Abstract
Objective
Elevated pre-operative C-reactive protein (CRP) serum values have been reported to be associated with poor overall survival for patients with pancreatic neuroendocrine neoplasms (pNEN). The aim of this study was to identify mechanisms linking CRP to poor prognosis in pNEN.
Methods
The malignant properties of pNENs were investigated using the human pNEN cell-lines BON1 and QGP1 exposed to CRP or IL-6. Analyses were performed by ELISA, Western blot, flow cytometry and immunocytochemistry as well as invasion and proliferation assays. To compare cytokine profiles and CRP levels, 76 serum samples of pNEN patients were analyzed using Luminex technology. In parallel, the expression of CRP and growth signaling pathway proteins was assessed on cell lines and paraffin-embedded primary pNEN.
Results
In BON1 and QGP1 cells, inflammation (exposure to IL-6) significantly upregulated CRP expression and secretion as well as migratory properties. CRP stimulation of BON1 cells increased IL-6 secretion and invasion. This was accompanied by activation/phosphorylation of the ERK, AKT and/or STAT3 pathways. Although known CRP receptors – CD16, CD32 and CD64 – were not detected on BON1 cells, CRP uptake of pNEN cells was shown after CRP exposure. In patients, increased pre-operative CRP levels (≥5 mg/L) were associated with significantly higher serum levels of IL-6 and G-CSF, as well as with an increased CRP expression and ERK/AKT/STAT3 phosphorylation in pNEN tissue.
Conclusion
The malignant properties of pNEN cells can be stimulated by CRP and IL-6 promoting ERK/AKT/STAT pathways activation as well as invasion, thus linking systemic inflammation and poor prognosis.
Keywords: CRP, pancreatic neuroendocrine neoplasms, pNEN, pNET, IL-6, inflammation, prognosis
Introduction
Pancreatic neuroendocrine neoplasm (pNEN), the most aggressive neuroendocrine malignancy, has an increasing incidence, with 3.2 cases per 1,000,000 inhabitants currently being diagnosed annually in the United States (1, 2, 3, 4). Since prognosis varies widely between individual pNENs (5), there is a critical need to better define the molecular features associated with prognosis in these tumors.
Recently, elevated pre-operative C-reactive protein (CRP) values have been shown to be prognostic for survival in patients with pNENs; patients with elevated pre-operative CRP levels had a significantly shorter overall survival (6). However, the molecular mechanisms for this phenomenon remained unclear.
CRP is an acute-phase protein which is produced mainly in the liver and rarely in atherosclerotic lesions, kidney, neurons and alveolar macrophages (7, 8, 9). CRP synthesis is triggered largely through secretion of interleukin 6 (IL-6) from macrophages and T-cells (10). Any type of inflammatory process is able to activate IL-6, thus causing increased concentration of CRP in systemic circulation (11). The function of CRP was initially considered to be related to its role in the innate immune system (12). CRP activates the complement system, which is then able to enhance the ability of antibodies and phagocytic cells to clear microbes and damaged cells from the organism. CRP binds to certain Fc-receptors, such as CD16, CD32 and CD64, whose names are derived from their binding specificity to the antibody region Fc (Fragment crystallizable). Additionally, it acts as opsonin for various pathogens. Interaction of CRP with Fc-receptors, for example internalization in human aortic endothelial cells (13), leads to the secretion of pro-inflammatory cytokines enhancing inflammatory response (14, 15).
Another protein that is involved in response to inflammatory products is STAT3 (signal transducer and activator of transcription 3). It is a crucial regulator of gene expression in response to pro-inflammatory cytokines (including IL-6) as it plays an important role in regulating growth, survival, differentiation and pathogen resistance (16, 17). Constitutive activation of STAT3 signaling pathway has been observed in a growing number of human cancers, such as breast cancer, multiple myeloma, head and neck tumors and both ovarian and prostate cancers (18, 19, 20, 21). Nothing is known about the role of STAT3 in pNEN. CRP and STAT3 may be part of the link between inflammation and cancer (22). Tumor-associated inflammatory cues in the tumor microenvironment, such as macrophages or tumor-associated fibroblasts and pro-inflammatory cytokines (including TNF-alpha and IL-6), contribute to genomic instability and are important tumor progression-promoting signals (23, 24).
Chronic inflammation is able to stimulate endocrine cells leading to their hyperplasia and neoplastic transformation (25) such as IL-1 was able to direct cancer cells into neuroendocrine differentiation. In other cancer entities, such as pancreatic ductal adenocarcinoma, squamous cell and adenocarcinoma of the esophagus, melanoma, soft tissue sarcoma and gastric cancer, circulating pro-inflammatory cytokines are also related to the pathogenesis and tumor progression (26, 27, 28, 29, 30). Additionally, in all the above-mentioned entities, elevated pre-operative CRP levels were also reported to be significantly associated with poor prognosis (31, 32, 33, 34, 35).
We, therefore, aimed to identify the mechanisms linking CRP and inflammation to poor prognosis in pancreatic neuroendocrine neoplasms.
Materials and methods
Clinical samples
For this study, snap-frozen tumor tissue (n = 14), paraffin-embedded tissue (n = 20) as well as serum (n = 76) from patients with pNEN were obtained from the Pancobank of the European Pancreas Center (EPZ/Department of Surgery, University Hospital Heidelberg; Ethical Approval Votes no. 301/2001 and 159/2002), a member of BMBH/Biomaterial Bank Heidelberg. A written informed consent was obtained from all patients after full explanation of the purpose and nature of all procedures used.
The histological examination of formalin-fixed, paraffin-embedded and H&E-stained pancreatic tissue sections was performed by an experienced pancreas pathologist (F B) and only tumor sections with more than 90% pNEN tissue were used.
Patients characteristics from which the clinical samples derived are summarized in Table 1.
Table 1.
Characteristics of the pNEN study population.
| Parameter | n |
|---|---|
| Total samplesa | 89 |
| Serum | 76 |
| Frozen tissue | 14 |
| FFPEb | 20 |
| Age at surgery: median (min–max) | 57 (23–83) |
| Sex | |
| Male | 52 |
| Female | 37 |
| Tumor localization | |
| Head | 39 |
| Body | 11 |
| Tail | 29 |
| Multifocal | 10 |
| ENETS stage | |
| I | 13 |
| II | 22 |
| III | 52 |
| IV | 2 |
| Grading (WHO 2010) | |
| G1 | 34 |
| G2 | 43 |
| G3 | 12 |
| Resection | |
| R0 | 67 |
| R1 | 16 |
| R2 | 6 |
aOverlapping samples; bFFPE, formalin-fixed paraffin-embedded tissue.
pNEN cell lines BON1 and QGP1
The human metastasized adherent pNEN cell line BON1 was obtained from Dr M Kidd, Yale University School of Medicine, as a gift and has been authenticated by STRS analysis. BON1 cells were cultured as a monolayer in T-75 flasks (Corning) in RPMI 1640:Ham’s F-12 medium in a 1:1 volume ratio (Invitrogen) supplemented with 10% fetal bovine serum (Life Technologies) and penicillin/streptomycin (100 IU/mL) at 37°C with 5% CO2 as previously described (36).
The human adherent pNEN cell line QGP1 was purchased from the Japanese Cancer Research Resources Bank (JCRB), which is part of the National Institutes of Biomedical Innovation, Health and Nutrition and was cultivated as a monolayer in T-75 flasks in RPMI 1640 supplemented with 10% fetal bovine serum (Life Technologies) and penicillin/streptomycin (100 IU/mL) at 37°C with 5% CO2.
To investigate the role of CRP as well as inflammation in pNEN cell proliferation and metastasis, BON1 and QGP1 cells were treated with the pro-inflammatory cytokine IL-6 (recombinant human IL-6, 25 ng/ml, Cell Signaling) or recombinant human CRP (20 µg/mL) (R&D Systems) for 48 h.
Protein extraction and Western blot analysis
Proteins from frozen pNEN tissue specimen (60–80 mg) pulverized in liquid nitrogen or from BON1 cells pellets were extracted using RIPA buffer containing complete protease inhibitors (Sigma-Aldrich) and supplemented with phosphatase inhibitor sets 1 and 2 (Sigma-Aldrich). Using an ultrasonic homogenizer (SonoPuls mini20 Bandelin®, Berlin, Germany), the suspensions were subjected to a 30-s sonication step on ice (ampl. 80%, 0.99 kJ) and subsequently centrifuged at 16,000 g for 10 min.
Supernatants were collected and divided into aliquots, and the total protein concentration was determined using a BCA Protein Determination Kit (Thermo Scientific) following the manufacturer’s instructions.
Western blot was performed as previously described (37) using the following monoclonal antibodies: anti-CRP, anti-IL-6, anti-total-STAT3 (all Abcam), anti-phospho-STAT3 (Tyr705), anti-total-ERK, anti-phospho-ERK (Thr202/Tyr204), anti-total-AKT and anti-phosho-AKT (Ser473) (all Cell Signaling). The secondary antibody used was goat anti-rabbit (R&D Systems). Quantification of protein expression was performed using ImageJ (EHD imaging, Damme, Germany).
Immunohistochemistry and immunocytochemistry
The expression of CRP and IL-6 was assessed on paraffin-embedded primary tumor samples by immunohistochemistry and on BON1 cells by immunocytochemistry as previously described (38, 39). Briefly, tissue sections were de-paraffinized and rehydrated with a graded ethanol series, and heat-based antigen retrieval was carried out in citrate buffer. Both tissue sections and BON1 cells were blocked with Universal Blocking Reagent (BioGenex, Fremont, CA, USA) and incubated with anti-CRP antibody (Abcam) or anti-IL-6 antibody (Abcam) before being washed and developed using goat anti-rabbit secondary antibody (CRP, Dako) or goat anti-mouse secondary antibody (IL-6, Dako) and the DAB kit (Dako). After the tissue sections were counterstained with hematoxylin and washed with water, they were incubated in graded alcohol solutions and roticlear (Roche) and then mounted. Optical imaging and analysis was performed using Zeiss Axioplan2 Microscope (Zeiss) with Axio Vision and K5400 Zeiss software.
Ten different high-power fields (400×) were selected for each slide and integrated optical density was used as the measure of staining intensity. Negative controls were processed in the absence of primary antibodies.
ELISA and PETIA
The determination of CRP and IL-6 levels in cell extracts and culture supernatants was performed using human C-Reactive-Protein ELISA Kit (Abcam) and human interleukin-6 ELISA Kit (Abcam) according to the manufacturer’s instructions.
CRP was additionally measured with PETIA (particle-enhanced turbidimetric immunoassay) using Dimension EXL (Siemens Healthineers) at the central clinical laboratory of the University Hospital of Heidelberg.
Multiplex cytokine analysis
A multiplex assay for quantitative determination of inflammatory mediators was applied to assess the concentrations of selected cytokines in serum. The analysis was performed using the Bio-Plex Pro Human Cytokine 17-Plex panel (Bio-Rad Laboratories GmbH). The following determinants were simultaneously detected: IL-1β, IL-2, IL-4, IL-5, IL6, IL-7, IL-8, IL-10, IL-12, IL-13, IL-17, G-CSF, GM-CSF, IFN-γ, MCP-1, MIP-1β and TNF-α. Multiplexing was performed according to the manufacturer’s instruction manual and analyzed on a Bio-Rad Bio-Plex 200 system. Values were calculated by the Bio-Plex™ software (Bio-Plex Manager, version 6.1, Bio-Rad). All serum samples were measured in triplicates.
Flow cytometry
BON1-cells were detached from the culture flasks by cell dissociation reagent (accutase, Thermo Fisher Scientific). 1 × 106 cells in 200 µL were incubated with the primary FcgRs antibodies (FcgRI-APC, clone 10.1; FcgRII-FITC, clone 8.26; FcgRIII-PE, clone 3G8; and isotype control (BD Pharmingen)) at RT for 1 h. After washing with PBS, labeled cells were analyzed using flow cytometry technique with FACS-Canto II flow cytometer (Becton Dickinson) and Flow Jo software 8.8. Cells incubated with only secondary antibody were used as negative control.
Proliferation assays
The proliferation capability of BON1 cells was firstly assessed by a MTS cell proliferation colorimetric assay kit (Biovision, Milpitas, CA, USA) according to the manufactures instructions. Briefly, BON1 cells (5 × 103/well) were seeded in 96-well plate and incubated for 3 days with IL-6 (25 ng/mL, Cell Signaling) or CRP (20 µg/mL, R&D Systems). Twenty microliters of MTS reagent were added to each well. After 3 h of incubation, optical density was read at 490 nm and 650 nm using a SYNERGY/HTX multi-mode reader (BioTek Instruments).
Secondly, we used the BrdU Cell Proliferation ELISA Kit (Abcam), in order to assess the same mechanism, according to the manufactures instructions.
RNA extraction and quantitative reverse transcriptase PCR (qRT-PCR)
Total mRNA extraction from cells as well as cDNA synthesis was performed using MPLC RNA IsoI. Kit (Roche) according to the manufacturer’s instructions. For PCR, the following primer pairs were used:
BCL2 5′-GGATGCCTTTGTGGAACTGT-3′, 5′-AGCCTGCAGCTTTGTTTCAT-3′;
MMP-9 5′-TTCTGCCCCAGCGAGAGA-3′, 5′-GTGCAGGCGGAGTAGGATTG-3′
GAPDH 5′-ATGTTCGTCATGGGTGTGAA-3′, 5′-GTCTTCTGGGTGGCAGTGAT-3′
Gene expression was quantified by qRT-PCR on a LightCycler 480 II using LightCycler® 480 SYBR green I master reagent (Roche). The quantitative PCR was performed in a 20 µL reaction solution containing 6 µL nuclease-free water, 2 µL cDNA templates, 10 µL Master Mix and 1 µL each of forward and reverse primer. The amplification conditions were initial denaturation at 95°C for 15 min followed by 45 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 30 s and elongation at 72°C for 20 s. The experiment was performed in triplicates. A qualitative PCR was also performed in order to confirm the presence of single and appropriate bands for each primer set. PCR data were analyzed using the ΔΔCT method as previously described (40).
Cell migration and invasion assays
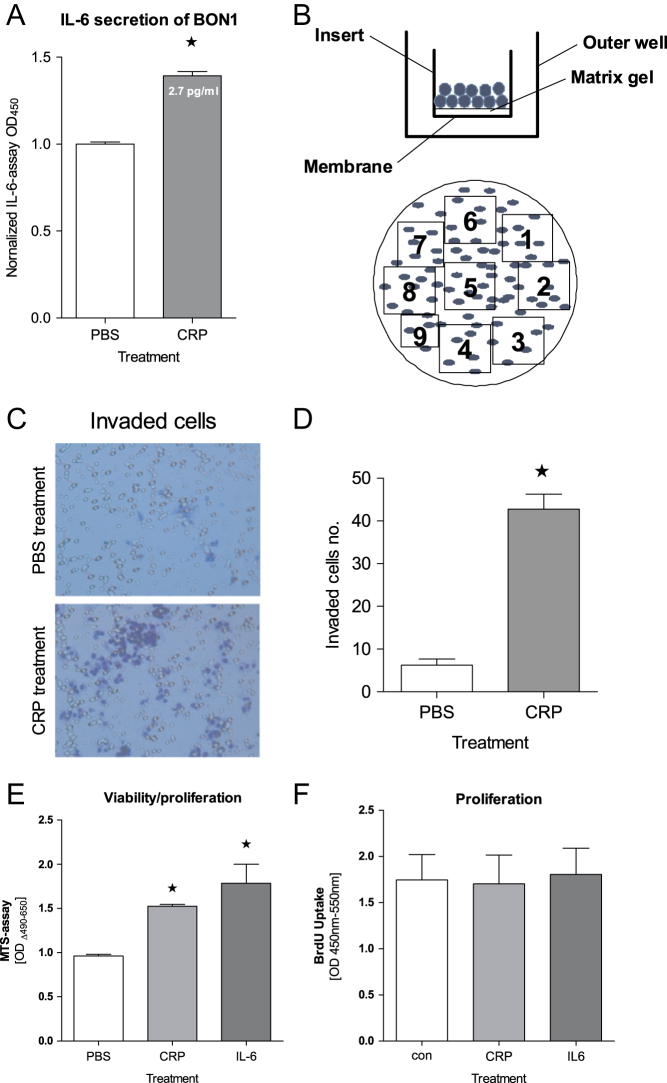
The migration and invasion potential of BON1 cells was assessed using a 24-well plate containing inserts with 12 µm pore size polycarbonate membranes (Merck Millipore) which was coated with a uniform layer of dried basement membrane matrix solution (1 mg/mL; Corning) on the upper surface of the inserts’ membrane. The lower compartment was filled with RPMI 1640/Ham’s-F12 medium containing 30% FBS. BON1 cell suspensions (3 × 105 cells/well in 300 µL serum-free medium) were added to the inside of each insert. After 48 h of incubation, inserts were removed and cells were fixed and stained with 0.05% crystal violet. Cells on the upper surface of the insert membrane (invasive cells) were removed with a cotton swab. Membranes were cut out. Cells that were able to invade and migrate (taken from the bottom of the membrane and the bottom of the outer well) were counted with a Zeiss Axioskop 2 Microscope with nine individual fields per membrane (Fig. 1B).
Figure 1.
Impact of CRP on BON1 cells. (A) IL-6 secretion of BON1 cells significantly increased upon CRP exposure (20 µg/mL, 48 h, ELISA, *P < 0.01). (B) Migration/invasion assay model: BON1 cells seeded in the inserts pass through basement membrane and cling to the bottom of insert’s membrane. Invasive cells are counted with nine individual fields per membrane. Blue dots = BON1 cells, clear circles = membrane pores. (C and D) Compared to normal non-treated BON1 cells, significantly more BON1 cells were capable of invading the gel and migrating through the membrane after CRP treatment (invasion assay, *P < 0.0001). (E) Proliferation capability of BON1 cells was strongly enhanced in the presence of CRP as well as IL-6 in MTS assay (*P < 0.001). (F) However, this effect cannot be seen in BrdU assay.
Inhibition of endocytosis and immunofluorescence staining
In order to investigate CRP internalization, 5 × 105 BON1 or QGP1 cells/mL were seeded in six-well plates on collagen-coated cover slides and cultured for 24 h. Both cell lines were treated with Dyngo 4a (Abcam), which inhibits clathrin-mediated endocytosis (IC50 Dyngo BON1 = 638, 1 nM, QGP-1 = 214, 1 nM). CRP was labeled with FITC using a KPL Surelink™ Fluorescein (FITC)-labeling Kit (SeraCare, Milford, MA, USA) following the manufacturer’s instructions.
After 24-h incubation, cells were additionally treated with the prepared FITC-conjugated CRP (20 µg/mL) for 24 h, harvested and seeded on collagen-coated glass plates. Then, cells were fixed in ice cold acetone for 10 min. Cells were washed three times with PBS for 5 min. Afterward, the cells were permeabilized using 0.3% Triton (Sigma-Aldrich) for 20 min. Cells were blocked in blocking buffer (0.2% FCS, 2% BSA, 0.2% gelatin fish skin in 100 mL 1× PBS) for 45 min at RT. Subsequently, cells were incubated with DAPI (1:10,000) for 1 h. After another washing step, the cells were incubated for 30 min at RT with PBS or DAPI and visualized using fluorescent microscopy (Zeiss Axioplan2 Microscope with Axio Vision and K5400 Zeiss software, Zeiss).
Statistical analysis
Statistical analyses were performed using Prism 5 (GraphPad Software). The Mann–Whitney test was used to determine the statistical difference between two groups. Comparisons between more than two groups were performed using the Kruskal–Wallis test, followed by the Dunn’s post hoc test where appropriate. Statistical significance is indicated by an asterisk. A P value <0.05 was considered statistically significant.
Results
Impact of CRP on IL-6 secretion, invasion and proliferation of BON1 and QGP1 cells
Given the fact that systemic elevated CRP is part of an inflammatory response and associated with poor prognosis in pNEN, we first examined the impact of the exposure of pNEN cells such as BON1 and QGP1 to CRP alone. Upon treatment with CRP (20 µg/mL for 48 h) IL-6 secretion by BON1 cells was significantly increased (Fig. 1A), while QGP1 cells did not secrete more IL-6 (data not shown).
Next, the functional significance of CRP with respect to essential malignant properties of cancer cells was examined. Invasion (Fig. 1C and D) as well as viability/proliferation measured by the MTS assay- (Fig. 1E) were both significantly increased upon exposure to CRP in comparison to unstimulated BON1 cells, while QGP1 cells did not show any differences (data not shown). Proliferation – measured by BrdU-ELISA – was not affected by CRP in BON1 (Fig. 1F) and QGP1 cells (data not shown).
Impact of pro-inflammatory IL-6 on CRP secretion, proliferation and invasion of BON1 and QGP1 cells
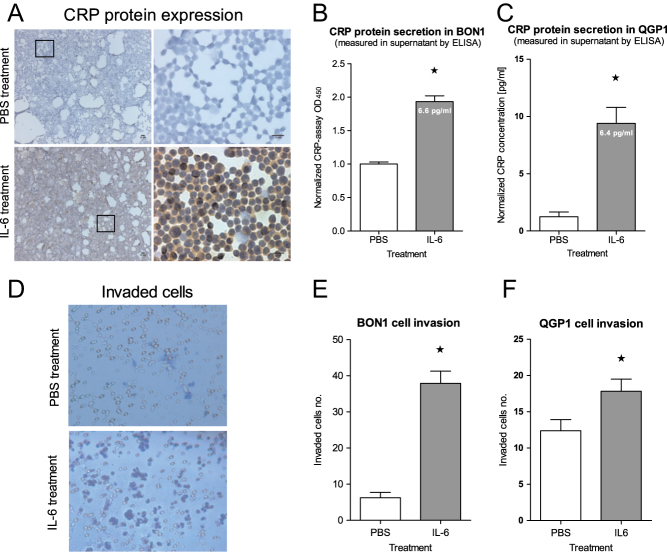
To investigate whether BON1 and QGP1 cells spontaneously express and secrete CRP under normal culture conditions, CRP concentrations were measured in cell supernatants using ELISA and the high-sensitive CRP test of the Heidelberg University Hospital Clinical Laboratory. Both assays revealed marginal amount of CRP in the supernatants of BON1 and QGP1 cells (control group Fig. 2B and C).
Figure 2.
Impact of pro-inflammatory IL-6 on BON1 and QGP cells. (A) Immunocytochemical analysis of BON1 cells and ELISA analysis of (B) BON1 and (C) QGP1 cell supernatant shows CRP expression/secretion being significantly increased upon IL-6 treatment (25 ng/mL, 48 h, *P < 0.01). (D and E) BON1 cells as well as (F) QGP1 cells were found to be more invasive after IL-6 exposure in comparison to control cells, (invasion assay, *P < 0.001).
Since CRP synthesis of hepatocytes is triggered by inflammatory mediators such as IL-6, IL-1β and TNF-α (11), BON1 and QGP1 cells were exposed to the pro-inflammatory cytokine IL-6. Three days after treatment with IL-6 (25 ng/mL), a strong intracellular CRP expression by BON1 cells was found (Fig. 2A). Concordantly, a two-fold increased CRP secretion of BON1 (OD) and a nine-fold increased CRP secretion of QGP1 (pg/mL) cells was measured (Fig. 2B and C).
We also sought to investigate the functional significance of IL-6 exposure with respect to invasion as key property of cancer cells, which was examined using a gel on top of a semi-permeable membrane with 12 µm pores. Invasion of cells after 48 h of IL-6 exposure was found to be increased seven-fold in BON1 and 1.5-fold QGP1 cells (Fig. 2E and F). A correlation between CRP secretion and cell invasion was not given since the high inflammatory effect of IL-6 exceeded the relative low CRP concentration in the supernatant (secretion). Another explanation for the lower invasiveness of QGP-1 cells may be the stronger QGP1-cell-cluster formation in the presence of IL-6.
Impact of IL-6 and CRP on signaling pathways in BON1 and QGP1 cells
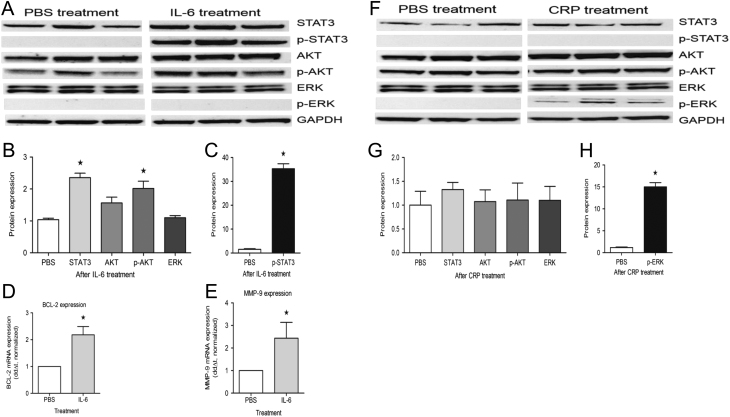
To examine the impact of pro-inflammatory IL-6 on signaling pathways in BON1 cells, we first determined STAT3 expression in IL-6-treated and PBS-treated BON1 cells as IL-6 treatment led to a robust STAT3 activation as reflected by increased phosphorylated STAT3 in IL-6-treated BON1 cells in comparison to control cells that did not show any STAT3 phosphorylation on the Western blot. The potential activation of the ERK and AKT pathways was also examined and a strong activation of AKT, but not of ERK, was found in these cells (Fig. 3A, B and C).
Figure 3.
Impact of pro-inflammatory IL-6 and CRP on signaling pathways in BON1 cells. (A) Activation/phosphorylation of AKT and STAT3, not ERK upon IL-6 treatment (Western blot, BON1 triplicates, 25 ng/mL, 48 h). (B and C) Quantitative analysis (normalized protein expression) showing STAT3, p-STAT3 and p-AKT are significantly increased upon IL-6 treatment (*P < 0.05). (D and E) mRNA expression of the STAT3 downstream genes BCL-2 and MMP-9 is significantly increased upon IL-6 treatment (qRT-PCR *P < 0.05). (F, G and H) Exposure of CRP to BON1 cells leads to phosphorylation of ERK, not AKT/STAT3 (Western blot (normalized protein expression), BON1 triplicates, *P < 0.001).
Whether the effects of IL-6 on STAT3 phosphorylation translated into active STAT3-mediated transcription was investigated as well. The expression of two STAT3-responsive genes, BCL2 and MMP-9, was examined by qRT-PCR. Both genes were strongly upregulated in IL-6-treated BON1 cells (Fig. 3D and E).
CRP triggered phosphorylation of ERK (growth signaling pathway). STAT3 and AKT were not affected by CRP treatment (Fig. 3F, G and H).
In QGP1 cells, none of those effects on signaling pathways were seen (data not shown).
CRP receptors in BON1 and QGP1 cells
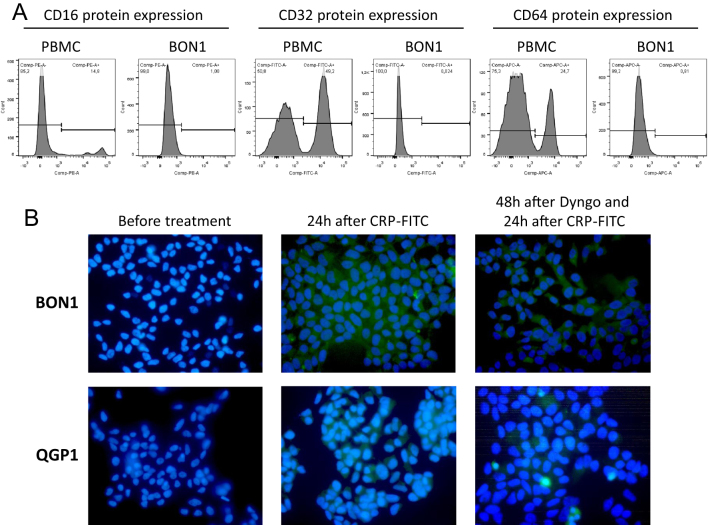
Having shown that CRP itself was able to directly promote proliferation and invasion by phosphorylation of ERK in BON1 cells, we next examined the presence of putative CRP receptors on BON1 and QGP1 cells. It has been previously demonstrated that CRP was capable of binding to three receptors: FcgRI (CD64), FcgRII (CD32) and probably FcgRIII (CD16) on neutrophils (41, 42, 43). Using FACS analysis, none of these FcgRs could be detected on BON1 cells (Fig. 4A).
Figure 4.
CRP internalization in BON1 and QGP1 cells. (A) Flow cytometry analysis showed no expression of previously described CRP receptors such as FcgRI (CD64), FcgRII (CD32) and FcgRIII (CD16) on the surface of BON1 cells. Normal human peripheral blood mononuclear cells (PBMC) carrying these receptors were analyzed as control cells. (B) Using FITC-labeled CRP, this protein was found in the cytoplasm of BON1 and QGP1 cells (middle), thus suggesting internalization of CPR. Inhibition of clathrin-mediated endocytosis using Dyngo seemed to result in less CRP internalization not excluding endocytosis a possible mechanism.
Therefore, FITC-labeled CRP was used in immunofluorescence microscopy in order to visualize CRP on neuroendocrine tumor cells. Indeed, FITC-labeled CRP was detectable within BON1 and QGP1 cells (Fig. 4B, middle), thereby suggesting an internalization of CRP either through endocytosis or another yet unknown receptor.
In order to specify this mechanism of internalization, clathrin-mediated endocytosis was inhibited by treatment with Dyngo. Subsequently, FITC-labeled CRP was given. Figure 4B (right) shows that this treatment visually tended to result in less CRP internalization. Endocytosis as possible mechanism is therefore not excluded.
CRP and IL-6 expression in human pNEN are associated with STAT3, AKT and ERK pathway activation and systemic cytokine levels
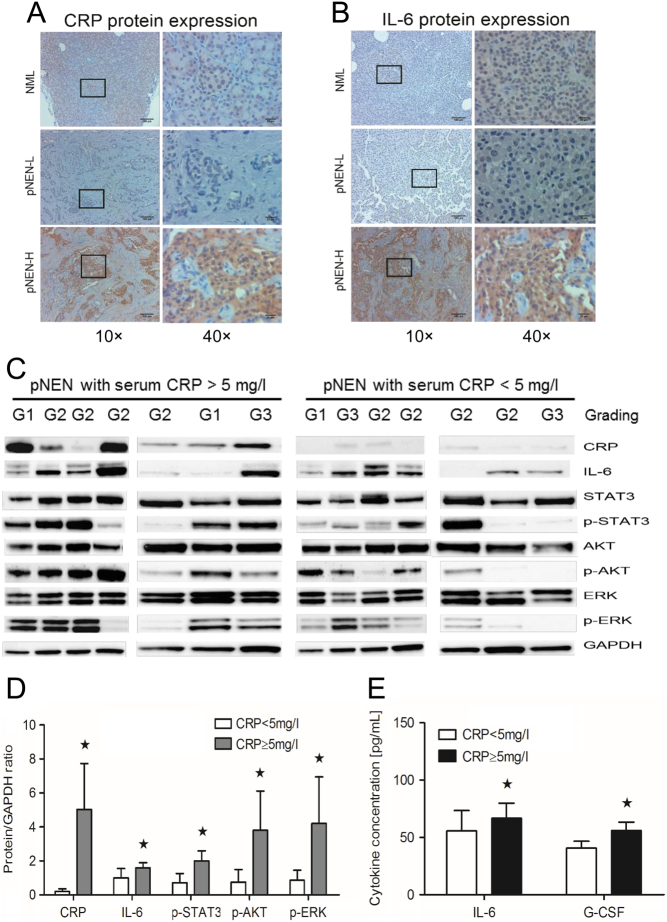
Given the impact of CRP on proliferation and invasion of BON1 cells, we examined CRP expression and growth signaling pathways in human pNEN tissue. Both CRP and IL-6 expression were significantly higher in immunohistochemistry and Western blot analysis in pNEN of patients with pre-operative CRP serum concentration above 5 mg/L compared to pNEN of patients with pre-operative CRP serum concentration below 5 mg/L (Fig. 5A, B and C). Additionally, high serum CRP concentrations were associated with increased phosphorylation of STAT3, AKT and ERK, as well as strongly increased IL-6 levels in comparison to low CRP serum concentrations (Fig. 5C and D). The increase of serum CRP was also associated with increased systemic levels of IL-6 and G-CSF (Fig. 5E), thus indicating a complex inflammatory response accompanying CRP secretion. These results confirm the link observed in vitro between pNEN and systemic inflammation in the human disease.
Figure 5.
Impact of CRP in pNENs. (A and B) Immunohistochemical and (C) Western blot analysis showing high CRP and IL-6 expression in tumor sections of patients with high pre-operative serum CRP concentrations (≥5 mg/L) in comparison to patients with low pre-operative serum CRP concentrations (<5 mg/L). There were no correlations between grading (3 NET G1, 8 NET G2 and 3 G3) and CRP. (C and D) High pre-operative serum CRP concentrations (≥5 mg/L) lead to significant phosphorylation of STAT3, ERK and AKT, as well as strongly increased IL-6 expression in tumor tissue in comparison to low pre-operative serum CRP concentrations (Western blot, *P < 0.05), thereby supporting CRP exposure to be relevant for neuroendocrine tumor cells with regards to proliferation and tumor biology. (E) Both IL-6 and G-CSF were significantly increased in serum from pNENs patients with high pre-operative serum CRP concentration (≥5 mg/L) (ELISA, *P < 0.05). NML, normal pancreas tissue; pNEN-L, pNEN patients with low pre-operative serum CRP concentrations; pNEN-H: pNEN patients with high pre-operative serum CRP concentrations.
Discussion
In this study, we showed that pancreatic neuroendocrine neoplasm cells (BON1 and QGP1) upregulate and secrete CRP when stimulated by inflammation (IL-6), Additionally, IL-6 is secreted by BON1 cells during exposure to CRP. Both CRP and IL-6 increased invasion of BON1 and QGP1 cells. Furthermore, the ERK, AKT and/or STAT3 signaling were delineated as critical effector pathways that are strongly associated with the essential malignant properties of cancer cells. Those in vitro observations were confirmed in human neuroendocrine disease. Increased CRP and IL-6 expression, as well as ERK/AKT/STAT3 phosphorylation in tissue of pNENs patients with elevated CRP serum levels, were identified.
Although the role of CRP in the innate immune response as opsonin is well established, its role in tumorigenesis and progression – particularly in pancreatic neuroendocrine neoplasms – is unknown. Although in breast cancer cell lines increased invasion was only observed in synergy with sphingosine-1-phosphate (44), elevated CRP levels at the time of diagnosis of breast cancer were found to be significantly correlated with stage, size and grade of the tumor and metastasis (45). Increased pre-operative serum CRP has also been demonstrated to enhance the metastasis of renal cell carcinoma (46). Consistent with these studies, we demonstrated that elevated CRP increased the invasion capability of BON1 and QGP1 cells. Low CRP concentration (<5 mg/L) was revealed to inhibit proliferation of vascular endothelium cells (47); however, high CRP concentration (≥5 mg/L) increased proliferation of multiple myeloma cells and macrophages (48, 49, 50). In this study, we show activation of growth signaling pathways in neuroendocrine tumor cells (BON1) and increased viability and proliferation using the MTS assay, while in the BrdU assay, high CRP concentrations were not shown to be pro-proliferative in the pNEN cell lines BON1 and QGP1.
It is remarkable that both pNEN cell lines BON1 and QGP1 reacted differently to the same stimuli as well as secreted CRP in different concentrations. The different behavior of pNEN cell lines may correspond to the biological heterogeneity (51, 52, 53) that is well known from human pNENs (5). Our study is limited by the small number of pNEN cell lines currently available for research. One important goal of future studies has to be the generation of new pNEN cell lines, especially from well-differentiated tumors. Another limitation of this study is that in vitro experiments were conducted with highly proliferative pNEN cell lines, whereas the patient cohort included a majority of G1/G2 pNENs (Table 1).
IL-6 has also been shown to stimulate the proliferation and metastasis of a diverse range of tumors (54, 55), in which signaling involves several distinct downstream intracellular signaling cascades, including the AKT/STAT3 pathway. This ultimately leads to transcriptional changes that have an impact on survival, proliferation, differentiation and migration (56). We examined the IL-6-secretion of BON1 cells and determined the effects of IL-6 exposure on proliferation and migration/invasion. The response to IL-6 is most likely mediated by STAT3 activation, which has been associated with poor prognosis in renal cell carcinoma, for example (57). In agreement with these findings, our data showed that the effects of IL-6 in augmenting both BON1 and QGP1 cells migration and proliferation are likely mediated via the AKT/STAT3 pathway.
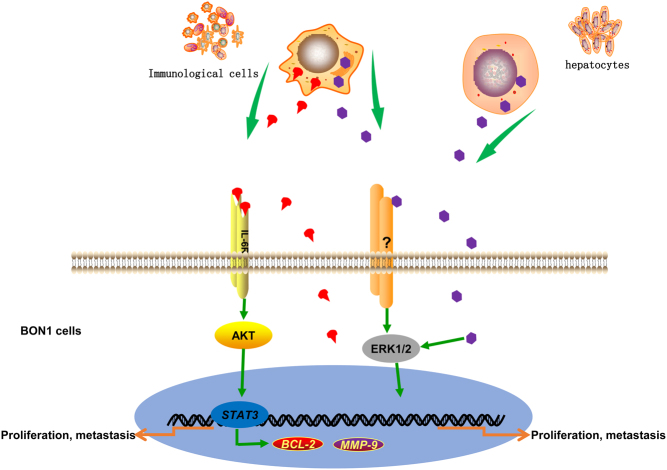
Interaction of CRP with monocytes/macrophages was reported to induce the secretion of IL-1 and TNF as well as IL-1-induced IL-6 production (58, 59). Moreover, monocytes/macrophages were shown to synthesize CRP (8) and IL-6 (59), thus suggesting a positive feedback mechanism between CRP and IL-6 in monocytes/macrophages in the promotion of systemic inflammation. The present study indicates that CRP may play a role in pNEN tumor progression using an IL-6/AKT/STAT-3/CRP signaling as shown in Fig. 6. CRP either binds to an unknown receptor or becomes internalized by endocytosis, triggers IL-6 production and activates the IL-6/AKT/STAT3/CRP axis in BON1 and QGP1 cells. The mutual stimulation between CRP and IL-6 in those cells may represent a positive feedback mechanism which promotes tumor progression as well.
Figure 6.
Schematic model of possible signal transduction stimulated by CRP in BON1 cells. In pNEN microenvironment, special immunological cells and hepatocytes are one source of CRP and/or IL-6. The other source of CRP and IL-6 is BON1 or QGP1 cells. The binding of IL-6 to its ligand-binding receptor initiates activation of AKT, which in turn phosphorylates and activates STAT3 transcription factors. Activated STAT3 binds to specific sites in the target gene promoters inducing transcription of BCL-2 and MMP-9, thus resulting in proliferation and metastasis (invasion/migration) of pancreatic neuroendocrine tumor cells, as well as the expression and secretion of CRP. Secreted CRP may exert its effects by internalization either through an unknown receptor (?) or via endocytosis. It activates ERK as well as IL-6 secretion, thereby leading to proliferation and metastasis in pancreatic neuroendocrine tumor cells.
In conclusion, systemic CRP is used by pNEN tumor cells to induce IL-6 expression and, by itself, may activate the IL-6/AKT/STAT3/CRP axis which promotes invasion and metastasis of pNEN.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
Simon Schimmack was supported by the Heidelberger Stiftung Chirurgie (Heidelberg Foundation of Surgery). Yongchao Yang was sponsored by the China Scholarship Council (CSC, 201506370096).
Author contribution statement
Funding acquisition: O S, S S and Y L: Conception and design of experiments: O S, S S and Y Y. Experiments were conducted by Y Y, K F, M H and S S. Data analysis: Y Y, M H, S S, F B, K F, M S and Y L. Manuscript was written by S S, Y Y and O S. Revision of manuscript: S S, Y Y, K F, M H, Y L, M S, F B, T H and O S.
References
- 1.Lawrence B, Gustafsson BI, Chan A, Svejda B, Kidd M, Modlin IM. The epidemiology of gastroenteropancreatic neuroendocrine tumors. Endocrinology and Metabolism Clinics of North America 2011. 40 , vii. ( 10.1016/j.ecl.2010.12.005) [DOI] [PubMed] [Google Scholar]
- 2.Metz DC, Jensen RT. Gastrointestinal neuroendocrine tumors: pancreatic endocrine tumors. Gastroenterology 2008. 135 . ( 10.1053/j.gastro.2008.05.047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Modlin IM, Oberg K, Chung DC, Jensen RT, de Herder WW, Thakker RV, Caplin M, Delle Fave G, Kaltsas GA, Krenning EP, et al Gastroenteropancreatic neuroendocrine tumours. Lancet: Oncology 2008. 9 . ( 10.1016/S1470-2045(07)70410-2) [DOI] [PubMed] [Google Scholar]
- 4.Yao JC, Hassan M, Phan A, Dagohoy C, Leary C, Mares JE, Abdalla EK, Fleming JB, Vauthey JN, Rashid A, et al One hundred years after ‘carcinoid’: epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the United States. Journal of Clinical Oncology 2008. 26 . ( 10.1200/JCO.2007.15.4377) [DOI] [PubMed] [Google Scholar]
- 5.Schimmack S, Svejda B, Lawrence B, Kidd M, Modlin IM. The diversity and commonalities of gastroenteropancreatic neuroendocrine tumors. Langenbeck’s Archives of Surgery 2011. 396 . ( 10.1007/s00423-011-0739-1) [DOI] [PubMed] [Google Scholar]
- 6.Wiese D, Kampe K, Waldmann J, Heverhagen AE, Bartsch DK, Fendrich V. C-reactive protein as a new prognostic factor for survival in patients with pancreatic neuroendocrine neoplasia. Journal of Clinical Endocrinology and Metabolism 2016. 101 . ( 10.1210/jc.2015-3114) [DOI] [PubMed] [Google Scholar]
- 7.Yasojima K, Schwab C, McGeer EG, McGeer PL. Human neurons generate C-reactive protein and amyloid P: upregulation in Alzheimer’s disease. Brain Research 2000. 887 . ( 10.1016/s0006-8993(00)02970-x) [DOI] [PubMed] [Google Scholar]
- 8.Dong Q, Wright JR. Expression of C-reactive protein by alveolar macrophages. Journal of Immunology 1996. 156 . [PubMed] [Google Scholar]
- 9.Yasojima K, Schwab C, McGeer EG, McGeer PL. Generation of C-reactive protein and complement components in atherosclerotic plaques. American Journal of Pathology 2001. 158 . ( 10.1016/S0002-9440(10)64051-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinhold B, Ruther U. Interleukin-6-dependent and -independent regulation of the human C-reactive protein gene. Biochemical Journal 1997. 327 . ( 10.1042/bj3270425) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pepys MB, Hirschfield GM. C-reactive protein: a critical update. Journal of Clinical Investigation 2003. 111 . ( 10.1172/JCI18921) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volanakis JE. Human C-reactive protein: expression, structure, and function. Molecular Immunology 2001. 38 . ( 10.1016/S0161-5890(01)00042-6) [DOI] [PubMed] [Google Scholar]
- 13.Devaraj S, Du Clos TW, Jialal I. Binding and internalization of C-reactive protein by Fcgamma receptors on human aortic endothelial cells mediates biological effects. Arteriosclerosis, Thrombosis, and Vascular Biology 2005. 25 . ( 10.1161/01.ATV.0000168573.10844.ae) [DOI] [PubMed] [Google Scholar]
- 14.Du Clos TW. Function of C-reactive protein. Annals of Medicine 2000. 32 . ( 10.3109/07853890009011772) [DOI] [PubMed] [Google Scholar]
- 15.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nature Reviews: Immunology 2008. 8 . ( 10.1038/nri2206) [DOI] [PubMed] [Google Scholar]
- 16.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nature Reviews: Molecular Cell Biology 2002. 3 . ( 10.1038/nrm909) [DOI] [PubMed] [Google Scholar]
- 17.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nature Reviews: Cancer 2009. 9 . ( 10.1038/nrc2734) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatterjee M, Jain S, Stuhmer T, Andrulis M, Ungethum U, Kuban RJ, Lorentz H, Bommert K, Topp M, Kramer D, et al. STAT3 and MAPK signaling maintain overexpression of heat shock proteins 90alpha and beta in multiple myeloma cells, which critically contribute to tumor-cell survival. Blood 2007. 109 . ( 10.1182/blood-2006-05-024372) [DOI] [PubMed] [Google Scholar]
- 19.Dhir R, Ni Z, Lou W, DeMiguel F, Grandis JR, Gao AC. Stat3 activation in prostatic carcinomas. Prostate 2002. 51 . ( 10.1002/pros.10079) [DOI] [PubMed] [Google Scholar]
- 20.Hambek M, Baghi M, Strebhardt K, May A, Adunka O, Gstottner W, Knecht R. STAT 3 activation in head and neck squamous cell carcinomas is controlled by the EGFR. Anticancer Research 2004. 24 . [PubMed] [Google Scholar]
- 21.Proietti CJ, Rosemblit C, Beguelin W, Rivas MA, Diaz Flaque MC, Charreau EH, Schillaci R, Elizalde PV. Activation of Stat3 by heregulin/ErbB-2 through the co-option of progesterone receptor signaling drives breast cancer growth. Molecular and Cellular Biology 2009. 29 . ( 10.1128/MCB.00853-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature 2008. 454 . ( 10.1038/nature07205) [DOI] [PubMed] [Google Scholar]
- 23.Balkwill FR, Mantovani A. Cancer-related inflammation: common themes and therapeutic opportunities. Seminars in Cancer Biology 2012. 22 . ( 10.1016/j.semcancer.2011.12.005) [DOI] [PubMed] [Google Scholar]
- 24.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011. 144 . ( 10.1016/j.cell.2011.02.013) [DOI] [PubMed] [Google Scholar]
- 25.Cigrovski Berkovic M, Cacev T, Catela Ivkovic T, Zjacic-Rotkvic V, Kapitanovic S. New insights into the role of chronic inflammation and cytokines in the etiopathogenesis of gastroenteropancreatic neuroendocrine tumors. Neuroendocrinology 2014. 99 . ( 10.1159/000362339) [DOI] [PubMed] [Google Scholar]
- 26.Ernst M, Najdovska M, Grail D, Lundgren-May T, Buchert M, Tye H, Matthews VB, Armes J, Bhathal PS, Hughes NR, et al. STAT3 and STAT1 mediate IL-11-dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. Journal of Clinical Investigation 2008. 118 . ( 10.1172/JCI34944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmer R, Goumas FA, Waetzig GH, Rose-John S, Kalthoff H. Interleukin-6: a villain in the drama of pancreatic cancer development and progression. Hepatobiliary and Pancreatic Diseases International 2014. 13 . ( 10.1016/S1499-3872(14)60259-9) [DOI] [PubMed] [Google Scholar]
- 28.Kavanagh ME, O’Sullivan KE, O’Hanlon C, O’Sullivan JN, Lysaght J, Reynolds JV. The esophagitis to adenocarcinoma sequence; the role of inflammation. Cancer Letters 2014. 345 . ( 10.1016/j.canlet.2013.08.017) [DOI] [PubMed] [Google Scholar]
- 29.Linnskog R, Jonsson G, Axelsson L, Prasad CP, Andersson T. Interleukin-6 drives melanoma cell motility through p38alpha-MAPK-dependent up-regulation of WNT5A expression. Molecular Oncology 2014. 8 . ( 10.1016/j.molonc.2014.05.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radons J. The role of inflammation in sarcoma. Advances in Experimental Medicine and Biology 2014. 816 . ( 10.1007/978-3-0348-0837-8_11) [DOI] [PubMed] [Google Scholar]
- 31.Szkandera J, Stotz M, Absenger G, Stojakovic T, Samonigg H, Kornprat P, Schaberl-Moser R, Alzoughbi W, Lackner C, Ress AL, et al. Validation of C-reactive protein levels as a prognostic indicator for survival in a large cohort of pancreatic cancer patients. British Journal of Cancer 2014. 110 . ( 10.1038/bjc.2013.701) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gockel I, Dirksen K, Messow CM, Junginger T. Significance of preoperative C-reactive protein as a parameter of the perioperative course and long-term prognosis in squamous cell carcinoma and adenocarcinoma of the oesophagus. World Journal of Gastroenterology 2006. 12 . ( 10.3748/wjg.v12.i23.3746) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang S, Wang Y, Sui D, Liu H, Ross MI, Gershenwald JE, Cormier JN, Royal RE, Lucci A, Schacherer CW, et al. C-reactive protein as a marker of melanoma progression. Journal of Clinical Oncology 2015. 33 . ( 10.1200/JCO.2014.58.0209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura T, Matsumine A, Matsubara T, Asanuma K, Uchida A, Sudo A. Clinical significance of pretreatment serum C-reactive protein level in soft tissue sarcoma. Cancer 2012. 118 . ( 10.1002/cncr.26353) [DOI] [PubMed] [Google Scholar]
- 35.Nozoe T, Iguchi T, Adachi E, Matsukuma A, Ezaki T. Preoperative elevation of serum C-reactive protein as an independent prognostic indicator for gastric cancer. Surgery Today 2011. 41 . ( 10.1007/s00595-009-4297-x) [DOI] [PubMed] [Google Scholar]
- 36.Schimmack S, Taylor A, Lawrence B, Alaimo D, Schmitz-Winnenthal H, Buchler MW, Modlin IM, Kidd M. A mechanistic role for the chromatin modulator, NAP1L1, in pancreatic neuroendocrine neoplasm proliferation and metastases. Epigenetics and Chromatin 2014. 7 15 ( 10.1186/1756-8935-7-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schimmack S, Lawrence B, Svejda B, Alaimo D, Schmitz-Winnenthal H, Fischer L, Buchler MW, Kidd M, Modlin I. The clinical implications and biologic relevance of neurofilament expression in gastroenteropancreatic neuroendocrine neoplasms. Cancer 2012. 118 . ( 10.1002/cncr.26592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartmann N, Giese NA, Giese T, Poschke I, Offringa R, Werner J, Ryschich E. Prevailing role of contact guidance in intrastromal T-cell trapping in human pancreatic cancer. Clinical Cancer Research 2014. 20 . ( 10.1158/1078-0432.CCR-13-2972) [DOI] [PubMed] [Google Scholar]
- 39.Reiser-Erkan C, Erkan M, Pan Z, Bekasi S, Giese NA, Streit S, Michalski CW, Friess H, Kleeff J. Hypoxia-inducible proto-oncogene Pim-1 is a prognostic marker in pancreatic ductal adenocarcinoma. Cancer Biology and Therapy 2008. 7 . ( 10.4161/cbt.7.9.6418) [DOI] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 2001. 25 . ( 10.1006/meth.2001.1262) [DOI] [PubMed] [Google Scholar]
- 41.Bharadwaj D, Stein MP, Volzer M, Mold C, Du Clos TW. The major receptor for C-reactive protein on leukocytes is fcgamma receptor II. Journal of Experimental Medicine 1999. 190 . ( 10.1084/jem.190.4.585) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khreiss T, Jozsef L, Hossain S, Chan JS, Potempa LA, Filep JG. Loss of pentameric symmetry of C-reactive protein is associated with delayed apoptosis of human neutrophils. Journal of Biological Chemistry 2002. 277 . ( 10.1074/jbc.M205378200) [DOI] [PubMed] [Google Scholar]
- 43.Crowell RE, Du Clos TW, Montoya G, Heaphy E, Mold C. C-reactive protein receptors on the human monocytic cell line U-937. Evidence for additional binding to Fc gamma RI. Journal of Immunology 1991. 147 . [PubMed] [Google Scholar]
- 44.Kim ES, Cha Y, Ham M, Jung J, Kim SG, Hwang S, Kleemann R, Moon A. Inflammatory lipid sphingosine-1-phosphate upregulates C-reactive protein via C/EBPbeta and potentiates breast cancer progression. Oncogene 2014. 33 . ( 10.1038/onc.2013.319) [DOI] [PubMed] [Google Scholar]
- 45.Onitilo AA, Engel JM, Stankowski RV, Liang H, Berg RL, Doi SA. High-sensitivity C-reactive protein (hs-CRP) as a biomarker for trastuzumab-induced cardiotoxicity in HER2-positive early-stage breast cancer: a pilot study. Breast Cancer Research and Treatment 2012. 134 . ( 10.1007/s10549-012-2039-z) [DOI] [PubMed] [Google Scholar]
- 46.Tatokoro M, Saito K, Iimura Y, Fujii Y, Kawakami S, Kihara K. Prognostic impact of postoperative C-reactive protein level in patients with metastatic renal cell carcinoma undergoing cytoreductive nephrectomy. Journal of Urology 2008. 180 . ( 10.1016/j.juro.2008.04.025) [DOI] [PubMed] [Google Scholar]
- 47.Guo S, Meng S, Chen B, Liu J, Gao L, Wu Y. C-reactive protein can influence the proliferation, apoptosis, and monocyte chemotactic protein-1 production of human umbilical vein endothelial cells. DNA and Cell Biology 2011. 30 . ( 10.1089/dna.2010.1093) [DOI] [PubMed] [Google Scholar]
- 48.Devaraj S, Yun JM, Duncan-Staley C, Jialal I. C-reactive protein induces M-CSF release and macrophage proliferation. Journal of Leukocyte Biology 2009. 85 . ( 10.1189/jlb.0808458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang YR, Huang LJ, Ma YP, Lu YJ, Yang LH, Zhou YA. Mechanism of C-reactive protein on proliferation of multiple myeloma U266 cells. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2009. 17 . [PubMed] [Google Scholar]
- 50.Yang J, Wezeman M, Zhang X, Lin P, Wang M, Qian J, Wan B, Kwak LW, Yu L, Yi Q. Human C-reactive protein binds activating Fcgamma receptors and protects myeloma tumor cells from apoptosis. Cancer Cell 2007. 12 . ( 10.1016/j.ccr.2007.08.008) [DOI] [PubMed] [Google Scholar]
- 51.Hofving T, Arvidsson Y, Almobarak B, Inge L, Pfragner R, Persson M, Stenman G, Kristiansson E, Johanson V, Nilsson O. The neuroendocrine phenotype, genomic profile and therapeutic sensitivity of GEPNET cell lines. Endocrine-Related Cancer 2018. 25 . ( 10.1530/ERC-17-0445) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vandamme T, Peeters M, Dogan F, Pauwels P, Van Assche E, Beyens M, Mortier G, Vandeweyer G, de Herder W, Van Camp G, et al. Whole-exome characterization of pancreatic neuroendocrine tumor cell lines BON-1 and QGP-1. Journal of Molecular Endocrinology 2015. 54 . ( 10.1530/JME-14-0304) [DOI] [PubMed] [Google Scholar]
- 53.Kim JT, Li J, Song J, Lee EY, Weiss HL, Townsend CM, Jr, Evers BM. Differential expression and tumorigenic function of neurotensin receptor 1 in neuroendocrine tumor cells. Oncotarget 2015. 6 . ( 10.18632/oncotarget.4745) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeh HH, Lai WW, Chen HH, Liu HS, Su WC. Autocrine IL-6-induced Stat3 activation contributes to the pathogenesis of lung adenocarcinoma and malignant pleural effusion. Oncogene 2006. 25 . ( 10.1038/sj.onc.1209464) [DOI] [PubMed] [Google Scholar]
- 55.Morikawa T, Baba Y, Yamauchi M, Kuchiba A, Nosho K, Shima K, Tanaka N, Huttenhower C, Frank DA, Fuchs CS, et al. STAT3 expression, molecular features, inflammation patterns, and prognosis in a database of 724 colorectal cancers. Clinical Cancer Research 2011. 17 . ( 10.1158/1078-0432.CCR-10-2694) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yun UJ, Park SE, Jo YS, Kim J, Shin DY. DNA damage induces the IL-6/STAT3 signaling pathway, which has anti-senescence and growth-promoting functions in human tumors. Cancer Letters 2012. 323 . ( 10.1016/j.canlet.2012.04.003) [DOI] [PubMed] [Google Scholar]
- 57.Guo C, Yang G, Khun K, Kong X, Levy D, Lee P, Melamed J. Activation of Stat3 in renal tumors. American Journal of Translational Research 2009. 1 . [PMC free article] [PubMed] [Google Scholar]
- 58.Galve-de Rochemonteix B, Wiktorowicz K, Kushner I, Dayer JM. C-reactive protein increases production of IL-1 alpha, IL-1 beta, and TNF-alpha, and expression of mRNA by human alveolar macrophages. Journal of Leukocyte Biology 1993. 53 . ( 10.1002/jlb.53.4.439) [DOI] [PubMed] [Google Scholar]
- 59.Helle M, Boeije L, Pascual-Salcedo D, Aarden L. Differential induction of interleukin-6 production by monocytes, endothelial cells and smooth muscle cells. Progress in Clinical and Biological Research 1991. 367 . [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a