Abstract
The CCAAT box-harboring proteins represent a family of heterotrimeric transcription factors which is highly conserved in eukaryotes. In fungi, one of the particularly important homologs of this family is the Hap complex that separates the DNA-binding domain from the activation domain and imposes essential impacts on regulation of a wide range of cellular functions. So far, a comprehensive summary of this complex has been described in filamentous fungi but not in the yeast. In this review, we summarize a number of studies related to the structure and assembly mode of the Hap complex in a list of representative yeasts. Furthermore, we emphasize recent advances in understanding the regulatory functions of this complex, with a special focus on its role in regulating respiration, production of reactive oxygen species (ROS) and iron homeostasis.
Keywords: Hap complex, CCAAT box, reactive oxygen species, iron homeostasis, yeast, cytotoxicity
Introduction
The CCAAT box, similar as the classic TATA-box, cap signal, and GC-box, is also a major eukaryotic promoter element (Bucher, 1990). It appears in about 30% of eukaryotic genes and is located at a conserved distance of −60/−100 bp from the transcription start sites (TSS) (Maity and De Crombrugghe, 1998; Mantovani, 1999; Dolfini et al., 2009), where normally the TATA box is present (Yang et al., 2007). The CCAAT box is usually found in TATA-less promoters (Dolfini et al., 2009), and sequences around the box are highly conserved when comparing with specific genes in different species (Dorn et al., 1987; Chodosh et al., 1988a). In S. cerevisiae, the CCAAT box is reported to be found at promoters of the genes encoding cytochromes and those who are specifically activated by non-fermentable carbon sources (McNabb et al., 1995). In higher eukaryotes, the CCAAT box exists in all kinds of promoters but is more prevalent in cell-cycle regulated genes (Berry et al., 1992; Ronchi et al., 1996; Marziali et al., 1997; Mantovani, 1998). Deletion or mutation of this pentanucleotide dramatically affects basal levels of gene transcripts (Myers et al., 1986), highlighting its role in transcriptional regulation.
A number of DNA-binding proteins, such as the CCAAT/enhancer binding protein (C/EBP), CCAAT transcription factor/nuclear factor 1 (CTF/NF1), and CCAAT displacement protein (CDP), were found to selectively recognize the CCAAT homologous sequences (Zorbas et al., 1992; Andrés et al., 1994; Aufiero et al., 1994; Harada et al., 1995; Osada et al., 1996). Among them, nuclear factor Y (NF-Y) was originally discovered by its ability to bind to the conserved Y box element present at the promoter of the mouse MHC Class II Ea and importantly, the results, based on a series of experiments such as gel retardation, saturation mutagenesis, methylation interference, and cross-competition, clearly demonstrate that NF-Y recognition strictly requires all the five nucleotides (CCAAT) in the region and may also benefit from additional flanking sequences (Dorn et al., 1987; Karsenty et al., 1988; Ronchi et al., 1995; Mantovani, 1998). A detailed analysis in more than 100 promoters has shown that the CCAAT box and NF-Y both play important and sometimes even indispensable roles in gene regulation (Myers et al., 1986; Dorn et al., 1987; Greene et al., 1987; Karsenty et al., 1988; Danilition et al., 1991; Skalnik et al., 1991; Pramson, 1993; Szabo et al., 1993; Hasan et al., 1994; Mantovani, 1998), confirming that NF-Y is the major protein recognizing the CCAAT box (Mantovani, 1999). Actually, a list of transcription factors belonging to the NF-Y family has been discovered in a wide range of organisms and designated the Hap complex (e.g., the budding yeast Saccharomyces cerevisiae, the fission yeast Schizosaccharomyces pombe, Kluyveromyces lactis, Arabidopsis thaliana, and Aspergillus species) and the CBF/NF-Y (e.g., Xenopus and mammals), respectively (Kato, 2005). In this review, we will focus on the Hap complex subunits in yeasts and summarize recent understanding of the protein structure, function, and related signaling pathways.
Hap Complex in Yeasts
NF-Y is a ubiquitous heteromeric protein complex composed of three subunits (NF-YA, NF-YB, and NF-YC), and is necessary for DNA binding and transcriptional regulation (Hatamochi et al., 1988; Chodosh et al., 1988a; Kim and Sheffery, 1990; Bellorini et al., 1997b). The Hap complex from S. cerevisiae represents the first identified factor in this family and its discovery was originated from the studies by Guarente and his colleagues. They found that this heteromeric complex contains three subunits (Hap2, Hap3, and Hap4) and was originally recognized as a transcriptional activator through its capacity of binding to the CCAAT box located in the UAS2 element of CYC1, a gene encoding iso-1-cytochrome C (Guarente and Ptashne, 1981; Guarente and Mason, 1983; Guarente et al., 1984; Olesen et al., 1987; Hahn and Guarente, 1988; Forsburg and Guarente, 1989; Xing et al., 2006). Interestingly, the subunit Hap4, although confers an essential activity in transcriptional activation, was found to be dispensable for the binding of the Hap complex to the CCAAT box (Olesen and Guarente, 1990; Xing et al., 1993) and a novel subunit, designated Hap5, constitutes another major component of the Hap complex and exhibits the DNA-binding capacity to the CCAAT box (McNabb et al., 1995), indicating that the domains conferring the DNA binding and activation could be different in this complex. Indeed, further studies clarified that the DNA-binding domain of the Hap complex contains three essential subunits, Hap2/3/5 (the three homologs of NF-YA, NF-YB, and NF-YC subunits), while the activation domain is composed of only Hap4 (Mantovani, 1999; McNabb and Pinto, 2005; Bolotin-Fukuhara, 2017). More importantly, gene activation in S. cerevisiae requires the factor Hap4, in addition to the Hap2/Hap3/Hap5 subunits (Forsburg and Guarente, 1989; McNabb et al., 1995). The Hap2/3/5 subcomplex, although constitutively expressed, needs to combine another regulatory subunit Hap4 to trigger gene expression and intriguingly, two distinct amino acid domains of Hap5 (residues 71–102 and 115–146) mediate the recruitment of Hap4 to the trimeric complex (McNabb et al., 1997).
Following characterization of the Hap complex in S. cerevisiae, this highly conserved factor was also discovered and analyzed in mammals including human and other vertebrates (Chodosh et al., 1988a). Three heterologous components, including NF-Y, CP1, and CBF, were found to share remarkable sequence similarities to the yeast homologs of Hap complex and studies have shown that they form a functionally conserved, multisubunit complex that is capable of binding to the CCAAT nucleotide sequences in a way similar to its ortholog in the budding yeast, although considerable divergence between yeast and mammalian complexes exists because of the varied sequences outside of the highly conserved core motifs (Chodosh et al., 1988b; Maity et al., 1990; Vuorio et al., 1990; Li et al., 1992b; Sinha et al., 1995; Becker et al., 2006; Hooft van Huijsduijnen et al., 2018). In addition to S. cerevisiae, homologs of the Hap complex components were also present in other yeasts such as Hap2/Hap3/Hap5 in Candida glabrata (Ueno et al., 2011; Thiébaut et al., 2017), Php2/Php3/Php5 in Schizosaccharomyces pombe (Olesen et al., 1991; McNabb et al., 1997), Hap2/Hap3/Hap5 in Cryptococcus neoformans (Jung et al., 2010), and Hap2/Hap3/Hap5 in Candida albicans (Table 1; Johnson et al., 2005; Baek et al., 2008; Hsu et al., 2011; Singh et al., 2011). Interestingly, unlike yeast and mammalian Hap complex components in which each subunit is encoded by a single gene, the Hap2/3/5 subunits in plants could be represented by multiple genes. For example, Arabidopsis genome harbors at least three isoforms of each of the Hap subunits and even more, searching the Plant Transcription Factor Database (PlantTFDB) identifies a total of 59 predicted genes coding for the three subunits of NF-Y in tomato, including 10 NF-YA, 29 NF-YB, and 20 NF-YC genes (Edwards et al., 2002; Siefers et al., 2008; Petroni et al., 2012; Li et al., 2016), suggesting that more diverse roles in gene transcription might be assigned to this complex in higher plants.
Table 1.
CCAAT-box binding proteins in this paper.
| Species | Name | Subunits | Reference |
|---|---|---|---|
| Saccharomyces cerevisiae | Hap | Hap2/3/4/5 | McNabb et al., 1997; McNabb and Pinto, 2005 |
| Schizosaccharomyces pombe | Php | Php2/3/4/5 | Olesen et al., 1991; McNabb et al., 1997 |
| Cryptococcus neoformans | Hap | Hap2/3/5/X | Jung et al., 2010 |
| Candida albicans | Hap | Hap2/31/32/43/5 | Johnson et al., 2005; Baek et al., 2008; Hsu et al., 2011; Singh et al., 2011 |
| Candida glabrata | Hap | Hap2/3/4/5 | Ueno et al., 2011; Thiébaut et al., 2017 |
| Mammals | NF-Y | NF-YA/B/C | Hatamochi et al., 1988; Chodosh et al., 1988a; Kim and Sheffery, 1990 |
The Hap complex in S. cerevisiae binds to the CCAAT sequence in the upstream activation sequence (UAS) of numerous cytochrome genes, acting as a master regulator of respiratory metabolism (Buschlen et al., 2003; Schüller, 2003), similar to its role in the mammalian cells (Chodosh et al., 1988a). Analysis of expression profiles in HAP2 or HAP3 gene deletion mutant cells after a diauxic shift identified those whose expressions were significantly up- or down-regulated, and actually the CCAAT box is clearly enriched in the UAS of these genes (Bonander et al., 2008). Sequence analyses showed the presence of a conserved CCAAT box at the promoters of almost all nuclear encoded respiratory genes and strong evidence supported that activation of these genes requires the Hap2/3/4/5 complex (Buschlen et al., 2003). However, different from the mammalian cells in which the trimeric complex (NF-Y, CP1, and CBF) is sufficient to regulate gene expression, gene activation in S. cerevisiae requires another factor (Hap4) in addition to the Hap2/Hap3/Hap5 (Forsburg and Guarente, 1989; McNabb et al., 1995). Intriguingly, only fungi contain the Hap4 subunit (Bourgarel et al., 1999; Kato, 2005; Mercier et al., 2008). Similar to the homologs in other higher eukaryotes, Hap4 in fungi has structurally split its activation function by incorporating into NF-YA and NF-YC, the two NF-Y subunits harboring activation motifs that are displayed as long glutamine-rich stretches in the respective N-terminal (NF-YA) or C-terminal (NF-YC) regions (Li et al., 1992a; Coustry et al., 1996; Di Silvio et al., 1999; Hortschansky et al., 2017). In addition, Hap4 homologs are expressed in other yeasts whereas the function is different (Bourgarel et al., 1999; Hortschansky et al., 2007; Mercier et al., 2008; Jung et al., 2010). For example, Hap4 homologs, including the S. pombe homolog Php4, the C. neoformans homolog HapX, and the C. albicans homolog Hap43, were all found to act as transcriptional repressors, instead of being activators (Hsu et al., 2013). Importantly, these homologs were found to be critical in regulation and maintenance of iron homeostasis, mainly because they negatively regulate expression of iron-consuming genes under low-iron conditions.
Architecture of the Hap Complex
NY-F has been identified in various eukaryotes, including yeasts, fungi, higher plants, and vertebrates, and it has been clear that the domains required for DNA and subunit binding in all three core subunits (NF-YA, NF-YB, and NF-YC) are evolutionarily conserved (Li et al., 1992b; Quan et al., 2018). The architecture of NY-F has been extensively studied (Mantovani, 1999; Matuoka and Yu Chen, 1999; Matuoka and Chen, 2002; Dolfini et al., 2012). With the characterization of the crystal structure of the NF-Y trimer bound to DNA (Huber et al., 2012; Nardini et al., 2013; Gnesutta et al., 2017; Nardone et al., 2017), it has been clear that the NF-YB and NF-YC interact with each other to form a tight heterodimer through a core region that belongs to the class of histone fold domain (HFD) proteins and is responsible for the non sequence-specific DNA binding. NF-YA could be divided into two domains hosting structurally extended α-helices, as the N-terminal region is required for the binding of NF-YB/NF-YC whereas the C-terminal region is involved in the recognition and binding of the CCAAT elements (Olesen and Guarente, 1990; Maity and De Crombrugghe, 1992; Xing et al., 1993, 2006; Mantovani et al., 1994; Nardini et al., 2013). In addition to the presence of the highly conserved core regions in these three subunits which are responsible for the formation of a trimer, both NF-YA and NF-YC also contain transcriptional activation domains that are glutamine rich and much less conserved (Coustry et al., 1996; Di Silvio et al., 1999). Importantly, the histone fold domain (HFD) present in the core region of NF-YB and NF-YC is not only responsible for the heterodimer interaction via a head-to-tail fashion that is similar to the H2A/H2B in the nucleosome (Luger et al., 1997; Romier et al., 2003) but also is needed for creating a molecular platform facilitating the binding and bending of DNA.
The architecture of the Hap complex has been extensively studied in fungi. A bifunctional lexA-HAP2 fusion experiment revealed that the 265-amino-acid sequence of Hap2 was composed of an essential core of 65 amino acids, a signature sequence which could be further divided into a DNA recognition sequence containing 21 amino acids, although it is devoid of the classical DNA-binding motif, and a subunit association sequence containing 44 amino acids (Olesen and Guarente, 1990). In comparison, the conserved domain in Php2 of S. pombe is a 60-amino-acid block (Olesen et al., 1991). In comparison, the Hap3 subunit harbors a short 7-amino-acid sequence that is responsible for DNA-binding and mutations of this motif fail to recruit Hap4 to the complex and were unable to interact with Hap2 (Xing et al., 1993). Moreover, an 87-amino-acid core domain was found to be conserved in Hap5/Php5 and required for assembly of the Hap2/3/4 complex (McNabb et al., 1997). Of importance, the relative position of each binding domain within different subunits varies and the flanking region of each domain shows no apparent sequence homology between yeast and higher eukaryotes. Actually, the predictions were further supported by recent studies about the crystal structures of the Hap complex subunits (Huber et al., 2012; Nardini et al., 2013; Nardone et al., 2017). In eukaryotes, intrinsic disorders (IDs) are highly abundant, and it is also no exception for NF-YA since a holistic disorder of 96.25% was observed in a general prevalence test for detecting intrinsic disorder of transcriptional regulation (Liu et al., 2006). However, the exact functions of these disordered regions in fungal species are not well understood. Moreover, Hap2, besides of the conserved core domain, harbors a second evolutionarily conserved nuclear localization signals (NLS) which is essential for nuclear localization (Steidl et al., 2004; Tüncher et al., 2005). Apparently, this structure is not limited in yeast species, HapB, the Hap2 homologs in the filamentous fungus Aspergillus nidulans, also contains two putative nuclear localization signal sequences, named as NLS1 and NLS2, respectively. Interestingly, only NLS2, but not NLS1, is required for the nuclear localization of HapB (Tüncher et al., 2005).
A characteristic histone fold domain (HFD) is commonly recognized as the structural marker for all Hap3 homologs, and structurally, this motif is composed of a long central α-helix flanked by two smaller α-helices and is dimerized through the extensive hydrophobic contact in a head-to-tail assembly mode (Arents et al., 1991; Gnesutta et al., 2013; Hortschansky et al., 2017). The C. albicans Hap3 contains two distinct homologs, namely Hap31 and Hap32, and studies have shown that Hap31 and Hap32 individually interact with Hap2 and Hap5 to form DNA-binding complexes under different iron conditions (Singh et al., 2011; Chakravarti et al., 2017).
Hap5 was discovered through a yeast two-hybrid screen, using the core sequence of Hap2 as the bait (Singh et al., 2011). DNA mobility assays showed that no CCAAT-binding activity was detected in the HAP5 depleting mutant, enforcing the role of Hap5 in gene activation (McNabb et al., 1995). Both in vitro and in vivo analyses of the evolutionarily conserved regions of S. cerevisiae Hap5 identified core domains (87 bp in the area of 154–242 amino acid residues) that are required for the assembly of the Hap2-Hap3-Hap5 heterotrimer (McNabb et al., 1995). Similarly, the core domains were also found in S. pombe Php5 and the mammalian homolog CBF-C. Moreover, a second, relatively smaller domain that is essential for the recruitment of Hap4 into the CCAAT-binding complex (CBC) was identified in Hap5 (115–146 amino acids) and Php5 (71–102 amino acids). However, this small domain alone is not sufficient for recruitment of Hap4 (McNabb et al., 1997).
Structural comparison of the Hap complex subunits in yeast, plant and mammalian cells suggests that the highly conserved subunits NF-YB and NF-YC were able to form a canonical HFD heterodimer that is mediated by DNA binding with the trimerization (Zemzoumi et al., 1999; Lee et al., 2003; Romier et al., 2003; Yamamoto et al., 2009; Huber et al., 2012; Nardini et al., 2013; Gnesutta et al., 2017). Nevertheless, significant differences also exist. For example, the corresponding amino acid residue that is located in helix α2 of the HFD structure and responsible for the association with the DNA phosphate backbone is very conserved and identical in both yeast and mammals, however, it differs in the plant Arabidopsis thaliana, as a crystal structure at 2.3 Å resolution of the core domains of the At-L1L/At-NF-YC3 dimer showed that the typical Lys/Arg present in yeast and mammalian NF-YBs was replaced with a negatively charged Asp (Asp84 in At-L1L and Asp55 in LEC1) (Gnesutta et al., 2017). This sequence diversity turns out be important for composition selectivity and sequence flexibility of GC or AT base pairs flanking the CCAAT box, and therefore reflecting functional differences in gene activation in various organisms. Moreover, a proline residue (Pro205 in A. thaliana NF-YA6 and Pro267 in Hap2/HapB subunit of S. cerevisiae and A. nidulans) located in A1A2 linker within the Hap/DNA complex was found to favor the correct preferred positioning of the A2 helix of the Hap2/HapB subunit toward the CCAAT box site, surprisingly, this amino acid somehow is lost in mammalian NF-YA (Nardini et al., 2013; Gnesutta et al., 2017).
Additionally, a structure-based study by Nardini et al. (2013) observed that the head-to-tail assembly of NF-YB and NF-YC subunits provides a stable and compact dimeric scaffold suitable for the interaction of NF-YA and DNA binding. Meanwhile, NF-YA was found to bind to the NF-YB/NF-YC dimer and incorporate an α helix deeply into the minor groove of DNA, and thus ensure the sequence-specific recognition to the CCAAT box. Although the crystal structures of the yeast Hap complex have yet to be solved, it is reasonable to assume the structure assembly and DNA recognition mode might be very similar, given the sequence and functional conservation of the Hap complex subunits in both yeast (Hap2/3/5) and mammals (NF-YA/NF-YB/NF-YC). This notion came from an indirect evidence showing the crystal structures of the HapB/HapC/HapE: DNA complex in the filamentous fungus Aspergillus nidulans (Huber et al., 2012). A sequence identity (~75%) of the histone fold motifs of HapC and HapE with their human homologs of NF-YB and NF-YC reflects a high structural conservation, as both HapC and HapE subunits adopt the similar tertiary structures and also display a nucleosome-like DNA bending by anchoring to the sugar-phosphate backbone in the minor groove of DNA (Bellorini et al., 1997a; Huber et al., 2012).
As mentioned above, Hap4 is only present in fungal species. Studies of Hap4-like protein in yeasts like C. neoformans (HapX) and C. albicans (Hap43) (Jung et al., 2010; Hsu et al., 2011) as well as in filamentous fungi, such as Aspergillus (HapX) and Fusarium (HapX) (Hortschansky et al., 2007; Schrettl et al., 2010; Lopez-Berges et al., 2012), revealed that this unique factor contains several highly conserved domains, including (1) an N-terminal CBC interaction domain; (2) a bZIP/coiled coil region mediating specific DNA binding and dimerization of basic region-leucine zipper (bZIP) transcription factors; (3) up to five cysteine-rich regions (CRR); and (4) a conserved C-terminal motif of approximate 25 amino acids (Brault et al., 2015). Similar domains also exist in the C-terminal domain of yeast bZIP regulators Yap5 and Yap7. It has been confirmed that once the stable Hap2-Hap3-Hap5 heterotrimer is formed, the highly conserved CCAAT sequence is required for the binding to Hap4 via the recruitment domain of Hap5 (McNabb and Pinto, 2005). In yeasts such as S. pombe and C. albicans, and also the filamentous fungus A. nidulans, the highly conserved 17-amino-acid HAP4-like (HAP4L) domain encompassed by the CBC interaction domain was found to contribute to interactions between the recruitment domain of Hap5 and Hap4 binding (Hortschansky et al., 2007; Singh et al., 2011; Brault et al., 2015). S. cerevisiae Hap4 displays a high sequence similarity to the N-terminal region of HapX; however, it lacks the bZIP domain and cysteine-rich regions (Tanaka et al., 2002). The lack of this conserved domain in CBF-C is coherent with the absence of Hap4 homologs in all mammalian complexes being analyzed so far (Mantovani, 1999).
The Assembly and DNA-Binding Modes of Hap Complex
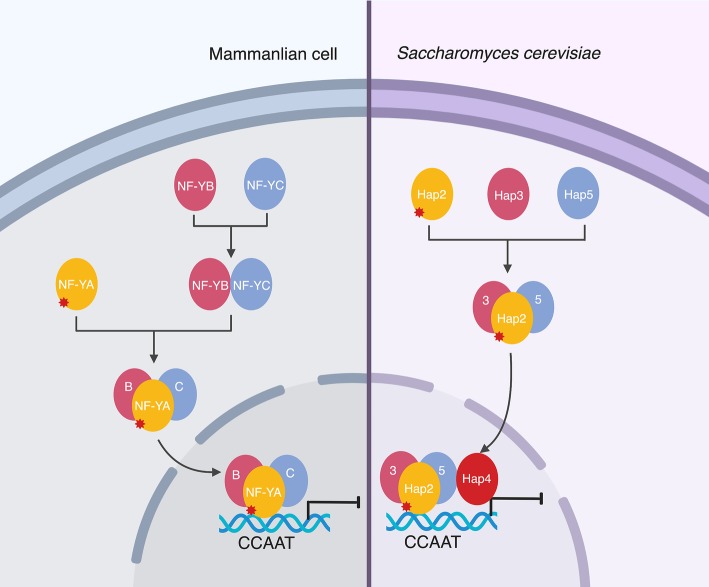
Although all three Hap complex subunits are required for DNA binding, only Hap2 homologs contain the nuclear localization signals (NLS). As such, an interesting question was raised: how do the other subunits transport into the nucleus? It has been generally accepted that a two-step strategy is utilized in mammalian cells. The subunits NF-YB/CBF-A and NF-YC/CBF-C form a heterodimer, afterward they interact with the third subunit, NF-YA/CBF-B, to form a heterotrimeric complex (Baxevanis et al., 1995; Sinha et al., 1995; Kato et al., 1998; Liang, 1998; Romier et al., 2003). Studies in plants like Arabidopsis revealed that the NF-YB/NF-YC heterodimer interacts with some specific transcription factors to form a complex (NF-YB-YC-TF) and regulates the expression of target genes through the binding of transcription factors to specific cis-elements located at the promoters (Wenkel et al., 2006; Yamamoto et al., 2009; Kumimoto et al., 2010). However, it is not the case in the yeasts. Actually, it has been found in S. cerevisiae that Hap2, Hap3, and Hap5 are assembled using a one-step strategy followed by the binding of Hap4 to this trimeric complex. Of importance, this recruitment only happens after Hap4 binds to the CCAAT-containing oligonucleotide (Figure 1; McNabb and Pinto, 2005). A comprehensive study using a GFP-tagged Hap subunit in both the wild-type and hap-deletion strains (ΔhapB, ΔhapC, and ΔhapE) demonstrated that the CBC is assembled in the cytoplasm first, and then the non-conserved NLS2 of Hap2 interacts with the nuclear import machinery and transports the CBC to the nucleus through a “piggy-back” mechanism (Steidl et al., 2004; Hortschansky et al., 2017).
Figure 1.
Comparison of the assembly mode of the Hap complex between yeast and mammalian cells. The symbol marked in red asterisk represents the NLS.
Gene Regulation of Hap Complex in Yeasts
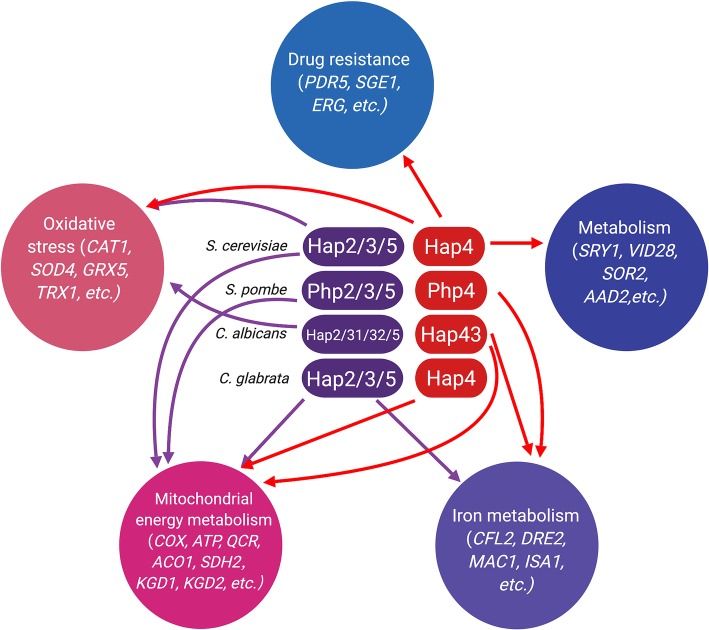
Experimental data obtained from S. cerevisiae indicate that the Hap complex, either directly or indirectly, controls the expression of hundreds of genes, highlighting its important role as a hierarchically highly placed transactivator. In S. cerevisiae, approximately 230 genes are positively regulated, while approximately 240 genes are negatively regulated by the Hap complex (Figure 2; Buschlen et al., 2003). In similar, about 16% of the C. albicans ORFs were differentially regulated in a Hap43-dependent manner under iron-depleted conditions (Singh et al., 2011). Notably, many of the identified genes were found to play roles in sensing oxidative stress and regulating iron homeostasis. Of course, many other pathways were also found to be regulated by the Hap complex. For example, the hap4 null mutant, in addition to its sensitivity to oxidative stress, also displayed chronological life span, decreased resistance to DNA damaging agents, smaller cell size, decreased utilization rate of nitrogen sources, etc. (Riego et al., 2002; Salmon et al., 2004; Longo et al., 2012; Bolotin-Fukuhara, 2017). In particular, the regulatory function of this complex in sensing oxidative stress and iron homeostasis will be addressed in more details.
Figure 2.
A summary of the Hap-mediated regulation in yeasts. Genes mainly regulated by the Hap2/3/5 homolog complex are marked with purple arrows, whereas genes mainly regulated by the Hap4 homolog are marked with red arrows.
Respiration and Reactive Oxygen Species
The observation that the Hap complex in S. cerevisiae binds to the CCAAT sequence located at the UAS of almost all cytochrome genes suggests that this complex may represent a master regulator in respiratory metabolism, since these target genes belong to the mitochondrial respiratory chain (Guarente and Ptashne, 1981; Guarente and Mason, 1983; Guarente and Hoar, 1984; Buschlen et al., 2003; Schüller, 2003). Indeed, when DeRisi et al. examined the global expression patterns linked with different growth phases of S. cerevisiae, they found a dramatic increase of expression in genes encoding the enzymes involving TCA cycle and pathways (DeRisi et al., 1997), and intriguingly, expression of these genes was dependent on the Hap complex (De Winde and Grivell, 1993; Dang et al., 1994). This conclusion was further supported by Buschlen et al. as they also observed that the Hap complex is responsible for the upregulation of genes involved in TCA cycle (Buschlen et al., 2003). Taken together, it is clear that the Hap complex controls the complete TCA cycle and related pathways.
The mitochondrion has long been known to serve as an important compartment participating in crucial cellular events such as energy generation and metabolism in most eukaryotes. Like nucleus, mitochondrion is also a genome-bearing organelle but its genome only encodes ~1% of the total mitochondrial proteins. The majority of mitochondrial proteins is actually encoded by nuclear genes and translocated from cytoplasm to the mitochondrial compartments after translated. This phenomenon raised an important question of how nuclear and mitochondrial genomes coordinate to maintain mitochondrial biosynthesis and functionality. A growing body of studies has demonstrated a mitochondrion-nuclei communication, which is mainly governed by anterograde (nuclear to mitochondrial) or retrograde (mitochondrial to nuclear) signaling, is present under normal and pathophysiological conditions (Ng et al., 2014; Quirós et al., 2016; Guaragnella et al., 2018). Meanwhile, the mitochondrial retrograde response was firstly described in S. cerevisiae where the nuclear RTGs (retrograde signaling genes) were able to sense the cellular changes such as perturbed respiratory function and trigger the expression of a variety of genes involved in alteration of metabolic adaptations. One example is the glyoxylate cycle and fatty acid beta oxidation (Liu and Butow, 2006). Evidence for the integration of the Hap complex in retrograde signaling has been reported, as it has been found that the expression of ACO1, a mitochondrial aconitase-encoding gene in S. cerevisiae, is co-regulated by the Hap2/3/5 complex and the retrograde genes Rtg1 and Rtg3 (Liu and Butow, 1999; Butow and Avadhani, 2004). Deletion of both HAP2 and RTG1 leads to a dramatic loss of mitochondrial DNA, however, this defect could be reversed by expressing an integrated single copy of ACO1, suggesting that the transcriptional regulation of ACO1 by the Hap complex and retrograde signaling pathway is able to directly affect mitochondrial DNA maintenance (Xin et al., 2005).
In addition, Buschlen et al. also found that the Hap complex regulates the expression of nuclear genes responsible for the mitochondrial processes (Galley, 2011) associated with the cellular metabolic state but not the replication and transcription of mitochondrial DNA (Buschlen et al., 2003). Interestingly, HAP4 gene appears to be dispensable in promoting the growth rate of either aerobiosis or anaerobiosis in a standard glucose medium. However, the growth pattern is changed in a low glucose condition where the cell is still able to respire in the absence of HAP4 gene, although its respiration capacity is significantly reduced. On the other hand, the growth rate is also related to the respiration regulation. Transcription of the respiration genes requires the Hap2/3/5 complex when the cells were grown below the intermediate specific growth rate, whereas the Hap4 protein is activated when they are above the intermediate specific growth rate (Raghevendran et al., 2006). Therefore, Hap4 plays an important role in regulating the expression of genes crucial for mitochondrial respiration and reductive pathways.
When the host is invaded by microbial pathogens, one of the major strategies that immune cells like macrophages and neutrophils act to clear the pathogen is to expose the microbe to toxic levels of reactive oxygen species (ROS) such as superoxides, peroxides, and hydroxyl radicals (Collette et al., 2014; Gazendam et al., 2014; Kaloriti et al., 2014; Dantas et al., 2015). Accordingly, in order to survive in the host environment, the pathogen has to evolve strategies to maintain the redox balance. Studies have shown that the Hap complex is one of the major redox sensors for yeast cells and a member of the fine-tuning mechanism that adapts to different levels of oxidative stress (Thön et al., 2007; Chevtzoff et al., 2010; Marinho et al., 2014). For example, the subunits Hap2, Hap31/32, and Hap5 in C. albicans were found to be actively involved in regulating the expression of oxidative stress genes (e.g., CAT1, SOD4, GRX5, and TRX1) in response to iron availability (Chakravarti et al., 2017).
Iron Homeostasis
Iron is an indispensable cofactor in various cellular processes including oxygen transport, amino acid metabolism, and DNA biosynthesis. However, excessive iron may catalyze the formation of reactive oxygen species that may destroy cells by damaging lipids, proteins, and nucleic acids, through a Fenton reaction (Stohs and Bagchi, 1995; Kalinowski, 2005). In fact, in order to prevent any potential hazards caused by iron limitation or excess, cells have to precisely regulate their iron uptake and storage. Recent studies have identified the GATA-type and the bZIP-type transcription factors as key players in regulating iron homeostasis in most fungal species, and found that these factors act through a negative transcriptional feedback loop (Hortschansky et al., 2017). When the environmental iron is sufficient, iron uptake genes are directly repressed by the GATA family transcription factors. By contrast, when the environmental iron is low, the Hap complex directly represses the iron utilization genes; meanwhile, the GATA factor-encoding gene was also inhibited in order to de-repress the iron uptake genes.
In the budding yeast S. cerevisiae, iron regulation depends on the activation and cytoplasm-to-nucleus translocation of two transcriptional activators, Aft1 and Aft2. Under iron-depletion conditions, Aft1 and Aft2 directly bind to the conserved promoter sequences of iron uptake genes and activate the transcription, thus inducing iron acquisition from external sources and releasing free iron deposited in intracellular vacuoles (Blaiseau et al., 2001; Rutherford et al., 2002; Courel et al., 2005; Philpott and Protchenko, 2008; Yatsyshyn et al., 2009). Interestingly, Aft2 appears to behave as a weaker transcriptional activator compared to Aft1, as mutants lacking Aft1 exhibit only poor growth under iron-depletion conditions whereas mutants depleting of both Aft1 and Aft2 almost completely abolish their growth, suggesting that differential expression of iron-uptake genes might be mediated by these two factors (Rutherford et al., 2003).
Unlike the iron-regulatory mode in S. cerevisiae, the expression of iron-responsive genes in the basidiomycete Ustilago maydis (Urbs1) (Voisard et al., 2015), the ascomycetes Penicillium chrysogenum (Srep) (Haas et al., 1997), and Neurospora crass (Sre) (Zhou et al., 1998), and some pathogenic fungi like Aspergillus fumigatus (SreA) (Schrettl et al., 2008), Histoplasma capsulatum (Sre1) (Chao et al., 2008), and Cryptococcus neoformans (Cir1) (Won et al., 2006), is controlled by GATA-type transcriptional repressors with are characterized with the conserved Cys2/Cys2-type zinc finger domains flanked by a cysteine-rich region (Haas et al., 2008) The GATA-type repressors, through a direct binding to the conserved sequence elements of promoter, negatively regulate the expression of iron uptake genes under iron-rich conditions.
In S. pombe, iron homeostasis is controlled by the GATA-type transcription factor Fep1 and the CCAAT-binding factor Php4 (Bird, 2007; Labbé et al., 2007). When the environmental iron is sufficient, Fep1 binds to the GATA cis-acting elements present in the promoter regions of iron acquisition genes and turns off gene expression to avoid the harmful consequences of iron overload (Pelletier et al., 2002, 2003). When the environmental iron is limited, Fep1 is dissociated from DNA, leading to transcriptional de-repression of iron acquisition genes (Pelletier et al., 2002, 2007; Jbel et al., 2009). Meanwhile, the Php4-encoding gene is induced and represses transcription of the PHP complex genes. Unlike the other orthologs, Php4 lacks the b (ZIP) domain and fails to directly bind to DNA. The Php4-dependent gene induction is mediated by the association of Php4 with the monothiol glutaredoxin Grx4. In an iron-replete condition, the [2Fe-2S]2+ cluster coordinates the binding of Grx4 and inactivates Php4, resulting in the nuclear exportation of Php4 in a Crm1-dependent manner (Mercier and Labbé, 2009). Moreover, a spectroscopic study by Dlouhy et al. found that Grx4 was able to bind to a [2Fe-2S]2+ cluster in a way that is similar to previously characterized CGFS glutaredoxins (Dlouhy et al., 2017). In an iron-depleting condition, the binding of Grx4 to the [2Fe-2S]2+ cluster was decomposed, resulting in the functional switch of Php4 as a repressor (Brault et al., 2015; Encinar del Dedo et al., 2015). For example, a genome-wide microarray analysis showed that Php4 is responsible for the transcriptional repression of 86 genes under the condition of iron starvation (Mercier et al., 2008; Mercier and Labbé, 2009) and among them, several genes encode proteins that execute their functions in the iron-dependent metabolic pathways, such as the iron-sulfur cluster assembly, mitochondrial respiration, tricarboxylic acid cycle, and heme biosynthesis. Therefore, the strict regulation of intracellular iron levels is through the interaction between Php4 and Fep1 where each of them controls the expression of its counterpart (Mercier et al., 2008).
Analogous to C. albicans are Sfu1 (GATA-type factor) and Hap43 (CCAAT-binding factor). Unlike the system of S. pombe, the opportunistic human fungal pathogen C. albicans has evolved a new transcription factor (namely Sef1) that could be integrated into the widely conserved regulatory circuit containing both Sfu1 and Hap43. Sfu1 was found to inhibit the expression of SEF1 and iron uptake genes in iron-replete conditions, however, under iron-depleting conditions, the expression of iron uptake genes and HAP43 who is responsible for the repression of iron utilization genes was specifically induced by Sef1 (Chen et al., 2011). More importantly, deletion of the newly identified iron responsive factor Sef1 substantially attenuated the virulence of C. albicans, more likely due to the composite alteration of gene expression related to both iron uptake and pathogenicity (Chen et al., 2011). This could also explain a lack of SEF1 in the non-pathogenic fungi such as S. pombe. In addition to Sef1, studies in the human fungal pathogen C. albicans also suggest that the Hap complex participates in virulence and a different mode of iron regulation might be involved (Chen et al., 2011; Hsu et al., 2011). Unlike those non-pathogenic fungi, the pathogenic fungus C. albicans lives in an iron-poor environment because the host sequesters most of the free iron by hemoglobin, ferritin, transferrin, and lactoferrin, thus urging the fungus to compete with the host and other microbes for the limited iron supply (Schaible and Kaufmann, 2004). By doing so, regulation of iron homeostasis by the Hap complex may evolve a different mode in C. albicans when compared to the nonpathogenic fungi, since the ability of this fungus to capture iron is critical for both its survival and pathogenesis (Sutak et al., 2008). Indeed, a previous study has revealed that the Hap43/Cap2 harbors an amino-terminal bipartite domain containing a fungal-specific Hap-4 like domain and a Yap-like bZIP domain and is responsible for the expression of about 16% of gene ORFs, supporting a critical role of Hap43 in regulation of iron homeostasis (Singh et al., 2011). Of noted, Hap43 directly executes its function in iron regulation. For example, a feed-forward loop exists and is required for the Hap43-dependent expression of Hap2/3/5 complex components, as Hap43 acts to maintain the mRNA level of HAP5 under iron-depleting conditions and mutation analysis further confirmed the functional requirement of both the Hap-4 like and bZIP domains present in Hap43 (Singh et al., 2011). Remarkably, the observation that iron-triggered activation of the Hap complex components requires only Hap43, instead of the core Hap2/3/5 subunits, highly suggests the presence of additions inputs of Hap43 (Singh et al., 2011). Consistent with this hypothesis, the Hap43 regulatory cascade was found to play a dual but contrasting role (both positive and negative) in maintenance of iron homeostasis in C. albicans. For example, a Hap32-dependent Hap5 recruitment on promoters of Hap43-induced or repressed genes (FRP1 and ACO1 respectively) was identified and characterized previously (Singh et al., 2011). This robust coordination of the regulatory machinery could define a unique role of the Hap complex in iron homeostasis and pathogenicity of C. albicans.
Earlier studies (Chakravarti et al., 2017) established the requirement of the Hap complex in connecting the iron acquisition to oxidative stress response, by regulating the expression of genes involved in the production of ROS under iron-overloaded conditions, and the genes include the catalase-encoding gene CAT1, the superoxide dismutase-encoding gene SOD4, the glutaredoxin-encoding gene GRX5, and the thioredoxin-encoding gene TRX1. Furthermore, high levels of catalase increase resistance to oxidative stress and affect iron homeostasis by enhancing cellular demand for iron, thereby reducing the resistance to iron limitation (Pradhan et al., 2017). In addition, both iron and oxidative stress are closely linked since high iron can catalyze production of reactive oxygen species via the Fenton reaction (Pierre et al., 2002). Our unpublished data demonstrated that Hap43 was indeed associated with the resistance to oxidative stress when iron is replete. The dual role is also found in Candida glabrata (Thiébaut et al., 2017). Hap5, together with Hap4, regulates cellular respiration and contributes to iron regulation by binding to Yap5, a transcription factor containing a HAP4-like domain. Interestingly, S. cerevisiae lacks an ortholog of the GATA factor and alternatively evolves new iron regulators that are absent in other fungal species. Moreover, although the CBC/Hap4 complex does exist in S. cerevisiae, it was found that this complex acts to activate gene transcription required for respiration, instead of iron regulation (Guarente and Hoar, 1984). A phylogenetic analysis indicates that the GATA factors are lost in Saccharomyces species undergoing a genome-wide replication like Saccharomyces cerevisiae (Merhej et al., 2015). In contrast, in Saccharomycotina species, such as C. albicans, which are not concerned with whole genome duplication, iron regulation depends on typical Fep1 and Php4 orthologs (Singh et al., 2011).
Similar to the yeasts like S. pombe, in the filamentous fungus A. fumigatus, SreA (GATA-type factor) inhibits the expression of iron acquisition genes during iron adequacy, so it is important to accommodate iron excess. HapX (a CCAAT-binding factor) acts through interaction with CBC and the CBC/HapX complex is critical for adapting to both iron starvation and iron overdose. During iron starvation, the CBC/HapX complex inhibits genes expression involved in the iron-dependent pathway, such as the heme biosynthesis, to reserve iron and activate the iron carrier system to increase iron availability (Hortschansky et al., 2007; Schrettl et al., 2010). However, this complex was found to activate the expression of genes required for vacuolar iron storage and some iron-dependent signaling pathways under iron-replete conditions (Gsaller et al., 2014).
Conclusion
In this review, we have described considerable progresses in understanding the structure and DNA-binding of the Hap complex in yeasts. Moreover, the Hap complex-dependent regulation network was evaluated. As stated, the Hap complex mainly regulates gene expression associated with the oxidative stress response, iron homeostasis, and virulence. However, our understanding of the HAP regulon is still incomplete and many important questions remain to be answered, such as specific mechanisms related to the DNA binding and iron sensing assigned by the Hap4/HapX, proteomic maps of the Hap complex and post-transcriptional/translational modification modes of Hap complex. Undoubtedly, deciphering these questions will greatly help our understanding of the Hap complex regulatory circuits in yeasts and other fungi.
Author Contributions
YM and CC conceived and designed this review, screened and selected the articles, analyzed and interpreted the information, drafted the manuscript, and read and approved the final version of the manuscript. YM designed the figures.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to all members of the Chen-labs, for their valuable suggestions, comments and discussion.
Footnotes
Funding. This research was financially supported by the Chinese National Natural Science Foundation (grants Nos. 31870141, 31570140, 31370174), the Key International collaboration project of the Chinese Academy of Sciences (No. 153831KYSB20170043) and the “100 Talent Program” awarded by the Chinese Academy of Sciences.
References
- Andrés V., Chiara M. D., Mahdavi V. (1994). A new bipartite DNA-binding domain: cooperative interaction between the cut repeat and homeo domain of the cut homeo proteins. Genes Dev. 8, 245–257. 10.1101/gad.8.2.245, PMID: [DOI] [PubMed] [Google Scholar]
- Arents G., Burlingame R. W., Wang B. C., Love W. E., Moudrianakis E. N. (1991). The nucleosomal core histone octamer at 3.1 a resolution: a tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. USA 88, 10148–10152. 10.1073/pnas.88.22.10148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufiero B., Neufeld E. J., Orkin S. H. (1994). Sequence-specific DNA binding of individual cut repeats of the human CCAAT displacement/cut homeodomain protein. Proc. Natl. Acad. Sci. USA 91, 7757–7761. 10.1073/pnas.91.16.7757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek Y. U., Li M., Davis D. A. (2008). Candida albicans ferric reductases are differentially regulated in response to distinct forms of iron limitation by the Rim101 and CBF transcription factors. Eukaryot. Cell 7, 1168–1179. 10.1128/EC.00108-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxevanis A. D., Arents G., Moudrianakis E. N., Landsman D. (1995). A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic Acids Res. 23, 2685–2691. 10.1093/nar/23.14.2685, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D. M., Fikes J. D., Guarente L. (2006). A cDNA encoding a human CCAAT-binding protein cloned by functional complementation in yeast. Proc. Natl. Acad. Sci. USA 88, 1968–1972. 10.1073/pnas.88.5.1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellorini M., Lee D. K., Dantonel J. C., Zemzoumi K., Roeder R. G., Tora L., et al. (1997a). CCAAT binding NF-Y-TBP interactions: NF-YB and NF-YC require short domains adjacent to their histone fold motifs for association with TBP basic residues. Nucleic Acids Res. 25, 2174–2181. 10.1093/nar/25.11.2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellorini M., Zemzoumi K., Farina A., Berthelsen J., Piaggio G., Mantovani R. (1997b). Cloning and expression of human NF-YC. Gene 193, 119–125. 10.1016/s0378-1119(97)00109-1 [DOI] [PubMed] [Google Scholar]
- Berry M., Grosveld F., Dillon N. (1992). A single point mutation is the cause of the Greek form of hereditary persistence of fetal haemoglobin. Nature 358, 499–502. 10.1038/358499a0, PMID: [DOI] [PubMed] [Google Scholar]
- Bird A. J. (2007). Metallosensors, the ups and downs of gene regulation. Adv. Microb. Physiol. 53, 231–267. 10.1016/S0065-2911(07)53004-3 [DOI] [PubMed] [Google Scholar]
- Blaiseau P. L., Lesuisse E., Camadro J. M. (2001). Aft2p, a novel iron-regulated transcription activator that modulates, with Aft1p, intracellular iron use and resistance to oxidative stress in yeast. J. Biol. Chem. 276, 34221–34226. 10.1074/jbc.M104987200, PMID: [DOI] [PubMed] [Google Scholar]
- Bolotin-Fukuhara M. (2017). Thirty years of the HAP2/3/4/5 complex. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 543–559. 10.1016/j.bbagrm.2016.10.011, PMID: [DOI] [PubMed] [Google Scholar]
- Bonander N., Ferndahl C., Mostad P., Wilks M. D. B., Chang C., Showe L., et al. (2008). Transcriptome analysis of a respiratory Saccharomyces cerevisiae strain suggests the expression of its phenotype is glucose insensitive and predominantly controlled by Hap4, Cat8 and Mig1. BMC Genomics 9:365. 10.1186/1471-2164-9-365, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgarel D., Nguyen C. C., Bolotin-Fukuhara M. (1999). HAP4, the glucose-repressed regulated subunit of the HAP transcriptional complex involved in the fermentation-respiration shift, has a functional homologue in the respiratory yeast Kluyveromyces lactis. Mol. Microbiol. 31, 1205–1215. 10.1046/j.1365-2958.1999.01263.x, PMID: [DOI] [PubMed] [Google Scholar]
- Brault A., Mourer T., Labbé S. (2015). Molecular basis of the regulation of iron homeostasis in fission and filamentous yeasts. IUBMB Life 67, 801–815. 10.1002/iub.1441, PMID: [DOI] [PubMed] [Google Scholar]
- Bucher P. (1990). Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 212, 563–578. 10.1016/0022-2836(90)90223-9, PMID: [DOI] [PubMed] [Google Scholar]
- Buschlen S., Amillet J. M., Guiard B., Fournier A., Marcireau C., Bolotin-Fukuhara M. (2003). The S. cerevisiae HAP complex, a key regulator of mitochondrial function, coordinates nuclear and mitochondrial gene expression. Int. J. Genomics 4, 37–46. 10.1002/cfg.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butow R. A., Avadhani N. G. (2004). Mitochondrial signaling: the retrograde response. Mol. Cell 14, 1–15. 10.1016/S1097-2765(04)00179-0, PMID: [DOI] [PubMed] [Google Scholar]
- Chakravarti A., Camp K., McNabb D. S., Pinto I. (2017). The iron-dependent regulation of the Candida albicans oxidative stress response by the CCAAT-binding factor. PLoS One 12:e0170649. 10.1371/journal.pone.0170649, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L. Y., Marletta M. A., Rine J. (2008). Sre1, an iron-modulated GATA DNA-binding protein of iron-uptake genes in the fungal pathogen Histoplasma capsulatum. Biochemistry 47, 7274–7283. 10.1021/bi800066s, PMID: [DOI] [PubMed] [Google Scholar]
- Chen C., Pande K., French S. D., Tuch B. B., Noble S. M. (2011). An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe 10, 118–135. 10.1016/j.chom.2011.07.005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevtzoff C., Yoboue E. D., Galinier A., Casteilla L., Daignan-Fornier B., Rigoulet M., et al. (2010). Reactive oxygen species-mediated regulation of mitochondrial biogenesis in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 285, 1733–1742. 10.1074/jbc.M109.019570, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh L. A., Baldwin A. S., Carthew R. W., Sharp P. A. (1988a). Human CCAAT-binding proteins have heterologous subunits. Cell 53, 11–24. 10.1016/0092-8674(88)90483-7 [DOI] [PubMed] [Google Scholar]
- Chodosh L. A., Olesen J., Hahn S., Baldwin A. S., Guarente L., Sharp P. A. (1988b). A yeast and a human CCAAT-binding protein have heterologous subunits that are functionally interchangeable. Cell 53, 25–35. 10.1016/0092-8674(88)90484-9 [DOI] [PubMed] [Google Scholar]
- Collette J. R., Zhou H., Lorenz M. C. (2014). Candida albicans suppresses nitric oxide generation from macrophages via a secreted molecule. PLoS One 9:e96203. 10.1371/journal.pone.0096203, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courel M., Lallet S., Camadro J.-M., Blaiseau P.-L. (2005). Direct activation of genes involved in intracellular iron use by the yeast iron-responsive transcription factor Aft2 without its paralog Aft1. Mol. Cell. Biol. 25, 6760–6771. 10.1128/MCB.25.15.6760-6771.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustry F., Maity S. N., Sinha S., De Crombrugghe B. (1996). The transcriptional activity of the CCAAT-binding factor CBF is mediated by two distinct activation domains, one in the CBF-B subunit and the other in the CBF-C subunit. J. Biol. Chem. 271, 14485–14491. 10.1074/jbc.271.24.14485, PMID: [DOI] [PubMed] [Google Scholar]
- Dang V. -D., Valens M., Bolotin-Fukuhara M., Daignan-Fornier B. (1994). A genetic screen to isolate genes regulated by the yeast CCAAT-box binding protein Hap2p. Yeast 10, 1273–1283. 10.1002/yea.320101004, PMID: [DOI] [PubMed] [Google Scholar]
- Danilition S. L., Frederickson R. M., Taylor C. Y., Miyamoto N. G. (1991). Transcription factor binding and spacing constraints in the human beta-actin proximal promoter. Nucleic Acids Res. 19, 6913–6922. 10.1093/nar/19.24.6913, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantas A. D. S., Day A., Ikeh M., Kos I., Achan B., Quinn J. (2015). Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomol. Ther. 5, 142–165. 10.3390/biom5010142, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Winde J. H., Grivell L. A. (1993). Global regulation of mitochondrial biogenesis in Saccharomyces cerevisiae. Prog. Nucleic Acid Res. Mol. Biol. 46, 51–91. 10.1016/S0079-6603(08)61018-1 [DOI] [PubMed] [Google Scholar]
- DeRisi J. L., Iyer V. R., Brown P. O. (1997). Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278, 680–686. 10.1126/science.278.5338.680, PMID: [DOI] [PubMed] [Google Scholar]
- Di Silvio A., Imbriano C., Mantovani R. (1999). Dissection of the NF-Y transcriptional activation potential. Nucleic Acids Res. 27, 2578–2584. 10.1093/nar/27.13.2578, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlouhy A. C., Beaudoin J., Labbé S., Outten C. E. (2017). Schizosaccharomyces pombe Grx4 regulates the transcriptional repressor Php4 via [2Fe-2S] cluster binding. Metallomics 9, 1096–1105. 10.1039/C7MT00144D, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfini D., Gatta R., Mantovani R. (2012). NF-Y and the transcriptional activation of CCAAT promoters. Crit. Rev. Biochem. Mol. Biol. 47, 29–49. 10.3109/10409238.2011.628970 [DOI] [PubMed] [Google Scholar]
- Dolfini D., Zambelli F., Pavesi G., Mantovani R. (2009). A perspective of promoter architecture from the CCAAT box. Cell Cycle 8, 4127–4137. 10.4161/cc.8.24.10240 [DOI] [PubMed] [Google Scholar]
- Dorn A., Bollekens J., Staub A., Benoist C., Mathis D. (1987). A multiplicity of CCAAT box-binding proteins. Cell 50, 863–872. 10.1016/0092-8674(87)90513-7, PMID: [DOI] [PubMed] [Google Scholar]
- Edwards D., Murray J. A. H., Smith A. G. (2002). Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiol. 117, 1015–1022. 10.1104/pp.117.3.1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinar del Dedo J., Gabrielli N., Carmona M., Ayté J., Hidalgo E. (2015). A cascade of iron-containing proteins governs the genetic iron starvation response to promote iron uptake and inhibit iron storage in fission yeast. PLoS Genet. 11:e1005106. 10.1371/journal.pgen.1005106, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. (1989). Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 3, 1166–1178. 10.1101/gad.3.8.1166, PMID: [DOI] [PubMed] [Google Scholar]
- Galley H. F. (2011). Oxidative stress and mitochondrial dysfunction in sepsis. Br. J. Anaesth. 107, 57–64. 10.1093/bja/aer093, PMID: [DOI] [PubMed] [Google Scholar]
- Gazendam R. P., Van Hamme J. L., Tool A. T. J., Van Houdt M., Verkuijlen P. J. J. H., Herbst M., et al. (2014). Two independent killing mechanisms of Candida albicans by human neutrophils: evidence from innate immunity defects. Blood 124, 590–597. 10.1182/blood-2014-01-551473, PMID: [DOI] [PubMed] [Google Scholar]
- Gnesutta N., Nardini M., Mantovani R. (2013). The H2A/H2B-like histone-fold domain proteins at the crossroad between chromatin and different DNA metabolisms. Transcription 4, 114–119. 10.4161/trns.25002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnesutta N., Saad D., Chaves-Sanjuan A., Mantovani R., Nardini M. (2017). Crystal structure of the Arabidopsis thaliana L1L/NF-YC3 histone-fold dimer reveals specificities of the LEC1 family of NF-Y subunits in plants. Mol. Plant 10, 645–648. 10.1016/j.molp.2016.11.006, PMID: [DOI] [PubMed] [Google Scholar]
- Greene J., Larin Z., Taylor I., Prentice H., Gwinn K., Kingston R. (1987). Multiple basal elements of a human hsp70 promoter function differently in human and rodent cell lines. Mol. Cell. Biol. 7, 3646–3655. 10.1128/MCB.7.10.3646, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsaller F., Hortschansky P., Beattie S. R., Klammer V., Tuppatsch K., Lechner B. E., et al. (2014). The Janus transcription factor HapX controls fungal adaptation to both iron starvation and iron excess. EMBO J. 33, 2261–2276. 10.15252/embj.201489468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guaragnella N., Coyne L. P., Chen X. J., Giannattasio S. (2018). Mitochondria-cytosol-nucleus crosstalk: learning from Saccharomyces cerevisiae. FEMS Yeast Res. 18. 10.1093/femsyr/foy088, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Hoar E. (1984). Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the “TATA box”. Proc. Natl. Acad. Sci. USA 81, 7860–7864. 10.1073/pnas.81.24.7860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Lalonde B., Gifford P., Alani E. (1984). Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell 36, 503–511. 10.1016/0092-8674(84)90243-5, PMID: [DOI] [PubMed] [Google Scholar]
- Guarente L., Mason T. (1983). Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell 32, 1279–1286. 10.1016/0092-8674(83)90309-4, PMID: [DOI] [PubMed] [Google Scholar]
- Guarente L., Ptashne M. (1981). Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 78, 2199–2203. 10.1073/pnas.78.4.2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas H., Angermayr K., Stöffler G. (1997). Molecular analysis of a Penicillium chrysogenum GATA factor encoding gene (sreP) exhibiting significant homology to the Ustilago maydis urbs1 gene. Gene 184, 33–37. 10.1016/S0378-1119(96)00570-7, PMID: [DOI] [PubMed] [Google Scholar]
- Haas H., Eisendle M., Turgeon B. G. (2008). Siderophores in fungal physiology and virulence. Annu. Rev. Phytopathol. 46, 149–187. 10.1146/annurev.phyto.45.062806.094338, PMID: [DOI] [PubMed] [Google Scholar]
- Hahn S., Guarente L. (1988). Yeast HAP2 and HAP3: transcriptional activators in a heteromeric complex. Science 240, 317–321. 10.1126/science.2832951, PMID: [DOI] [PubMed] [Google Scholar]
- Harada R., Bérubé G., Tamplin O. J., Denis-Larose C., Nepveu A. (1995). DNA-binding specificity of the cut repeats from the human cut-like protein. Mol. Cell. Biol. 15, 129–140. 10.1128/MCB.15.1.129, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S., Koda T., Kakinuma M. (1994). An upstream NF-Y-binding site is required for transcriptional activation from the hst promoter in F9 embryonal carcinoma cells. J. Biol. Chem. 269, 25042–25048. PMID: [PubMed] [Google Scholar]
- Hatamochi A., Golumbek P. T., Van Schaftingen E., de Crombrugghe B. (1988). A CCAAT DNA binding factor consisting of two different components that are both required for DNA binding. J. Biol. Chem. 263, 5940–5947. PMID: [PubMed] [Google Scholar]
- Hooft van Huijsduijnen R., Li X. Y., Black D., Matthes H., Benoist C., Mathis D. (2018). Co-evolution from yeast to mouse: cDNA cloning of the two NF-Y (CP-1/CBF) subunits. EMBO J. 9, 3119–3127. 10.1002/j.1460-2075.1990.tb07509.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortschansky P., Eisendle M., Al-Abdallah Q., Schmidt A. D., Bergmann S., Thön M., et al. (2007). Interaction of HapX with the CCAAT-binding complex—a novel mechanism of gene regulation by iron. EMBO J. 26, 3157–3168. 10.1038/sj.emboj.7601752, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortschansky P., Haas H., Huber E. M., Groll M., Brakhage A. A. (2017). The CCAAT-binding complex (CBC) in Aspergillus species. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 560–570. 10.1016/j.bbagrm.2016.11.008 [DOI] [PubMed] [Google Scholar]
- Hsu P.-C., Chao C.-C., Yang C.-Y., Ye Y.-L., Liu F.-C., Chuang Y.-J., et al. (2013). Diverse Hap43-independent functions of the Candida albicans CCAAT-binding complex. Eukaryot. Cell 12, 804–815. 10.1128/EC.00014-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. C., Yang C. Y., Lan C. Y. (2011). Candida albicans Hap43 is a repressor induced under low-iron conditions and is essential for iron-responsive transcriptional regulation and virulence. Eukaryot. Cell 10, 207–225. 10.1128/EC.00158-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber E. M., Scharf D. H., Hortschansky P., Groll M., Brakhage A. A. (2012). DNA minor groove sensing and widening by the ccaat-binding complex. Structure 20, 1757–1768. 10.1016/j.str.2012.07.012, PMID: [DOI] [PubMed] [Google Scholar]
- Jbel M., Mercier A., Pelletier B., Beaudoin J., Labbe S. (2009). Iron activates in vivo DNA binding of Schizosaccharomyces pombe transcription factor Fep1 through its amino-terminal region. Eukaryot. Cell 8, 649–664. 10.1128/EC.00001-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. C., Cano K. E., Kroger E. C., McNabb D. S. (2005). Novel regulatory function for the CCAAT-binding factor in Candida albicans. Eukaryot. Cell 4, 1662–1676. 10.1128/EC.4.10.1662-1676.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W. H., Saikia S., Hu G., Wang J., Fung C. K., D’Souza C., et al. (2010). HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathog. 6:e1001209. 10.1371/journal.ppat.1001209, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski D. S. (2005). The evolution of iron chelators for the treatment of iron overload disease and cancer. Pharmacol. Rev. 57, 547–583. 10.1124/pr.57.4.2, PMID: [DOI] [PubMed] [Google Scholar]
- Kaloriti D., Jacobsen M., Yin Z., Patterson M., Tillmann A., Smith D. A., et al. (2014). Mechanisms underlying the exquisite sensitivity of Candida albicans to combinatorial cationic and oxidative stress that enhances the potent fungicidal activity of phagocytes. MBio 5, e01334–e01314. 10.1128/mBio.01334-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G., Golumbek P., De Crombrugghe B. (1988). Point mutations and small substitution mutations in three different upstream elements inhibit the activity of the mouse alpha 2(I) collagen promoter. J. Biol. Chem. 263, 13909–13915. PMID: [PubMed] [Google Scholar]
- Kato M. (2005). An overview of the CCAAT-box binding factor in filamentous fungi: assembly, nuclear translocation, and transcriptional enhancement. Biosci. Biotechnol. Biochem. 69, 663–672. 10.1271/bbb.69.663 [DOI] [PubMed] [Google Scholar]
- Kato M., Aoyama A., Naruse F., Tateyama Y., Hayashi K., Miyazaki M., et al. (1998). The Aspergillus nidulans CCAAT-binding factor AnCP/AnCF is a heteromeric protein analogous to the HAP complex of Saccharomyces cerevisiae. Mol. Gen. Genet. 257, 404–411. 10.1007/s004380050664 [DOI] [PubMed] [Google Scholar]
- Kim G., Sheffery M. (1990). Physical characterization of the purified CCAAT transcription factor, alpha-CP1. J. Biol. Chem. 265, 13362–13369. PMID: [PubMed] [Google Scholar]
- Kumimoto R. W., Zhang Y., Siefers N., Holt B. F. (2010). NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 63, 379–391. 10.1111/j.1365-313X.2010.04247.x, PMID: [DOI] [PubMed] [Google Scholar]
- Labbé S., Pelletier B., Mercier A. (2007). Iron homeostasis in the fission yeast Schizosaccharomyces pombe. Biometals 20, 523–537. 10.1007/s10534-006-9056-5, PMID: [DOI] [PubMed] [Google Scholar]
- Lee H., Fischer R. L., Goldberg R. B., Harada J. J. (2003). Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. USA 100, 2152–2156. 10.1073/pnas.0437909100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Li K., Ju Z., Cao D., Fu D., Zhu H., et al. (2016). Genome-wide analysis of tomato NF-Y factors and their role in fruit ripening. BMC Genomics 17:36. 10.1186/s12864-015-2334-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Y., Mantovani R., Hooft Van Huijsduijnen R., Andre I., Benoist C., Mathis D. (1992b). Evolutionary variation of the CCAAT-binding transcription factor NF-Y. Nucleic Acids Res. 20, 1087–1091. 10.1093/nar/20.5.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Y., Van Huijsduijnen R. H., Mantovani R., Benoist C., Mathis D. (1992a). Intron-exon organization of the NF-Y genes. Tissue-specific splicing modifies an activation domain. J. Biol. Chem. 267, 8984–8990. [PubMed] [Google Scholar]
- Liang S. G. (1998). Pathway of complex formation between DNA and three subunits of CBF/NF-Y. J. Biol. Chem. 273, 31590–31598. 10.1074/jbc.273.47.31590, PMID: [DOI] [PubMed] [Google Scholar]
- Liu Z., Butow R. A. (1999). A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol. Cell. Biol. 19, 6720–6728. 10.1128/MCB.19.10.6720, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Butow R. A. (2006). Mitochondrial retrograde signaling. Annu. Rev. Genet. 40, 159–185. 10.1146/annurev.genet.40.110405.090613, PMID: [DOI] [PubMed] [Google Scholar]
- Liu J., Perumal N. B., Oldfield C. J., Su E. W., Uversky V. N., Dunker A. K. (2006). Intrinsic disorder in transcription factors. Biochemistry 45, 6873–6888. 10.1021/bi0602718, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo V. D., Shadel G. S., Kaeberlein M., Kennedy B. (2012). Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 16, 18–31. 10.1016/j.cmet.2012.06.002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Berges M. S., Capilla J., Turra D., Schafferer L., Matthijs S., Jochl C., et al. (2012). HapX-mediated iron homeostasis is essential for rhizosphere competence and virulence of the soilborne pathogen fusarium oxysporum. Plant Cell 24, 3805–3822. 10.1105/tpc.112.098624, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260. 10.1038/38444, PMID: [DOI] [PubMed] [Google Scholar]
- Maity S. N., De Crombrugghe B. (1992). Biochemical analysis of the B subunit of the heteromeric CCAAT-binding factor. A DNA-binding domain and a subunit interaction domain are specified by two separate segments. J. Biol. Chem. 267, 8286–8292. PMID: [PubMed] [Google Scholar]
- Maity S. N., De Crombrugghe B. (1998). Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci. 23, 174–178. 10.1016/S0968-0004(98)01201-8, PMID: [DOI] [PubMed] [Google Scholar]
- Maity S. N., Vuorio T., de Crombrugghe B. (1990). The B subunit of a rat heteromeric CCAAT-binding transcription factor shows a striking sequence identity with the yeast Hap2 transcription factor. Proc. Natl. Acad. Sci. USA 87, 5378–5382. 10.1073/pnas.87.14.5378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. (1998). A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 26, 1135–1143. 10.1093/nar/26.5.1135, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. (1999). The molecular biology of the CCAAT-binding factor NF-Y. Gene 239, 15–27. 10.1016/S0378-1119(99)00368-6, PMID: [DOI] [PubMed] [Google Scholar]
- Mantovani R., Li X. Y., Pessara U., Hooft Van Huisjduijnen R., Benoist C., Mathis D. (1994). Dominant negative analogs of NF-YA. J. Biol. Chem. 269, 20340–20346. PMID: [PubMed] [Google Scholar]
- Marinho H. S., Real C., Cyrne L., Soares H., Antunes F. (2014). Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2, 535–562. 10.1016/j.redox.2014.02.006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marziali G., Perrotti E., Ilari R., Testa U., Coccia E. M., Battistini A. (1997). Transcriptional regulation of the ferritin heavy-chain gene: the activity of the CCAAT binding factor NF-Y is modulated in heme-treated friend leukemia cells and during monocyte-to-macrophage differentiation. Mol. Cell. Biol. 17, 1387–1395. 10.1128/MCB.17.3.1387, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuoka K., Chen K. Y. (2002). Transcriptional regulation of cellular ageing by the CCAAT box-binding factor CBF/NF-Y. Ageing Res. Rev. 1, 639–651. 10.1016/S1568-1637(02)00026-0, PMID: [DOI] [PubMed] [Google Scholar]
- Matuoka K., Yu Chen K. (1999). Nuclear factor Y (NF-Y) and cellular senescence. Exp. Cell Res. 253, 365–371. 10.1006/excr.1999.4605, PMID: [DOI] [PubMed] [Google Scholar]
- McNabb D. S., Pinto I. (2005). Assembly of the Hap2p/Hap3p/Hap4p/Hap5p-DNA complex in Saccharomyces cerevisiae. Eukaryot. Cell 4, 1829–1839. 10.1128/EC.4.11.1829-1839.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb D. S., Tseng K. A., Guarente L. (1997). The Saccharomyces cerevisiae Hap5p homolog from fission yeast reveals two conserved domains that are essential for assembly of heterotetrameric CCAAT-binding factor. Mol. Cell. Biol. 17, 7008–7018. 10.1128/MCB.17.12.7008, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb D. S., Xing Y., Guarente L. (1995). Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev. 9, 47–58. 10.1101/gad.9.1.47, PMID: [DOI] [PubMed] [Google Scholar]
- Mercier A., Labbé S. (2009). Both Php4 function and subcellular localization are regulated by iron via a multistep mechanism involving the glutaredoxin Grx4 and the exportin Crm1. J. Biol. Chem. 284, 20249–20262. 10.1074/jbc.M109.009563, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier A., Watt S., Bähler J., Labbé S. (2008). Key function for the CCAAT-binding factor Php4 to regulate gene expression in response to iron deficiency in fission yeast. Eukaryot. Cell 7, 493–508. 10.1128/EC.00446-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhej J., Delaveau T., Guitard J., Palancade B., Hennequin C., Garcia M., et al. (2015). Yap7 is a transcriptional repressor of nitric oxide oxidase in yeasts, which arose from neofunctionalization after whole genome duplication. Mol. Microbiol. 96, 951–972. 10.1111/mmi.12983, PMID: [DOI] [PubMed] [Google Scholar]
- Myers R. M., Tilly K., Maniatis T. (1986). Fine structure genetic analysis of a beta-globin promoter. Science 232, 613–618. 10.1126/science.3457470, PMID: [DOI] [PubMed] [Google Scholar]
- Nardini M., Gnesutta N., Donati G., Gatta R., Forni C., Fossati A., et al. (2013). Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 152, 132–143. 10.1016/j.cell.2012.11.047, PMID: [DOI] [PubMed] [Google Scholar]
- Nardone V., Chaves-Sanjuan A., Nardini M. (2017). Structural determinants for NF-Y/DNA interaction at the CCAAT box. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 571–580. 10.1016/j.bbagrm.2016.09.006, PMID: [DOI] [PubMed] [Google Scholar]
- Ng S., De Clercq I., Van Aken O., Law S. R., Ivanova A., Willems P., et al. (2014). Anterograde and retrograde regulation of nuclear genes encoding mitochondrial proteins during growth, development, and stress. Mol. Plant 7, 1075–1093. 10.1093/mp/ssu037, PMID: [DOI] [PubMed] [Google Scholar]
- Olesen J. T., Fikes J. D., Guarente L. (1991). The Schizosaccharomyces pombe homolog of Saccharomyces cerevisiae HAP2 reveals selective and stringent conservation of the small essential core protein domain. Mol. Cell. Biol. 11, 611–619. 10.1128/MCB.11.2.611, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen J. T., Guarente L. (1990). The HAP2 subunit of yeast CCAAT transcriptional activator contains adjacent domains for subunit association and DNA recognition: model for the HAP2/3/4 complex. Genes Dev. 4, 1714–1729. 10.1101/gad.4.10.1714, PMID: [DOI] [PubMed] [Google Scholar]
- Olesen J., Hahn S., Guarente L. (1987). Yeast HAP2 and HAP3 activators both bind to the CYC1 upstream activation site, UAS2, in an interdependent manner. Cell 51, 953–961. 10.1016/0092-8674(87)90582-4, PMID: [DOI] [PubMed] [Google Scholar]
- Osada S., Yamamoto H., Nishihara T., Imagawa M. (1996). DNA binding specificity of the CCAAT/enhancer-binding protein transcription factor family. J. Biol. Chem. 271, 3891–3896. 10.1074/jbc.271.7.3891, PMID: [DOI] [PubMed] [Google Scholar]
- Pelletier B., Beaudoin J., Mukai Y., Labbé S. (2002). Fep1, an iron sensor regulating iron transporter gene expression in Schizosaccharomyces pombe. J. Biol. Chem. 277, 22950–22958. 10.1074/jbc.M202682200 [DOI] [PubMed] [Google Scholar]
- Pelletier B., Beaudoin J., Philpott C. C., Labbé S. (2003). Fep1 represses expression of the fission yeast Schizosaccharomyces pombe siderophore-iron transport system. Nucleic Acids Res. 31, 4332–4344. 10.1093/nar/gkg647, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier B., Mercier A., Durand M., Peter C., Jbel M., Beaudoin J., et al. (2007). Expression of Candida albicans Sfu I in fission yeast complements the loss of the iron-regulatory transcription factor Fep I and requires Tup co-repressors. Yeast 24, 883–900. 10.1002/yea.1539, PMID: [DOI] [PubMed] [Google Scholar]
- Petroni K., Kumimoto R. W., Gnesutta N., Calvenzani V., Fornari M., Tonelli C., et al. (2012). The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24, 4777–4792. 10.1105/tpc.112.105734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott C. C., Protchenko O. (2008). Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot. Cell 7, 20–27. 10.1128/EC.00354-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre J. L., Fontecave M., Crichton R. R. (2002). Chemistry for an essential biological process: the reduction of ferric iron. Biometals 15, 341–346. 10.1023/A:1020259021641, PMID: [DOI] [PubMed] [Google Scholar]
- Pradhan A., Herrero-de-Dios C., Belmonte R., Budge S., Lopez Garcia A., Kolmogorova A., et al. (2017). Elevated catalase expression in a fungal pathogen is a double-edged sword of iron. PLoS Pathog. 13:e1006405. 10.1371/journal.ppat.1006405, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramson P. (1993). A serum response element and a binding site for NF-Y mediate the serum response of the human thrombospondin 1 Gene. J. Biol. Chem. 268, 4989–4996. [PubMed] [Google Scholar]
- Quan S., Niu J., Zhou L., Xu H., Ma L., Qin Y. (2018). Identification and characterization of NF-Y gene family in walnut (Juglans regia L.). BMC Plant Biol. 18:255. 10.1186/s12870-018-1459-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirós P. M., Mottis A., Auwerx J. (2016). Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 17, 213–226. 10.1038/nrm.2016.23, PMID: [DOI] [PubMed] [Google Scholar]
- Raghevendran V., Patil K. R., Olsson L., Nielsen J. (2006). Hap4 is not essential for activation of respiration at low specific growth rates in Saccharomyces cerevisiae. J. Biol. Chem. 281, 12308–12314. 10.1074/jbc.M512972200, PMID: [DOI] [PubMed] [Google Scholar]
- Riego L., Avendaño A., DeLuna A., Rodríguez E., González A. (2002). GDH1 expression is regulated by GLN3, GCN4, and HAP4 under respiratory growth. Biochem. Biophys. Res. Commun. 293, 79–85. 10.1016/S0006-291X(02)00174-2, PMID: [DOI] [PubMed] [Google Scholar]
- Romier C., Cocchiarella F., Mantovani R., Moras D. (2003). The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 278, 1336–1345. 10.1074/jbc.M209635200, PMID: [DOI] [PubMed] [Google Scholar]
- Ronchi A., Bellorini M., Mongelli N., Mantovani R. (1995). CCAAT-box binding protein NF-Y (CBF, CP1)recognizes the minor groove and distorts DNA. Nucleic Acids Res. 23, 4565–4572. 10.1093/nar/23.22.4565, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchi A., Berry M., Raguz S., Imam A., Yannoutsos N., Ottolenghi S., et al. (1996). Role of the duplicated CCAAT box region in gamma-globin gene regulation and hereditary persistence of fetal haemoglobin. EMBO J. 15, 143–149. 10.1002/j.1460-2075.1996.tb00342.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford J. C., Jaron S., Ray E., Brown P. O., Winge D. R. (2002). A second iron-regulatory system in yeast independent of Aft1p. Proc. Natl. Acad. Sci. USA 98, 14322–14327. 10.1073/pnas.261381198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford J. C., Jaron S., Winge D. R. (2003). Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J. Biol. Chem. 278, 27636–27643. 10.1074/jbc.M300076200, PMID: [DOI] [PubMed] [Google Scholar]
- Salmon T. B., Evert B. A., Song B., Doetsch P. W. (2004). Biological consequences of oxidative stress-induced DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res. 32, 3712–3723. 10.1093/nar/gkh696, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaible U. E., Kaufmann S. H. E. (2004). Iron and microbial infection. Nat. Rev. Microbiol. 2, 946–953. 10.1038/nrmicro1046, PMID: [DOI] [PubMed] [Google Scholar]
- Schrettl M., Beckmann N., Varga J., Heinekamp T., Jacobsen I. D., Jöchl C., et al. (2010). HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 6:e1001124. 10.1371/journal.ppat.1001124, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M., Kim H. S., Eisendle M., Kragl C., Nierman W. C., Heinekamp T., et al. (2008). SreA-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 70, 27–43. 10.1111/j.1365-2958.2008.06376.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller H.-J. (2003). Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 43, 139–160. 10.1007/s00294-003-0381-8, PMID: [DOI] [PubMed] [Google Scholar]
- Siefers N., Dang K. K., Kumimoto R. W., Bynum W. E., Tayrose G., Holt B. F. (2008). Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 149, 625–641. 10.1104/pp.108.130591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. P., Prasad H. K., Sinha I., Agarwal N., Natarajan K. (2011). Cap2-HAP complex is a critical transcriptional regulator that has dual but contrasting roles in regulation of iron homeostasis in Candida albicans. J. Biol. Chem. 286, 25154–25170. 10.1074/jbc.M111.233569, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S., Maity S. N., Lu J., De Crombrugghet B. (1995). Recombinant rat CBF-C, the third subunit of CBF/NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Biochemistry 92, 1624–1628. 10.1073/pnas.92.5.1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalnik D. G., Strauss E. C., Orkin S. H. (1991). CCAAT displacement protein as a repressor of the myelomonocytic-specific gp91-phox gene promoter. J. Biol. Chem. 266, 16736–16744. PMID: [PubMed] [Google Scholar]
- Steidl S., Tüncher A., Goda H., Guder C., Papadopoulou N., Kobayashi T., et al. (2004). A single subunit of a heterotrimeric CCAAT-binding complex carries a nuclear localization signal: piggy back transport of the pre-assembled complex to the nucleus. J. Mol. Biol. 342, 515–524. 10.1016/j.jmb.2004.07.011, PMID: [DOI] [PubMed] [Google Scholar]
- Stohs S. J., Bagchi D. (1995). Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 18, 321–336. 10.1016/0891-5849(94)00159-H, PMID: [DOI] [PubMed] [Google Scholar]
- Sutak R., Lesuisse E., Tachezy J., Richardson D. R. (2008). Crusade for iron: iron uptake in unicellular eukaryotes and its significance for virulence. Trends Microbiol. 16, 261–268. 10.1016/j.tim.2008.03.005, PMID: [DOI] [PubMed] [Google Scholar]
- Szabo S. J., Gold J. S., Murphy T. L., Murphy K. M. (1993). Identification of cis-acting regulatory elements controlling interleukin-4 gene expression in T cells: roles for NF-Y and NF-ATc. Mol. Cell. Biol. 13, 4793–4805. 10.1128/MCB.13.8.4793, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A., Kato M., Nagase T., Kobayashi T., Tsukagoshi N. (2002). Isolation of genes encoding novel transcription factors which interact with the Hap complex from Aspergillus species. Biochim. Biophys. Acta Gene Struct. Expr. 1576, 176–182. 10.1016/S0167-4781(02)00286-5, PMID: [DOI] [PubMed] [Google Scholar]
- Thiébaut A., Delaveau T., Benchouaia M., Boeri J., Garcia M., Lelandais G., et al. (2017). The CCAAT-binding complex controls respiratory gene expression and iron homeostasis in Candida glabrata. Sci. Rep. 7:3531. 10.1038/s41598-017-03750-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thön M., Al-Abdallah Q., Hortschansky P., Brakhage A. A. (2007). The thioredoxin system of the filamentous fungus Aspergillus nidulans: impact on development and oxidative stress response. J. Biol. Chem. 282, 27259–27269. 10.1074/jbc.M704298200, PMID: [DOI] [PubMed] [Google Scholar]
- Tüncher A., Spröte P., Gehrke A., Brakhage A. A. (2005). The CCAAT-binding complex of eukaryotes: evolution of a second NLS in the HapB subunit of the filamentous fungus Aspergillus nidulans despite functional conservation at the molecular level between yeast, A. nidulans and human. J. Mol. Biol. 352, 517–533. 10.1016/j.jmb.2005.06.068, PMID: [DOI] [PubMed] [Google Scholar]
- Ueno K., Matsumoto Y., Uno J., Sasamoto K., Sekimizu K., Kinjo Y., et al. (2011). Intestinal resident yeast Candida glabrata requires Cyb2p-mediated lactate assimilation to adapt in mouse intestine. PLoS One 6:e24759. 10.1371/journal.pone.0024759, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisard C., Wang J., McEvoy J. L., Xu P., Leong S. A. (2015). urbs1, a gene regulating siderophore biosynthesis in Ustilago maydis, encodes a protein similar to the erythroid transcription factor GATA-1. Mol. Cell. Biol. 13, 7091–7100. 10.1128/mcb.13.11.7091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuorio T., Maity S. N., De Crombrugghe B. (1990). Purification and molecular cloning of the “A” chain of a rat heteromeric CCAAT-binding protein. Sequence identity with the yeast Hap3 transcription factor. J. Biol. Chem. 265, 22480–22486. PMID: [PubMed] [Google Scholar]
- Wenkel S., Turck F., Singer K., Gissot L., Le Gourrierec J., Samach A., et al. (2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18, 2971–2984. 10.1105/tpc.106.043299, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H. J., Sham A., White R., Kronstad J. W. (2006). Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 4:e410. 10.1371/journal.pbio.0040410, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin J. C., Wang X., Kaufman B. A., Butow R. A. (2005). Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science 307, 714–717. 10.1126/science.1106391 [DOI] [PubMed] [Google Scholar]
- Xing Y., Fikes J. D., Guarente L. (1993). Mutations in yeast HAP2/HAP3 define a hybrid CCAAT box binding domain. EMBO J. 12, 4647–4655. 10.1002/j.1460-2075.1993.tb06153.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Zhang S., Olesen J. T., Rich A., Guarente L. (2006). Subunit interaction in the CCAAT-binding heteromeric complex is mediated by a very short alpha-helix in HAP2. Proc. Natl. Acad. Sci. USA 91, 3009–3013. 10.1073/pnas.91.8.3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Kagaya Y., Toyoshima R., Kagaya M., Takeda S., Hattori T. (2009). Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 58, 843–856. 10.1111/j.1365-313X.2009.03817.x, PMID: [DOI] [PubMed] [Google Scholar]
- Yang C., Bolotin E., Jiang T., Sladek F. M., Martinez E. (2007). Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene 389, 52–65. 10.1016/j.gene.2006.09.029, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsyshyn V. Y., Ishchuk O. P., Voronovsky A. Y., Fedorovych D. V., Sibirny A. A. (2009). Production of flavin mononucleotide by metabolically engineered yeast Candida famata. Metab. Eng. 11, 163–167. 10.1016/j.ymben.2009.01.004, PMID: [DOI] [PubMed] [Google Scholar]
- Zemzoumi K., Frontini M., Bellorini M., Mantovani R. (1999). NF-Y histone fold α1 helices help impart CCAAT specificity. J. Mol. Biol. 286, 327–337. 10.1006/jmbi.1998.2496, PMID: [DOI] [PubMed] [Google Scholar]
- Zhou L. W., Haas H., Marzluf G. A. (1998). Isolation and characterization of a new gene, sre, which encodes a GATA-type regulatory protein that controls iron transport in Neurospora crassa. Mol. Gen. Genet. 259, 532–540. 10.1007/s004380050845 [DOI] [PubMed] [Google Scholar]
- Zorbas H., Rein T., Krause A., Hoffmann K., Winnacker E. L. (1992). Nuclear factor I (NF I) binds to an NF I-type site but not to the CCAAT site in the human α-globin gene promoter. J. Biol. Chem. 267, 8478–8484. PMID: [PubMed] [Google Scholar]