The peptidoglycan in Gram-negative bacteria is a dynamic structure in constant remodeling. This dynamism, achieved through synthesis and degradation, is essential because the peptidoglycan is necessary to maintain the structure of the cell but has to have enough plasticity to allow the transport and assembly of macromolecular complexes in the periplasm and outer membrane. In addition, this remodeling has to be coordinated with the division process.
KEYWORDS: Brucella, inhibitor, peptidoglycan hydrolases
ABSTRACT
The peptidoglycan in Gram-negative bacteria is a dynamic structure in constant remodeling. This dynamism, achieved through synthesis and degradation, is essential because the peptidoglycan is necessary to maintain the structure of the cell but has to have enough plasticity to allow the transport and assembly of macromolecular complexes in the periplasm and outer membrane. In addition, this remodeling has to be coordinated with the division process. Among the multiple mechanisms bacteria have to degrade the peptidoglycan are the lytic transglycosidases, enzymes of the lysozyme family that cleave the glycan chains generating gaps in the mesh structure increasing its permeability. Because these enzymes can act as autolysins, their activity has to be tightly regulated, and one of the mechanisms bacteria have evolved is the synthesis of membrane bound or periplasmic inhibitors. In the present study, we identify a periplasmic lytic transglycosidase inhibitor (PhiA) in Brucella abortus and demonstrate that it inhibits the activity of SagA, a lytic transglycosidase we have previously shown is involved in the assembly of the type IV secretion system. A phiA deletion mutant results in a strain with the incapacity to synthesize a complete lipopolysaccharide but with a higher replication rate than the wild-type parental strain, suggesting a link between peptidoglycan remodeling and speed of multiplication.
INTRODUCTION
The complex structure of the envelope of Gram-negative bacteria requires highly regulated mechanisms for its synthesis, as well as for the control of its homeostasis. In particular, regulation of the synthesis and degradation of the peptidoglycan, a polymer of sugars and amino acids that form a mesh-like layer outside the plasma membrane, has to be tightly regulated. The peptidoglycan is composed of linear chains of N-acetylglucosamine and N-acetylmuramic acid (MurNAc), and the alternating sugars are connected by β-(1,4)-glycosidic bonds. Each MurNAc is attached to a short (4- to 5-residue) amino acid chain containing, in the majority of Gram-negative bacteria, d-alanine and d-glutamic acid (1). Because of its structure, the peptidoglycan imposes a physical barrier for the transport and assembly of outer membrane components. For this reason, Gram-negative bacteria encode muramidases, enzymes that degrade the peptidoglycan in order to allow, for example, the assembly of type III or type IV secretion systems essential in virulence, motility, or conjugation (1). Muramidases are specialized enzymes of the lysozyme superfamily that generate, locally, gaps in the peptidoglycan meshwork (1). Since the peptidoglycan is an essential component, the activity of these muramidases has to be tightly regulated to avoid overcleavage and, eventually, lethality. To regulate these enzymes, bacteria have a battery of inhibitors that modulate their activity through their specific spatiotemporal inhibition (2, 3).
Brucellaceae are highly adapted intracellular pathogens that cause brucellosis, a widespread zoonosis that affects animal production and inflicts important human health problems in regions of endemicity (4, 5). The capacity of Brucella to modulate the host cellular and immune response is completely dependent on several virulence factors that have to be either assembled in the inner and outer membrane or secreted through the peptidoglycan (6–11).
We have recently identified and characterized a muramidase in Brucella abortus (SagA) involved in the early stages of the intracellular replication process, necessary for the proper assembly of the type IV secretion system and thus the secretion of several of its substrates (10). Here, we describe the identification of an inhibitor (phiA) of SagA and characterize its role in the virulence process. We determined that overexpression of SagA affects the synthesis of the lipopolysaccharide (LPS) and results in a strain with some membrane defects, indicating that the lack of control of the muramidase activity can be highly detrimental for the bacterium. The development of an in vivo activity assay for SagA allowed us to demonstrate that phiA encodes a periplasmic muramidase inhibitor and that SagA is, at least, one of its targets. A phiA deletion mutant results in a rough phenotype but has a higher multiplication rate than the wild-type parental strain, indicating that the protein is necessary for the homeostasis of the periplasm.
RESULTS
Development of an in vivo assay to determine the peptidoglycanase activity of SagA.
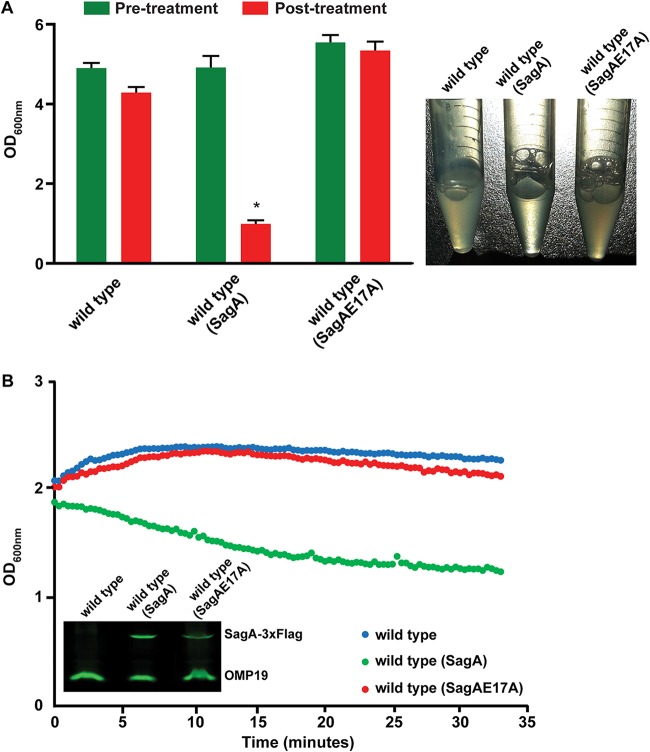
We have previously identified and characterized a protein with peptidoglycanase activity (SagA) necessary for the correct assembly of the type IV secretion system and, consequently, for the secretion of several of its substrates and for the early stages of the intracellular replication cycle (10). One interesting observation we made was that the Brucella abortus SagA-overexpressing strain seemed to be more sensitive to membrane-destabilizing agents than the wild-type strain, suggesting that the noncontrolled activity of SagA could be detrimental for the bacterium (unpublished results). To further characterize SagA, we determined its localization in the cell. As shown in Fig. 1, SagA has a predictive carboxy-terminal transmembrane segment, and when we performed periplasmic extractions, as well as membrane preparations, the results indicated that the protein is located in the inner membrane. Even though we were able to measure the activity of SagA in vitro (10), the phenotype of the overexpressing strain prompted us to evaluate the activity of SagA in vivo. For this, we developed an assay that consisted of growing the strains to a defined optical density at 600 nm (OD600) and resuspending them in a buffer containing a low concentration of Zwittergent 3-16 (see Materials and Methods) and monitored the decrease or lack of decrease in OD600 over time. The rationale was that generating a mild destabilization of the membrane would result in lysis of the SagA-overexpressing strain. Figure 2 shows the results of the assay with the wild-type strain, the wild-type strain overexpressing SagA, and the wild-type strain overexpressing a catalytic inactive SagA (SagAE17A) (10). As can be observed, overexpression of the active, but not the inactive, SagA induced a significant lysis of bacterial cells that could be monitored over time and resulted in a final OD600 significantly lower in the strain with the active SagA. These results demonstrate that the activity of SagA can be measured in vivo and that the lack of control of its activity can be highly detrimental for the bacterium.
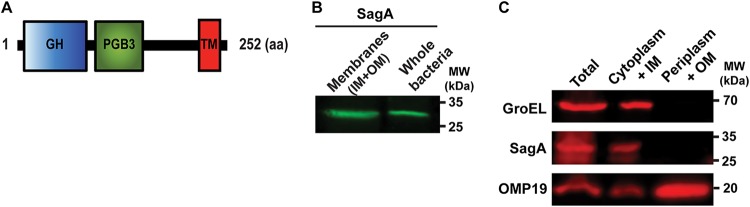
FIG 1.
SagA is an inner membrane-bound protein. (A) Schematic diagram of the domain structure of SagA. GH, glycosyl hydrolase domain; PGB3, peptidoglycan-binding domain; TM, transmembrane domain; aa, amino acids. (B) Western blot with anti-Flag monoclonal antibody of total membranes and whole cells of the B. abortus SagA-3×FLAG strain. (C) Western blot with anti-Flag monoclonal antibody of periplasmic and cytoplasmic fractions after a periplasmic fractionation of the B. abortus SagA-3×FLAG strain. Cytoplasmic control, GroEL; periplasmic/outer membrane control, OMP19; IM, inner membrane; OM, outer membrane.
FIG 2.
The activity of SagA can be measured in vivo. (A) OD600 at 15 min after incubation of the strains in a buffer containing 0.25% Zwittergent 3-16 (posttreatment). Pretreatment refers to the OD600 before adding the Zwittergent. A representative picture of bacterial suspensions at the end of the treatment (right) is shown. *, P < 0.0001. (B) Optical density determination over a 35-min period after incubation of the strains in a buffer containing 0.25% Zwittergent 3-16. A decrease in the optical density indicates lytic activity of SagA. Inset gel, expression levels of both proteins.
bab1_0102 encodes a periplasmic peptidoglycanase inhibitor.
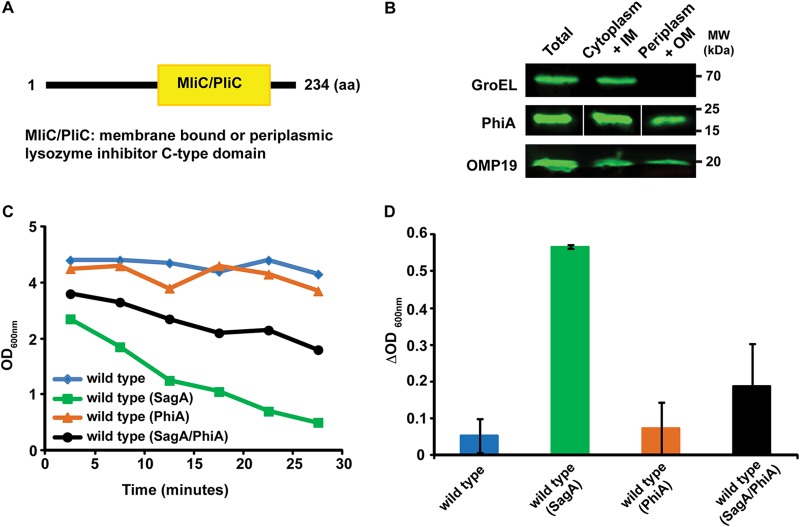
The results shown above indicated that the activity of SagA has to be highly regulated in order to avoid autolysis of the bacterium. Regulation can be achieved either at the transcriptional/translational level or by controlling the activity of the enzyme, for example, by specific inhibitors (2). To determine whether this second mechanism operates on SagA, we searched the genome of B. abortus for genes encoding putative lysozyme inhibitors and identified bab1_0102 and bab1_0466. bab1_0466 is predicted to encode a periplasmic lysozyme inhibitor and it has been crystalized alone as well as in complex with human lysozyme (12). bab1_0102 encodes a predictive protein with a periplasmic or membrane lysozyme C-type inhibitor domain (Fig. 3A). To determine the localization of Bab1_0102, we Flag tagged the gene, expressed it in B. abortus, and performed periplasmic extractions. As can be observed in Fig. 3B, the protein product of bab1_0102 localizes to the periplasmic space, strongly suggesting that it might act on proteins present in this compartment or in the inner membrane but with its active domain facing to the periplasm. To determine whether Bab1_0102 inhibits the muramidase activity of SagA, we exploited the in vivo activity assay developed, coexpressed both genes in B. abortus and performed the assay as indicated above. Figure 3C and D show that expression of Bab1_0102 significantly inhibited SagA-mediated autolysis, demonstrating that Bab1_0102 is an inhibitor of SagA's peptidoglycanase activity. For this reason, we renamed bab1_0102 “phiA” for peptidoglycan hydrolase inhibitor A.
FIG 3.
PhiA is a periplasmic inhibitor of SagA. (A) Schematic diagram of the domain structure of PhiA. (B) Western blot with anti-Flag monoclonal antibody of periplasmic and cytoplasmic fractions after a periplasmic fractionation of the B. abortus PhiA-3×FLAG strain. Cytoplasmic control, GroEL; periplasmic/outer membrane control, OMP19. (C) Optical density determination over a 30-min period after incubation of the strains in a buffer containing 0.25% Zwittergent 3-16. (D) The change (Δ value) in OD600 after a 15-min treatment with 0.25% Zwittergent 3-16 was determined as a direct measure of the activity of SagA. Coexpression of SagA and PhiA significantly inhibits SagA-mediated autolysis.
Deletion of phiA impairs the synthesis of the LPS and is detrimental for the virulence process.
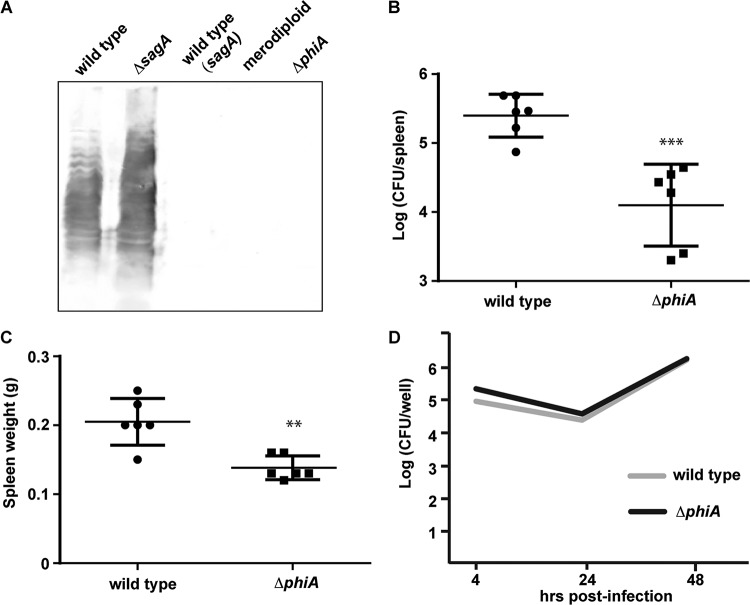
During the study of SagA, we observed that overexpression of the gene from a multicopy plasmid and a strong promoter results in a rough strain (Fig. 4A, third lane). Interestingly, a merodiploid strain with only one extra copy of the sagA gene in the chromosome also exhibits this phenotype (Fig. 4A, fourth lane). To explore whether the lack of control of the activity of SagA has an impact on the bacterium, we generated a gene deletion mutant and analyzed the ability of the ΔphiA strain to synthesize a complete LPS. As seen in Fig. 4A, deletion of phiA generated a strain with no detectable assembled O antigen, as for the SagA-overexpressing or merodiploid strains. As expected for a rough strain, the mutant was significantly less virulent in mouse infections, measured as the capacity to colonize the spleens at 14 days postinfection (Fig. 4B and C). When we determined the capacity of the strain to multiply intracellularly in HeLa cells, the mutant showed a replication rate identical to that of the wild-type parental strain (Fig. 4D), indicating that the diminished virulence observed in mice is not the consequence of the incapacity to replicate intracellularly.
FIG 4.
Deletion of phiA affects the synthesis of the LPS and virulence in mice. (A) Western blot analysis of total cell extracts of the B. abortus wild-type strain, ΔsagA mutant strain, B. abortus wild-type strain with a plasmid expressing SagA [wild type (sagA)], a B. abortus merodiploid strain carrying one extra copy of sagA in the chromosome, and a ΔphiA mutant, developed with an anti O-antigen monoclonal antibody (M84). (B) Spleen bacterial load of intraperitoneally infected BALB/c mice (six animals per group) inoculated with 105 CFU of B. abortus wild-type or B. abortus ΔphiA mutant (ΔphiA) strains at 2 weeks postinfection. ***, P < 0.001. (C) Spleen weight of intraperitoneally infected BALB/c mice inoculated with 105 CFU of wild-type (B. abortus) or B. abortus ΔphiA (ΔphiA) strains at 2 weeks postinfection. **, P < 0.05. (D) An antibiotic protection assay was performed to determine the intracellular replication of B. abortus wild-type and ΔphiA strains in HeLa cells.
phiA is involved in the control of the division process.
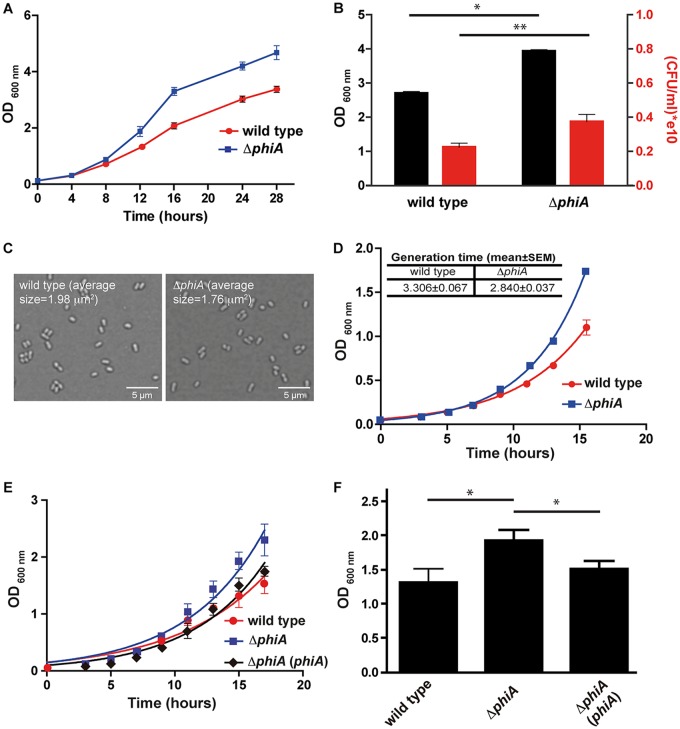
During the manipulation of the B. abortus ΔphiA mutant, we observed that the strain grew faster in plates and that it consistently reached higher OD600 values in liquid cultures than the wild-type parental strain. To determine whether the mutant has an altered division rate, we performed in vitro replication curves and determined the rate of multiplication, as well as the final OD600 that the strains reached at the end of the exponential phase. As shown in Fig. 5A, the ΔphiA mutant strain showed a faster replication in rich medium that was evidenced as early as at 10 h from the initiated culture and was maintained throughout the complete culture, reaching a higher final OD600 . Because the OD600 measures light dispersion and this is proportional to the number of bacteria or to their size, it is possible that the mutant has a significantly bigger size and that this results in a higher OD600. To determine whether this is the case, we measured the OD600, as well as the viable numbers of bacteria (measured in CFU), of the wild-type and mutant strains at 20 h postinoculation of the culture. As shown in Fig. 5B, the ΔphiA mutant showed a statistically significant higher final OD600, as well as viable CFU, indicating that the strain multiplies with faster kinetics. Consistent with this, Fig. 5C shows microscopy images of the wild-type and ΔphiA mutant strains confirming that the sizes and forms of both strains are equivalent. To determine the generation time of both strains, we assessed growth curves until 16 h postinoculation and calculated the generation time, as indicated in Materials and Methods. The results of these experiments (Fig. 5D) demonstrate a statistically significant decrease in the generation time of the ΔphiA mutant (3.36 h in the wild-type strain versus 2.84 h in the mutant). Figure 5E and F show that the addition of a plasmid carrying phiA in the mutant complemented the growth phenotypes. Altogether, these results demonstrate that PhiA is involved in the regulation of the generation time in Brucella.
FIG 5.
The ΔphiA mutant has a higher multiplication rate. (A) Growth curve in TSB of B. abortus wild-type and ΔphiA mutant strains over 28 h. (B) OD600 and CFU at 20 h of the growth curve. *, P < 0.0001; **, P < 0.05. (C) Bright-field microscopy images of the B. abortus wild-type and ΔphiA mutant strains grown in TSB at 24 h of the growth curve. The average size (100 bacteria measured) of the bacteria is indicated in each image. (D) Growth curve in TSB of B. abortus wild-type and ΔphiA mutant strains over 16 h. In this curve, the generation time of each strain was calculated (inset table). (E) Growth curve in TSB of B. abortus wild-type, ΔphiA mutant, and complemented strains over 18 h. (F) OD600 at 15 h of the growth curve. *, P < 0.05.
DISCUSSION
The composition and topology of the outer membranes and periplasmic spaces of Gram-negative bacteria are highly regulated features. The peptidoglycan imposes a physical barrier for macromolecules going either in or out of the cytoplasm, and for this reason, it has to have a highly controlled permeability. Muramidases or lytic-transglycosidases regulate this permeability by locally degrading the N-acetylglucosamine and N-acetylmuramicacid mesh-like structure and allowing the transport and assembly of high-molecular-weight complexes that need to be positioned in the outer membrane. The activity of these enzymes has to be exquisitely controlled since they act as double-edged knives: too much activity will be lethal, but no activity will be detrimental for the structure of the bacterium. One of the mechanisms to control the activity of these muramidases is their specific inhibition through periplasmic or membrane bound proteins that act as inhibitors of their enzymatic activity (2). Here, we have identified and characterized a new muramidase inhibitor that targets SagA, a peptidoglycanase that we have previously found to be important for the virulence of B. abortus (10). SagA is a peptidoglycanase necessary for the proper assembly of the type IV secretion system of Brucella, an essential virulence factor for the intracellular multiplication of the bacterium (13, 14). The absence of SagA affects the secretion of several substrates of the type IV secretion system and negatively impacts the capacity of the bacteria to replicate intracellularly. Cellular localization studies indicated that SagA is an inner membrane-bound enzyme. Interestingly, overexpression of SagA results in a rough strain, demonstrating that the lack of control of its activity is detrimental for the bacterium. Moreover, we took advantage of this and developed an in vivo assay to monitor the activity of SagA, measuring the lysis of the overexpressing strain after a mild disruption of the outer membrane. Using this technique, we were able to measure the in vivo activity of SagA and confirmed that mutation of the glutamic acid 17 to alanine renders the enzyme inactive (negative control). The fact that the activity of SagA has to be tightly regulated led us to search and identify phiA, a gene encoding a periplasmic peptidoglycanase inhibitor. In vivo SagA activity assays in a SagA/PhiA-coexpressing strain demonstrated that phiA encodes an inhibitor of SagA. Deletion of phiA resulted in a rough bacterium, the same phenotype as the SagA-overexpressing strain, indicating that the lack of control of SagA's activity significantly affects the synthesis or assembly of the LPS. It is becoming increasingly accepted in the last few years that the periplasmic spaces and the outer membranes of bacteria are highly organized structures with a defined topology. To achieve this organization, bacteria have evolved mechanisms to compartmentalize the synthetic and/or translocation machinery of the different components of these structures, such as the LPS (15), the peptidoglycan (15, 16), secretion systems (17–19), or adhesins (20, 21). For this reason, the assembly of these structures has to be extremely coordinated. We hypothesize that the overactivity of SagA, either due to its overexpression or because its inhibitor is absent, alters the homeostasis of the periplasm, affecting the capacity to coordinate the synthesis and assembly of several of its components, such as the LPS synthetic machinery.
When growing the ΔphiA strain, we consistently observed that the mutant grew faster than the wild-type parental strain. To corroborate this observation, we performed in vitro growth curves and measured the generation time, confirming that the mutant divides significantly faster than does the wild type. Because the division process requires rupture and reassembly of the peptidoglycan, this remodeling, particularly degradation, could act as a bottleneck in the generation time. Because the activity of SagA and probably other peptidoglycan hydrolases is upregulated in the ΔphiA mutant, this could increase the division rate due to a more efficient degradation of the peptidoglycan. If so, this would imply that at least part of the replication rate could be modulated by the activity of lytic transglycosidases or glycosyl hydrolases. Remodeling of the peptidoglycan is a central feature during the division process and can occur through the activity of three types of enzymes: N-acetylmuramyl-l-alanine amidases, endopeptidases, or lytic transglycosidases (3). In any of these cases, the phenotypes of single or multiple mutations in these enzymes are always associated with the capacity of the bacteria to complete the septum and separate the cells after division (3, 22, 23). The hypothesis that the activity of peptidoglycan hydrolases could alter the speed of division implies that the multiplication rate and proper permeability of the peptidoglycan, as well as the assembly of macromolecular complexes in the outer membrane, are processes in a constant “toggle war” and for this reason have to be finely tuned. Although this is purely speculative at this stage, it is an interesting hypothesis to test in the future.
MATERIALS AND METHODS
Media and culture conditions.
Brucella strains were grown at 37°C in tryptic soy broth (TSB) or tryptic soy agar (TSA). E. coli strains were grown at 37°C in Luria-Bertani media. If necessary, media were supplemented with the appropriate antibiotics at the indicated final concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; and nalidixic acid, 5 μg/ml.
Recombinant DNA techniques.
(i) Construction of plasmids pLF, pLF/SagA, and pLF/SagA(E17A). The plasmid pLF was generated from the vector pLFC (24). In order to eliminate the Cya sequence and replace it with the 3×FLAG sequence, pLFC was digested with SpeI and XbaI. A fragment coding for 3×FLAG epitope was amplified from pBAD24-3×FLAG (25) using the primers 3FLAG·SpeI and 3FLAG·XbaI (24), and the PCR product was digested with SpeI and XbaI and ligated to the vector pLFC into the corresponding sites. The resulting plasmid was named pLF.
To generate the plasmid pLF/SagA, coding for in-frame fusion 3×FLAG-SagA, a DNA fragment was amplified from B. abortus 2308 genomic DNA using primers MDG20 (5′-GCTCTAGAGCCGAAAGGAAATCACCAATG-3′) and MDG23 (5′-CGAGCTCTCATGCCTCTTTCCTCACCC-3′). The PCR product was digested with SpeI and XbaI and ligated to the pLF vector digested with the same enzymes.
In the same way, the sagA(E17A) sequence was amplified from pQE30/SagA(E17A) (10) and cloned into pLF vector.
(ii) Construction of wild type (SagA) and wild type (SagAE17A). To construct the strain overexpressing sagA, the plasmid pLF/SagA was introduced into Brucella abortus 2308 wild-type strain by biparental mating using the E. coli S17-λpir strain. In the same way, the plasmid pLF/SagA(E17A) was introduced into B. abortus 2308 wild-type strain, and the resulting strain was named wild type (SagAE17A).
(iii) Construction of the ΔphiA mutant strain. To construct a B. abortus 2308 ΔphiA mutant strain, the regions flanking the phiA gene were amplified and ligated using the recombinant PCR technique (26). The resulting fragment was digested with EcoRI and BamHI and ligated to the pK18mobSacB plasmid digested with the same enzymes. The primers used for PCR amplification were Fw-UP-102 (5′-CCGGAATTCGCCGCGAAAATCGATCCG-3′) and Rv-UP-102 (5′-TTATTGGGGATTTCAAGTGG-3′) to amplify a 400-bp upstream region and Fw-DW-102 (5′-CTTGAAATCCCCAATAAGCCCGTTGCGGTTTTGC-3′) and Rv-DW-102 (5′-CGCGGATCCAAGGCGCAGCTTGAACGGC-3′) to amplify a 400-bp downstream region. Fw-UP-102 and Rv-DW-102 were used in an overlapping PCR. The resulting fragment was digested with EcoRI and BamHI and ligated to the pK18mobSacB plasmid digested with the same enzymes. The plasmid pK18mobSacBΔphiA was introduced into the B. abortus 2308 wild-type strain by biparental mating using the E. coli S17-λpir strain. Double recombination events (Kms Sacr) were selected, and gene knockout was confirmed by genomic PCR.
(iv) Construction of plasmid pBBR2/phiA-3×FLAG. In order to generate the in-frame fusion phiA-3×FLAG, a DNA fragment containing the phiA gene was amplified from B. abortus 2308 genomic DNA using the primers MDG38 (5′-CTAGCTAGCACTTGAAATCCCCAATAAGTG-3′) and MDG102rv (5′-CCGGAATTCTTCTTCGGCGGTGTCTGCGCG-3′). This fragment was digested with NheI and EcoRI and ligated into the corresponding sites of pBBR4-3×FLAG vector (26). The resulting plasmid was digested with BamHI and HindIII, and the in-frame fusion phiA-3×FLAG was subcloned in the pBBR1-MCS2 (27) vector in the same restriction sites.
(v) Construction of wild type (PhiA), wild type (SagA/PhiA), and complemented ΔphiA (phiA) strains. To construct the strains overexpressing phiA, the plasmid pBBR2/phiA-3×FLAG was introduced into Brucella abortus 2308 wild type, wild type (SagA), and ΔphiA strains by biparental mating using the E. coli S17-λpir strain.
(vi) Construction of a sagA merodiploid strain. To construct a B. abortus 2308 sagA merodiploid strain, the 3×FLAG-tagged sagA gene was amplified from pBBR4/sagA-3×FLAG plasmid using the primers CC7-Flag (CGCGGATCCACTCTAGAGATCGTCATCCTTGTAA) and MDG16 (10). The resulting fragment was digested with BamHI and ligated to the pK18mobSacB plasmid digested with the same enzyme. The resulting plasmid was introduced into B. abortus 2308 by biparental mating using the E. coli S17-λpir strain. Simple recombination events (Kmr Sacs) were selected, and the presence of the integrated plasmid was confirmed by genomic PCR.
Analysis of protein expression and subcellular localization.
(i) Whole bacteria. Brucella whole-cell extracts were resuspended in Laemmli sample buffer and heated to 100°C for 5 min. Samples were submitted to SDS-PAGE (10 or 12.5%, depending on the assay) and transferred to nitrocellulose membranes. The presence of 3×FLAG-tagged proteins was assessed by immunoblot analysis with mouse anti-Flag M2 monoclonal antibody (dilution, 1:5,000) and IRDye secondary anti-mouse antibody (LI-COR, Inc.).
(ii) Membrane proteins. Brucella membrane extracts were obtained as described previously (10), subjected to SDS-PAGE (10%), and transferred to nitrocellulose membranes. The presence of SagA-3×FLAG protein was assessed by immunoblot analysis with anti-Flag M2 monoclonal antibody (dilution, 1:5,000).
(iii) Periplasmic and cytoplasmic proteins. Analysis of periplasmic and cytoplasmic localization was performed as described previously (26). Briefly, 2.5 × 1010 bacterial cells were centrifuged for 10 min at 3,300 × g. The pellets were treated with 0.2 M Tris-HCl (pH 7.6), 1 M sucrose, and 0.25% Zwittergent 3-16 solution and then incubated for 10 min at room temperature. The samples were centrifuged for 30 min at 8,000 × g, and the resultant fractions were processed for Western blotting. The presence of SagA-3×FLAG and PhiA-3×FLAG proteins was assessed using anti-Flag M2 monoclonal antibody (dilution, 1:5,000) (10). Anti-GroEL polyclonal antibody (1:2,000; our laboratory) and anti-OMP-19 monoclonal antibody (1:2,000; Axel Cloeckaert) were used as cytoplasmic and periplasmic controls, respectively.
All antibodies were diluted in Tris-buffered saline (TBS), 1% nonfat milk, and 0.1% Tween solution, and detection was performed using an IRDye secondary anti-mouse antibody (1:20,000) and the Odyssey Imaging System (LI-COR, Inc.).
LPS analysis.
Brucella whole-cell extracts were subjected to SDS-PAGE (10%) and transferred to nitrocellulose membranes. The presence of O-polysaccharide was evaluated by immunoblot analysis with monoclonal antibody M84 (dilution 1:3,000) and diluted in TBS–1% nonfat milk–0.1% Tween solution, and detection was performed using an IRDye secondary anti-mouse antibody (1:20,000) and the Odyssey Imaging System (LI-COR, Inc.).
In vivo activity assay of SagA.
B. abortus strains were grown in TSB overnight, and 2.5 × 1010 bacterial cells were centrifuged for 10 min at 3,300 × g. The pellets were washed with 0.2 M Tris-HCl (pH 7.6) and treated with 0.2 M Tris-HCl (pH 7.6) and 0.25% Zwittergent 3-16 solution for 15 min. The OD600 was measured pre- and posttreatment or monitored during treatment.
Mouse infections.
Mouse infections were performed as described previously (28). Groups of six 8- to 9-week-old female BALB/c mice were intraperitoneally inoculated with 105 CFU of B. abortus 2308 wild-type or B. abortus ΔphiA strains in phosphate-buffered saline (PBS). At 2 weeks postinfection, spleens from infected mice were removed and homogenized in 2 ml of PBS. Serial dilutions from individual spleens were plated on TSA to quantify the recovered CFU. All mice were bred in accordance with institutional animal guidelines under specific-pathogen-free conditions in the local animal facility (biosafety level 3 [BSL3]; Institute for Research in Biotechnology) of the University of San Martín. Mouse studies were approved by the local regulatory agencies (CICUAE-UNSAM).
HeLa cell infection.
A standard antibiotic protection assay was performed in HeLa cells. Cells were seeded in 24-well plates in suitable culture medium at 105 cells/ml and incubated overnight at 37°C. Brucella strains were grown in TSB with the appropriate antibiotics for 24 h and diluted in culture medium prior to infection. The suspension was added at a multiplicity of infection of 500:1 and centrifuged at 300 × g for 10 min. After 1 h of incubation at 37°C, the cells were washed, and fresh medium containing 100 μg/ml streptomycin and 50 μg/ml gentamicin was added. At 4, 24, or 48 h postinfection, the cells were washed and lysed with 0.1% Triton X-100. The intracellular CFU were determined by direct plating on TSB agar plates.
In vitro growth assay.
Brucella growth curves were done in TSB supplemented with 5 μg/ml nalidixic acid. Overnight cultures were diluted to an OD600 of 0.1 or 0.045 and grown at 37°C and 250 rpm with shaking. At the times indicated in Fig. 5, aliquots were taken, and the OD600 and CFU/ml were determined. The growth curves were done in triplicate. The generation time of each strain was determined by calculating the slope of the linear regression of the OD600 as a function of time.
Confocal microscopy and determination of bacterial size.
For characterization of live cell shape and size, 3-μl portions of the bacterial cell culture were collected and spotted onto a fresh 1% agarose pad, covered with a coverslip, and sealed with VALAP (a 1:1:1 mixture of vaseline, lanolin, and paraffin) as previously described (29). Bright-field microscopy of bacterial cells was performed using a microscope Olympus IX81 (confocal microscope) with a 60× oil immersion objective. The bacterial size was calculated with ImageJ software using the Analyze Particles tool.
ACKNOWLEDGMENTS
We thank members of the J.E.U. laboratory for useful discussions. We especially thank Alfonso Soler Bistue for sharing knowledge about bacterial growth and Francisco Guaimas for help with the confocal microscopy image acquisition and processing.
This study was supported by grants PICT-1028-2014 and PICT-2717-2016 to J.E.U. and M.G.D.G., respectively. C.C. and J.E.U. are members of the National Research Council of Argentina (CONICET). A.M.R. is a fellow of CONICET, and M.G.D.G. is a fellow of ANPCyT (Argentina).
REFERENCES
- 1.Koraimann G. 2003. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell Mol Life Sci 60:2371–2388. doi: 10.1007/s00018-003-3056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Callewaert L, Van Herreweghe JM, Vanderkelen L, Leysen S, Voet A, Michiels CW. 2012. Guards of the great wall: bacterial lysozyme inhibitors. Trends Microbiol 20:501–510. doi: 10.1016/j.tim.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Typas A, Banzhaf M, Gross CA, Vollmer W. 2011. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbel MJ. 1997. Brucellosis: an overview. Emerg Infect Dis 3:213–221. doi: 10.3201/eid0302.970219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pappas G, Akritidis N, Bosilkovski M, Tsianos E. 2005. Brucellosis. N Engl J Med 352:2325–2336. doi: 10.1056/NEJMra050570. [DOI] [PubMed] [Google Scholar]
- 6.Briones G, Inon de Iannino N, Roset M, Vigliocco A, Paulo PS, Ugalde RA. 2001. Brucella abortus cyclic β-1,2-glucan mutants have reduced virulence in mice and are defective in intracellular replication in HeLa cells. Infect Immun 69:4528–4535. doi: 10.1128/IAI.69.7.4528-4535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byndloss MX, Tsolis RM. 2016. Brucella spp. virulence factors and immunity. Annu Rev Anim Biosci 4:111–127. doi: 10.1146/annurev-animal-021815-111326. [DOI] [PubMed] [Google Scholar]
- 8.Comerci DJ, Martinez-Lorenzo MJ, Sieira R, Gorvel JP, Ugalde RA. 2001. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell Microbiol 3:159–168. doi: 10.1046/j.1462-5822.2001.00102.x. [DOI] [PubMed] [Google Scholar]
- 9.Czibener C, Ugalde JE. 2012. Identification of a unique gene cluster of Brucella spp. that mediates adhesion to host cells. Microbes Infect 14:79–85. doi: 10.1016/j.micinf.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Giudice MG, Ugalde JE, Czibener C. 2013. A lysozyme-like protein in Brucella abortus is involved in the early stages of intracellular replication. Infect Immun 81:956–964. doi: 10.1128/IAI.01158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myeni S, Child R, Ng TW, Kupko JJ III, , Wehrly TD, Porcella SF, Knodler LA, Celli J. 2013. Brucella modulates secretory trafficking via multiple type IV secretion effector proteins. PLoS Pathog 9:e1003556. doi: 10.1371/journal.ppat.1003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Um SH, Kim JS, Kim K, Kim N, Cho HS, Ha NC. 2013. Structural basis for the inhibition of human lysozyme by PliC from Brucella abortus. Biochemistry 52:9385–9393. doi: 10.1021/bi401241c. [DOI] [PubMed] [Google Scholar]
- 13.O’Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli ML, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol 33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 14.Sieira R, Comerci DJ, Sanchez DO, Ugalde RA. 2000. A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J Bacteriol 182:4849–4855. doi: 10.1128/jb.182.17.4849-4855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vassen V, Valotteau C, Feuillie C, Formosa-Dague C, Dufrene YF, De Bolle X. 2019. Localized incorporation of outer membrane components in the pathogen Brucella abortus. EMBO J 38:e100323. doi: 10.15252/embj.2018100323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cameron TA, Anderson-Furgeson J, Zupan JR, Zik JJ, Zambryski PC. 2014. Peptidoglycan synthesis machinery in Agrobacterium tumefaciens during unipolar growth and cell division. mBio 5:e01219-14. doi: 10.1128/mBio.01219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aguilar J, Cameron TA, Zupan J, Zambryski P. 2011. Membrane and core periplasmic Agrobacterium tumefaciens virulence type IV secretion system components localize to multiple sites around the bacterial perimeter during lateral attachment to plant cells. mBio 2:e00218-11. doi: 10.1128/mBio.00218-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aguilar J, Zupan J, Cameron TA, Zambryski PC. 2010. Agrobacterium type IV secretion system and its substrates form helical arrays around the circumference of virulence-induced cells. Proc Natl Acad Sci U S A 107:3758–3763. doi: 10.1073/pnas.0914940107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geiger T, Pazos M, Lara-Tejero M, Vollmer W, Galán JE. 2018. Peptidoglycan editing by a specific ld-transpeptidase controls the muramidase-dependent secretion of typhoid toxin. Nat Microbiol 3:1243–1254. doi: 10.1038/s41564-018-0248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Czibener C, Merwaiss F, Guaimas F, Del Giudice MG, Serantes DA, Spera JM, Ugalde JE. 2016. BigA is a novel adhesin of Brucella that mediates adhesion to epithelial cells. Cell Microbiol 18:500–513. doi: 10.1111/cmi.12526. [DOI] [PubMed] [Google Scholar]
- 21.Posadas DM, Ruiz-Ranwez V, Bonomi HR, Martín FA, Zorreguieta A. 2012. BmaC, a novel autotransporter of Brucella suis, is involved in bacterial adhesion to host cells. Cell Microbiol 14:965–982. doi: 10.1111/j.1462-5822.2012.01771.x. [DOI] [PubMed] [Google Scholar]
- 22.Heidrich C, Ursinus A, Berger J, Schwarz H, Holtje JV. 2002. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J Bacteriol 184:6093–6099. doi: 10.1128/jb.184.22.6093-6099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Priyadarshini R, Popham DL, Young KD. 2006. Daughter cell separation by penicillin-binding proteins and peptidoglycan amidases in Escherichia coli. J Bacteriol 188:5345–5355. doi: 10.1128/JB.00476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchesini MI, Herrmann CK, Salcedo SP, Gorvel JP, Comerci DJ. 2011. In search of Brucella abortus type IV secretion substrates: screening and identification of four proteins translocated into host cells through VirB system. Cell Microbiol 13:1261–1274. doi: 10.1111/j.1462-5822.2011.01618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spano S, Ugalde JE, Galan JE. 2008. Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe 3:30–38. doi: 10.1016/j.chom.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Dohmer PH, Valguarnera E, Czibener C, Ugalde JE. 2014. Identification of a type IV secretion substrate of Brucella abortus that participates in the early stages of intracellular survival. Cell Microbiol 16:396–410. doi: 10.1111/cmi.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 28.Czibener C, Del Giudice MG, Spera JM, Fulgenzi FR, Ugalde JE. 2016. Delta-pgm, a new live-attenuated vaccine against Brucella suis. Vaccine 34:1524–1530. doi: 10.1016/j.vaccine.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 29.Brown PJ, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X, Fuqua C, Brun YV. 2012. Polar growth in the alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci U S A 109:1697–1701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]