In silico analyses of Treponema pallidum subsp. pallidum genomes and predicted proteomes to search for homologs of known bacterial outer membrane proteins (OMPs) led to the identification of tp0126 as a gene encoding a putative member of the OmpW family of porins/virulence factors. Our previous investigations on the role of Tp0126 in T. pallidum biology and syphilis pathogenesis showed that Tp0126 is fully conserved among T. pallidum strains and that transcription of tp0126 is driven by σ70.
KEYWORDS: OmpW, Tp0126, Treponema pallidum, functional redundancy, phase variation
ABSTRACT
In silico analyses of Treponema pallidum subsp. pallidum genomes and predicted proteomes to search for homologs of known bacterial outer membrane proteins (OMPs) led to the identification of tp0126 as a gene encoding a putative member of the OmpW family of porins/virulence factors. Our previous investigations on the role of Tp0126 in T. pallidum biology and syphilis pathogenesis showed that Tp0126 is fully conserved among T. pallidum strains and that transcription of tp0126 is driven by σ70. These initial results pointed to a housekeeping function for this protein. We also demonstrated that a guanosine homopolymer of various lengths located between the −10 and −35 consensus sequences in the tp0126 promoter modulates transcription consistently with phase variation, a mechanism that we also previously described for other T. pallidum genes encoding putative OMPs/virulence factors and that is often employed as a strategy for immune evasion. Circular dichroism spectra of recombinant Tp0126 also supported its structural homology with OmpW. Here we further investigated the humoral and cellular responses to Tp0126 during experimental and natural syphilis and the ability of Tp0126 to confer protection against syphilis in immunized rabbits. B-cell epitope mapping showed that compared to sera from experimentally infected animals, immunizations enhanced humoral immunity to sequences located in the putative Tp0126 surface-exposed loops, while phagocytosis assays showed that postimmunization sera opsonized T. pallidum. Despite such promising results, no significant protection was seen following infectious challenge in immunized animals versus controls. Functional redundancy and phase variation might explain the lack of effectiveness of this vaccine candidate and/or design.
INTRODUCTION
Syphilis is a sexually transmitted infection caused by the spirochete Treponema pallidum subsp. pallidum (1). This disease still represents a public health concern, given a global prevalence that ranges from 18 million to 56 million cases and an estimated incidence of 6 million to 11 million new episodes every year (2–6). Although the majority of these cases occur in developing countries, syphilis has also become resurgent in industrialized Asian, European, and American nations, including the United States, where its incidence has risen steadily since 2000 (7–11). Syphilis is a multistage, systemic, and chronic infection that, if left untreated, has the potential to cause significant morbidity and mortality. Syphilis might in fact progress to affect the cardiovascular and central nervous systems of infected individuals, potentially leading to aortic aneurism, hearing or vision loss, dementia, paralysis, and stroke-like syndrome (1, 12). Additionally, vertical transmission of T. pallidum during pregnancy accounts for a high proportion of perinatal morbidity and mortality in underprivileged settings (13–15) and up to 50% of stillbirths in sub-Saharan Africa (15). The development of an effective syphilis vaccine could significantly curtail the spread of this infection. In fact, despite being an easily treatable infection (16), past public health initiatives to eliminate syphilis and congenital syphilis did not fully achieve their intended goals (17, 18). Because T. pallidum immune clearance from early lesions is mediated by phagocytosis of opsonized pathogen cells by macrophages (19–21), T. pallidum outer membrane (OM) proteins (OMPs) are the most suitable candidates for vaccine development. However, OMP identification has been hindered by several limitations associated with working with T. pallidum in a laboratory setting, including the unusual fragility of the pathogen’s OM and the inability to genetically manipulate this bacterium (1). Analysis of annotated proteomes of T. pallidum strains using bioinformatics tools has therefore been of pivotal importance to identify proteins bearing sequence/structural homology to known prokaryotic OMPs (22–24). Continued research efforts to deepen our knowledge on these proteins are, however, necessary, particularly because sufficient experimental data to label most of these in silico-identified proteins as bona fide OMPs are still lacking, and independent studies have sometimes led to discordant conclusions on the location and/or structure of some OMP candidates (25–29). Among the newly identified putative T. pallidum OMPs, Tp0126 is annotated as a possible member of the OmpW family of proteins, known to be comprised of small, eight-stranded β-barrel monomeric porins that are widespread among Gram-negative bacteria. Previous work conducted in our laboratory supported that Tp0126 is indeed an OMP. We showed that in the initial T. pallidum Nichols strain genome (24), the tp0126 open reading frame (ORF) was incorrectly annotated as a longer polypeptide and was therefore not predicted to have a cleavable signal sequence for sorting to the outer membrane (24). A cleavable signal peptide could, however, be easily predicted after we experimentally identified the gene transcriptional start site (TSS) and most likely start codon (24). Moreover, analysis of circular dichroism (CD) spectra produced by recombinant Tp0126 following protein refolding suggested that the percentages of β-barrel (43%), α-helix (30%), and random-coil (27%) components were compatible with a β-barrel OMP, further supporting the structural homology of Tp0126 to OmpW (24). Evidence that Tp0126 might be an excellent vaccine candidate came from the observations that the protein sequence is fully conserved among T. pallidum strains and isolates and that the Tp0126 promoter is recognized by the housekeeping factor σ70 (24), which could support steady expression of this target in the pathogen. However, we later showed that tp0126 transcription undergoes on/off switching depending on the length of a poly(G) repeat of various lengths located between the −10 and −35 σ70 consensus sequences. This mechanism, known as phase variation, was previously observed by us to modulate the transcription of other T. pallidum genes encoding putative OMPs with porin/adhesin function (30). To test our hypothesis that Tp0126 could be a possible vaccine candidate for syphilis, we evaluated whether immunization with recombinant refolded Tp0126 (rTp0126) and a modified emulsion-based Sigma adjuvant system (mSAS) elicited antibodies able to opsonize T. pallidum and protected rabbits against syphilis infection. To this end, immunized and control animals were challenged with T. pallidum, and correlates of protection, such as attenuation of lesion development and treponemal burden in primary chancres and animal seroconversion following challenge, were measured. The biological and immunological bases of the outcome of the protection experiment were further investigated by performing enzyme-linked immunosorbent assay (ELISA)-based Pepscan B-cell epitope mapping to compare the humoral responses to Tp0126 that developed in immunized rabbits to those that developed in experimentally infected ones and in syphilis patients, and we identified Tp0126 T-cell epitopes recognized during experimental infection by performing a lymphocyte proliferation assay. Finally, we investigated how the tp0126 expression level and length of the poly(G) tracts associated with the tp0126 promoter changed in immunized and control animals following infectious challenge.
RESULTS
Humoral response to Tp0126 following rabbit immunization and T. pallidum opsonophagocytosis assay using anti-Tp0126 antiserum.
Rabbits immunized with recombinant rTp0126 and mSAS developed specific humoral immunity to the antigen, which was detectable 3 weeks after the third immunization (data not shown). Prechallenge sera had a titer of 1:20,000 (Fig. 1A) for each of the three rabbits used in this pilot protection experiment. Preimmunization sera had no reactivity to Tp0126 (not shown). At the end of the five-dose immunization cycle, we performed B-cell epitope mapping using overlapping synthetic peptides (20-mers overlapping by 10 amino acids [aa]) (Table 1). B-cell epitope identification using prechallenge sera from the immunized animals (1:5,000) showed that reactivity to predicted Tp0126 surface-exposed loops was limited to L1 (peptide 3), L2 (peptide 9), and possibly a small portion of L4 (peptide 16), while no reactivity was seen toward peptides encompassing L3 (Fig. 1B). Reactivity to peptides mapping onto the predicted protein scaffolding (peptides 1, 2, 7, and 8) was also seen in immunized animals (Fig. 1B). No reactivity was detected against the T. pallidum antigen Tp0574, used here as a control. Figure 1C provides a graphical view (in red) of the Tp0126 regions recognized by sera from immunized rabbits. Despite this limited reactivity to putative surface-exposed sequences, the phagocytosis assay showed that pooled sera from immunized animals were opsonic for T. pallidum and promoted significantly higher pathogen ingestion than did noninfected rabbit serum (NRS) (P = 0.039) (Fig. 1D).
FIG 1.
(A) Titration of prechallenge sera from each rabbit. (B) B-cell epitope mapping performed with postimmunization sera. Bars relative to peptides containing putative surface-exposed amino acids are in gray, while bars relative to peptides that map onto the predicted Tp0126 β-scaffolding are in white. Gray/white bars contain amino acids that map to both the predicted surface-exposed loops and the scaffolding. FL, full-length peptide. (C) Tp0126 sequences recognized by prechallenge rabbit immune sera, highlighted in red in the Tp0126 structural model. (D) Opsonophagocytosis assay performed using pooled prechallenge sera from Tp0126-immunized rabbits. IRS, day 90 immune serum from rabbits infected i.t. with the T. pallidum Nichols strain; NRS, noninfected rabbit serum.
TABLE 1.
Tp0126 synthetic peptide sequences
| Peptide | Sequencea | Peptide coordinatesb |
|---|---|---|
| 1 | DPWDTTAAGRSTIRLSAMGA | 29–49 |
| 2 | STIRLSAMGAVPLFQVDWCN | 39–59 |
| 3 | VPLFQVDWCNSGRGDDRNAN | 49–69 |
| 4 | SGRGDDRNANAQTNGHKYIY | 59–79 |
| 5 | AQTNGHKYIYPAFSAALGFE | 69–89 |
| 6 | PAFSAALGFEHFVCRGLSLG | 79–99 |
| 7 | HFVCRGLSLGIDASVQYHCS | 89–109 |
| 8 | IDASVQYHCSYPNNTYSPTT | 99–119 |
| 9 | YPNNTYSPTTPYYYLAIPVA | 109–129 |
| 10 | PYYYLAIPVALTAGYTVAFW | 119–139 |
| 11 | LTAGYTVAFWRIRLPLTVGA | 129–149 |
| 12 | RIRLPLTVGAGFNYQHYYTS | 139–159 |
| 13 | GFNYQHYYTSTYYGLVLKAA | 149–169 |
| 14 | TYYGLVLKAAAGCYFQLTEH | 159–179 |
| 15 | AGCYFQLTEHWSLGVSATYS | 169–189 |
| 16 | WSLGVSATYSGVPRSCEKII | 179–199 |
| 17 | GVPRSCEKIIEEDRQQTNTR | 189–209 |
| 18 | EEDRQQTNTRTAQFIAAGVD | 199–219 |
| 19 | TAQFIAAGVDVRYHL | 209–223 |
Sequences are listed from N to C termini. The underlined amino acids are in predicted external loops.
According to Tp0126 annotation described previously (24).
Protection against experimental syphilis infection in Tp0126-immunized animals.
Rabbits immunized with rTp0126 and mSAS were challenged intradermally (i.d.) at 8 sites on their shaved backs with 106 freshly harvested T. pallidum (Nichols) cells per injection site. Three unimmunized control rabbits were also challenged. As shown by previous work with different vaccine candidates (25, 31–33), syphilitic lesion development (measured as lesion diameter) is attenuated in animals immunized with protective antigens compared to unimmunized controls, while the treponemal burden within lesions that develop at challenge sites is significantly lower in immunized/protected animals than in controls. Additionally, if an antigen is protective, fewer lesions undergo ulceration in immunized animals (25, 31–33) than in unimmunized controls. Measurement of lesion diameters over a 30-day period postchallenge, however, did not show significant differences between immunized and control animals (Fig. 2A), and eventually, all lesions ulcerated in both groups of animals (Fig. 2B). Dark-field microscopy (DFM) analysis of needle aspirates from all of the challenge sites (Fig. 2C) as well as quantification of treponemal mRNA using reverse transcription-quantitative PCR (RT-qPCR) as parallel approaches to evaluate treponemal burden within lesions (Fig. 2D) showed a slightly decreased burden in samples from immunized animals, albeit not significant (P > 0.05), compared to controls, overall suggesting that immunization with recombinant Tp0126 failed to induce any protection against T. pallidum. At the end of the 30-day observation period, all immunized and control animas were VDRL and fluorescent treponemal antibody absorption (FTA-ABS) positive, further confirming the lack of protection.
FIG 2.
(A) Measurement of indurated lesion diameters at challenge sites. (B) Percentage of ulcerating lesions. Open circles represent Tp0126-immunized rabbits; solid circles are unimmunized control rabbits. (C) Dark-field microscopy counts of treponemal cells in aspirates from challenge lesions. Needle aspirates were obtained from each lesion at day 13 postchallenge. (D) Molecular evaluation of treponemal burden within challenge lesions. Burden was measured by quantification of T. pallidum tp0574 mRNA normalized to the rabbit HPRT message.
Humoral and cellular responses to Tp0126 during experimental syphilis infection.
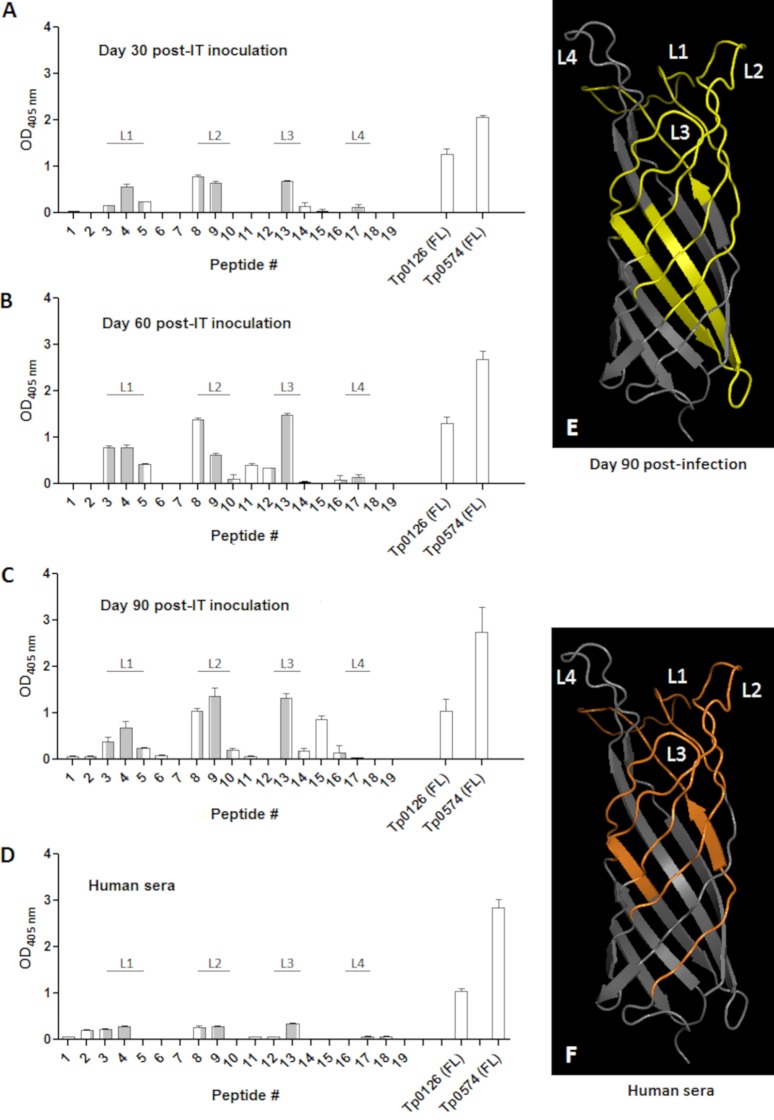
Previous work showed that rabbits infected with T. pallidum developed a humoral response to Tp0126 as soon as 4 weeks after intratesticular (i.t.) inoculation and that animal infection with different syphilis strains led to differential levels of Tp0126-specific antibodies (24). Furthermore, analysis of sera from human patients with latent syphilis showed that all samples had a detectable humoral response against Tp0126, although it was significantly weaker than the reactivity to the control antigen Tp0574, the 47-kDa lipoprotein, which is one of the most abundantly expressed and immunogenic antigens of the syphilis spirochete (24). To expand on these early studies and understand which Tp0126 sequences preferentially elicit a humoral response during experimental and natural infection, we performed B-cell epitope mapping using overlapping synthetic peptides (Table 1). Sera for this investigation were obtained from new animals experimentally infected i.t. with T. pallidum (Nichols strain) at days 30, 60, and 90 postinoculation and from patients (n = 7) with latent syphilis. Pepscan mapping of B-cell epitopes showed that in all sera from experimentally infected animals, humoral reactivity was mainly directed toward peptides entirely encompassed in the predicted Tp0126 surface-exposed loops (peptides 3, 4, 9, and 13) (Fig. 3A to C), while only a few peptides mapping predominantly (peptides 5 and 8) or entirely (peptide 15) to the predicted Tp0126 β-barrel scaffolding were recognized. Reactivity to peptides was generally lower at day 30 postinoculation and higher in sera collected at days 60 and 90 (Fig. 3A to C). Reactivity in human sera was also mainly directed toward peptides mapping onto predicted Tp0126 surface loops (peptides 3, 4, 9, and 13) (Fig. 3D), rather than peptides mapping onto the predicted protein scaffolding (peptides 2 and 8) (Fig. 3D). Figures 3E and F provide a graphical view of the Tp0126 regions recognized by day 90 rabbit sera (in yellow) and human sera (in orange), respectively, showing that although three (L1, L2, and L3) out of four of the putative protein surface loops appear to be targets of the humoral response, the L4 loop is consistently not recognized by either group of sera. Additionally, because we did not conduct any prior investigation to assess whether Tp0126 harbored T-cell epitopes, splenic lymphocytes harvested from rabbits euthanized at day 90 postinfection were used in proliferation assays also using synthetic peptides to identify Tp0126 T-cell epitopes. Splenic lymphocytes proliferated mainly upon exposure to peptides encompassing the predicted Tp0126 β-scaffolding (peptides 2, 7, and 11) compared to control wells (Fig. 4A) but also in response to two peptides mapping to the predicted L2 and L3 loops (peptides 9 and 13, respectively). Figure 4B shows a structural model of Tp0126, with the sequences eliciting proliferation in green.
FIG 3.
B-cell epitope mapping using synthetic peptides. (A to C) Peptides (20-mers overlapping by 10 aa) based on the Tp0126 sequence (Table 1) were used along with sera collected at day 30 (A), day 60 (B), and day 90 (C) from rabbits infected i.t. with the Nichols strain of T. pallidum. Bars represent the mean OD readings after background (from no-antigen wells) subtraction. (D) Reactivity of sera (n = 7) from latent syphilis patients. Bars relative to peptides that contain putative surface-exposed amino acids are in gray, while bars relative to peptides that map onto the predicted Tp0126 β-scaffolding are in white. Gray/white bars contain amino acids that map to both the predicted surface-exposed loops and the scaffolding. (E) Tp0126 sequences recognized by day 90 immune rabbit sera, highlighted in yellow in the Tp0126 three-dimensional (3D) model. (F) Tp0126 sequences recognized by human sera. Sequences are highlighted in orange in the Tp0126 3D model. The Tp0126 3D model was generated using I-TASSER as described previously (24). FL, full-length peptide.
FIG 4.
(A) T-cell proliferation assay with splenic lymphocytes. Splenocytes were harvested from rabbits at day 90 after experimental i.t. infection using test antigens (rTp126 and Tp0126-derived synthetic peptides), the control antigen concanavalin A (ConA), and no antigen (No Ag). Bars represent the geometric means ± standard errors of means of quadruplicate experimental values for each antigen for three rabbits. Significant differences (P ≤ 0.05) compared to the no-antigen value are indicated by an asterisk. (B) Tp0126 sequences that induced proliferation. Sequences are highlighted in green in the Tp0126 3D model. The Tp0126 3D model was generated using I-TASSER as described previously (24).
Expression of tp0126 mRNA in challenged rabbits and correlation with length of the poly(G) tract in the tp0126 promoter.
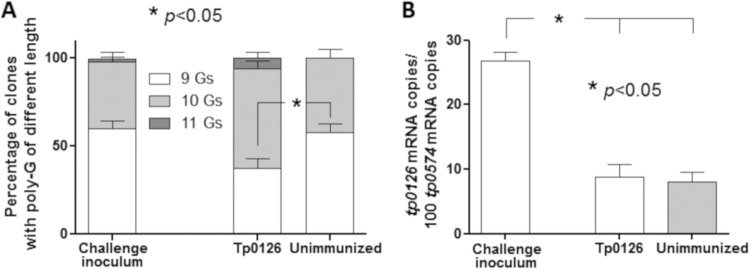
In addition to evaluation of treponemal burden (Fig. 2D), RNA extracted both from treponemes of the challenge inoculum and from lesion biopsy specimens from all immunized and control animals was used to evaluate the level of tp0126 expression. In parallel, DNA extracted from the same samples was used to amplify the region containing the variable poly(G) tract previously shown to influence tp0126 promoter activity by changing the spacing between the −35 and −10 consensus sequences in the gene promoter (24). More specifically, a poly(G) length of 7 or 8 G’s was associated with high promoter activity, while lengths of ≥9 G’s were associated with decreased gene transcription (24). Analysis of the poly(G) length profile in these samples showed that compared to both the treponemes in the challenge inoculum, a significantly low proportion of poly(G) tracts with 9 G residues (mostly replaced by 10-G tracts) was found in treponemes from Tp0126-immunized animals (Fig. 5A), while treponemes present in lesions from unimmunized animals had a poly(G) length profile not dissimilar from that of the challenge inoculum (Fig. 5A). tp0126 message quantification showed that expression levels were significantly higher in treponemes from the challenge inoculum than in treponemes present in lesions from Tp0126-immunized animals (Fig. 5B). However, low tp0126 expression was also detected in treponemes from unimmunized (control) animals (Fig. 5B).
FIG 5.
(A) Distribution of poly(G) lengths. Distributions were determined in the inoculum treponemes as well as in treponemes in lesions from Tp0126-immunized and control rabbits. G length percentages are 61.2% (9 G’s), 37.1% (10 G’s), and 1.7% (11 G’s) in the challenge inoculum; 36.8% (9 G’s), 58.2% (10 G’s), and 5.0% (11 G’s) in Tp0126-immunized rabbits; and 54.6% (9 G’s) and 45.4% (10 G’s) in control samples. (B) tp0126 mRNA levels. Message levels are normalized to the tp0574 message in the T. pallidum challenge inoculum and in treponemes from postchallenge lesions of unimmunized and Tp0126-immunized rabbits.
DISCUSSION
The feasibility of developing a fully protective syphilis vaccine is supported by early evidence that sterile immunity to infection was induced in rabbits after repeated immunizations with γ-irradiated T. pallidum as well as by human inoculation studies showing that patients with late latent syphilis were resistant to symptomatic reinfection with heterologous T. pallidum strains (34, 35). Given the role of opsonophagocytosis in T. pallidum immune clearance during natural infection, putative OMPs like Tp0126 have naturally become the most suitable candidates for a syphilis vaccine. The outcome of our protection experiment was therefore disappointing, as lesion development and treponemal burden in immunized animals were not significantly attenuated or reduced compared to controls. The Pepscan analysis using rabbit prechallenge sera suggested that despite having achieved good antibody titers to Tp0126, such reactivity was not uniformly directed toward all the putative Tp0126 surface loops but was directed toward just a subset of them. This result was somewhat in contrast to the epitope mapping outcome using sera from experimentally infected animals or naturally infected patients, where reactivity to a wider array of putative surface sequences was seen. In immunized animals, for example, no reactivity was ever detected to peptide 13 (putative external loop 3), which, on the contrary, was consistently recognized by sera from both experimentally infected animals and patients. Partial reactivity to surface loop sequences might play a role in the outcome of our immunization/challenge experiment. The plausibility of this hypothesis is also supported by the limited opsonic ability (although significant compared to the negative control) of the postimmunization Tp0126 antiserum shown by the opsonophagocytosis assay. Future work will focus on understanding the contribution of humoral immunity to the single putative Tp0126 loops (and in particular loop 3) in treponemal clearance. In parallel, work should be done to identify with more certainty the structure of Tp0126. Our CD data, although compatible with a small β-barrel protein, suggest that there might also be an α-helix component not predicted in silico. Whether such a difference is due to our refolding process or to a genuine feature of the Tp0126 structure is unknown. Refolding of rTp0126 to a structure not exactly identical to the native one might also have had a role in the lack of reactivity to peptide 13 mentioned above.
Although, for the time being, our results do not support Tp0126 as a good vaccine candidate, alternative immunization strategies could improve humoral reactivity toward surface sequences, which in turn could lead to better protection with this antigen. Possible strategies currently being considered in our laboratory include production of the antigen in a heterologous system, such as the nonpathogenic Borrelia burgdorferi B314 strain, as we reported previously for the TprK antigen (36), and purification of OM vesicles for immunization, rather than using the whole organism. Such a design would also bypass the issue of refolding a denatured antigen prior to immunization, a practice that has been adopted today with several syphilis vaccine candidates, including ours, to induce antibodies to conformational epitopes in addition to linear ones. Pepscan mapping performed using patient sera, however, which showed generally low reactivity to linear peptides compared to the full-length antigen, might suggest that our refolding approach for Tp0126 might indeed be able to reconstitute some conformational epitopes not present in the peptides. An alternative approach also being considered in our laboratory is immunization with HIV- or human papillomavirus (HPV)-derived viruslike particles (VLPs) carrying the whole antigen embedded in the envelope (if HIV VLPs are used) or sequences corresponding to the surface loops of the protein mounted on a capsid protein (e.g., L1 in the case of HPV VLPs).
Furthermore, our data seem to partially support a role for phase variation in the outcome of our experiment. Within treponemes present in lesions of immunized animals, the majority of the Tp0126-associated poly(G) tracts harbored 10 G residues, compared to the 9 seen in controls (both inoculum treponemes and treponemes from unimmunized animals). We previously showed that a 10-residue poly(G) tract was associated with a lower transcription level than with shorter lengths, which is consistent with the evidence that upon performing message quantification, significantly higher tp0126 mRNA levels were seen in the inoculum treponemes than in those from Tp0126-immunized animals. Therefore, our results suggest that immunization might have selected against a subset of treponemes expressing higher Tp0126 levels but not necessarily against those expressing the antigen at lower levels. At this point, however, it is not clear how to interpret the low tp0126 message level seen in treponemes from lesions harvested from unimmunized controls, which was expected to be as high as that in the inoculum treponemes, at least based on the distribution of the poly(G) lengths. Too little is known on the transcriptional regulation of this T. pallidum gene aside from our previous work on phase variation, but one could hypothesize that the different anatomical district (skin) that the treponemes were harvested from compared to the inoculum (testes) or the longer infection period (a 10-day period in testes versus a 13-day period in skin) could also influence tp0126 gene expression.
It should also be considered that the T. pallidum genome encodes a second putative OmpW homolog, Tp0733. In this context, functional redundancy with the Tp0733 protein could have played a role in ensuring that the biological role of Tp0126, which supposedly is to facilitate the transport of small hydrophobic molecules across the OM, would nonetheless be maintained in the presence of immune pressure toward Tp0126. Future work in the laboratory will aim at studying tp0733 expression in our samples to further evaluate this hypothesis.
A limitation of our study was the lack of an adjuvant-only control rabbit group. In our case, however, providing data from adjuvant-only controls would have been more critical if the animals immunized with Tp0126 plus mSAS had shown signs of protection. Since there were no significant differences in treponemal burden and lesion progression between immunized and naive control animals, we can infer that the adjuvant was not sufficient to induce any protective response in the immunized animals. Another limitation was the challenge dose used (106 cells/injection site), which is orders of magnitude higher than the infective dose responsible for natural syphilis transmission. The use of a lower, more “natural” challenge dose could have led to a different outcome for our experiment. Nonetheless, previous work done with other T. pallidum vaccine candidates (25, 31–33) showed that differences in lesion development and treponemal burden are evident even with high challenge doses if the immunization induces partial protection. Future experiments will be designed to include lower challenge doses and an adjuvant-only control group.
Despite our preliminary results, we believe that given the very high degree of conservation of this protein among T. pallidum strains and subspecies, further investigations and approaches to improve and/or better evaluate the protective potential of Tp0126 might be worthy before ruling out this antigen as a possible vaccine candidate. Such future endeavors will likely go far beyond the pairing of one or more recombinant antigens with adjuvants but will necessarily involve a structure-guided antigen design and advances in the isolation of human monoclonal antibodies, next-generation sequencing, and nanoparticle biology. The application of these approaches will eventually facilitate the goal of obtaining an effective syphilis vaccine to stem the spread of this important infection.
MATERIALS AND METHODS
Ethics statement.
Outbred adult male New Zealand White rabbits ranging from 3.0 to 4.5 kg were purchased from R&R Research (Stanwood, WA). While the animals were in our vivarium, care was provided according to the Guide for the Care and Use of Laboratory Animals (37). All procedures performed on the animals were approved by the University of Washington Institutional Animal Care and Use Committee (IACUC) (protocol number 4243-01; principal investigator, L.G.). Deidentified sera from human syphilis patients were used in this study, and this research has been determined by the University of Washington Human Subjects Division not to meet the federal regulatory definition of human subject research.
Strain propagation and rabbit experimental infection to evaluate humoral and cellular responses to Tp0126.
Prior to entry into the study, each rabbit was bled, and individual sera were tested with both a treponemal test (FTA-ABS) and a nontreponemal test (VDRL test; BD, Franklin Lakes, NJ) to rule out infection with Treponema paraluiscuniculi, the rabbit syphilis agent. Only rabbits seronegative by both tests were used for T. pallidum propagation, experimental infections to evaluate humoral and cellular immune responses to Tp0126, or immunization/challenge experiments to evaluate Tp0126 protective ability. The Nichols strain of T. pallidum was propagated by means of intratesticular (i.t.) inoculation as previously described (38) and always harvested at peak orchitis, generally after 10 to 12 days from the inoculation date. Briefly, on harvest day, the rabbit was euthanized, testes were removed, and bacteria were extracted from minced testes in sterile saline supplemented with 10% heat-inactivated normal rabbit serum (NRS). The testicular extract was then collected in sterile 50-ml tubes and spun twice at 1,000 rpm (180 × g) for 10 min in a 5810R centrifuge (Eppendorf, Hauppauge, NY) to remove rabbit tissue debris. Treponemes were counted by dark-field microscopy (DFM), and extracts were diluted in NRS-saline to the desired concentration depending on the downstream application (either long-term experimental infection or infectious challenge). Long-term experimental infections were conducted to evaluate the development of humoral and cellular responses to Tp0126 and to perform B- and T-cell epitope mapping using synthetic peptides and recombinant Tp0126 (rTp0126). To this end, three rabbits were infected i.t. with a total of 5 × 106 freshly harvested T. pallidum cells per testis. Following i.t. inoculation, treponemal motility was assessed by DFM to ensure that the time elapsed between harvest and reinoculation did not affect treponemal viability. Establishment of infection in these rabbits was assessed by monitoring the development of orchitis during the following weeks as well as by performing FTA-ABS and VDRL tests on sera collected at day 30 postinoculation. Sera from i.t.-infected animals were collected at days 30, 60, and 90 postinfection to assess humoral immunity to Tp0126 and to obtain infected rabbit serum (IRS) for the opsonophagocytosis assay (described below). At day 90, animals were terminally bled and euthanized to isolate splenic lymphocytes to perform T-cell proliferation assays (as described below). Extracted sera were heat inactivated at 56°C for 30 min and stored in aliquots at −20°C until used for ELISAs or opsonophagocytosis assays.
Expression and purification of recombinant Tp0126.
An rTp0126 protein devoid of the signal peptide was expressed following ORF cloning into the pEXP-5-NT TOPO vector (Life Technologies, Carlsbad, CA), which added to the Tp0126 mature coding sequence 22 vector-encoded NH2-terminal amino acids, including the 6×His tag. No additional residues were added at the COOH terminus, as the tp0126 stop codon was included in the reverse primer. Amplification of the tp0126 ORF from Nichols strain DNA was performed in our laboratory, as were ORF cloning into the expression vector and confirmation of the correct sequence/orientation as previously described (24). For protein expression, a starter culture of Escherichia coli Rosetta cells (Sigma, St. Louis, MO) transformed with the recombinant plasmid was prepared by inoculating 2 ml of LB-ampicillin (100 μg/ml) broth with a single E. coli colony and cultured at 37°C in a shaking incubator at 250 rpm until the optical density at 600 nm (OD600) reached 1.0 absorbance units (AU). The starter culture was then diluted to 20 ml in fresh broth. When the OD600 of the new culture again reached 1.0 AU, it was then diluted into 2 liters of fresh broth and split in two 4-liter flasks. The culture was induced with 1 mM (final concentration) isopropyl-β-d-thiogalactopyranoside (IPTG) when it reached an OD600 of 0.6 AU and then left in the shaking incubator for 5 h at 30°C. Subsequently, cultures were centrifuged at 3,500 × g for 15 min at 4°C in a Sorvall RC-5B centrifuge. Cells were resuspended in 100 ml of sterile phosphate-buffered saline (PBS) and centrifuged again. Following pellet resuspension in 5 ml of binding buffer (50 mM NaH2PO4, 8 M urea, 10 mM imidazole [pH 8.0]) per g of culture, cells were sonicated in ice with 100 pulses of 6 s, each separated by 10-s intervals. Insoluble components were precipitated by centrifugation at 24,000 × g for 30 min at 4°C, and the cell lysate was saved. For affinity chromatography, 2 ml of Ni-nitrilotriacetic acid (NTA) agarose (Qiagen, Germantown, MD) was packed into a 1.5- by 12-cm column and washed with 3 resin volumes of molecular-grade H2O and 6 resin volumes of binding buffer. The cell lysate was then loaded, and the flow was adjusted to 1 ml/min. Washes were performed by using 10 resin volumes of binding buffer, followed by 6 resin volumes of wash buffer (50 mM NaH2PO4, 8 M urea, 20 mM imidazole [pH 8.0]). Recombinant Tp0126 was eluted with 10 ml of elution buffer (50 mM NaH2PO4, 8 M urea, 300 mM imidazole [pH 8.0]) in 1-ml fractions. Eluates were analyzed for purity and yield by SDS-PAGE. Prior to each immunization, rTp0126 was refolded using a Profoldin (Hudson, MA) column containing lysophosphatidylcholine (5 mM), arginine (150 mM), glycerol (10%), dodecyl maltoside (0.7 mM), and Tris-HCl (0.1 mM) (pH 7.5) according to the manufacturer’s instructions. The concentration of refolded rTp0125 was determined using a Micro-BCA protein assay kit (Thermo Fisher, Waltham, MA). CD spectra (190 to 260 nm) were acquired in triplicate at room temperature using 0.5 mg/ml of recombinant, refolded Tp0126 in a Jasco-1500 high-performance CD spectrometer. CD spectra were analyzed using Dichroweb (http://dichroweb.cryst.bbk.ac.uk/html/home.shtml), and the spectra from buffer alone were used for background subtraction. CD analysis results were consistent with those previously reported (24).
Rabbit immunization and challenge with rTp0126.
Rabbits (n = 3) were injected with 50 μg of refolded rTp0126 every 3 weeks for a total of five immunizations. Prior to injection, the antigen was mixed with a Sigma adjuvant system (SAS; Sigma) supplemented with 0.5 mg/ml of N-acetylmuramyl-l-alanyl-d-isoglutamine (MDP [muramyl dipeptide]; Sigma) and 0.1 mg/ml of FSL-1 (Pam2CGDPKHPKSF from InvivoGen, San Diego, CA) as a Toll-like receptor 2/6 (TLR2/6) agonist, hence referred to as modified SAS (mSAS). Immunogen-adjuvant preparation was performed according to the manufacturer’s instructions by mixing 2 ml of the antigen solution in each vial of mSAS. Immunizations were performed by administering 300 μl of the antigen-adjuvant emulsion intradermally (i.d.) (50 μl in six different sites), 600 μl intramuscularly (i.m.) into each hind leg, and 100 μl subcutaneously (s.c.) in the neck region. During the immunization cycle, animals were bled prior to the first immunization and 3 weeks after each injection, prior to the subsequent one, to evaluate the development of humoral immunity to Tp0126. Humoral immunity to Tp0126 was evaluated by an ELISA using both rTp0126 for serum titration and synthetic peptides for B-cell epitope mapping. Animals were then challenged with freshly harvested Nichols strain cells on eight sites on their shaved backs. A group (n = 3) of naive animals was also challenged as described above to serve as unimmunized controls. For the infectious challenge, a total of 106 T. pallidum cells were inoculated at each of the inoculation sites. Aliquots of the challenge inoculum were spun, and pellets were resuspended in either 400 μl of Ultraspec RNA reagent (reagent now discontinued; Biotex Laboratories, Houston, TX) or 400 μl of DNA lysis buffer (10 mM Tris [pH 8.0], 0.1 M EDTA [pH 8.0], 0.5% SDS). DNA/RNA extraction was performed on these samples to allow analysis of tp0126 expression and length of the poly(G) tract associated with the tp0126 promoter in the challenge inoculum (as described below). Following challenge, cutaneous lesion development was monitored by recording the diameter of indurated lesions and progression (from erythematous to indurated, ulcerated, and healed lesions) every day for 30 days. Lesion aspirates were collected at day 13 postchallenge from each lesion and resuspended in 18 μl of sterile saline on a microscope slide to be examined by DFM. Treponemes were counted in a blind manner in a total of 100 fields per aspirate. Concurrent with lesion aspirates, at day 13 postinoculation, two cutaneous lesion biopsy specimens were obtained from each immunized and control animal using a 4-mm biopsy punch to evaluate treponemal burden by quantification of T. pallidum mRNA and to compare tp0126 expression levels and poly(G) tract lengths in these samples to those in the inoculum. To this end, biopsy specimens were split and minced with a sterile scalpel. Half of the biopsy sample was homogenized in 400 μl of Ultraspec, while the remaining half was resuspended in 400 μl of DNA lysis buffer. Samples were stored at −80°C until use. For all samples in Ultraspec, the RNA extraction, DNase treatment, and reverse transcription procedures were performed as reported previously (39). Treponemal burden in lesions from immunized and control animals was determined using a previously developed RT-qPCR approach that targets the tp0574 gene mRNA (encoding the 47-kDa lipoprotein) and normalizes it to the message for rabbit hypoxanthine phosphoribosyltransferase (HPRT) (39). Construction of the tp0574 standard and amplification conditions were also previously reported (39). All samples resuspended in DNA lysis buffer were extracted using the Qiagen DNA minikit. Data from message quantification and treponemal counts from aspirates were analyzed with Student’s unpaired two-tailed t test, and significance was set at a P value of ≤0.05. At the end of the 30-day observation period, rabbits were bled to test sera by VDRL and FTA-ABS tests and then euthanized.
Quantification of tp0126 mRNA.
The tp0126 message level was analyzed using a relative quantification protocol with external standards that normalizes tp0126 message to tp0574 reference gene mRNA levels as previously described (24). To obtain the tp0126 standard, a 201-bp target sequence within the tp0126 ORF was amplified from Nichols strain DNA using primers (5′-CGCAAGTGTGCAGTACCATT and 5′-GCCTTGAGCACAAGACCGTA) and cloned into the pCR IV-TOPO vector (Life Technologies) as previously described (24). The same primers were used for message quantification. For each sample analyzed, quadruplicate amplifications were performed for both the tp0126 and tp0574 genes. Data from message quantification were analyzed with Student’s unpaired two-tailed t test, and significance was set at a P value of ≤0.05.
Fluorescent fragment length analysis of the tp0126-associated poly(G) repeat.
A 275-bp amplicon containing the tp0126-associated poly(G) tract was obtained to perform fluorescent fragment length analysis (FLA) as previously described (24) and to assess the different lengths of the poly(G) tracts and their relative proportions. Briefly, amplification was performed using a 6-carboxyfluorescein (FAM)-labeled sense primer (5′-FAM-ACGACATACCCCGCAGTC) and an antisense primer (5′-GTTTCTTGTGCAGCTCCCCACACTC) with no modifications but carrying a terminal “pigtail” sequence (GTTTCTT) to ensure the uniform addition of nontemplated A overhangs to all amplicons. Amplification products were purified using the QIAquick PCR purification kit (Qiagen), and concentrations were measured using an ND-1000 instrument (NanoDrop Technologies, Wilmington, DE). Samples were diluted to a 0.2-ng/μl final concentration. For FLA, 1 μl of each sample was mixed with 15.4 μl of Hi-Di formamide and 0.1 μl of an HD400 ROX-labeled DNA size marker (Life Technologies). Samples were transferred to a 96-well plate, denatured at 94°C for 2 min, stored on ice for 1 min, and loaded onto an ABI3730xl DNA analyzer (Life Technologies). Output data were analyzed using GeneMapper 4.0 software (Life Technologies). Amplicon length was determined by comparison to the ROX-labeled marker, and intensity was measured as the area under a peak. For each DNA sample, FLA was performed in triplicate using three independent amplification products obtained with the same template. For data analysis, the sum of the area underneath all peaks generated by amplicons with the same number of G’s was divided by the total area underneath all peaks.
Analysis of the humoral response to Tp0126 during experimental/natural infection and following rabbit immunization using rTp0126 and synthetic peptides.
Overlapping synthetic peptides (20-mers overlapping by 10 aa) (Table 1) were designed based upon the Tp0126 sequence from the Nichols strain and our previous work to identify the most likely start codon of the protein (24). Only peptide 19, which covers the protein COOH terminus, was synthesized as 15-mers (Table 1). Peptides were produced by GenScript (Piscataway, NJ). Upon reception, lyophilized peptides were rehydrated in sterile PBS plus 0.1% sodium azide to a stock solution of 200 μg/ml. The solubility of hydrophobic peptides was increased by adding dimethyl sulfoxide (DMSO) (4%, vol/vol) when needed. Reconstituted peptides were stored at −20°C until use. Recombinant Tp0126, rTp0574 (the T. pallidum 47-kDa lipoprotein), and synthetic peptides were used for ELISAs. Recombinant Tp0126 was obtained as described above, while rTp0574 (the 47-kDa lipoprotein) was purchased from ViroStat (Portland, ME) to be used as a positive-control antigen with sera from experimentally infected rabbits. Recombinant antigens and peptides were used to coat the wells of a 96-well ELISA EIA II Plus microplate (ICN, Irvine, CA). Plates containing 0.5 μg/well of rTp0126, rTp0574, or a synthetic peptide in 100 μl of sterile PBS–0.1% sodium azide were incubated at 37°C for 2 h and subsequently at 4°C overnight to ensure antigen binding to the test wells. Plates were then washed three times with PBS plus 0.05% Tween 20 and blocked with 200 μl of 3% nonfat dry milk (NFM) in sterile PBS for 1 h. At this step, NFM was used to block no-antigen wells for background signals. Wells were then washed again as described above, and primary antibody was applied immediately thereafter. Sera obtained from infected rabbits at the three time points (days 30, 60, and 90) postinoculation were pooled to obtain three serum specimens, each representative of one time point. Pooled sera were diluted 1:20 in PBS plus 1% NFM, and 100 μl of sera was added to peptide-containing or no-antigen wells and incubated at room temperature overnight. Sera obtained from immunized animals 3 weeks after the last boost were also pooled and adsorbed against a sonicated E. coli lysate to minimize reactivity to possible contaminants carried over during rTP0126 purification. Sera were first titrated against rTp0126 and subsequently used with synthetic peptides at the first dilution (1:5,000) that generated a signal not exceeding the maximum detectable range of the instrument with full-length rTp0126. Human sera (n = 7; kindly provided by Francesco Drago, University of Genoa, Italy) were used at a 1:20 dilution. After incubation with the primary antibody, plates were washed again as described above, and 100 μl of a goat anti-rabbit IgG(H+L) alkaline phosphatase conjugate (Sigma) or goat anti-human IgG (also from Sigma) diluted 1:2,000 in PBS–1% NFM was added to each well and incubated for 3 h at room temperature. Plates were washed and developed with 50 μl/well of 1 mg/ml para-nitrophenyl phosphate (pNPP) substrate (Sigma), dissolved in glycine buffer (0.1 M glycine [pH 10.4], 1 mM MgCl2, and 1 mM ZnCl2) for 1 h according to the manufacturer’s instructions. Absorbance was measured as the OD405 using a Molecular Devices SpectraMax Plus microplate reader (Molecular Devices, San Jose, CA). The mean for triplicate experimental wells minus the mean for no-antigen wells tested with the same serum was calculated for all proteins/peptides tested. Plotted data represent the means ± standard errors of the means (SEM) for triplicate wells tested. Results were analyzed using Student’s t test, with significance set at a P value of ≤0.05.
Opsonophagocytosis assay.
The opsonic activity of the antiserum raised against rTp0126 was tested as previously described (40). Briefly, 2 × 106 proteose peptone-induced rabbit peritoneal macrophages were incubated for 4 h on coverslips with 107 freshly extracted Nichols treponemes and 10% (final concentration) NRS. Additionally, 1% (final concentration) anti-Tp0126 test antiserum (obtained from Tp0126-immunized animals) was added to cultures to assess its opsonic ability. Pooled sera from three rabbits infected long-term (90 days) with T. pallidum (IRS) were used as positive controls to promote treponemal phagocytosis. Following incubation with treponemes and test/control sera, macrophages were washed, fixed with ethanol, stained by indirect immunofluorescence for T. pallidum, and examined in a blind manner by fluorescence microscopy for the presence of fluorescein-labeled antigens within phagocytic vacuoles. Triplicate specimens were prepared under each condition, and 100 macrophages were counted for each coverslip. The results presented represent data from three separate experiments, each using a different macrophage donor rabbit. Mean values (±SEM) for the percentages of macrophages ingesting T. pallidum were calculated and compared for different antisera by Student’s t test, with significance set at a P value of ≤0.05.
T-cell proliferation assay.
Splenic lymphocytes were harvested, as previously described (41), from rabbits at day 90 after experimental i.t. infection. Approximately 5 × 105 cells in 200 μl of culture medium were plated in quadruplicate for each animal and each recombinant antigen/synthetic peptide in flat-bottomed 96-well plates (Corning, Corning, NY) and incubated at 37°C in a 5% CO2 atmosphere. rTp0126 (2.5 μg) or synthetic peptides (2.5 μg) were added as test antigens, while 0.5 μg/well concanavalin A (ConA; ICN Pharmaceuticals, Costa Mesa, CA) was used as a positive control. PBS alone (no-antigen wells) was used to measure background reactivity. Three days after exposure to rTp0126, synthetic peptides, or ConA, T cells were pulsed with 0.5 μCi [3H]thymidine per well and harvested after 24 h to measure tritiated thymidine incorporation using a Wallac Microbeta Trilux microplate scintillation analyzer (Perkin-Elmer, Waltham, MA). The geometric mean for quadruplicate wells with no antigens (determined as the background value) was compared to the geometric means for the quadruplicate experimental wells. Data represent the geometric mean (±SEM) log10 counts per minute for each antigen for three rabbits. Data were analyzed with Student’s unpaired two-tailed t test, and significance was set at a P value of ≤0.05.
ACKNOWLEDGMENTS
This work was supported by the University of Washington Royalty Research Fund grant (to L.G.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We are grateful to Kerry Laing and Barbara Molini for assistance with some of the experimental procedures and to Francesco Drago (University of Genoa, Italy) for providing deidentified patient sera.
We remember Arturo Centurion-Lara, who passed away prematurely in July 2018, for the conversations on the topic of the manuscript. We miss you greatly.
REFERENCES
- 1.Radolf JD, Lukehart SA (ed). 2006. Pathogenic Treponema: molecular and cellular biology. Caister Academic Press, Norfolk, England. [Google Scholar]
- 2.World Health Organization. 2011. Prevalence and incidence of selected sexually transmitted infections Chlamydia trachomatis, Neisseria gonorrhoeae, syphilis and Trichomonas vaginalis: methods and results used by WHO to generate 2005 estimates. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Newman L, Kamb M, Hawkes S, Gomez G, Say L, Seuc A, Broutet N. 2013. Global estimates of syphilis in pregnancy and associated adverse outcomes: analysis of multinational antenatal surveillance data. PLoS Med 10:e1001396. doi: 10.1371/journal.pmed.1001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerbase AC, Rowley JT, Mertens TE. 1998. Global epidemiology of sexually transmitted diseases. Lancet 351(Suppl 3):2–4. [DOI] [PubMed] [Google Scholar]
- 5.Anonymous. 2017. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390:1211–1259. doi: 10.1016/S0140-6736(17)32154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. 2016. Report on global sexually transmitted infection surveillance 2015. WHO Document Production Services, Geneva, Switzerland. [Google Scholar]
- 7.Savage EJ, Marsh K, Duffell S, Ison CA, Zaman A, Hughes G. 2012. Rapid increase in gonorrhoea and syphilis diagnoses in England in 2011. Euro Surveill 17(29):pii=20224 https://www.eurosurveillance.org/content/10.2807/ese.17.29.20224-en. [PubMed] [Google Scholar]
- 8.Savage EJ, Hughes G, Ison C, Lowndes CM. 2009. Syphilis and gonorrhoea in men who have sex with men: a European overview. Euro Surveill 14(47):pii=19417 https://www.eurosurveillance.org/content/10.2807/ese.14.47.19417-en. [DOI] [PubMed] [Google Scholar]
- 9.Simms I, Fenton KA, Ashton M, Turner KM, Crawley-Boevey EE, Gorton R, Thomas DR, Lynch A, Winter A, Fisher MJ, Lighton L, Maguire HC, Solomou M. 2005. The re-emergence of syphilis in the United Kingdom: the new epidemic phases. Sex Transm Dis 32:220–226. doi: 10.1097/01.olq.0000149848.03733.c1. [DOI] [PubMed] [Google Scholar]
- 10.Tucker JD, Cohen MS. 2011. China’s syphilis epidemic: epidemiology, proximate determinants of spread, and control responses. Curr Opin Infect Dis 24:50–55. doi: 10.1097/QCO.0b013e32834204bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2018. Sexually transmitted disease surveillance 2016. Centers for Disease Control and Prevention, Atlanta, GA: https://demystifyingmedicine.od.nih.gov/dm18/m01d23/reading01.pdf. [Google Scholar]
- 12.LaFond RE, Lukehart SA. 2006. Biological basis for syphilis. Clin Microbiol Rev 19:29–49. doi: 10.1128/CMR.19.1.29-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamb ML, Newman LM, Riley PL, Mark J, Hawkes SJ, Malik T, Broutet N. 2010. A road map for the global elimination of congenital syphilis. Obstet Gynecol Int 2010:312798. doi: 10.1155/2010/312798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan NX, Rydzak C, Yang LG, Vickerman P, Yang B, Peeling RW, Hawkes S, Chen XS, Tucker JD. 2013. Prioritizing congenital syphilis control in south China: a decision analytic model to inform policy implementation. PLoS Med 10:e1001375. doi: 10.1371/journal.pmed.1001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldenberg RL, Thompson C. 2003. The infectious origins of stillbirth. Am J Obstet Gynecol 189:861–873. doi: 10.1067/S0002-9378(03)00470-8. [DOI] [PubMed] [Google Scholar]
- 16.Workowski KA, Berman SM. 2011. Centers for Disease Control and Prevention sexually transmitted disease treatment guidelines. Clin Infect Dis 53(Suppl 3):S59–S63. doi: 10.1093/cid/cir694. [DOI] [PubMed] [Google Scholar]
- 17.Centers for Disease Control and Prevention. 1999. The national plan to eliminate syphilis in the United States. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 18.World Health Organization. 2007. The global elimination of congenital syphilis: rationale and strategy for action. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 19.Baker-Zander SA, Lukehart SA. 1992. Macrophage-mediated killing of opsonized Treponema pallidum. J Infect Dis 165:69–74. doi: 10.1093/infdis/165.1.69. [DOI] [PubMed] [Google Scholar]
- 20.Lukehart SA. 1982. Activation of macrophages by products of lymphocytes from normal and syphilitic rabbits. Infect Immun 37:64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukehart SA, Miller JN. 1978. Demonstration of the in vitro phagocytosis of Treponema pallidum by rabbit peritoneal macrophages. J Immunol 121:2014–2024. [PubMed] [Google Scholar]
- 22.Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG, Dodson R, Gwinn M, Hickey EK, Clayton R, Ketchum KA, Sodergren E, Hardham JM, McLeod MP, Salzberg S, Peterson J, Khalak H, Richardson D, Howell JK, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton MD, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith HO, Venter JC. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 23.Cox DL, Luthra A, Dunham-Ems S, Desrosiers DC, Salazar JC, Caimano MJ, Radolf JD. 2010. Surface immunolabeling and consensus computational framework to identify candidate rare outer membrane proteins of Treponema pallidum. Infect Immun 78:5178–5194. doi: 10.1128/IAI.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giacani L, Brandt SL, Ke W, Reid TB, Molini BJ, Iverson-Cabral S, Ciccarese G, Drago F, Lukehart SA, Centurion-Lara A. 2015. Transcription of TP0126, Treponema pallidum putative OmpW homolog, is regulated by the length of a homopolymeric guanosine repeat. Infect Immun 83:2275–2289. doi: 10.1128/IAI.00360-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Centurion-Lara A, Castro C, Barrett L, Cameron C, Mostowfi M, Van Voorhis WC, Lukehart SA. 1999. Treponema pallidum major sheath protein homologue Tpr K is a target of opsonic antibody and the protective immune response. J Exp Med 189:647–656. doi: 10.1084/jem.189.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giacani L, Molini BJ, Kim EY, Godornes BC, Leader BT, Tantalo LC, Centurion-Lara A, Lukehart SA. 2010. Antigenic variation in Treponema pallidum: TprK sequence diversity accumulates in response to immune pressure during experimental syphilis. J Immunol 184:3822–3829. doi: 10.4049/jimmunol.0902788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hazlett KRO, Sellati TJ, Nguyen TT, Cox DL, Clawson ML, Caimano MJ, Radolf JD. 2001. The TprK protein of Treponema pallidum is periplasmic and is not a target of opsonic antibody or protective immunity. J Exp Med 193:1015–1026. doi: 10.1084/jem.193.9.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Centurion-Lara A, Giacani L, Godornes C, Molini BJ, Brinck Reid T, Lukehart SA. 2013. Fine analysis of genetic diversity of the tpr gene family among treponemal species, subspecies and strains. PLoS Negl Trop Dis 16:e2222. doi: 10.1371/journal.pntd.0002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anand A, LeDoyt M, Karanian C, Luthra A, Koszelak-Rosenblum M, Malkowski MG, Puthenveetil R, Vinogradova O, Radolf JD. 2015. Bipartite topology of Treponema pallidum repeat proteins C/D and I: outer membrane insertion, trimerization, and porin function require a C-terminal beta-barrel domain. J Biol Chem 290:12313–12331. doi: 10.1074/jbc.M114.629188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giacani L, Lukehart S, Centurion-Lara A. 2007. Length of guanosine homopolymeric repeats modulates promoter activity of subfamily II tpr genes of Treponema pallidum ssp. pallidum. FEMS Immunol Med Microbiol 51:289–301. doi: 10.1111/j.1574-695X.2007.00303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun ES, Molini BJ, Barrett LK, Centurion-Lara A, Lukehart SA, Van Voorhis WC. 2004. Subfamily I Treponema pallidum repeat protein family: sequence variation and immunity. Microbes Infect 6:725–737. doi: 10.1016/j.micinf.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Morgan CA, Lukehart SA, Van Voorhis WC. 2002. Immunization with the N-terminal portion of Treponema pallidum repeat protein K attenuates syphilitic lesion development in the rabbit model. Infect Immun 70:6811–6816. doi: 10.1128/IAI.70.12.6811-6816.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan CA, Lukehart SA, Van Voorhis WC. 2003. Protection against syphilis correlates with specificity of antibodies to the variable regions of Treponema pallidum repeat protein K. Infect Immun 71:5605–5612. doi: 10.1128/IAI.71.10.5605-5612.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller JN. 1973. Immunity in experimental syphilis. VI. Successful vaccination of rabbits with Treponema pallidum, Nichols strain, attenuated by g-irradiation. J Immunol 110:1206–1215. [PubMed] [Google Scholar]
- 35.Cameron CE, Lukehart SA. 2014. Current status of syphilis vaccine development: need, challenges, prospects. Vaccine 32:1602–1609. doi: 10.1016/j.vaccine.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parveen N, Fernandez MC, Haynes AM, Zhang RL, Godornes BC, Centurion-Lara A, Giacani L. 2019. Non-pathogenic Borrelia burgdorferi expressing Treponema pallidum TprK and Tp0435 antigens as a novel approach to evaluate syphilis vaccine candidates. Vaccine 37:1807–1818. doi: 10.1016/j.vaccine.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 38.Lukehart SA, Marra CM. 2007. Isolation and laboratory maintenance of Treponema pallidum. Curr Protoc Microbiol Chapter 12:Unit 12A.1. doi: 10.1002/9780471729259.mc12a01s7. [DOI] [PubMed] [Google Scholar]
- 39.Giacani L, Molini B, Godornes C, Barrett L, Van Voorhis WC, Centurion-Lara A, Lukehart SA. 2007. Quantitative analysis of tpr gene expression in Treponema pallidum isolates: differences among isolates and correlation with T-cell responsiveness in experimental syphilis. Infect Immun 75:104–112. doi: 10.1128/IAI.01124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaffer JM, Baker-Zander SA, Lukehart SA. 1993. Opsonization of Treponema pallidum is mediated by immunoglobulin G antibodies induced only by pathogenic treponemes. Infect Immun 61:781–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukehart SA, Baker-Zander SA, Sell S. 1980. Characterization of lymphocyte responsiveness in early experimental syphilis. I. In vitro response to mitogens and Treponema pallidum antigens. J Immunol 124:454–460. [PubMed] [Google Scholar]