Brucella is an intracellular bacterial pathogen that causes chronic systemic infection in domesticated livestock and poses a zoonotic infectious risk to humans. The virulence of Brucella is critically dependent on its ability to replicate and survive within host macrophages. Brucella modulates host physiological pathways and cell biology in order to establish a productive intracellular replicative niche. Conversely, the host cell presumably activates pathways that limit infection.
KEYWORDS: Brucella neotomae, MAP kinase, autophagy, brucellosis, intracellular growth, p38, phagosome, type IV secretion system
ABSTRACT
Brucella is an intracellular bacterial pathogen that causes chronic systemic infection in domesticated livestock and poses a zoonotic infectious risk to humans. The virulence of Brucella is critically dependent on its ability to replicate and survive within host macrophages. Brucella modulates host physiological pathways and cell biology in order to establish a productive intracellular replicative niche. Conversely, the host cell presumably activates pathways that limit infection. To identify host pathways contributing to this yin and yang during host cell infection, we performed a high-throughput chemical genetics screen of known inhibitors and agonists of host cell targets to identify host factors that contribute to intracellular growth of the model pathogen Brucella neotomae. Using this approach, we identified the p38 mitogen-activated protein (MAP) kinase pathway and autophagy machinery as both a linchpin and an Achilles’ heel in B. neotomae’s ability to coopt host cell machinery and replicate within macrophages. Specifically, B. neotomae induced p38 MAP kinase phosphorylation and autophagy in a type IV secretion system-dependent fashion. Both p38 MAP kinase stimulation and an intact autophagy machinery in turn were required for phagosome maturation and intracellular replication. These findings contrasted with those for Legionella pneumophila, where chemical inhibition of the p38 MAP kinase pathway and autophagy factor depletion failed to block intracellular replication. Therefore, results from a chemical genetics screen suggest that intersections of the MAP kinase pathways and autophagy machinery are critical components of Brucella’s intracellular life cycle.
INTRODUCTION
Brucella is a facultative, intracellular, Gram-negative pathogen that causes chronic systemic infection in mammals. Humans become infected through ingestion of contaminated milk, percutaneous blood exposure, or inhalation of aerosolized organisms from birthing farm animals. Infection is associated with prolonged and debilitating fever, hepatosplenomegaly, lymphadenopathy, and potentially life-threatening sequelae, including endocarditis, osteomyelitis, and meningitis (1).
The VirB-based type IV secretion system (T4SS) is a major virulence determinant of Brucella spp. (2–5) and is required for intracellular replication in host macrophages. VirB mutants also show dramatically reduced virulence in experimental models. The VirB operon encodes a multiprotein molecular syringe, induced by phagosomal acidification, that translocates effector proteins from the bacterial cytoplasm across the phagosomal membrane into the host cell cytoplasm. Effectors have multiple effects on the host cell that ultimately result in altered maturation of the bacterial phagosome and establishment of a productive intracellular growth niche.
After internalization, Brucella-containing vacuoles (BCVs) mature along an endosomal route marked by the recruitment of late endosomal markers, Rab7 and lysosome-associated membrane protein 1 (LAMP-1) (6, 7). The BCV then diverges from canonical endosomal maturation, taking on properties of the endoplasmic reticulum (ER), where intracellular replication begins (3, 8, 9). In contrast, the BCVs of T4SS mutants fail to avoid end-stage endosomal maturation, and bacteria are degraded within mature lysosomal compartments.
The relationship between autophagy and host cell infection remains to be fully defined. For example, Brucella melitensis 16M was found to induce the maturation of microtubule-associated protein 1A/1B light chain 3-I (LC3-I) to LC3-II and the corresponding formation of LC3 puncta (10), markers of autophagosome formation. Furthermore, early BCVs were enclosed in multiple membranes, a morphological feature of autophagosomes, and 3-methyladenine, an indirect autophagy inhibitor, blocked intracellular replication. Similarly, Brucella abortus strain 1308 was found to colocalize, early during infection, with the autophagosome marker monodansylcadaverine (11), and intracellular replication was similarly inhibited by 3-methyladenine (6, 12). However, although B. abortus BCVs were noted to have multilaminar features suggestive of autophagosomes late during infection (9), an association with LC3 was not evident. Furthermore, deletion or small interfering RNA (siRNA) knockdown of many but not all autophagy machinery components still supported formation of multilaminar BCVs, and intracellular replication was not affected (9).
We reasoned that we could use a chemical genetics screen to identify host pathways coopted by Brucella to orchestrate these curious cellular events. For this screen, we turned to the model Brucella pathogen Brucella neotomae. B. neotomae, which natively infects desert pack rats, is highly related to select-agent Brucella species (B. melitensis, B. abortus, and B. suis) and also shares similar host cell infection dynamics and type IV secretion system-dependent intracellular replication and pathogenesis (5, 13). However, in contrast to B. melitensis, B. abortus, and B. suis, it is not a select agent and is categorized as a biosafety level 2 pathogen, making it a useful model for high-throughput experiments. Therefore, B. neotomae was used to screen a large number of known bioactive agents to identify host cell pathways relevant to intracellular infection. Analysis of screening hits led to the identification of the p38 mitogen-activated protein (MAP) kinase (MAPK) pathway as a critical mediator of T4SS-dependent cell intracellular replication at least in part through regulation of autophagy-dependent BCV formation.
RESULTS
Chemical genetics screen identifies pathways that inhibit intracellular growth of B. neotomae.
A high-throughput screen was developed to identify inhibitors of B. neotomae intracellular growth in THP-1 human macrophages. Although direct bacterial growth inhibition assays were initially considered, they did not offer sufficient robustness in a high-throughput screening (HTS) format. Instead, we took advantage of the novel observation of the ability of B. neotomae to kill and permeabilize THP-1 macrophages after a productive intracellular growth cycle. To enable real-time, nondestructive quantification of host cell death, we incorporated the impermeant nucleic acid binding dye SYTOX green in the assay medium, as we previously described (14–16). SYTOX green permeates dead cells and binds to nuclear DNA. The large increase in fluorescence upon DNA binding provided a real-time, quantitative readout of host cell death and, therefore, an indirect measure of B. neotomae intracellular replicative capacity.
Assay optimization in a 384-well plate format resulted in a mean Z′ of 0.70 ± 0.05 comparing azithromycin-treated wells (positive growth inhibition control, i.e., no cytotoxicity) with untreated control wells (negative control) at 48 h postinfection in three separate experiments (see Fig. S1A in the supplemental material). A total of 9,685 known bioactive compounds were then screened in duplicate for anticytotoxicity activity indicative of reduced bacterial growth. Z-scores between two replicates showed high correlation (R2 = 0.83) (Fig. S1B). Compound libraries included known kinase inhibitors, G-protein-coupled receptor inhibitors and agonists, antimicrobials, chemotherapeutics, ion-channel-active agents, and FDA-approved drugs with a variety of mechanisms of action.
Hits (n = 1,717) were categorized based on the average replicate Z-scores as strong (Z ≥ 7) (n = 364), medium (5 ≤ Z < 7) (n = 543), or weak (3 ≤ Z < 5) (n = 810). After removal of duplicates, the remaining 236 strong hits were divided into seven broad functional categories based on mechanism of action (Table S1 and Fig. S1C). For the purposes of our study, we excluded known direct antimicrobials from further analysis, although we noted an unexpected activity of many prototypically Gram-positive antimicrobials (e.g., fusidic acid), a characteristic shared with other fastidious intracellular pathogens such as Legionella pneumophila (15). Furthermore, in contrast to L. pneumophila, we also noted a relative efficacy of polar antibiotics (aminoglycosides and β-lactams among strong hits), potentially suggesting increased access to Brucella intracellular compartments. We instead focused subsequent investigation on hits that may potentially limit intracellular growth through action on the host cell. Forty-nine remaining compounds with previously described host cell targets were advanced to secondary screening (Table S1).
Hits were first tested for their ability to inhibit intracellular growth using a recombinant B. neotomae strain constitutively expressing a bacterial luminescence operon. In previous studies, luminescence was found to correlate highly with intracellular CFU (5). However, the dynamic range of the luminescence assay was compressed relative to CFU counts (5). Although the luminescence assay in general provides an underestimate of the magnitude of intracellular replication compared with CFU data, it was used preferentially for most experiments described below as it provided a facile, real-time, non-host-cell-destructive way to examine large numbers of experimental variables.
Eleven compounds were confirmed as strong hits in a secondary orthogonal screen using this bacterial luminescence assay (Z-scores less than or equal to −7) and were the subject of further study in this work (Table S1). These hits consisted of eight kinase inhibitors, two ion channel modulators, and a proton pump inhibitor. Known activities of hits are as follows: CDK4/cyclin D1 and calcium/calmodulin-dependent protein kinase II inhibitor (arcyriaflavin A), PDK1 inhibitor (BX912), IκB kinase inhibitor (IKK-16), cyclin-dependent kinase (CDK)/glycogen synthase kinase 3β (GSK-3β) inhibitor (indirubin-3′-monoxime [I3M]), CSF1R inhibitor (Ki20227), JAK inhibitor (CYT387), Rho kinase inhibitor (Rhodblock6), Jun N-terminal protein kinase (JNK) inhibitor (SP600125), H+/K+-ATPase proton pump inhibitor (esomeprazole), small-conductance Ca2+-activated K+ channel blocker (NS8593), and ATP-sensitive K+ channel opener (pinacidil). Several additional hits were also confirmed (Table S1), in particular multiple adenosine receptor agonists in G-protein-coupled receptor libraries, and will be the subject of future work.
For subsequent analysis, J774A.1 murine macrophage host cells, except where otherwise indicated, were used preferentially for analysis based on simpler protocols for propagation that did not require differentiation using exogenous agents (calcitriol for THP-1 cells). Levels of intracellular growth inhibition in J774A.1 cells determined by CFU and luminescence measurements were highly correlated (Fig. 1A). As expected, all compounds significantly decreased both bacterial replication, as assessed by luminescence, and host cell cytotoxicity, as assessed by SYTOX green fluorescence, in the same assay wells (48 h postinfection; P < 0.05 by analysis of variance [ANOVA] and Dunnett’s post hoc comparisons to dimethyl sulfoxide [DMSO]-infected controls) during infection of both THP-1 and J774A.1 cells (Fig. S1D), using the same inhibitor concentrations as the ones in Fig. 1A. These linked observations for luminescence and fluorescence readouts are presumptively based on decreased intracellular replication-associated cytotoxicity, which was the foundation of the original high-throughput, chemical genetics screening assay. Similar significant inhibition of intracellular growth by all inhibitors was also observed during infection of primary murine bone marrow-derived macrophages (BMDM) (Fig. 1B) (P < 0.0001 for ANOVA and all post hoc comparisons), indicating that observations extend beyond continuous macrophage cell lines. To rule out effects on initial phagocytosis as an alternative explanation for reduced intracellular bacterial counts at 48 h postinfection, we assessed microscopically the median number of bacteria taken up by infected J774A.1 cells at 4 h postinfection in the presence of the same concentration of compounds (Fig. S1F). There was no significant difference in intracellular bacterial counts compared to DMSO controls (P > 0.99 in post hoc comparisons), except for a significant increase in uptake with arcyriaflavin A and pinacidil treatments (P = 0.0001 and P = 0.001, respectively). Therefore, decreased phagocytosis did not appear to contribute to observed intracellular growth defects.
FIG 1.
Chemical genetics screen identifies pathways that inhibit intracellular growth of B. neotomae. (A) Inhibitory effects of selected small molecules on intracellular replication in J774A.1 host cells at 48 h postinfection. Both B. neotomae (Bn) luminescence and CFU were measured in parallel. Data shown are means ± SD from at least two independent experiments, 3 replicates for luminescence data and 2 replicates for CFU data. Concentrations of the following tested compounds were in general above the IC50 (Table 1) and substantially below the CC50: arcyriaflavin A (3.6 μM), BX912 (3.6 μM), IKK16 (3.2 μM), I3M (3.6 μM), Ki20227 (2.0 μM), CYT387 (1.4 μM), Rhodblock6 (6.9 μM), SP600125 (10.2 μM), esomeprazole (10.2 μM), NS8593 (4.7 μM), and pinacidil (10.2 μM). (B) Inhibitory effects of selected small molecules on intracellular replication in primary murine bone marrow-derived macrophages (BMDM) at 48 h postinfection based on luminescence signals. Data shown are means ± SD from 4 replicates. Saponin (0.2%) and 0.3% DMSO in the absence of infection were used as positive and negative cytotoxicity controls, respectively. Significant suppression of luminescence was observed for all inhibitors in comparison with the DMSO control (P < 0.0001 by ANOVA and Dunnett’s post hoc multiple-comparison tests). (C) Effect of kinase target siRNA on intracellular growth of B. neotomae in J774A.1 host cells as assessed by luminescence. Data points represent means ± SD from at least three independent experiments. Significant suppression of luminescence (*, P < 0.001) was observed for siRNA knockdowns compared to the NTsi control. (D) Effect of siRNA knockdowns on mRNA expression assayed using same experimental protocol as the one described for panel C. Data points represent means ± SD from three independent samples and were normalized to the β-actin level within each sample and then to expression in untreated controls.
Hits showed various degrees of relative selectivity (Table 1) for intracellular growth inhibition compared with toxicity toward host cells based on ratios of the 50% cytotoxicity concentration (CC50) to the 50% intracellular growth inhibitory concentration (IC50). Furthermore, hits were also relatively selective for inhibiting intracellular bacterial growth compared with extracellular (axenic) replication in bacterial growth medium, with the exception of esomeprazole, SP600125, and NS8593, which had detectable axenic inhibitory activity at concentrations only 2- to 4-fold above the intracellular IC50. Complete axenic bacterial growth inhibition (i.e., MIC) was observed for only two compounds (NS8593 and indirubin-3-monoxime) at the highest concentration tested (100 μM). Therefore, inhibition of intracellular growth could not be accounted for by either host cell destruction (thereby eliminating the intracellular growth niche) or direct antibacterial activity, with rare exceptions. Instead, intracellular growth limitation was presumably caused by nonlethal perturbations to the host cell.
TABLE 1.
Compound potency, cytotoxicity, and selectivity
| Compound | Target | Mean IC50 (μM) ± SD in J774 cellsa | Mean IC50 (μM) ± SD in THP cellsa | Mean IC50 (μM) ± SD, axenicb | MIC (μM)d | Mean CC50 (μM) ± SD in J774 cellsa | Selectivityc
|
|
|---|---|---|---|---|---|---|---|---|
| Intracellular vs J774 cells | Axenic vs intracellular | |||||||
| Ki20227 | CSF1R | 0.3 ± 0.1 | 1.2 ± 0.5 | >100 | >100 | 45.8 ± 1.7 | 147.7 | >333 |
| CYT387 | JAK | 0.8 ± 0.3 | 0.5 ± 0.04 | >100 | >100 | 5.5 ± 0.3 | 6.8 | >125 |
| Arcyriaflavin A | CDK4 | 2.2 ± 0.3 | 2.1 ± 0.2 | >100 | >100 | >100 | >44.6 | >45.5 |
| Indirubin-3-monoxime | CDK/GSK-3β | 2.4 ± 0.1 | 0.13 ± 0.07 | 27.8 ± 4.5 | 100 | >100 | >42.6 | 11.6 |
| IKK16 | IκB kinase | 2.4 ± 0.4 | 5.0 ± 0.1 | >100 | >100 | 12.7 ± 0.3 | 5.2 | >41.7 |
| BX912 | PDK1 | 4.3 ± 1.1 | 1.5 ± 0.1 | >100 | >100 | 53.7 ± 9.0 | 12.6 | >23.3 |
| Rhodblock6 | Rho kinase | 11 ± 5.8 | 3.6 ± 1.8 | >100 | >100 | >100 | >9.2 | >9.1 |
| SP600125 | JNK | 9.3 ± 1.1 | 8.4 ± 4.9 | 19.3 ± 4.3 | >100 | >100 | >10.7 | 2.1 |
| NS8593 | K+ channel | 13.1 ± 0.4 | 3.8 ± 0.4 | 43.2 ± 6.1 | 100 | >100 | >7.6 | 3.3 |
| Pinacidil | K+ channel | >15 | 11.5 ± 2.2 | >100 | >100 | >100 | >6.7 | >6.7 |
| Esomeprazole | H+/K+-ATPase | 4.5 ± 0.1 | 3.8 ± 1.0 | 24.6 ± 2.9 | >100 | 54.8 ± 6.9 | 12 | 5.5 |
IC50, concentration that leads to 50% bacterial growth inhibition by a luminescence assay within either THP or J774A.1 cells; CC50, concentration that leads to 50% death of J774A.1 host cells as measured by a SYTOX green cytotoxicity assay.
Concentration that leads to bacterial growth inhibition of 50% by a luminescence assay in bacterial growth medium.
Selectivity is the CC50/IC50 ratio (relative effect on intracellular growth of B. neotomae compared to cytotoxicity for J774A.1 host cells) or the IC50 axenic/IC50 J774 ratio (relative effect on intracellular growth in J774A.1 host cells compared to axenic, extracellular growth).
MIC where there was completion inhibition of B. neotomae luminescence compared with the positive control (100 μg/ml gentamicin).
We reasoned that the kinase inhibitor hits, in particular, modulate host cell signaling pathways to limit intracellular growth. To verify the involvement of putative kinase inhibitor targets (our most selective hits), we tested effects of siRNA-mediated inhibition of CDK1, CDK4, CSF1R, and JAK1 expression on intracellular replication. These siRNA kinase knockdowns led to a significant 1.9- to 3.2-fold decrease (P < 0.001) in B. neotomae luminescence signals at 48 h postinfection (Fig. 1C) in comparisons to treatment with the commercially available nontargeting Dicer siRNA control (NTsi) (Table S2), which was determined bioinformatically by the manufacturer not to interact with the mouse or human transcriptome. Efficient suppression of mRNA expression by siRNA constructs was confirmed by quantitative reverse transcription-PCR (Fig. 1D). Screening hits such as proton pump inhibitors, e.g., esomeprazole, were initially intriguing and were repeatedly identified as weak to strong hits (data not shown). However, these drugs are concentrated in acidic phagosomes and presumptive direct bacterial inhibitors, as described for Helicobacter pylori (17), and were therefore not further studied.
MAPK p38 is induced by and modulates intracellular infection.
Based on the master regulatory role of several targeted kinases, we questioned whether their signaling would converge on common mitogen-activated protein kinase (MAPK) pathways that in turn would modulate B. neotomae infection. We first explored the relationship of MAP kinase activation and B. neotomae infection in J774A.1 cells. In particular, phosphorylation of p38 MAPK was stimulated at 4 h postinfection during wild-type (wt) B. neotomae infection, with further incremental stimulation at 24 h and 48 h (Fig. 2A), in comparison to uninfected controls. Phosphorylation of extracellular signal-regulated kinase (ERK), however, was not similarly stimulated. In contrast, ΔvirB4 mutant infection did not induce p38 phosphorylation.
FIG 2.
Effect of p38 MAPK inhibition on B. neotomae trafficking and intracellular growth. (A) Phosphospecific Western blots were performed after mock infection (uninfected) or infection of J774A.1 cells with the B. neotomae wild-type (B. neotomae) or ΔvirB4 mutant strain for the indicated times. (B) MAP kinase phosphorylation observed 48 h after infection of J774A.1 cells in the presence of target siRNA or NTsi. Results shown are representative of data from two independent experiments (see also Fig. S1 in the supplemental material). (C) Effects of SB203580 on MAPK signaling in B. neotomae-infected J774A.1 cells at 48 h postinfection. (D to F) For J774A.1 cells infected with B. neotomae (D), J774A.1 cells infected with L. pneumophila (Lp) (E), and THP-1 cells infected with B. neotomae (F), effects of p38 inactivation by SB203580 on intracellular growth were assessed based on bacterial luminescence. Data points represent means ± SD from assays performed in triplicate. (G) LAMP-1 colocalization with BCVs in J774A.1 cells treated with DMSO or SB203580. Data shown represent means ± SD obtained from two experiments, with at least 100 BCVs scored under each condition per experiment (P < 0.05 for 24-h and 48-h comparisons). (H) Colocalization of BCVs (red) with LAMP-1 (green) and mTurquiose2::calreticulin (blue) after treatment with SB203580 at the indicated times after infection of J774A.1 cells. Bars, 5 μm. For all panels, * designates a significant difference between treatment conditions and the DMSO control (see also Fig. S2 in the supplemental material).
To examine potential connections with inhibitor targets, we measured MAPK activity in the presence or absence of siRNA inhibition of targeted genes. At 48 h postinfection, B. neotomae markedly induced the phosphorylation of p38 (Fig. 2B) in host cells transfected with the NTsi control. In contrast, in host cells transfected with siRNAs targeting kinase target expression, B. neotomae-induced p38 phosphorylation was suppressed, while conversely, ERK phosphorylation was stimulated. We also examined the effects of the direct p38 MAPK pathway inhibitor SB203580. Similar to observed effects of high-throughput screening target inhibition, SB203580 suppressed infection-induced p38 phosphorylation, effects observable at concentrations of as low as 1 μM, with almost complete suppression at 10 μM, as assessed by Western blotting (Fig. 2C). ERK phosphorylation was also similarly stimulated at low concentrations of the inhibitor, while MEK phosphorylation was stimulated only at higher concentrations.
Based on p38 pathway stimulation during T4SS-competent infection, we tested whether the p38 inhibitor SB203580 would block intracellular bacterial replication (Fig. 2D). As little as 1 μM SB203580 was observed to inhibit intracellular growth by approximately 3-fold, as assessed by luminescence (P = 0.0001), and 100-fold, as assessed by CFU enumeration (Fig. S2A). Furthermore, by confocal microscopy, SB203580 decreased the mean area occupied by BCVs in confocal microscopy images by ∼10-fold at 48 h postinfection (Fig. S2B and S2D). However, at 24 h postinfection, the mean percentages of infected J774A.1 cells treated with either SB203580 or DMSO were not statistically different (Fig. S3C) (P = 0.86). These results suggested that inhibitor effects resulted from interference with intracellular replication rather than a defect in bacterial uptake.
Furthermore, growth inhibition appeared pathogen specific, as up to 10 μM SB203580 did not similarly affect intracellular growth of L. pneumophila in J774A.1 cells (Fig. 2E). THP-1 cells were less sensitive to inhibition, as 1 or 2 μM SB203580 did not affect intracellular growth, whereas 5 μM and 10 μM significantly decreased luminescence by ∼53% and 67%, respectively (Fig. 2F), perhaps explaining the nondetection of SB203580 during the chemical genetics high-throughput screening effort using the THP-1 cell line. Similar inhibition of luminescence was observed in J774A.1 cells using the structurally related inhibitor SB202190 (Fig. S2E) (IC50 of 4 μg/ml).
We next characterized phagosome maturation in the presence of SB203580. Previously, we noted that later during maturation, but before extensive replication, B. neotomae phagosomes excluded the late endosomal and lysosome-associated marker LAMP-1 and increased the association with the endoplasmic reticulum-associated marker calreticulin (5). Larger replicative BCVs (≥2 μm in diameter) at 24 and 48 h postinfection, however, demonstrated higher associations with LAMP-1, as discussed further below. SB203580 treatment increased the colocalization of small BCVs (<2 μm in diameter) with LAMP-1 at 24 and 48 h (Fig. 2G). At 48 h postinfection, SB203580 also reduced the colocalization of BCVs with calreticulin, a marker of the endoplasmic reticulum (Fig. 2H). These alterations in phagosome maturation were associated with suppression of intracellular replication such that BCVs contained one or only small numbers of organisms (Fig. 2H). Taken together, these observations suggested a potential link between eukaryotic signal transduction, particularly p38 stimulation, and T4SS-dependent phagosome maturation and intracellular growth of B. neotomae.
Autophagy is required for B. neotomae intracellular replication and trafficking.
As other Brucella species were noted to either induce autophagy or reside in phagosomes with morphological properties of autophagosomes (9, 10, 18), we next questioned whether screening hits may affect the relationship, if any, of B. neotomae with autophagy. Autophagy is associated with the processing of microtubule-associated protein 1A/1B light chain 3 (LC3) into a form that associates with autophagosomal membranes and coalescences into LC3 puncta observed by immunofluorescence microscopy (19, 20). Notably, B. neotomae infection significantly induced LC3 puncta at 6 h postinfection relative to either uninfected macrophages or host cells infected with the ΔvirB4 T4SS mutant (P < 0.0001) (Fig. 3A). In addition, although almost all BCVs overlapped LC3B at 6 h postinfection, induction of LC3 puncta appeared as a more global response, as many additional LC3 puncta were present throughout the cytoplasm that were not associated with BCVs (Fig. 3B). Interestingly, most confirmed hits significantly reduced the formation of LC3 puncta (P < 0.0001), with the exception of arcyriaflavin A, NS5893, and pinacidil (Fig. 3A and B). These results suggested the general convergence of kinase inhibitor hits on B. neotomae-induced autophagosome formation.
FIG 3.
Autophagy is required for B. neotomae intracellular replication and trafficking. (A) Mean numbers of LC3 puncta per infected J774A.1 host cell ± SD at 6 h postinfection, scored across at least 50 macrophages under each condition in two independent experiments. ui, uninfected; B. neotomae, cells infected with wild-type bacteria; ΔvirB4, cells infected with the T4SS mutant. All other conditions include cells infected with B. neotomae in the presence of the indicated compounds. * indicates a P value of 0.0001 compared to B. neotomae infection alone. (B) Colocalization of LC3 (green) and B. neotomae (red) at 6 h postinfection in J774A cells. Host cells were treated with DMSO or the indicated compounds, except for the single panel showing infection with the ΔvirB4 mutant. Bars, 5 μm. (C and D) For B. neotomae (C) or L. pneumophila (D), J774A.1 host cells treated with the indicated siRNAs were infected with the respective luminescent bacterial species. Data shown are means ± SD of relative bioluminescence for 4 replicates per data point. (E) Colocalization of B. neotomae (red) and LAMP-1 (green) 24 and 48 h after infection of J774A.1 cells. The inset shows a region, designated by arrows, enlarged to highlight colocalization events. Bars, 5 μm. (F) Time course of colocalization of B. neotomae in phagosomes of <2 μm in diameter with LAMP-1 during infection of siRNA-transfected J774A.1 host cells expressing LAMP-1::mTurquoise2. * designates a significant difference between all siRNA treatments and NTsi. (G) Colocalization of B. neotomae wild-type or ΔvirB4 BCVs (red) with LAMP-1 (green) and LC3B (blue) in J774A.1 host cells transfected with targeting siRNA or NTsi at the indicated times postinfection. Bars, 5 μm. (H) Mean percentages of J774A.1 cells ± SD containing any replicative BCVs (defined as phagosomes of ≥2 μm in diameter) colocalizing with LAMP-1 at 48 h postinfection, based on 100 scored host cells under each condition from quadruplicate independent experiments. (I) Mean percentage of replicative BCVs colocalizing with both LAMP-1 and LC3B ± SD 48 h after infection of J774A.1 cells, based on a minimum of 41 BCVs scored under each condition in triplicate independent experiments. * designates significant differences between siRNA and NTsi treatments (see also Fig. S3 in the supplemental material).
We next considered whether autophagy is required for intracellular replication. We transfected J774A.1 cells with siRNAs targeting ATG5, ATG12, LC3B, and Sequestome-1/p62, whose gene products play critical roles in autophagosome formation (21, 22). The first three are involved in elongation of the phagophore; LC3 plays additional roles in closure of the autophagosomal membrane and as a cargo adaptor (23); and p62, a docking protein, captures and delivers cellular content to autophagosomes (24) and has been further noted for its crucial role in targeting bacterial pathogens, including Salmonella enterica serovar Typhimurium, Shigella flexneri, and Listeria monocytogenes, to autophagosomes (25–27). Treatment with 30 nM each siRNA depleted the respective proteins to nearly undetectable levels by Western blotting (Fig. S3A). In these siRNA knockdowns, intracellular bacterial luminescence was significantly suppressed at 48 h postinfection by 2.65- to 3.16-fold (P = 0.0001) in comparisons with the NTsi control (Fig. 3C). Furthermore, the area occupied by BCVs was ∼10 times larger in the NTsi control than in autophagy knockdown host cells at 48 h postinfection (P < 0.001) (Fig. S3C). Importantly, at 4 h postinfection, the mean number of BCVs in infected J774A.1 hosts did not significantly differ between autophagy knockdown and the NTsi control (Fig. S3D) (P > 0.9), nor did the percentage of infected J774 cells differ at 24 h postinfection (P = 0.3 by ANOVA) (data not shown). Taken together, these results suggested that autophagy knockdown interfered with intracellular replication rather than initial infection of J774A host cells. In contrast, intracellular growth of L. pneumophila did not appear to be significantly affected at 48 h postinfection by the same siRNA knockdowns (Fig. 3D) (P = 0.3 by ANOVA), suggesting that the findings were largely specific to B. neotomae.
To determine whether autophagy deficiency altered subsequent phagosome maturation, we performed several assessments. Colocalization of B. neotomae wt and ΔvirB4 strains with LAMP-1 and LC3B was assessed by confocal microscopy over a 48-h time course. Under control conditions, LAMP-1 colocalization decreased, starting at 4 h postinfection, in nonreplicative or prereplicative BCVs. In contrast, in autophagy siRNA knockdowns, high LAMP-1 colocalization was relatively maintained at all time points (inclusive of nonreplicative/prereplicative BCVs), notable at the 24- and 48-h time points (Fig. 3E and F).
siRNA inhibition was also associated with the absence of induction of LC3 puncta induction and a lack of association of LC3B with BCVs at all time points (Fig. 3G). We also found that the percentage of J774A. 1 cells containing at least one replicative BCV (rBCV) (phagosomes of ≥2 μm) with LAMP-1 colocalization (9) was significantly lower in siRNA-treated host cells at 48 h postinfection (Fig. 3H and Fig. S3E). The frequency of colocalization of both LC3B and LAMP-1 with BCVs at 48 h postinfection was significantly reduced (Fig. 3I), associated with the corresponding loss of replicative capacity.
Linkage between p38 and autophagy pathways.
As T4SS-dependent growth of B. neotomae was similarly affected by inhibition of both autophagy and the p38 pathway, we next questioned whether these two processes might cooperatively enable intracellular infection. We first examined the effect of autophagy inhibition on MAPK activation. In siRNA autophagy-inhibited host cells, wild-type B. neotomae infection failed to induce p38 phosphorylation, while ERK and MEK phosphorylation was stimulated (48 h postinfection) (Fig. 4A). In contrast, phosphorylation of MAPK was unaffected by autophagy inhibition in the ΔvirB4 mutant and uninfected controls. Furthermore, the modest stimulation of p38 phosphorylation by tumor necrosis factor alpha (TNF-α) did not appear to be blocked by siRNA autophagy knockdowns (Fig. S4E). The data suggested that these autophagy-deficient host cells are not defective in response to all stimuli but defective in response to B. neotomae specifically. Both B. neotomae wt and ΔvirB4 infections modestly stimulated Akt serine 473 phosphorylation. However, autophagy suppression did not further modulate Akt phosphorylation levels. Therefore, autophagy suppression by siRNA and abolition of T4SS function both inhibited B. neotomae-induced p38 phosphorylation and intracellular growth, suggesting connections among p38 MAP kinase signaling, autophagy, and virulence.
FIG 4.

Linkage between p38 and autophagy pathways. (A) Changes in MAPK signaling in autophagy-defective J774A.1 cells at 48 h postinfection. (B) Colocalization of B. neotomae (red), LAMP-1::mTurquoise2 (pseudocolored green), and LC3B (pseudocolored blue) in J774A.1 cells at the indicated times postinfection with DMSO treatment or at 48 h postinfection with SB203580 treatment. Arrowheads highlight BCVs colocalizing with LC3B and LAMP-1. Arrows indicate regions magnified in insets. Bars, 5 μm. (C) Quantification of dual colocalization of LC3B and LAMP-1 with BCVs in J774A.1 host cells. Data points are the means ± SD from three separate experiments, and 100 colocalization events were scored under each condition per experiment. Colocalization was significantly reduced (*) by SB203580 treatment at all concentrations tested in post hoc comparisons with the DMSO control (P ≤ 0.0002).
We then examined the formation of LC3B puncta in the presence of the p38 inhibitor SB203580. p38 inhibition blocked the formation of LC3B puncta and decreased colocalization of B. neotomae with LC3B at all time points examined (Fig. 4B and C). Similar findings were observed in primary BMDM from BALB/c mice. LC3 puncta were induced by B. neotomae at 4 h postinfection (Fig. S4A and B), and LC3 was found to colocalize with BCVs at both early and late time points (Fig. S4A and D). In turn, intracellular bacterial growth, formation of LC3 puncta, mean BCV area, and LC3B and LAMP-1 colocalization with BCVs were significantly suppressed by treatment of BMDM with 10 μM SB203850 (Fig. S4A to D). Therefore, linkage of intracellular growth, autophagy, and the p38 MAP kinase pathways was also maintained during infection of primary macrophages.
Rab7 is required for BCV maturation, p38 phosphorylation, and intracellular growth.
Previously, Rab7, a GTPase involved in late endosome and autophagosome maturation (28), was implicated as a required factor in maturation of the B. abortus BCV into a replicative phagosome (8). Therefore, we examined the relationship of Rab7 with B. neotomae infection. High Rab7 colocalization with BCV was noted at 6 h postinfection (∼75% ± 8%), as assessed by confocal microscopy (Fig. 5A and B and Fig. S5A). However, treatment with 2 μM SB203580 reduced this colocalization by half (Fig. 5B) (P = 0.0001). There was similarly high Rab7 colocalization with ΔvirB4 BCVs at 6 h postinfection (P = 0.99) and a not significantly different level of inhibition by SB203580 (P = 0.3). Therefore, T4SS status did not appear to affect Rab7 recruitment.
FIG 5.
Rab7 is required for BCV maturation, p38 phosphorylation, and intracellular growth. (A) Representative confocal microscopy images of Rab7 (green) colocalization with BCVs (red) at 6 h postinfection in J774A.1 cells treated with DMSO or SB203580. Bars, 5 μm. (B) Mean percent Rab7 colocalization with BCVs ± SD in J774A.1 cells from three to four independent experiments, with at least 70 BCVs scored under each condition per experiment. Horizontal bars indicate the two groups of conditions, each of whose members are significantly different (P ≤ 0.001 by Tukey’s multiple-comparison test [*]) in pairwise comparisons to each member of the second group, i.e., for all pairwise comparisons of colocalization between DMSO and SB203580 treatment conditions during either B. neotomae or ΔvirB4 mutant infection. In contrast, there was no significant difference in colocalization between B. neotomae and ΔvirB4 mutant infections under either DMSO or SB203580 treatment conditions. (C) Mean bacterial luminescence ± SD from three independent experiments using J774A.1 host cells treated with different concentrations of Rab7si. (D) Representative confocal images of BCVs (red) colocalizing with LC3B (green) and mTurquiose2::calreticulin (blue) in J774A.1 host cells transfected with 50 nM NTsi or Rab7si. Bars, 5 μm. (E) Representative images of BCVs (red) colocalizing with LAMP-1 (green) and mTurquiose2::calreticulin (blue) at 48 h postinfection in host cells transfected with 50 nM NTsi or Rab7si. Bars, 5 μm. (F) Mean percent colocalization of LAMP-1 with BCVs ± SD 48 h after infection of J774A.1 host cells from three independent experiments, with >100 BCVs analyzed under each condition per experiment (*, P < 0.01 for Rab7 siRNA compared with the control). (G) Effect of Rab7si transfection on MAPK signaling and autophagy protein expression 48 h after infection of J774A.1 host cells. NTsi was transfected at 50 nM (see also Fig. S4 in the supplemental material).
To investigate the contribution of Rab7 to intracellular replication, we infected J774A host cells transfected with Rab7 siRNA (Rab7si) and the NTsi-treated control. Rab7si transfection substantially reduced Rab7 protein, as assessed by Western blotting (Fig. 5G), and in turn significantly suppressed B. neotomae intracellular replication at 48 h postinfection by up to 70%, as assessed by luminescence (P < 0.01 for all comparisons with NTsi) (Fig. 5C). By confocal microscopy, Rab7 siRNA treatment suppressed the formation of LC3 puncta during both early and late infection (Fig. 5D), the association of BCVs with LC3 at all time points (Fig. S5B and D), and the high association of replicative BCVs with calreticulin notable at 24 h postinfection (Fig. 5D). Conversely, Rab7 siRNA treatment significantly enhanced (P = 0.01) colocalization with LAMP-1 at late time points (Fig. 5E and F). It further reduced p38 phosphorylation and reciprocally stimulated ERK and MEK phosphorylation (Fig. 5G). It also induced the accumulation of p62 and depletion of the 56-kDa ATG5-ATG12 conjugate (detected with anti-ATG5 antibody), associated with autophagy inhibition (29, 30). Therefore, Rab7, p38, and autophagy proteins appear to act in a mutually cross-stimulatory and dependent fashion to promote phagosome maturation and intracellular growth of B. neotomae.
DISCUSSION
A chemical genetics screening approach identified host pathways important for intracellular replication of B. neotomae. Several classes of small molecules with known biological function were identified that significantly inhibited intracellular replication without affecting the viability of host cells. Inhibitor targets potentially could act at the host-pathogen interface and therefore might provide insight into the Brucella intracellular life cycle.
The identification of several kinase inhibitors suggested potential convergence of common signal transduction pathways on Brucella biology. In particular, we found that B. neotomae both stimulated phosphorylation of p38 throughout infection and was dependent on p38 activation for intracellular growth. Furthermore, induction of p38 phosphorylation by B. neotomae was dependent on an intact T4SS, suggesting links among T4SS, p38, and intracellular growth.
Our results are consistent with the previously observed dependence of intracellular growth of B. melitensis on p38 and JNK pathways in primary human monocytes based on suppressive effects of the inhibitors SB203850 and SP600125, respectively (31), although we also noted here direct antibacterial activity of SP600125, which may have been contributory. Notably, several strains of B. abortus, B. melitensis, and B. suis were also noted previously to stimulate p38 phosphorylation in J774 cells (32, 33), although activation was examined only during the first 4 h postinfection for all species and up to 24 h postinfection for B. suis, rather than throughout an extended 48-h infection period as in our studies. These previous investigations noted that the lipopolysaccharide O-chain was not responsible for this stimulation, while the dependence on T4SS was not examined. Nevertheless, the stimulation of p38 across the genus suggests potential generalizability of our findings, a presumption that would need to be confirmed in future experiments with highly pathogenic, zoonotic species.
Of particular interest was the additional tripartite association of p38 phosphorylation, autophagy, and intracellular growth. B. neotomae was noted to induce LC3 puncta within 4 h postinfection. This was similar to previous descriptions of induction of LC3 puncta for B. melitensis and B. abortus. Here we also found that p38 inhibition; treatment with kinase inhibitors, with the exception of arcyriaflavin A; and infection with a T4SS mutant all abrogated the formation of LC3 puncta, suggesting that autophagy induction was a downstream cell biological link between T4SS-dependent host signaling and maturation of a replicative intracellular growth niche. Furthermore, supporting autophagy as a key process intertwined with B. neotomae pathogenesis, siRNA knockdowns of autophagy proteins substantially inhibited intracellular growth.
However, contradictory evidence for the importance of autophagy during Brucella infection was described previously. Specifically, B. abortus was shown to form replicative phagosomes in HeLa cells depleted for autophagy proteins, ATG5, LC3, and ULK1, using siRNA knockdowns; in bone marrow-derived macrophages isolated from cre-floxed C57BL/6J mice deleted for ATG5 and ATG16; and in ATG4B-null mice (9). Furthermore, intracellular growth of B. melitensis in mouse embryonic fibroblasts was unaffected by deletion of ATG5, with a concomitant absence of the formation of LC3 puncta at the 10-h time point reported, as assessed by visualization of green fluorescent protein (GFP)-LC3 (18).
The differing potential contributions of autophagy to successful infection, both positive for B. neotomae, B. melitensis (10), and B. abortus (6, 12) and indifferent for B. abortus (9) and B. melitensis (18), ultimately may relate to a number of factors, including bacterial species or strain, host cell type, and experimental variables. For example, the intracellular biology of Brucella in a cervical carcinoma cell line (HeLa) and embryonic fibroblasts may be very different from that in macrophages, presumably the predominant site of organism multiplication in mammals. In macrophages isolated from floxed mice, cre expression was induced from the lysosome M locus that is turned on during macrophage differentiation (34). However, it is possible that Atg5 and Atg16 were deleted from only a subset of the macrophage population (9), without reported assessment of deletion efficiency, potentially affecting observations. Furthermore, Atg4B knockouts (Atg4B is one of four related Atg4 paralogs) are only partially defective for autophagy (35). In addition, siRNA inhibition in this and previous studies may also have been partial and transient, especially during later time points using transiently transfected host cells. Of note, we observed both recapitulation of p38-dependent induction of LC3 puncta and abrogation of intracellular growth with p38 inhibition during infection of primary bone marrow-derived macrophages from BALB/c mice, indicating that our observations were not limited to continuous cell lines. Our observations are in contrast to those of Starr et al., who observed what were called autophagosomal BCVs (aBCVs) at later stages of infection with B. abortus 2308 (9). These phagosomes colocalized with LAMP-1 and were associated with multilaminar membranes morphologically suggestive of autophagosomes (9). However, in contrast to our findings, these aBCVs did not colocalize with LC3B, generally considered a defining feature of autophagosomes (36). Taken together, these data indicate that the role of autophagy in Brucella infection may be nuanced.
Mechanistically, our observations of p38-dependent autophagy induction by B. neotomae are consistent with previously observed relationships of p38 and autophagy. For example, in RAW macrophages, interferon gamma (IFN-γ)-induced autophagy was blocked by treatment with SB203850 and also, interestingly, by JAK inhibition, reminiscent of our JAK inhibitor screening hit, CYT397 (37). Furthermore, p38 itself was found to phosphorylate LAMP-2A in lysosomes and thereby activate autophagic machinery (38). In addition, ER stress-induced autophagy induction was found to be a p38-dependent process (38–40). The JNK pathway was also found to stimulate autophagy (41) and may coordinately activate autophagy along with p38 in response to stress. Although not investigated here, the identification of the screening hit SP600125, a known JNK inhibitor, is potentially suggestive of an additional contributory role of this alternative MAP kinase pathway in B. neotomae-induced autophagy, with caveats of potential direct antibacterial activity mentioned above.
We believe that the chemical genetics screen proved to be a powerful approach to identify pathways operating at the pathogen-host interface. There were additional pathways of interest that will be the subject of future studies and were not investigated here in the interest of focus, for example, the strong hit sirolimus that inhibits the mTOR pathway, further potential biology of interest related to the several cell cycle hits identified, and multiple adenosine G-protein-coupled receptor agonistics. One potential implication of the observed intersection with major cellular signaling pathways is that B. neotomae replication is somehow tethered to and perhaps takes advantage of the physiological state of the host cell.
The contrast with the relative insusceptibility of L. pneumophila to cell pathway inhibition is instructive. Although both organisms replicate within an endoplasmic reticulum-associated intracellular niche, the pathways to that niche and the niches themselves are markedly different. L. pneumophila avoids passage through an endosomal, low-pH compartment, and L. pneumophila-containing vacuoles (LCVs) depart from canonical endosomal maturation at the point of uptake (42). In contrast, Brucella requires acidified endosomal passage for T4SS induction and subsequent phagosomal maturation (2). Perhaps reflective of these differences, we noted contrasting physiological dependencies in this study, notably that siRNA-mediated autophagy protein inhibition and p38 inhibition by SB203850 had minimal to no effect on L. pneumophila replication, in marked contrast to their effects on B. neotomae. Our findings are consistent with previous observations indicating that L. pneumophila produces effectors that inhibit rather than promote autophagy (43). Furthermore, Rab7si did not affect intracellular replication of L. pneumophila (42), in contrast to the substantial inhibition noted for B. neotomae and previously for B. abortus (8). The latter observations may merely reflect a delay or reduced progression to a late endosomal niche that induces the VirB T4SS (44) rather than a direct role in phagosome maturation; however, it is possible that known additional contributions of Rab7 to autophagosome formation may also be contributory (28). Finally, intracellular B. neotomae appeared more sensitive to polar antibiotics than L. pneumophila, based on screening results for aminoglycosides and β-lactams, which essentially have no activity against intracellular growth of L. pneumophila (15). These results suggest that these antibiotics have greater access to intracellular Brucella, perhaps based on a prolonged association of phagosomes with endosomal compartments during Brucella’s intracellular life cycle.
Additionally, differences in outputs from chemical genetics screens between B. neotomae and L. pneumophila are also worthy of comment. We performed a screen for inhibition of L. pneumophila intracellular growth against the same set of known bioactives (16). In contrast to the current B. neotomae screen, we identified essentially no compound other than known antimicrobials that inhibited intracellular growth in the L. pneumophila screen (L. Chiaraviglio and J. E. Kirby, unpublished data). These observations likely reflect the extreme degeneracy in effector function previously noted for L. pneumophila, in which any one blow to a participatory host pathway is compensated for by redundant mechanisms (45). These differences also likely explain the capacity of L. pneumophila to complement T4SS-defective B. neotomae but not the converse (5). Lacking this degeneracy, B. neotomae provided an excellent model for discovery at the pathogen-host interface using a chemical genetic approach. A larger screen of chemical libraries with compounds of unknown function may potentially identify novel therapeutics that would disrupt infection by this high-consequence genus and could potentially be leveraged to reduce the prolonged treatment course required for organism eradication.
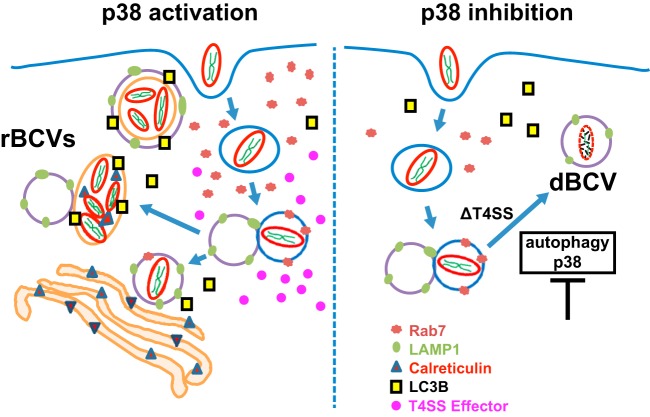
Based on observations in this study, we propose a model for the B. neotomae intracellular life cycle (Fig. 6). Specifically, B. neotomae traffics through the early and late endosomes, the latter associated with Rab7 recruitment. During this transit, B. neotomae presumptively induces a VirB4-dependent BCV transformation associated with p38 phosphorylation and autophagosome formation. MAP kinase induction and autophagosomal maturation appear to be at least partially mutually dependent events that are required for the formation of a productive intracellular replicative niche in macrophages. Taken together, these results highlight B. neotomae as a tractable model whose intracellular replicative capacity is inherently sensitive to host cell biology and signal transduction dynamics.
FIG 6.
Model of B. neotomae intracellular infection. BCVs initially mature along the endocytic pathway, a process dependent on Rab7 and associated with accumulation of LAMP-1. Endosomal maturation induces the expression of the VirB T4SS. The ensuing translocation of virulence factors into the host cell stimulates p38 phosphorylation and autophagy. MAP kinase induction and autophagolysosomal maturation are associated with the formation of a replicative BCV (rBCV). Association with endoplasmic reticulum compartments occurs at an early stage of rBCV maturation but for B. neotomae appears to be superseded by autophagosomal transformation of the fully developed rBCV. If such maturation is inhibited either by blocking p38 phosphorylation either directly (SB203580) or indirectly (depleting Rab7 [interference with upstream signal transduction events or T4SS deletion]) or by blocking autophagy either directly (autophagy protein knockdown) or indirectly (p38 inhibition or T4SS deletion), terminal endocytic maturation into a lysosomal, degradative BCV (dBCV) occurs instead. Core signal transduction pathways downstream of several hits identified in a chemical genetics screen link Brucella replicative capacity with the host cell physiological state.
MATERIALS AND METHODS
Bacterial strains, cell lines, and reagents.
The tdTomato fluorescent B. neotomae strain 5K33 (5) or luminescent strains of B. neotomae (5) and Legionella pneumophila (46) were used for the detection of intracellular growth of B. neotomae or L. pneumophila in macrophages. B. neotomae and L. pneumophila strains were passaged at 37°C in a humidified 5% CO2 incubator on Trypticase soy broth (TSB) medium (BD, Franklin Lakes, NJ) supplemented with 50 μg/ml of nourseothricin and on buffered charcoal yeast extract (BCYE) medium (47) including 100 μg/ml of thymidine, respectively.
THP-1 human monocyte and J774A.1 murine macrophage cell lines, both obtained from the American Type Culture Collection (Manassas, VA), were grown in RPMI 1640 (Corning, Tewksbury, MA) supplemented with 10% fetal bovine serum (FBS) (Gemini Bio-Products, West Sacramento, CA) and iron-supplemented calf serum (Gemini Bio-Products), respectively.
For all studies, infections were performed at an estimated multiplicity of infection (MOI) of 1 bacterium per plated macrophage. Key reagents are listed in Table 2.
TABLE 2.
Strains and key resources used in this studya
| Reagent or resource | Source or reference | Catalog no. or identifier |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-p38 antibody | Cell Signaling Technology | 9212 |
| Rabbit monoclonal anti-p-p38 antibody (Thr180/Tyr182) | Cell Signaling Technology | 9211 |
| Rabbit monoclonal anti-p44/42 (ERK1/2) antibody | Cell Signaling Technology | 9102 |
| Rabbit monoclonal anti-p-p44/42 (ERK1/2) antibody (Thr202/Tyr204) | Cell Signaling Technology | 9101 |
| Rabbit monoclonal anti-MEK1/2 antibody | Cell Signaling Technology | 9122 |
| Rabbit monoclonal anti-p-MEK1/2 antibody (Ser217/221) | Cell Signaling Technology | 9121 |
| Rabbit monoclonal anti-Akt antibody | Cell Signaling Technology | 9272 |
| Rabbit monoclonal anti-p-Akt antibody (Ser473) | Cell Signaling Technology | 9271 |
| Rabbit monoclonal anti-ATG5 antibody | Cell Signaling Technology | 2630 |
| Rabbit monoclonal anti-ATG12 antibody | Cell Signaling Technology | 2011 |
| Rabbit monoclonal anti-LC3 antibody | Cell Signaling Technology | 2775 |
| Rabbit monoclonal anti-p62 antibody | Cell Signaling Technology | 5114 |
| Rabbit monoclonal anti-β-actin antibody | Cell Signaling Technology | 4967 |
| Rat monoclonal anti-LAMP-1 antibody (1DB4) | Santa Cruz | sc-19992 |
| Mouse monoclonal anti-Rab7 antibody (B-3) | Santa Cruz | sc-376362 |
| Mouse monoclonal anti-cathepsin D antibody (D-7) | Santa Cruz | sc-377299 |
| Goat anti-rabbit IgG(H+L)-Alexa Fluor 488 | Invitrogen, Thermo Fisher | A-11070 |
| Goat anti-mouse IgG(H+L)-Alexa Fluor 488 | Invitrogen, Thermo Fisher | A-11017 |
| Chicken anti-rat IgG(H+L)-Alexa Fluor 488 | Invitrogen, Thermo Fisher | A-21470 |
| Goat anti-rabbit IgG(H+L)-Alexa Fluor 350 | Invitrogen, Thermo Fisher | A-11069 |
| Goat anti-rabbit IgG(H+L) secondary antibody-HRP | Thermo Fisher | 31460 |
| Goat anti-mouse IgG(H+L) cross-adsorbed secondary antibody-HRP | Thermo Fisher | 31432 |
| Protease/phosphatase inhibitor cocktail (100×) | New England Biolabs | 5872 |
| Small molecules | ||
| Arcyriaflavin A | Santa Cruz | sc-202470 |
| BX912 | Sigma | SML1086-5MG |
| IKK16 (IKK inhibitor VII) | APExBIO | B1586 |
| Indirubin-3-monoxime | Enzo | BML-CC207-0001 |
| KI20227 | Enzo | ENZ-CHM204-0001 |
| CYT387 (momelotinib) | APExBIO | A4143 |
| Rhodblock6 | Sigma | R1283-5MG |
| SP600125 | Sigma | S5567-10MG |
| Esomeprazole | Fisher | 50-148-627 |
| NS8593 | Alomone Labs | N-196 |
| Pinacidil | Santa Cruz | sc-203198 |
| SB202190 | Cayman | 10010399 |
| TNF-α | ProSpec | Cyt-252 |
| Critical commercial kits | ||
| Pierce BCA protein assay kit | Thermo Fisher | 23225 |
| SuperSignal West Femto maximum-sensitivity substrate | Thermo Fisher | 34095 |
| Lipofectamine RNAiMax transfection reagent | Thermo Fisher | 13778100 |
| Bacterial strains | ||
| B. neotomae | BEI Resources | 5K33 |
| B. neotomae-Lux | 5 | NA |
| B. neotomae-TdTomato | 5 | NA |
| Legionella pneumophila | ||
| L. pneumophila 02fla-Lux | 46 | NA |
| Experimental models: cell lines | ||
| J774A.1 | ATCC | ATCC TIB-67 |
| J774A.1(mTurquoise2::calreticulin) | 5 | NA |
| J774A.1(LAMP-1::mTurquoise2) | 5 | NA |
| THP-1 | ATCC | ATCC TIB-202 |
| L-929 | ATCC | ATCC CCL-1 |
| Software | ||
| GraphPad Prism 7.0 | GraphPad Software | |
| Image Lab | Bio-Rad | |
| Inkscape 0.92.3 | Inkscape | |
| Zen2.1 | Zeiss | |
| Illustrator CS6 | Adobe | |
See Tables S2 and S3 in the supplemental material for detailed oligonucleotide lists. NA, not applicable.
High-throughput screening.
The day before infection, 15,000 THP-1 cells, treated with 100 nM calcitriol (Cayman Chemical, Ann Arbor, MI), were seeded into 384-well, white microplates (catalog number 3570; Corning) in a 30-μl well volume. Immediately preceding infection, robotic pin transfer of 100 nl of screening compounds dissolved in DMSO stock solutions was performed using a custom Epson robotic platform in duplicate, with final compound concentrations usually at either 4 μg/ml or 20 μM. Library arrays were obtained from the ICCB-Longwood collection of known bioactive compounds from Biomol (now Enzo Life Sciences, Farmingdale, NY), Microsource (Gaylordsville, CT), Sigma-Aldrich (St. Louis, MO), Biofocus (Saffron Walden, UK), Enzo Life Sciences (Farmingdale, NY), Prestwick (Illkirch, France), and Chembridge (San Diego, CA).
Host cells were then infected with B. neotomae at an estimated MOI of 1 for a total well volume of 60 μl with a final SYTOX green concentration of 200 nM and incubated for 2 days. A 50-μg/ml final concentration of azithromycin was used as a positive control for complete B. neotomae growth suppression. The negative control was medium with 0.3% DMSO instead of test compounds.
Host cell viability was determined at 2 days postinfection by quantifying SYTOX green (Thermo Fisher Scientific, Waltham, MA) fluorescence using an Envision multiplate reader (PerkinElmer, Waltham, MA), with excitation and emission filters of 485/14 and 535/25 nm, respectively, using a dichroic mirror with a cutoff of 505 nM, as described previously (15). The Z′ statistic was calculated as Z′ factor = 1 − [3 × (σp + σn)]/|μp − μn|, with the mean (μ) and standard deviation (SD) (σ) calculated for 20 positive-control (p) and negative-control (n) wells, respectively, from three independent experiments. A Z′ value of between 0.5 and 1.0, as observed during our assay validation, is generally considered to indicate a robust screening assay (48). The Z-score was calculated as Z-score = (fluorescence of test well − μp)/σp, with μp and σp determined from 16 positive controls on the same screening plate and averaged for duplicate assays to prioritize compounds for further analysis.
For secondary screening using cherry picks of library compounds, plates were set up similarly except that intracellular replication was monitored directly by measurement of bacterial luminescence rather than indirectly through effects on host cell viability. Percent intracellular growth inhibition (see Table S1 in the supplemental material) was normalized to values for positive (azithromycin) and negative (DMSO) growth controls, and Z-scores were calculated as described above for the primary high-throughput screen.
Dose-dependent growth inhibitor activity or cytotoxicity for J774A.1 cells.
Dose-response analysis was performed using 2-fold serial dilutions prepared with the previously validated HP D300 digital dispensing system (49). Final concentrations of each compound ranged from 0.075 μM to 100 μM. Bacterial growth was measured using luminescence, and cytotoxicity was measured using SYTOX green fluorescence, as previously described (14).
B. neotomae or L. pneumophila infection of cultured macrophages for microwell assays.
J774A.1 cells were plated in a 50-μl volume at 105 cells/well in 96-well plates (Greiner Bio-One, Monroe, NC) in RPMI 1640 medium containing 9% iron-supplemented calf serum and incubated for 24 h. Wells were washed two times with phosphate-buffered saline (PBS) without Ca2+ or Mg2+. Luminescent B. neotomae or L. pneumophila cells were added to an MOI of 1. After 4 h, 100 μg/ml of gentamicin was added for 1 h to kill extracellular bacteria. Wells were then washed with PBS three times to remove antibiotic, and fresh RPMI medium was added to each well. Intracellular growth was monitored by measurement of luminescence periodically or by lysis of host cells with 0.1% saponin and CFU enumeration after plating serial dilutions on solid medium.
Colocalization of B. neotomae with organelle markers.
To investigate BCV maturation, macrophages were plated in 12-well plates containing 1.5 thickness, 12-mm-round coverslips (Warner Instruments, Hamden, CT) at a density of 2 × 105 cells per well. After 24 h, host cells were infected with B. neotomae constitutively expressing the tdTomato fluorescent protein. Plates were immediately centrifuged at 930 × g for 10 min to synchronize bacterial uptake and incubated at 37°C with 5% CO2. Gentamicin treatment was performed as described above, and host cells were incubated for the indicated times and then fixed with 4% paraformaldehyde for 30 min at room temperature. Cell membranes were permeabilized with PBS containing 0.3% Triton X-100 for 15 min at room temperature and then blocked with PBS containing 2% goat serum (Gemini Bio-Products, West Sacramento, CA). The permeabilized cells were probed with primary antibodies (1:100) and then incubated with Alexa Fluor-conjugated secondary antibody (1:300) for 1 h at 37°C. Coverslips were mounted with ProLong gold (Invitrogen, Carlsbad, CA) and visualized with a Zeiss LSM 880 confocal microscope (Carl Zeiss Microscopy, Thornwood, NY) using enhanced green fluorescent protein (eGFP) settings for detecting Alexa Fluor 488, tdTomato settings for tdTomato, DAPI (4′,6-diamidino-2-phenylindole) settings for Alexa Fluor 350, and enhanced cyan fluorescent protein (eCFP) settings for mTurquoise2. The image analysis program Zen 2.1 (Zeiss, Jena, Germany) was used for colocalization analysis. For rBCV (replicative BCV) quantification, only BCVs of >2 μm in diameter were scored in order to standardize comparisons.
Isolation and culture of mouse bone marrow-derived macrophages.
BMDM were isolated from femurs of 6-week-old BALB/c mice (Charles River Laboratories, Wilmington, MA) as previously described (50). Briefly, bone marrow was initially cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 9% FBS, glutamine, nonessential amino acids, and 20% L-929 cell culture supernatant for 6 days in non-tissue-culture-treated petri dishes. BMDM were collected in chilled PBS and seeded in 12-well plates at 5 × 105 cells per well. After overnight incubation, cells were infected for colocalization experiments as described above.
siRNA-mediated silencing in J774A.1 cells.
For siRNA knockdown, specific siRNA oligonucleotides (listed in Table S2) were designed on the Integrated DNA Technologies (IDT) website, except for the negative control, which was purchased from Integrated DNA Technologies (Skokie, IL), and Rab7si, which was purchased from Thermo Fisher Scientific. J774A.1 cells were plated in 6-well plates for Western blotting, 12-well plates for immunofluorescence, or 96-well plate for intracellular growth experiments at a density of 5 × 105, 2 × 105, or 105 cells per well, respectively. Cells were treated with 1 to 50 nM siRNA using Lipofectamine RNAiMax transfection reagent (Invitrogen, CA) according to the manufacturer’s protocol. After 72 h, transfected J774A.1 cells were washed with PBS three times and infected with B. neotomae expressing tdTomato or the bacterial luminescence operon.
Alternatively, RNA was purified from transfected cells using the RNeasy minikit (Qiagen, Germantown, MD) for reverse transcriptase quantitative PCR (qPCR) analysis. Equivalent amounts of cellular RNA were analyzed based on Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA) measurements. cDNA synthesis was performed using Moloney murine leukemia virus (M-MuLV) reverse transcriptase with a random primer mix (New England Biolabs, New Bedford, MA). The DyNAmo HS SYBR green qPCR kit (Thermo Fisher Scientific) was used for qPCR reactions. Cycling and detection were performed on a CFX 384 real-time PCR detection system (Bio-Rad, Hercules, CA), with an initial 95°C denaturation step for 5 min followed by 40 cycles of 95°C for 15 s and 60°C for 40 s. The relative expression level of mRNA in transfectants compared to controls was estimated from the cycle threshold (CT) values using the 2−ΔΔCT method (51), which includes normalization to the β-actin mRNA expression level within each sample.
Western blotting.
J774A.1 macrophage cells were seeded in 6-well plates at a density of 106 cells per well and then infected. After 4 h, extracellular bacteria were killed with gentamicin as described above. At 48 h postinfection, the host cell lysate was prepared in 1× radioimmunoprecipitation assay (RIPA) buffer (Cell Signaling Technology, Danvers, MA) containing 1× protease/phosphatase inhibitor cocktail (Cell Signaling Technology). Cell lysates were stored for 30 min on ice and centrifuged at 12,000 × g for 15 min at 4°C to remove insoluble material. Protein was quantified with the Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific). Whole-cell lysates were then boiled for 5 min in sample loading buffer, and 5 to 10 μg of cellular protein per well was loaded onto a 8 to 16% precast SDS-PAGE gradient gel (Bio-Rad, Hercules, CA), electrophoresed at 100 V for 90 min, and then transferred using a Mini-Transblot cell (Bio-Rad) to a 0.45-μm nitrocellulose membrane (Bio-Rad) at 50 to 100 mA for 2 h.
Membranes were blocked with 5% bovine serum albumin (BSA) (Santa Cruz Biotechnology, Dallas, TX) in Tris-buffered saline with 0.1% Tween 20 for 1 h at room temperature and incubated with primary antibodies (1:1,000 in BSA blocking buffer) at 4°C overnight. Following incubation with horseradish peroxidase (HRP)-conjugated secondary antibody (1:10,000 in 5% nonfat dried milk blocking buffer) for 1.5 h at room temperature, membranes were immersed in SuperSignal West Femto maximum-sensitivity substrate (Thermo Fisher Scientific) for 5 min and imaged on a Bio-Rad ChemiDoc system.
Statistical analysis.
Graphing and statistical analysis were performed using GraphPad Prism 7 (GraphPad Software, La Jolla, CA). Unpaired Student’s t test was used for two-group comparisons, analysis of variance (ANOVA) followed by Dunnett’s post hoc test was used for all multigroup comparisons to control conditions, and ANOVA followed by Tukey’s post hoc test was used for pairwise multigroup comparisons (used only in Fig. 5B). When ANOVA and post hoc methods were used, significant P values for post hoc comparisons are reported only if the ANOVAs were also significant. Statistical details can be found in relevant sections of Results and in the figure legends. A P value of <0.05 was considered statistically significant.
Ethics statement.
Murine experimental work was approved by the Beth Israel Deaconess Medical Center IACUC, protocol number 097-2017. All vertebrate animal experiments were performed according to the NIH Guide for the Care and Use of Laboratory Animals (52).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01AI099122 to J.E.K. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank all staff at the ICCB-Longwood Screening Facility with their help and guidance in performing high-throughput screening work. We thank Katelyn E. Zulauf, Kenneth P. Smith, and Lucius Chiaraviglio for helpful comments on the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00044-19.
REFERENCES
- 1.Young EJ. 1995. An overview of human brucellosis. Clin Infect Dis 21:283–289; quiz, 290. doi: 10.1093/clinids/21.2.283. [DOI] [PubMed] [Google Scholar]
- 2.Boschiroli ML, Ouahrani-Bettache S, Foulongne V, Michaux-Charachon S, Bourg G, Allardet-Servent A, Cazevieille C, Liautard JP, Ramuz M, O’Callaghan D. 2002. The Brucella suis virB operon is induced intracellularly in macrophages. Proc Natl Acad Sci U S A 99:1544–1549. doi: 10.1073/pnas.032514299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Celli J, de Chastellier C, Franchini DM, Pizarro-Cerda J, Moreno E, Gorvel JP. 2003. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J Exp Med 198:545–556. doi: 10.1084/jem.20030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanna N, Jiménez de Bagüés MP, Ouahrani-Bettache S, El Yakhlifi Z, Köhler S, Occhialini A. 2011. The virB operon is essential for lethality of Brucella microti in the BALB/c murine model of infection. J Infect Dis 203:1129–1135. doi: 10.1093/infdis/jiq163. [DOI] [PubMed] [Google Scholar]
- 5.Kang YS, Kirby JE. 2017. Promotion and rescue of intracellular Brucella neotomae replication during coinfection with Legionella pneumophila. Infect Immun 85:e00991-16. doi: 10.1128/IAI.00991-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pizarro-Cerdá J, Méresse S, Parton RG, van der Goot G, Sola-Landa A, Lopez-Goñi I, Moreno E, Gorvel JP. 1998. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect Immun 66:5711–5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaves-Olarte E, Guzmán-Verri C, Méresse S, Desjardins M, Pizarro-Cerdá J, Badilla J, Gorvel JP, Moreno E. 2002. Activation of Rho and Rab GTPases dissociates Brucella abortus internalization from intracellular trafficking. Cell Microbiol 4:663–676. doi: 10.1046/j.1462-5822.2002.00221.x. [DOI] [PubMed] [Google Scholar]
- 8.Starr T, Ng TW, Wehrly TD, Knodler LA, Celli J. 2008. Brucella intracellular replication requires trafficking through the late endosomal/lysosomal compartment. Traffic 9:678–694. doi: 10.1111/j.1600-0854.2008.00718.x. [DOI] [PubMed] [Google Scholar]
- 9.Starr T, Child R, Wehrly TD, Hansen B, Hwang S, López-Otin C, Virgin HW, Celli J. 2012. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe 11:33–45. doi: 10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo F, Zhang H, Chen C, Hu S, Wang Y, Qiao J, Ren Y, Zhang K, Wang Y, Du G. 2012. Autophagy favors Brucella melitensis survival in infected macrophages. Cell Mol Biol Lett 17:249–257. doi: 10.2478/s11658-012-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biederbick A, Kern HF, Elsasser HP. 1995. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur J Cell Biol 66:3–14. [PubMed] [Google Scholar]
- 12.Pizarro-Cerdá J, Moreno E, Sanguedolce V, Mege JL, Gorvel JP. 1998. Virulent Brucella abortus prevents lysosome fusion and is distributed within autophagosome-like compartments. Infect Immun 66:2387–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang YS, Brown DA, Kirby JE. 2019. Brucella neotomae recapitulates attributes of zoonotic human disease in a murine infection model. Infect Immun 87:e00255-18. doi: 10.1128/IAI.00255-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiaraviglio L, Kang YS, Kirby JE. 2016. High throughput, real-time, dual-readout testing of intracellular antimicrobial activity and eukaryotic cell cytotoxicity. J Vis Exp 2016:e54841. doi: 10.3791/54841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiaraviglio L, Kirby JE. 2015. High-throughput intracellular antimicrobial susceptibility testing of Legionella pneumophila. Antimicrob Agents Chemother 59:7517–7529. doi: 10.1128/AAC.01248-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiaraviglio L, Kirby JE. 2014. Evaluation of impermeant, DNA-binding dye fluorescence as a real-time readout of eukaryotic cell toxicity in a high throughput screening format. Assay Drug Dev Technol 12:219–228. doi: 10.1089/adt.2014.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sjøstrøm JE, Kühler T, Larsson H. 1997. Basis for the selective antibacterial activity in vitro of proton pump inhibitors against Helicobacter spp. Antimicrob Agents Chemother 41:1797–1801. doi: 10.1128/AAC.41.8.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamer I, Goffin E, De Bolle X, Letesson JJ, Jadot M. 2014. Replication of Brucella abortus and Brucella melitensis in fibroblasts does not require Atg5-dependent macroautophagy. BMC Microbiol 14:223. doi: 10.1186/s12866-014-0223-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshimori T. 2004. Autophagy: a regulated bulk degradation process inside cells. Biochem Biophys Res Commun 313:453–458. doi: 10.1016/j.bbrc.2003.07.023. [DOI] [PubMed] [Google Scholar]
- 21.Davis S, Wang J, Ferro-Novick S. 2017. Crosstalk between the secretory and autophagy pathways regulates autophagosome formation. Dev Cell 41:23–32. doi: 10.1016/j.devcel.2017.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaur J, Debnath J. 2015. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol 16:461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- 23.Schaaf MBE, Keulers TG, Vooijs MA, Rouschop KMA. 2016. LC3/GABARAP family proteins: autophagy-(un)related functions. FASEB J 30:3961–3978. doi: 10.1096/fj.201600698R. [DOI] [PubMed] [Google Scholar]
- 24.Lee YK, Lee JA. 2016. Role of the mammalian ATG8/LC3 family in autophagy: differential and compensatory roles in the spatiotemporal regulation of autophagy. BMB Rep 49:424–430. doi: 10.5483/BMBRep.2016.49.8.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, van der Goot FG, Sansonetti PJ, Lafont F. 2009. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe 6:137–149. doi: 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, Kakizuka A, Sztul E, Chakraborty T, Sasakawa C. 2009. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol 11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 27.Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. 2009. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol 183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 28.Hyttinen JM, Niittykoski M, Salminen A, Kaarniranta K. 2013. Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim Biophys Acta 1833:503–510. doi: 10.1016/j.bbamcr.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 29.Bjorkoy G, Lamark T, Pankiv S, Overvatn A, Brech A, Johansen T. 2009. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol 452:181–197. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 30.Sharifi MN, Mowers EE, Drake LE, Macleod KF. 2015. Measuring autophagy in stressed cells. Methods Mol Biol 1292:129–150. doi: 10.1007/978-1-4939-2522-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dimitrakopoulos O, Liopeta K, Dimitracopoulos G, Paliogianni F. 2013. Replication of Brucella melitensis inside primary human monocytes depends on mitogen activated protein kinase signaling. Microbes Infect 15:450–460. doi: 10.1016/j.micinf.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Jiménez de Bagüés MP, Gross A, Terraza A, Dornand J. 2005. Regulation of the mitogen-activated protein kinases by Brucella spp. expressing a smooth and rough phenotype: relationship to pathogen invasiveness. Infect Immun 73:3178–3183. doi: 10.1128/IAI.73.5.3178-3183.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reyes AW, Arayan LT, Simborio HL, Hop HT, Min W, Lee HJ, Kim DH, Chang HH, Kim S. 2016. Dextran sulfate sodium upregulates MAPK signaling for the uptake and subsequent intracellular survival of Brucella abortus in murine macrophages. Microb Pathog 91:68–73. doi: 10.1016/j.micpath.2015.10.024. [DOI] [PubMed] [Google Scholar]
- 34.Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, Schmidt RE, Mizushima N, Deretic V, Sibley LD, Virgin HW. 2008. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cabrera S, Marino G, Fernandez AF, Lopez-Otin C. 2010. Autophagy, proteases and the sense of balance. Autophagy 6:961–963. doi: 10.4161/auto.6.7.13065. [DOI] [PubMed] [Google Scholar]
- 36.Tanida I, Ueno T, Kominami E. 2008. LC3 and autophagy. Methods Mol Biol 445:77–88. doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- 37.Matsuzawa T, Kim B-H, Shenoy AR, Kamitani S, Miyake M, MacMicking JD. 2012. IFN-γ elicits macrophage autophagy via the p38 MAPK signaling pathway. J Immunol 189:813–818. doi: 10.4049/jimmunol.1102041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W, Zhu J, Dou J, She H, Tao K, Xu H, Yang Q, Mao Z. 2017. Phosphorylation of LAMP2A by p38 MAPK couples ER stress to chaperone-mediated autophagy. Nat Commun 8:1763. doi: 10.1038/s41467-017-01609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonnella R, Granato M, Farina A, Santarelli R, Faggioni A, Cirone M. 2015. PKC theta and p38 MAPK activate the EBV lytic cycle through autophagy induction. Biochim Biophys Acta 1853:1586–1595. doi: 10.1016/j.bbamcr.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 40.Kim D-S, Kim J-H, Lee G-H, Kim H-T, Lim JM, Chae S-W, Chae H-J, Kim H-R. 2010. p38 mitogen-activated protein kinase is involved in endoplasmic reticulum stress-induced cell death and autophagy in human gingival fibroblasts. Biol Pharm Bull 33:545–549. doi: 10.1248/bpb.33.545. [DOI] [PubMed] [Google Scholar]
- 41.Zhou YY, Li Y, Jiang WQ, Zhou LF. 2015. MAPK/JNK signalling: a potential autophagy regulation pathway. Biosci Rep 35:e00199. doi: 10.1042/BSR20140141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoffmann C, Finsel I, Otto A, Pfaffinger G, Rothmeier E, Hecker M, Becher D, Hilbi H. 2014. Functional analysis of novel Rab GTPases identified in the proteome of purified Legionella-containing vacuoles from macrophages. Cell Microbiol 16:1034–1052. doi: 10.1111/cmi.12256. [DOI] [PubMed] [Google Scholar]
- 43.Sherwood RK, Roy CR. 2016. Autophagy evasion and endoplasmic reticulum subversion: the yin and yang of Legionella intracellular infection. Annu Rev Microbiol 70:413–433. doi: 10.1146/annurev-micro-102215-095557. [DOI] [PubMed] [Google Scholar]
- 44.Altamirano-Silva P, Meza-Torres J, Castillo-Zeledon A, Ruiz-Villalobos N, Zuniga-Pereira AM, Chacon-Diaz C, Moreno E, Guzman-Verri C, Chaves-Olarte E. 2018. Brucella abortus senses the intracellular environment through the BvrR/BvrS two-component system, which allows B. abortus to adapt to its replicative niche. Infect Immun 86:e00713-17. doi: 10.1128/IAI.00713-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Connor TJ, Boyd D, Dorer MS, Isberg RR. 2012. Aggravating genetic interactions allow a solution to redundancy in a bacterial pathogen. Science 338:1440–1444. doi: 10.1126/science.1229556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coers J, Vance RE, Fontana MF, Dietrich WF. 2007. Restriction of Legionella pneumophila growth in macrophages requires the concerted action of cytokine and Naip5/Ipaf signalling pathways. Cell Microbiol 9:2344–2357. doi: 10.1111/j.1462-5822.2007.00963.x. [DOI] [PubMed] [Google Scholar]
- 47.Pasculle AW, Feeley JC, Gibson RJ, Cordes LG, Myerowitz RL, Patton CM, Gorman GW, Carmack CL, Ezzell JW, Dowling JN. 1980. Pittsburgh pneumonia agent: direct isolation from human lung tissue. J Infect Dis 141:727–732. doi: 10.1093/infdis/141.6.727. [DOI] [PubMed] [Google Scholar]
- 48.Zhang JH, Chung TD, Oldenburg KR. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 49.Smith KP, Kirby JE. 2016. Verification of an automated, digital dispensing platform for at-will broth microdilution-based antimicrobial susceptibility testing. J Clin Microbiol 54:2288–2293. doi: 10.1128/JCM.00932-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanson MS, Isberg RR. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun 63:3609–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.