Abstract
Fibroblast growth factor homologous factor 1 (FHF1) is an intracellular protein that does not bind to cell surface fibroblast growth factor receptor. Here, we report that FHF1 is abundantly present in Leydig cells with up‐regulation during its development. Adult male Sprague Dawley rats were intraperitoneally injected with 75 mg/kg ethane dimethane sulphonate (EDS) to ablate Leydig cells to initiate their regeneration. Then, rats daily received intratesticular injection of FHF1 (0, 10 and 100 ng/testis) from post‐EDS day 14 for 14 days. FHF1 increased serum testosterone levels without affecting the levels of luteinizing hormone and follicle‐stimulating hormone. FHF1 increased the cell number staining with HSD11B1, a biomarker for Leydig cells at the advanced stage, without affecting the cell number staining with CYP11A1, a biomarker for all Leydig cells. FHF1 did not affect PCNA‐labelling index in Leydig cells. FHF1 increased Leydig cell mRNA (Lhcgr, Scarb1, Star, Cyp11a1, Hsd3b1, Cyp17a1, Hsd17b3, Insl3, Nr5a1 and Hsd11b1) and their protein levels in vivo. FHF1 increased preadipocyte biomarker Dlk1 mRNA level and decreased fully differentiated adipocyte biomarker (Fabp4 and Lpl) mRNA and their protein levels. In conclusion, FHF1 promotes Leydig cell regeneration from stem cells while inhibiting the differentiation of preadipocyte/stem cells into adipocytes in EDS‐treated testis.
Keywords: differentiation, FHF1, Leydig cells, proliferation, regeneration, testosterone
1. INTRODUCTION
Adult Leydig cells (ALCs) are the testicular testosterone (T)‐producing cells, which play a vital role in the male reproductive system. The postnatal development of ALCs is required for the initiation and maintenance of spermatogenesis as well as for the promotion of the male secondary sexual characteristics.1 ALCs are derived from stem Leydig cells (SLCs), which exist near peritubular myoid cells.1 SLCs are spindle‐shaped cells and they differentiate into ALCs when ALCs wear off.1 When ALCs are dramatically damaged such as in the condition of ethane dimethane sulphonate (EDS)‐treated induction of ALC depletion, SLCs rapidly amplify their number via mitosis and then differentiate into ALCs, a process called Leydig cell (LC) regeneration.2, 3, 4, 5 When SLCs enter the LC lineage, they acquire luteinizing hormone (LH) receptor (LHCGR) for receiving LH trophic stimulation and express cholesterol transport proteins, high‐density lipoprotein receptor (SCARB1) and steroidogenic acute regulatory protein (STAR), as well as steroidogenic enzymes, including cytochrome P450 cholesterol side chain cleavage enzyme (CYP11A1), 3β‐hydroxysteroid dehydrogenase isoform 1 (HSD3B1), cytochrome P450 17α‐hydroxylase/17,20‐lyase (CYP17A1) and 17β‐hydroxysteroid dehydrogenase isoform 3 (HSD17B3).1 ALCs also secrete insulin‐like 3 to regulate spermatogenesis.6 The LC regeneration is very similar to the LC developmental process during puberty.7 SLCs begin to commit into progenitor Leydig cells (PLCs) on post‐EDS day 14 and then they further differentiate into immature Leydig cells (ILCs) on post‐EDS day 28, when the biomarker 11β‐hydroxysteroid dehydrogenase isoform 1 (HSD11B1) begins to be expressed in these advanced cells.7
The proliferation and differentiation of SLCs in the LC lineage is controlled by a set of growth factors and hormones.1 Although several critical growth factors such as platelet‐derived growth factor AA,8 dessert hedgehog,9 and kit ligand,10 have been reported, the regulatory growth factors are largely unknown.
By re‐analyzing the transcriptome of LC lineage cells,11 we identify fibroblast growth factor homologous factor 1 (FHF1), which is significantly up‐regulated from SLCs/PLCs into ILCs and further into ALCs, indicating that this peptide might exert autocrine effects on LC development. FHFs form a subfamily of proteins, which have sequences and structures similar to fibroblast growth factors (FGFs).12 FHFs are sometimes named according to FGF nomenclature (FHF1 = FGF12, FHF2 = FGF13, FHF3 = FGF11, FHF4 = FGF14). However, FHFs and FGFs have unrelated functions. FGFs bind to the cell surface receptor with tyrosine kinase activity.13 However, FHFs are expressed as intracellular peptides that bind to some intracellular proteins14 or cytoplasmic tails of some ion channels.15, 16 FHF members play a critical role in developmental cell processes and neuron excitability.15, 17 FHF1 is one of the FHF members that are widely expressed in many tissues such as cartilaginous skeleton, neuron, heart and testis, suggesting a role in the development of these tissues.14, 18 Here, we report that FHF1 stimulates LC development in rats.
2. RESULTS
2.1. FHF1 increases serum T levels in vivo
After re‐analyzing the transcriptome of SLCs, PLCs, ILCs and ALCs,11 we found that the main expression levels (arbitrary unit) of Fhf1 (Fgf12) in SLCs, PLCs, ILCs and ALCs were 46.13, 40.62, 130.12 and 213.18 respectively, suggesting that FHF1 is up‐regulated during LC development. We used an EDS‐treated LC regeneration model to study the function of FHF1 in vivo. Seven days after EDS, all LCs in the testis were eliminated, whereas SLCs were present.19 On post‐EDS day 14, PLCs reappeared, which were formed from the commitment of SLCs4 and on post‐EDS day 28, the PLCs differentiated into ILCs.20, 21 We intratesticularly injected FHF1 (0, 10 or 100 ng/testis/day) starting on post‐EDS day 14 up to day 28 (Figure 1A). After the treatment, FHF1 did not affect bodyweights and testis weights when compared to the control (Table S1). FHF1 dose‐dependently increased serum T levels with significance recorded at 100 ng/testis on post‐EDS day 28 (Figure 1B). However, it did not alter serum LH (Figure 1C) and FSH (Figure 1D) levels. These data suggest that FHF1 promotes LC regeneration primarily via direct action within the testis.
Figure 1.
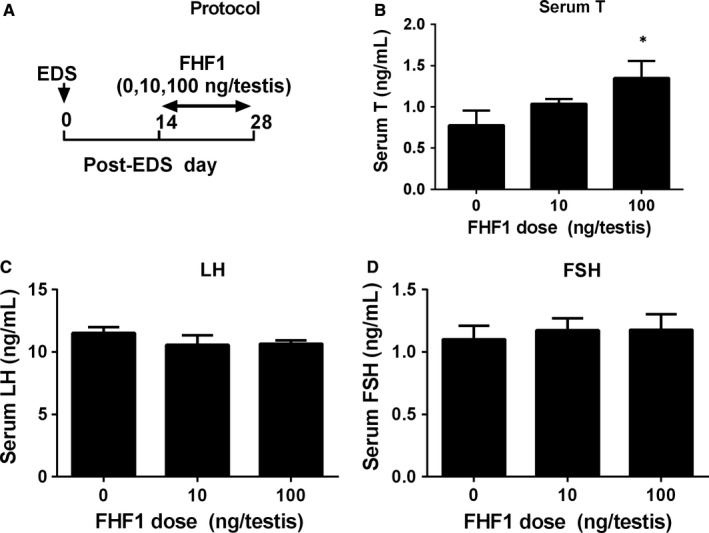
FHF1 experimental protocol and serum testosterone (T), LH and FSH levels after in vivo FHF1 treatment. A, Experimental protocol; B‐D, Serum T, LH, and FSH levels. Mean ± SEM, n = 8. Asterisks (*) designate significant difference from the control (FHF1, 0 ng/testis) at P < 0.05 respectively
2.2. FHF1 increases HSD11B1‐positive LC number in vivo
Elevation of serum T levels could be contributed by the increase of LC number at the advanced stage. We used two biomarkers to label LCs: CYP11A1 (representing all LCs) and HSD11B1 (representing LCs at the advanced stage).7, 20 As shown in Figure 2, FHF1 did not change the number of CYP11A1‐positive LCs. However, FHF1 significantly increased the number of HSD11B1‐positive LCs at 100 ng/testis. This indicates that more LCs reach the advanced stage in the LC lineage after FHF1 treatment. We also measured SOX9‐positive cells and we found that FHF1 did not alter SOX9‐positive Sertoli cell number (Figure 2L).
Figure 2.

Leydig cell (LC) and Sertoli cell (SC) numbers in the testes after in vivo FHF1 treatment Immunohistochemical staining of CYP11A1 (Panels A‐C), HSD11B1 (Panels E‐G) and SOX9 (Panels I‐K) of the testes from the rats treated with 0, 10 and 100 ng/testis FHF1 on post‐EDS day 28. Panels A, E and I: the control; Panels B, F and J: 10 ng/testis FHF1; Panels C, G and K: 100 ng/testis FHF1; Panel D, H and L: quantitative data. Black arrow indicates CYP11A1‐ and HSD11B1‐positive LCs and black arrowhead indicates SOX9‐positive SCs. Bar = 50 mm. Mean ± SEM, n = 8, * P < 0.05 when compared to the control
2.3. FHF1 does not increase the PCNA labelling index in LCs in vivo
We used PCNA to label the proliferating cells and CYP11A1 to stain all LCs. As shown in Figure S1, we did not find that FHF1 increased the ratio of PCNA labelling in CYP11A1‐positive LCs. These results indicate that the increased number of HSD11B1‐positive cells is not contributed by LC proliferation but possibly by its differentiation.
We asked whether the increased number of HSD11B1‐positive LCs came from the proliferation of PLCs. PLCs have a higher capacity of cell division.22 We isolated PLCs and performed cell cycle analysis after in vitro FHF1 treatment. FHF1 did not affect the percentage of cells entering the S and G2 phases (Figure S2). The result indicates that FHF1 does not alter PLC mitosis.
2.4. FHF1 promotes LC differentiation in vivo
We performed RNA sequencing analysis to explore the effects of FHF1 (100 ng/testis) on LC gene expression. We sequenced 14,028 transcripts in the testis of two groups (0 and 100 ng/testis FHF1). Among these transcripts, 197 transcripts were significantly up‐regulated (P < 0.05) and 99 transcripts were significantly down‐regulated (P < 0.05) in the FHF1 group when compared to the control (Figure 3A,B). GO analysis showed that most up‐regulated genes are related to the categories including response to gonadotropin, dioxin metabolic process and steroid metabolic process (Figure 3C) and most down‐regulated genes are related to the categories including positive regulation of T cell differentiation, negative regulation of protein processing and regulation of cell development (Figure 3D).
Figure 3.

RNA‐seq analysis of FHF1‐treated testis. A, Heatmap of mRNAs between FHF1 (F, F1‐4) and control (C, C1‐4) samples; Red colour = up‐regulated genes, Green colour = down‐regulated genes; B, Scatter analysis of mRNAs between FHF1 and control (CON) samples; C, up‐regulated GO; D, down‐regulated GO. Mean ± SEM, n = 4
In the LC steroidogenic pathway, gene expression of LC genes (Star, Cyp11a1, Hsd3b1, Cyp17a1 and Insl3) was up‐regulated by ≥2‐fold (Figure 4). We further verified the LC gene expression and compared it to the Sertoli cell genes using qPCR. As shown in Figure 5, all these LC genes were up‐regulated whereas Sertoli cell genes (Sox9, Fshr, Amh and Dhh) were not altered.
Figure 4.

Gene pathway of Leydig cell (LC) steroidogenesis in FHF1‐treated testis. Red colour = up‐regulated genes at ≥2‐fold, green colour = down‐regulated genes at ≤2‐folds; grey colour = unchanged genes, white colour = unmapped genes; the digital number is the ratio of control over FHF1. Mean ± SEM, n = 4
Figure 5.

QPCR measurement of mRNA levels in the testes after in vivo FHF1 treatment Leydig cell genes: (A) Lhcgr, (B) Scarb1, (C) Star, (D) Cyp11a1, (E) Hsd3b1, (F) Cyp17a1, (G) Hsd17b3, (H) Hsd11b1, (I) Nr5a1, and (J) Insl3. Sertoli cell genes: (K) Sox9, (L) Dhh, (M) Amh, and (N) Fshr. Proliferaring gene: (O) Pcna. Mean ± SEM, n = 8. Asterisks (*, **) designate significant differences from the control (FHF1, 0 ng/testis) at P < 0.05 and 0.01 respectively
We measured the levels of these LC gene products using Western blot. We found that these proteins had similar changes in their respective mRNA levels (Figure 6). In addition, we also used the semi‐quantitative measurement of CYP11A1, HSD11B1, and SOX9 densities in the testis and we found that their densities were similar to the Western blotting data (Figure 7).
Figure 6.

Protein levels of Leydig cells after in vivo FHF1 treatment Leydig cell (LC) proteins: A, Western blot band of LC proteins; B, Quantification of protein levels. Mean ± SEM, n = 8. Asterisks (*, **) designate significant differences from the control (FHF1, 0 ng/testis) at P < 0.05 and 0.01 respectively
Figure 7.

Semi‐quantitative measurement of CYP11A1, HSD11B1 and SOX9 levels and Leydig cell (LC) metrics in the testes after in vivo FHF1 treatment Immunohistochemical staining of CYP11A1 (Panels A‐C), HSD11B1 (Panels F‐H) and SOX9 (Panels K‐M) of the testes from the rats treated with 0, 10 and 100 ng/testis FHF1 on post‐EDS day 28. Panels A, F and K: the control (0 ng/testis FHF1); Panels B, G and L: 10 ng/testis FHF1; Panels C, H, and M: 100 ng/testis FHF1; Panels D, I, and N: quantitative data of protein density; Panels E, J and O: LC size, cytoplasmic size and nuclear size. Black arrow indicates CYP11A1, HSD11B1‐positive LCs and SOX9‐positive Sertoli cells. Mean ± SEM, n = 8, Asterisks (*, **) designate significant differences from the control (FHF1, 0 ng/testis) at P < 0.05 and 0.01 respectively
When SLCs and PLCs differentiate into ILCs, they increase cell size and cytoplasmic size.23 We used CYP11A1 to stain LCs to measure the LC metrics. As shown in Figure 7E,J,O, FHF1 significantly increased LC size and cytoplasmic size without affecting the nuclear size in the 100 ng/kg group, indicating that FHF1 stimulates the growth of LCs. These data suggest that FHF1 promotes LC regeneration.
2.5. FHF1 prevents the transition of stem cells into adipocyte in vivo
Our previous study demonstrated that SLCs are multipotent stem cells and can differentiate into adipocytes.24 We performed a transcriptome analysis to check whether FHF1 prevents the differentiation of stem cells into the adipocyte lineage. We found that the inhibitor of preadipocyte to adipocyte transition, Dlk1, was significantly up‐regulated, whereas the markers of fully differentiated adipocytes (Fabp4 and Lpl) were significantly down‐regulated (Figure 8A). We used qPCR to confirm the mRNA levels of these genes (Dlk1, Fabp4 and Lpl) and Western blotting to measure the protein levels of their products. The results showed that Dlk1 mRNA level was up‐regulated and Fabp4 and Lpl mRNA levels were significantly down‐regulated (Figure 8B). The protein levels were in parallel with their respective mRNA levels (Figure 8C). These results indicate that FHF1 promotes the differentiation of SLCs into the LC lineage by inhibiting the differentiation of preadipocyte/stem cells into adipocytes.
Figure 8.

Gene expression in preadipocyte differentiation after FHF1 treatment. A, Signalling pathway analysis: the expression of inhibitor of preadipocyte to adipocytes' transition (Dlk1) was significantly up‐regulated whereas the markers of fully differentiated adipocytes (Fabp4 and Lpl) were significantly down‐regulated. Red colour = up‐regulated genes at ≥2‐fold, green colour = down‐regulated genes at ≥2‐fold; grey colour = unchanged genes; white colour = unmapped genes. B, The expression of genes (Dlk1, Fabp4, and Lpl) was analysed using qPCR FHF1‐treated testes. C, Protein levels of DLK1, FABP4 and LPL were measured using Western blot. Mean ± SEM, n = 8. Asterisks (*, **) designate significant differences from the control (FHF1, 0 ng/testis) at P < 0.05 and 0.01 respectively
2.6. FHF1 regulates the signalling pathways
We measured the levels of total proteins (SIRT1, PGC‐1α and AKT1) and the phosphorylated protein (pAKT1) in the testis after FHF1 treatment. AKT1 level was not changed, whereas phosphorylated AKT1 (pAKT1) levels were significantly increased at 100 ng/testis FHF1 group (Figure 9). FHF1 significantly increased SIRT1 and PGC‐1α levels at 100 ng/testis (Figure 9). We further treated PLCs with 10 and 100 ng/mL FHF1 for 24 hours and found that FHF1 significantly increased pAKT1 and SIRT1 and PGC‐1α levels at 100 ng/mL (Figure 10). This indicates that phosphorylated AKT1 and SIRT1/PGC‐1α signalling pathways are involved in the FHF1‐mediated action in LC regeneration.
Figure 9.
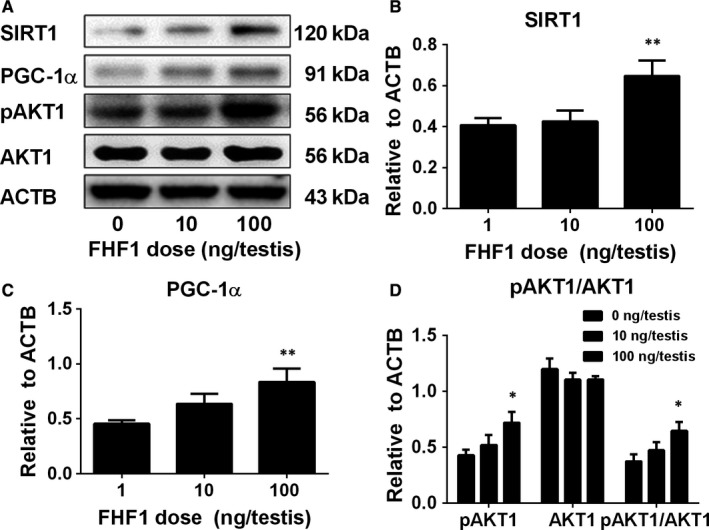
Signalling protein levels after in vivo FHF1 treatment Signalling proteins: A, Western blot bands; B, Quantification of protein levels. Mean ± SEM, n = 5. Asterisks (*, **) designate significant differences from the control (FHF1, 0 ng/testis) at P < 0.05 and 0.01 respectively
Figure 10.
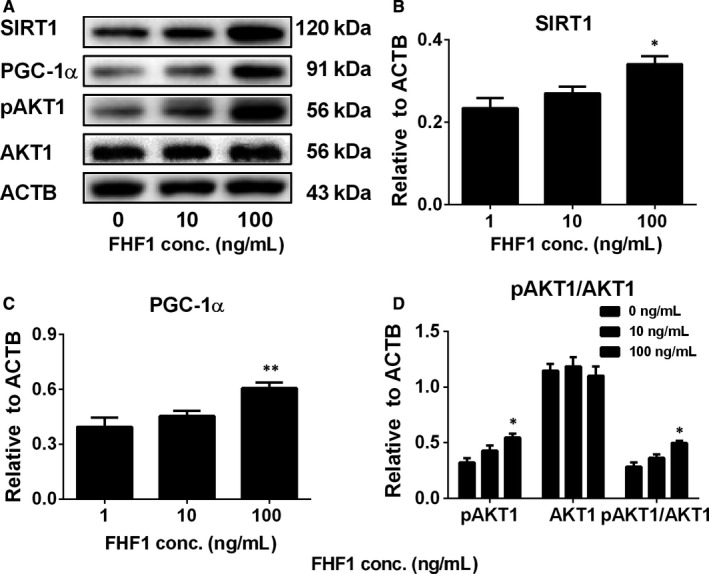
Signalling protein levels after in vitro FHF1 treatment to PLCs Signalling proteins: A, Western blot bands; B, Quantification of protein levels. Mean ± SEM, n = 5. Asterisks (*, **) designate significant differences from the control (FHF1, 0 ng/mL) at P < 0.05 and 0.01 respectively
3. DISCUSSION
Although classic FGFs belong to a family of signalling proteins that bind to the cell surface FGF receptors and play diverse roles in cell growth, differentiation, morphogenesis and developmental processes,25, 26, 27, 28 FHFs are not exact FGF members and are intracellular non‐secretory proteins.29 FHFs lack a signal sequence and cannot be released from cells30 to activate FGF receptors.31 FHF1 binds to islet brain‐2 and voltage‐gated sodium channels and plays a critical role in the membrane targeting and ion channel function.14 Here, we report that FHF1 is developmentally up‐regulated in the LC lineage and promotes LC regeneration by inducing their differentiation.
Serum androgen levels depend on the steroidogenic capacity of LCs and the number of these cells. In the adult rat testis, about 25 million ALCs reside in each testis and they respond to the stimulation of LH to produce T.32 LC number is achieved by the mitosis of SLCs and PLCs and their subsequent differentiation.1 We used the LC regeneration model after EDS injection to examine FHF1 function and its mechanisms. EDS depletes all LCs in adult rat testis without affecting SLCs,5 thus initiating LC regeneration.33, 34, 35 CD90‐positive SLCs isolated from the surface of seminiferous tubules after EDS treatment can be induced to differentiate into LCs in vitro.9 This suggests that SLCs in the EDS‐treated model are CD90‐positive cells and are homogenous. We treated rats with 10 and 100 ng/testis FHF1 on post‐EDS day 14 when SLCs begin to commit into the LC lineage. We selected these doses of FHF1 based on previous publication for doses of FGF136 and FGF16.37 Since the microarray showed that rat testis expressed FGF1, FHF1, and FGF16 with equivalent amounts, the doses of FHF1 might be comparable to those of FGF1 and FGF16. Apparently, FHF1 increased serum T level without affecting serum LH and FSH levels at 100 ng/mL (Figure 1). Previous studies demonstrated that several classic members of the FGF family, FGF2 and FGF16, were able to bind to FGF receptors, thus stimulating SLC and PLC mitosis and inhibiting LC steroidogenesis and regeneration.9, 38, 39 Unlike these classic FGFs, FHF1 instead did not affect LC proliferation, as shown by the unchanged number of CYP11A1‐positive LCs (Figure 2), unaltered PCNA‐labelling index in LCs (Figure S1) and unchanged cell cycle in PLCs (Figure S2). However, FHF1 increased HSD11B1‐positive LC number (Figure 2). This increase could be contributed by the differentiation of SLCs/PLCs into LCs at the advanced stage. Indeed, FHF1 increased the expression of Lhcgr, Scarb1, Star, Cyp11a1, Hsd3b1, Cyp17a1, Hsd17b3, Nr5a1 and Hsd11b1 and their proteins (Figure 5 and Figure 6).
Unlike the classic FGFs, FHF1 should enter the cells via endocytosis. Although the endocytosis efficiency of FHF1 is unknown, the previous study showed that FHF1 showed the dose‐dependent increase of endocytosis to intestinal epithelial cells when 10, 100 and 1000 ng/mL FHF1 were added and after 24‐48 hours, the endocytosis reached a maximum and about 96% of cells showed FHF1 endocytosis.40
The previous study demonstrated that SLCs are multipotent stem cells and are able to differentiate into either the LC lineage or the adipocyte lineage. 24 Using RNA sequencing and qPCR as well as Western blotting, we found that biomarkers of mature adipocytes (Fabp4 and Lpl) were significantly down‐regulated whereas Dlk1 (the biomarker of preadipocytes) and its protein levels were remarkably up‐regulated (Figure 8). DLK1, encoded by Dlk1, is a non‐canonical NOTCH1 ligand that inhibits NOTCH1 signalling in a dose‐dependent manner and modulates the adipogenesis process of 3T3‐L1 preadipocytes.41 FHF1 inhibits the differentiation of preadipocyte/stem cells into adipocytes, whereas it promotes the differentiation of SLCs into the LC lineage.
We further explored the possible pathways that FHF1 might be involved in. Previous studies demonstrated that the AKT1 signalling pathway is involved in LC development.42 AKT1 regulates numerous cellular processes, including cell differentiation, proliferation and apoptosis.43 AKT1 knockout in mice causes morphological abnormalities of mouse testis.44 Another growth factor, insulin‐like growth factor 1 (IGF‐1) also exerts to stimulate LC differentiation via increasing the phosphorylation of AKT1.45 Indeed, IGF‐1 null mice exhibited significant down‐regulation of LC genes, such as Star, Cyp11a1, Hsd3b1 and Cyp17a1 and the reduced T synthesis.46, 47
The SIRT1/PGC‐1α signalling may also be involved in FHF1‐mediated action. SIRT1 is an NAD‐dependent class III histone deacetylase and plays significant roles in many biological activities, including development, gene modification and metabolism via deactivation.48 SIRT1 targets PGC‐1α, a transcriptional co‐activator, which promotes β‐oxidation of fatty acids for the generation of energy.49 SIRT1 interacts with PGC‐1α and deacetylates it to increase PGC‐1α activity, thus promoting the biogenesis of mitochondria, which not only provide energy but also provide the space for steroidogenic enzyme CYP11A1.50 SIRT1 and PGC‐1α are present in the LCs of rodents, indicating that they regulate the mitochondrial biogenesis and steroidogenesis.49 Indeed, the present study demonstrated that FHF1 increased pAKT1, SIRT1 and PGC‐1α levels both in vivo (Figure 9) and in vitro (Figure 10), suggesting that FHF1 regulates Leydig cell development by increasing the levels of these proteins.
NR5A1 may also be involved in FHF1‐mediated action. FGF1 in vivo significantly increased NR5A1 mRNA and protein levels. NR5A1 is a ligand‐free nuclear receptor and is a critical transcription factor for promoting LC development. NR5A1 can bind to the promoters of many LC‐specific genes such as Star, Cyp11a1, Cyp17a1 and Hsd3b1.51, 52, 53, 54 Null mutation of NR5A1 caused LC agenesis.55 Forced expression of NR5A1 can even convert stem cells or fibroblasts into steroidogenic LC‐like cells by transcriptionally promoting the expression of LHCGR and other steroidogenic enzymes (CYP11A1, HSD11B1, CYP17A1 and HSD17B3).56
In conclusion, unlike classic FGFs, which inhibit LC differentiation and stimulate LC proliferation, FHF1 promotes LC differentiation without affecting their proliferation. FHF1 is possibly involved in several signalling pathways, including SIRT1/PGC‐1α and NR5A1 signalling.
4. MATERIALS AND METHODS
4.1. Chemicals and kits
FHF1 was purchased from GenScript (Catalogue number Z03129‐50; Piscataway, NJ). Immulite2000 Total T kit was purchased from Sinopharm (Hangzhou, Zhejiang, China). EDS was purchased from Pterosaur Biotech Co. (Hangzhou, China).
4.2. Animals and treatments
Twenty‐four adult (60‐day‐old) male Sprague Dawley rats were purchased from Shanghai Laboratory Animal Center (Shanghai, China). After one‐week adjustment, each rat received a single intraperitoneal injection of EDS (75 mg/kg bodyweight/once), which was dissolved in a mixture of DMSO: H2O (1:3, v/v), to eliminate LCs from rat testis. LC‐depleted rats were randomly divided into three groups with eight rats per group. FHF1 was dissolved in normal saline for injection. Each rat daily received intratesticular injection (20 μL) of 0, 10 or 100 ng/testis FHF1 for 14 days, starting on post‐EDS day 14. This time‐course of administration regimen was adopted because PLCs begin to emerge from SLCs on post‐EDS day 14.7 On post‐EDS day 28, rats were killed and the blood samples were collected. The serum sample was stored at −20°C to investigate T, LH and FSH. One testis per rat was frozen in −80°C for mRNA and protein expression study. The contralateral testis was punched and fixed in Bouin's solution for immunohistochemical and immunofluorescent staining processes. All animal procedures were performed in accordance with the protocol approved by the Animal Care and Use Committee of Wenzhou Medical University.
4.3. Serum T measurement
Immulite2000 Total T kit was employed to measure the serum T concentrations. The lower detection limit of serum T concentrations was 0.2 ng/mL.
4.4. ELISA for serum LH and FSH levels
According to the manufacturer's instructions (Chemicon CA), each ELISA kit was used to detect the serum levels of LH and FSH. Briefly, sample and assay diluent was mixed, washed and incubated with peroxidase‐conjugated IgG anti‐LH or anti‐FSH. Then, the conjugated complex was washed and substrate was added for the reaction. The levels of LH and FSH were measured at 550 nm using a microplate reader with a correction wavelength at 450 nm.
4.5. RNA sequencing
Sequencing analysis and base calling were conducted using Solexa pipeline v1.8 (Off‐Line Base Caller software, v1.8, Illumina, Foster City, CA). Sequence quality was examined using the FastQC software.57 The trimmed reads (trimmed 5′, 3′‐adaptor bases) were aligned to a reference genome using Hisat2 software.58 The transcript abundance for each sample was estimated with StringTie59 and the FPKM60 value for gene and transcript levels were calculated with R package Ballgown.61 The differentially expressed genes and transcripts were filtered using R package Ballgown.61 The novel genes and transcripts were predicted from assembled results by comparing to the reference annotation using StringTie and Ballgown and the coding potential of those sequences was then assessed using CPAT.60 Principle Component Analysis and correlation analysis were performed according to gene expression level. Hierarchical Clustering, Gene Ontology, Pathway analysis, scatter plots and volcano plots were performed with the differentially expressed genes in R, Python or shell environment for statistical computing and graphics.
4.6. Biological pathway analysis
Biological pathway analysis was performed as previously described.62 GenMAPP2.1 (San Francisco, CA) was used to map the signal pathways of potential pathways. We illustrated the biological pathways containing differentially expressed genes by importing our statistical results into the program. The results of differential gene expression profiles were confirmed using qPCR.
4.7. qPCR
Total RNAs were isolated from testes using Trizol. The concentrations of total RNAs were measured using NanoDrop 2000 (Thermo Scientific, Shanghai, China). The first‐strand cDNA was synthesized and used as the template for qPCR as previously described.63 SYBR Green qPCR kit (Roche, Basel, Switzerland) was used to analyse the LC (Lhcgr, Scarb1, Star, Cyp11a1, Hsd3b1, Cyp17a1, Hsd17b3, Hsd11b1 and Nr5a1) and the Sertoli cell (Fshr, Sox9, Amh and Dhh) mRNA levels. The PCR reaction mixture consisted of SYBR Green mix, primers, cDNAs and RNase‐free water. The procedure for qPCR was set as following: 95°C for 5 minutes, followed by 40 cycles of 95°C for 10 seconds and 60°C for 30 seconds. Ribosomal protein S16 (Rps16) was used as the internal control, which was the house‐keeping gene. The mRNA level of each gene was read as the Ct value and calculated using a standard curve method and was normalized to Rps16 as previously described.64 The primers and gene names are listed in Table S2.
4.8. Western blot
Western blot was performed as previously described.65 Proteins were prepared from FHF1‐injected testes and PLCs. Tissues were homogenized and put into lysis buffer (Bocai Biotechnology, China) to obtain protein samples. The total protein concentrations of samples were measured using the BCA Protein Assay Kit (Takara, Japan). Total protein (30 μg) was loaded and electrophoresed on 10% polyacrylamide gels and then the separated proteins were transferred onto the nitrocellulose membranes. The membranes were blocked with 5% non‐fat milk in TBST buffer for 2 hours and incubated with primary antibodies against LHCGR, SCARB1, STAR, CYP11A1, HSD3B1, CYP17A1, HSD17B3, NR5A1, HSD11B1 and ACTB at 4°C overnight. After that, the membranes were washed and incubated with HRP‐conjugated anti‐rabbit (1:2000; Bioword, St. Louis Park, MN) for 2 hours at room temperature. The band was visualized by chemiluminescence using an ECL kit (Amersham, Arlington Heights, IL). The house‐keeping protein, ACTB, serves as a control. The density of target protein calculated by using J‐Software was normalized to ACTB. All the antibodies used were listed in Table S3.
4.9. Immunohistochemical staining of the testis and enumeration of LCs
Immunohistochemical staining was performed to investigate the effects of FHF1 on LC and Sertoli cell number. One testis from each rat was used for immunohistochemical staining (Vector Laboratories, Inc, Burlingame, CA) according to the manufacturer's instructions. Eight testes per group were randomly prepared as testis samples and then embedded in paraffin as a tissue array. Tissue array samples were dehydrated in ethanol and xylene and then embedded in paraffin. Six micrometre‐thick transverse sections were cut and mounted on glass slides. Antigen retrieval was conducted by microwave irradiation in 10 mM (pH 6.0) citrate buffer for 10 minutes. After that, endogenous peroxidase was blocked with 0.5% of H2O2 in methanol for 30 minutes. The sections were then incubated with CYP11A1 or HSD11B1 or SOX9 polyclonal antibody diluted 1:200 for 1 hour at room temperature. Diaminobenzidine was used for visualizing the antibody‐antigen complexes, positively labelling LCs by brown cytoplasmic staining or Sertoli cells by brown nuclear staining. Mayer haematoxylin was applied in the counterstaining. The sections were dehydrated in graded concentrations of alcohol and cover‐slipped with resin (Thermo Fisher Scientific, Waltham, UK). Non‐immune rabbit IgG was used in the incubation of negative control sections with working dilution the same as the primary antibody.
In order to enumerate CYP11A1‐ or HSD11B1‐positive LC numbers or SOX9‐positive Sertoli cell number, sampling of the testis was performed according to a fractionator technique as previously described.66 Briefly, each testis was cut in eight discs and two discs were randomly selected. Then, discs were cut into four pieces and one piece was randomly selected from a total of eight pieces. These pieces of testis were embedded in paraffin in a tissue array as above. Paraffin blocks were sectioned into 6‐μm‐thick sections. Ten sections were randomly sampled from each testis per rat. Sections were used for immunohistochemical staining. Images were taken using a digital camera, under a 10 × objective and total microscopic fields per section were counted. The histochemical staining was performed as above. The total number of LCs or Sertoli cells was calculated by multiplying the number of LCs or Sertoli cells counted in a known fraction of the testis by the inverse of the sampling probability.
4.10. Immunofluorescent staining of the testis
Immunofluorescent staining was performed to investigate the effects of FHF1 on the proliferation of LCs. Sections were incubated with the primary antibody of PCNA for 60 minutes and then washed and incubated with the CYP11A1 antibody for double staining. Fluorescent secondary antibody (Alexa‐conjugated anti‐rabbit or anti‐mouse IgG, 1:500) were used after the primary antibody. Sections were counterstained with mounting medium containing DAPI. Sections were visualized under a fluorescent microscope (Olympus, Tokyo, Japan). The CYP11A1 (green colour) was used to label LCs and PCNA (red colour) was used to label proliferating cell nucleus.
4.11. PLC isolation
PLCs were isolated as described previously.67 Eighteen 21‐day‐old Sprague Dawley rats were killed by asphyxiation with CO2. In brief, the removed testis was digested with collagenase and DNase (Sigma‐Aldrich, St. Louis, MO) for 15 minutes. Then, the cell suspension was filtered using a 100 μm nylon mesh to remove the tissue debris and the cells were separated under Percoll gradient. The cells with a density of 1.07‐1.088 g/mL were transferred into a new tube and washed. Purified cells were evaluated using histochemical staining for HSD3B1 activity, with 0.4 mM etiocholanolone as the steroid substrate.68 More than 95% of PLCs were stained, indicating that the purity of PLCs was high.
4.12. PLC culture and cell cycle assay
PLCs were seeded into the 6‐well culture plates after isolation with a cell density of 106 cells/well. PLCs were treated with 0, 10 and 100 ng/mL FHF1 in 2.0 mL DMEM: F12 medium for 24 hours. Cells were harvested for analysis of the cell cycle using cytometry and the measurement of AKT1, pAKT1, SIRT and PGC‐1α levels using Western blotting. Then, the cells were harvested and fixed with 75% cold ethanol overnight at 4°C. The fixed cells were washed once with phosphate‐buffered saline (PBS) and stained darkly with propidium iodide (PI) for 30 minutes at room temperature. The stained cells were analysed using a flow cytometer (BD FACSAria, San Diego, CA).
4.13. Statistical analysis
Data are expressed as mean ± SEM. P < 0.05 was considered statistically significant. The differences among groups were evaluated by unpaired student t test when two groups were compared or by one‐way ANOVA followed by ad hoc Dunnett's multiple comparisons to compare with the control when three or more groups were compared. GraphPad version 6 software was used for statistical analysis (GraphPad Inc, CA).
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest to declare.
Supporting information
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (81730042, 81601266) and the Health Department of Zhejiang Province (2017KY483, 11‐CX29), Department of Science and Technology of Zhejiang Province (2019C03035), Wenzhou Science and Technology Bureau (ZS2017009).
Mo J, Chen X, Ni C, et al. Fibroblast growth factor homologous factor 1 stimulates Leydig cell regeneration from stem cells in male rats. J Cell Mol Med. 2019;23:5618–5631. 10.1111/jcmm.14461
Jiaying Mo and Xiuxiu Chen equally contributed to this work.
REFERENCES
- 1. Ye L, Li X, Li L, Chen H, Ge RS. Insights into the development of the adult leydig cell lineage from stem Leydig cells. Front Physiol. 2017;8:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O'Shaughnessy PJ, Morris ID, Baker PJ. Leydig cell re‐generation and expression of cell signaling molecules in the germ cell‐free testis. Reproduction. 2008;135:851‐858. [DOI] [PubMed] [Google Scholar]
- 3. Teerds KJ. Regeneration of Leydig cells after depletion by EDS: a model for postnatal Leydig cell renewal In: Payne AH, Hardy MP, Russell LD, eds. The Leydig cell. Vienna, IL: Cache River Press; 1996:203‐220. [Google Scholar]
- 4. Guo J, Zhou B, Chen H, et al. Comparison of cell types in the rat leydig cell lineage after ethane dimethanesulfonate treatment. In: Annual Meeting American Society of Andrology. San Antonio, Texas, USA: J. of Andrology; 2013. [DOI] [PubMed] [Google Scholar]
- 5. Stanley E, Lin CY, Jin S, et al. Identification, proliferation, and differentiation of adult Leydig stem cells. Endocrinology. 2012;153:5002‐5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anand‐Ivell R, Heng K, Hafen B, Setchell B, Ivell R. Dynamics of INSL3 peptide expression in the rodent testis. Biol Reprod. 2009;81:480‐487. [DOI] [PubMed] [Google Scholar]
- 7. Guo J, Zhou H, Su Z, et al. Comparison of cell types in the rat Leydig cell lineage after ethane dimethanesulfonate treatment. Reproduction. 2013;145:371‐380. [DOI] [PubMed] [Google Scholar]
- 8. Odeh HM, Kleinguetl C, Ge R, Zirkin BR, Chen H. Regulation of the proliferation and differentiation of leydig stem cells in the adult testis. Biol Reprod. 2014;90:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li X, Wang Z, Jiang Z, et al. Regulation of seminiferous tubule‐associated stem Leydig cells in adult rat testes. Proc Natl Acad Sci USA. 2016;113:2666‐2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu S, Chen X, Wang Y, et al. A role of KIT receptor signaling for proliferation and differentiation of rat stem Leydig cells in vitro. Mol Cell Endocrinol. 2017;444:5618‐8. [DOI] [PubMed] [Google Scholar]
- 11. Stanley EL, Johnston DS, Fan J, et al. Stem Leydig cell differentiation: gene expression during development of the adult rat population of Leydig cells. Biol Reprod. 2011;85:1161‐1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goldfarb M. Fibroblast growth factor homologous factors: evolution, structure, and function. Cytokine Growth Factor Rev. 2005;16:215‐220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brewer JR, Mazot P, Soriano P. Genetic insights into the mechanisms of Fgf signaling. Genes Dev. 2016;30:751‐771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Olsen SK, Garbi M, Zampieri N, et al. Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. J Biol Chem. 2003;278:34226‐34236. [DOI] [PubMed] [Google Scholar]
- 15. Liu CJ, Dib‐Hajj SD, Renganathan M, Cummins TR, Waxman SG. Modulation of the cardiac sodium channel Nav1.5 by fibroblast growth factor homologous factor 1B. J Biol Chem. 2003;278:1029‐1036. [DOI] [PubMed] [Google Scholar]
- 16. Wittmack EK, Rush AM, Craner MJ, Goldfarb M, Waxman SG, Dib‐Hajj SD. Fibroblast growth factor homologous factor 2B: association with Nav1.6 and selective colocalization at nodes of Ranvier of dorsal root axons. J Neurosci. 2004;24:6765‐6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cotton L, O'Bryan M, Hinton B. Cellular signaling by fibroblast growth factors (FGFs) and their receptors (FGFRs) in male reproduction. Endocr Rev. 2008;29:193‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Siekierska A, Isrie M, Liu Y, et al. Gain‐of‐function FHF1 mutation causes early‐onset epileptic encephalopathy with cerebellar atrophy. Neurology. 2016;86:2162‐2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo‐Xin H, Qing‐Quan L, Bing‐Bing C, et al. 7alpha‐hydroxytestosterone affects 1 beta‐hydroxysteroid dehydrogenase 1 direction in rat Leydig cells. Endocrinology. 2010;151:748‐754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Phillips DM, Lakshmi V, Monder C. Corticosteroid 11β‐dehydrogenase in rat testis. Endocrinology. 1989;125:209‐216. [DOI] [PubMed] [Google Scholar]
- 21. Ge RS, Hardy DO, Catterall JF, Hardy MP. Developmental changes in glucocorticoid receptor and 11beta‐hydroxysteroid dehydrogenase oxidative and reductive activities in rat Leydig cells. Endocrinology. 1997;138:5089‐5095. [DOI] [PubMed] [Google Scholar]
- 22. Ge RS, Hardy MP. Decreased cyclin A2 and increased cyclin G1 levels coincide with loss of proliferative capacity in rat Leydig cells during pubertal development. Endocrinology. 1997;138:3719‐3726. [DOI] [PubMed] [Google Scholar]
- 23. Shan LX, Hardy MP. Developmental changes in levels of luteinizing hormone receptor and androgen receptor in rat Leydig cells. Endocrinology. 1992;131:1107‐1114. [DOI] [PubMed] [Google Scholar]
- 24. Zhang M, Wang J, Deng C, et al. Transplanted human p75‐positive stem Leydig cells replace disrupted Leydig cells for testosterone production. Cell Death Dis. 2017;8:e3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. And W, Maciag T. The Heparin‐Binding (Fibroblast) growth factor family of proteins. Annu Rev Biochem. 2003;58:575‐606. [DOI] [PubMed] [Google Scholar]
- 26. Rifkin DB, Moscatelli D. Recent developments in the cell biology of basic fibroblast growth factor. J Cell Biol. 1989;109:5618‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yamaguchi TP, Rossant J. Fibroblast growth factors in mammalian development. Curr Opin Genet Dev. 1995;5:485‐491. [DOI] [PubMed] [Google Scholar]
- 28. Guillemot F, Zimmer C. From cradle to grave: the multiple roles of fibroblast growth factors in neural development. Neuron. 2011;71:574‐588. [DOI] [PubMed] [Google Scholar]
- 29. Zhang X, Bao L, Yang L, Wu Q, Li S. Roles of intracellular fibroblast growth factors in neural development and functions. Sci China Life Sci. 2012;55:1038‐1044. [DOI] [PubMed] [Google Scholar]
- 30. Pablo JL, Pitt GS. Fibroblast growth factor homologous factors: new roles in neuronal health and disease. Neuroscientist. 2016;22:19‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Olsen SK, Meirav G, Niccolo Z, et al. Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. J Biol Chem. 2003;278:34226‐34236. [DOI] [PubMed] [Google Scholar]
- 32. Hardy MP, Zirkin BR, Ewing LL. Kinetic studies on the development of the adult population of Leydig cells in testes of the pubertal rat. Endocrinology. 1989;124:762‐770. [DOI] [PubMed] [Google Scholar]
- 33. Rommerts FF, Teerds KJ, Hoogerbrugge JW. In vitro effects of ethylene‐dimethane sulfonate (EDS) on Leydig cells: inhibition of steroid production and cytotoxic effects are dependent on species and age of rat. Mol Cell Endocrinol. 1988;55:87‐94. [DOI] [PubMed] [Google Scholar]
- 34. Teerds KJ, De Rooij DG, Rommerts FF, Wensing CJ. The regulation of the proliferation and differentiation of rat Leydig cell precursor cells after EDS administration or daily HCG treatment. J Androl. 1988;9:343‐351. [DOI] [PubMed] [Google Scholar]
- 35. Smith LB, O'Shaughnessy PJ, Rebourcet D. Cell‐specific ablation in the testis: what have we learned? Andrology. 2015;3:1035‐1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen L, Li X, Wang Y, et al. Fibroblast growth factor 1 promotes rat stem Leydig cell development. Front Endocrinol. 2019;10:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duan Y, Wang Y, Li X, et al. Fibroblast growth factor 16 stimulates proliferation but blocks differentiation of rat stem Leydig cells during regeneration. J Cell Mol Med. 2019;23:2632‐2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu H, Yang Y, Zhang L, et al. Basic fibroblast growth factor promotes stem Leydig cell development and inhibits LH‐stimulated androgen production by regulating microRNA expression. J Steroid Biochem Mol Biol. 2014; 144 Pt B: 483–491. [DOI] [PubMed] [Google Scholar]
- 39. Xiao YC, Hardy DO, Sottas CM, Li XK, Ge RS. Inhibition of LH‐stimulated androgen production in rat immature Leydig cells: effects on nuclear receptor steroidogenic factor 1 by FGF2. Growth Factors. 2009;28:5618–9. [DOI] [PubMed] [Google Scholar]
- 40. Nakayama F, Yasuda T, Umeda S, et al. Fibroblast growth factor‐12 (FGF12) translocation into intestinal epithelial cells is dependent on a novel cell‐penetrating peptide domain: involvement of internalization in the in vivo role of exogenous FGF12. J Biol Chem. 2011;286:25823–25834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nueda ML, Gonzalez‐Gomez MJ, Rodriguez‐Cano MM, et al. DLK proteins modulate NOTCH signaling to influence a brown or white 3T3‐L1 adipocyte fate. Sci Rep. 2018;8:16923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Colon E, Zaman F, Axelson M, et al. Insulin‐like growth factor‐I is an important antiapoptotic factor for rat leydig cells during postnatal development. Endocrinology. 2007;148:128–139. [DOI] [PubMed] [Google Scholar]
- 43. Lawlor MA, Alessi DR. PKB/Akt: a key mediator of cell proliferation, survival and insulin responses? J Cell Sci. 2001;114:2903–2910. [DOI] [PubMed] [Google Scholar]
- 44. Chen WS, Xu PZ, Gottlob K, et al. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 2001;15:2203–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tai P, Shiraishi K, Ascoli M. Activation of the lutropin/choriogonadotropin receptor inhibits apoptosis of immature Leydig cells in primary culture. Endocrinology. 2009;150:3766–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hu GX, Lin H, Chen GR, et al. Deletion of the Igf1 gene: suppressive effects on adult Leydig cell development. J Androl. 2010;31:379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Baker J, Hardy MP, Zhou J, et al. Effects of an Igf1 gene null mutation on mouse reproduction. Mol Endocrinol. 1996;10:903–918. [DOI] [PubMed] [Google Scholar]
- 48. Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD‐dependent histone deacetylase. Nature. 2000;403:795–800. [DOI] [PubMed] [Google Scholar]
- 49. Patti ME, Butte AJ, Crunkhorn S, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc Natl Acad Sci USA. 2003;100:8466–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Higashida K, Kim SH, Jung SR, Asaka M, Holloszy JO, Han DH. Effects of resveratrol and SIRT1 on PGC‐1alpha activity and mitochondrial biogenesis: a reevaluation. PLoS Biol. 2013;11:e1001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen S, Shi H, Liu X, Segaloff DL. Multiple elements and protein factors coordinate the basal and cyclic adenosine 3′,5′‐monophosphate‐induced transcription of the lutropin receptor gene in rat granulosa cells. Endocrinology. 1999;140:2100–2109. [DOI] [PubMed] [Google Scholar]
- 52. Hu MC, Hsu NC, Pai CI, Wang CK, Chung B. Functions of the upstream and proximal steroidogenic factor 1 (SF‐1)‐binding sites in the CYP11A1 promoter in basal transcription and hormonal response. Mol Endocrinol. 2001;15:812–818. [DOI] [PubMed] [Google Scholar]
- 53. Sandhoff TW, Hales DB, Hales KH, McLean MP. Transcriptional regulation of the rat steroidogenic acute regulatory protein gene by steroidogenic factor 1. Endocrinology. 1998;139:4820–4831. [DOI] [PubMed] [Google Scholar]
- 54. Schimmer BP, Tsao J, Cordova M, Mostafavi S, Morris Q, Scheys JO. Contributions of steroidogenic factor 1 to the transcription landscape of Y1 mouse adrenocortical tumor cells. Mol Cell Endocrinol. 2011;336:85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sadovsky Y, Crawford PA, Woodson KG, et al. Mice deficient in the orphan receptor steroidogenic factor 1 lack adrenal glands and gonads but express P450 side‐chain‐cleavage enzyme in the placenta and have normal embryonic serum levels of corticosteroids. Proc Natl Acad Sci USA. 1995;92:10939–10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang Y, Li Z, Wu X, et al. Direct reprogramming of mouse fibroblasts toward Leydig‐like cells by defined factors. Stem Cell Reports. 2017;8:39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brown J, Pirrung M, McCue LA. FQC Dashboard: integrates FastQC results into a web‐based, interactive, and extensible FASTQ quality control tool. Bioinformatics. 2017; 33(19):3137–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Daehwan K, Ben L, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mihaela P, Pertea GM, Antonescu CM, Tsung‐Cheng C, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA‐seq reads. Nat Biotechnol. 2015;33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA‐Seq. Nat Methods. 2008;5:621–628. [DOI] [PubMed] [Google Scholar]
- 61. Frazee AC, Geo P, Jaffe AE, Ben L, Salzberg SL, Leek JT. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol. 2015;33: 243–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang Y, Yuan K, Li X, et al. Leukemia inhibitory factor stimulates steroidogenesis of rat immature Leydig cells via increasing the expression of steroidogenic acute regulatory protein. Growth Factors. 2016;34:166–176. [DOI] [PubMed] [Google Scholar]
- 63. Zhang L, Wang H, Yang Y, et al. NGF induces adult stem Leydig cells to proliferate and differentiate during Leydig cell regeneration. Biochem Biophys Res Commun. 2013;436:300–305. [DOI] [PubMed] [Google Scholar]
- 64. Lin H, Ge RS, Chen GR, et al. Involvement of testicular growth factors in fetal Leydig cell aggregation after exposure to phthalate in utero. Proc Natl Acad Sci USA. 2008;105:7218–7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wu X, Guo X, Wang H, et al. A brief exposure to cadmium impairs Leydig cell regeneration in the adult rat testis. Sci Rep. 2017;7:6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mendis‐Handagama SM, Keeney DS, Hardy MP, Ewing LL. Application of the disector method to enumerate cells in the testis. Ann N Y Acad Sci. 2010;564:86–98. [DOI] [PubMed] [Google Scholar]
- 67. Ge RS, Hardy MP. Variation in the end products of androgen biosynthesis and metabolism during postnatal differentiation of rat Leydig cells. Endocrinology. 1998;139:3787–3795. [DOI] [PubMed] [Google Scholar]
- 68. Payne AH, Wong KL, Vega MM. Differential effects of single and repeated administrations of gonadotropins on luteinizing hormone receptors and testosterone synthesis in two populations of Leydig cells. J Biol Chem. 1980;255:7118–7122. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


