Abstract
Glioma is the most common primary intracranial carcinoma with extremely poor prognosis. The significances of long non‐coding RNA (lncRNA) involved in glioma have been started revealed. However, the expression, roles and molecular mechanisms of most lncRNAs in glioma are still unknown. In this study, we identified a novel lncRNA LINC00526, which is significantly low expressed in glioma. Low expression of LINC00526 is correlated with aggravation and poor survival in glioma. Functional assays revealed that ectopic expression of LINC00526 inhibits glioma cell proliferation, migration, and invasion. LINC00526 silencing promotes glioma cell proliferation, migration and invasion. Mechanistically, we found that LINC00526 directly interacts with EZH2, represses the binding of EZH2 to AXL promoter, attenuates the transcriptional activating roles of EZH2 on AXL, and therefore represses AXL expression. Via repressing AXL, LINC00526 further represses PI3K/Akt/NF‐κB signalling. Intriguingly, we identified that NFKB1 and NFKB2 directly binds LINC00526 promoter and represses LINC00526 transcription. We further found that via activating NF‐κB signalling, AXL represses LINC00526 transcription. Therefore, LINC00526/EZH2/AXL/PI3K/Akt/NF‐κB form a feedback loop in glioma. Analysis of the TCGA data revealed that the expression of LINC00526 is inversely correlated with that of AXL in glioma tissues. In addition, functional rescue assays revealed that the tumour suppressive roles of LINC00526 are dependent on the negative regulation of AXL. Collectively, our data identified LINC00526 as a tumour suppressor in glioma via forming a double negative feedback loop with AXL. Our data also suggested LINC00526 as a potential prognostic biomarker and therapeutic candidate for glioma.
Keywords: AXL, feedback loop, glioma, long non‐coding RNA, progression
1. INTRODUCTION
Glioma is the most prevalent type of intracranial neoplasm with high morbidity and mortality.1 Although multiple therapeutic strategies have emerged, including surgical resection, radiotherapy, chemotherapy and so on, the outcomes of glioma patients are still very poor due to the excessive aggressive growth of glioma and resistance to conventional therapies.2, 3 Therefore, revealing the in‐depth molecular pathogenesis of glioma is of great importance to develop efficient therapies for glioma.
Accumulating genome and transcriptome high‐throughput sequencings have confirmed that most of human genome is transcribed for non‐coding RNAs.4, 5 For example, Iyer et al identified 58,648 genes transcribing for long non‐coding RNAs, and only 21,313 genes encoding for proteins in human cells.6 Long non‐coding RNA (lncRNA) is a class of transcripts with limited protein‐coding potential and more than 200 nucleotides in length.7 Increasing evidences have demonstrated that many lncRNAs are frequently dysregulated in numerous diseases, including cancers.8, 9, 10 Among these dysregulated lncRNAs, several lncRNAs are further verified to be potential diagnostic and prognostic biomarkers for a variety of cancers.11, 12, 13 Furthermore, many lncRNAs have important roles in various physiological and pathological processes.14, 15, 16 As to cancers, numerous lncRNAs have been shown to influence cell proliferation, cell cycle, cell apoptosis, cell senescence, cell migration and invasion, drug‐resistance, in vivo tumour growth and metastasis and so on.17, 18, 19, 20 Therefore, several lncRNAs were identified to be potential therapeutic targets for various cancers.21 LncRNAs have various and complex regulatory roles in control gene expression, particular for the expressions of critical oncogenes and tumour suppressors.22, 23, 24 LncRNAs modulate gene expression at transcriptional and post‐transcriptional levels via interacting with DNAs, proteins, mRNAs and/or microRNAs in different cellular contexts.25, 26, 27
The expression, roles and clinical significances of several lncRNAs in glioma have also been reported.28 Fu et al reported that lncRNA PVT1 is highly expressed in glioma and facilitates glioma tumorigenesis and progression.29 Wang et al revealed that lncRNA‐135528 inhibits glioma progression.30 Liu et al found that lncRNA LINC00470 promoted glioma progression.31 Xu et al identified that lncRNA AC003092.1 promotes glioma temozolomide chemosensitivity.32 Although several lncRNAs have been investigated in glioma, the expression, roles and functional mechanisms of most other lncRNAs in glioma are still unknown.
In a previous report, Zhou et al compared the differentially expressed lncRNAs in 14 glioma tissues and five normal tissues using lncRNA microarray analysis.33 Among these differentially expressed lncRNAs, we noted lncRNA LINC00526, which is markedly down‐regulated in glioma tissues. LINC00526 locates at chromosome 18p11.31, with 1322 nucleotides in length. Open reading frame (ORF) Finder from NCBI (https://www.ncbi.nlm.nih.gov/orffinder/) and Coding Potential Calculator (http://cpc.cbi.pku.edu.cn/programs/run_cpc.jsp) both confirmed that LINC00526 has no protein coding capability. In this study, we further investigated the express pattern of LINC00526 in larger glioma tissue samples, analysed its correlation with clinical features and prognosis, confirmed its biological roles in glioma progression and explored the potential molecular mechanisms mediating the dysregulation of LINC00526 in glioma and the mechanisms responding for the roles of LINC00526 in glioma.
2. MATERIALS AND METHODS
2.1. Tissue samples
Human glioma tissues were obtained from 41 glioma patients who received surgery at the first affiliated hospital of Nanchang university (Nanchang, Jiangxi, China). Normal brain tissues were obtained from 11 patients who received brain tissue resection for craniocerebral injury at the first affiliated hospital of Nanchang university (Nanchang, Jiangxi, China). All tissues were pathologically diagnosed. Tissue samples were preserved at −80°C. The uses of tissue samples were reviewed and approved by the ethics committee of the first affiliated hospital of Nanchang university (Nanchang, Jiangxi, China). Written informed consents were obtained from all patients.
2.2. Cell culture and treatment
Human normal glia cell line NHA, and glioma cell lines LN18, U87, U251 and U138 were obtained from cell banks of the Chinese Academy of Sciences (Shanghai, China). NHA, LN‐18 and U251 cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) (Invitrogen, Carlsbad, CA, USA) with 10% foetal bovine serum (FBS) (Invitrogen). U87 and U138 cells were cultured in Eagle's Minimum Essential Medium (MEM) (Invitrogen) with 10% FBS. All the cells were maintained in an incubator containing 5% CO2 at 37°C. Where indicated, glioma cells were treated with 10 μM Andrographolide (MedChemExpress, Monmouth Junction, NJ, USA) or 1 μM R428 (MedChemExpress) for indicated time.
2.3. RNA extraction, reverse transcription and quantitative real‐time polymerase chain reaction (qRT‐PCR)
Total RNA was extracted from indicated tissues or cells using the RNAiso Plus (Takara, Dalian, China), followed by treatment with DNase I (Takara) to remove genomic DNA. The extracted RNA was further used for reverse transcription with PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara). The generated cDNA was then used for quantitative real‐time polymerase chain reaction (qRT‐PCR) with SYBR® Premix Ex Taq™ II kit (Takara) on ABI7500 Real‐Time PCR System (Applied Biosystems, Foster City, CA, USA) following the standard SYBR Green protocol. The primers’ sequences were as follows: for LINC00526, 5′‐TTCAGGCTTCTGGGTCTC‐3′ (sense) and 5′‐TTCACGGTTGGTATTTCGG‐3′ (antisense); for AXL, 5′‐CACCTCCCTGCAGCTTTC‐3′ (sense) and 5′‐CTCAGGTTGAAGGGGGTG‐3′ (antisense)34; and for GAPDH, 5′‐GGTCTCCTCTGACTTCAACA‐3′ (sense) and 5′‐GTGAGGGTCTCTCTCTTCCT‐3′ (antisense).35 GAPDH was employed as an endogenous control. The quantification of the expression of RNAs was calculated by the 2−ΔΔCt method.
2.4. Plasmids construction
LINC00526 full‐length sequences were PCR‐amplified by the PfuUltra II Fusion HS DNA Polymerase (Agilent Technologies, Santa Clara, CA, USA) and the primers 5′‐CGGGATCCGCGGACTCCGCGGACAAG‐3′ (sense) and 5′‐GGAATTCCAAAATGCATCTTGTTTATTTGGC‐3′ (antisense). Then, the PCR products were cloned into the BamH I and EcoR I sites of pcDNATM3.1(+) (Invitrogen) and pSPT19 (Roche, Mannheim, Germany) plasmids, named as pcDNA‐LINC00526 and pSPT19‐LINC00526, respectively. The cDNA oligonucleotides silencing LINC00526 were synthesized by GenePharma (Shanghai, China) and inserted into the GenePharma SuperSilencingTM shRNA expression plasmid pGPU6/Neo. The shRNAs target sites were 5′‐GCTCAATGTCTCATAGCTACA‐3′ (shLinc‐1) and 5′‐GGTCCTCCAAGATGAGCTTAA‐3′ (shLinc‐2). AXL ORF expression plasmid (Catalog EX‐Z7835‐M68) was purchased from GeneCopoeia (Guangzhou, China).
2.5. Small interfering RNA (siRNA) synthesis and transfection
AXL specific and control Stealth siRNAs (siRNA ID HSS100897) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). The transfection of plasmids and siRNAs was carried out by the Lipofectamine 3000 (Invitrogen) following the protocol.
2.6. Stable cell lines construction
To obtain LINC00526 stably ectopically expressed glioma cells, pcDNA‐LINC00526 or pcDNA empty plasmids were transfected into U87 and U251 cells. The cells were treated with neomycin 48 hours after transfection for 4 weeks to select LINC00526 stably ectopically expressed cells. To obtain LINC00526 stably silenced glioma cells, shLinc‐1 or shLinc‐2 was transfected into U87 and U251 cells. The cells were treated with neomycin 48 hours after transfection for 4 weeks to select LINC00526 stably silenced cells. To obtain LINC00526 and AXL concurrently stably ectopically expressed glioma cells, AXL ORF expression plasmid (Catalog EX‐Z7835‐M68) was transfected into LINC00526 stably ectopically expressed U87 cells. The cells were treated with puromycin 48 hours after transfection for 4 weeks to select LINC00526 and AXL concurrently stably ectopically expressed cells.
2.7. Cell proliferation assays
Cell proliferation was evaluated by Cell Counting Kit‐8 (CCK‐8) and Ethynyl deoxyuridine (EdU) incorporation assays. For CCK‐8 assay, indicated glioma cells were seeded into 96‐well plates at a density of 2000 cells per well. After culture for indicated time, 10 μL CCK‐8 solution (Dojindo, Kumamoto, Japan) was added into per well. After incubation for 1 hour, the absorbance at 450 nm was measured to indicate cell proliferation rate. EdU incorporation assay was performed using the EdU kit (RiboBio, Guangzhou, China) in accordance with the protocol. The results were acquired by the Zeiss Photomicroscope (Carl Zeiss, Oberkochen, Germany) and analysed by counting at least five random fields.
2.8. Cell migration and invasion assays
Transwell migration and invasion assays were undertaken to evaluate the cell migration and invasion potential. Indicated glioma cells resuspended in serum‐free medium were seeded into the upper chamber of 24‐well inserts with 8 μm pores (Millipore, Billerica, MA, USA). For invasion assay, the upper chamber was pre‐coated with Matrigel (BD Biosciences, San Jose, CA, USA). Medium containing 10% FBS was added to lower chamber. After culture at 37°C for 48 hours, the cells remaining on the upper chamber were scraped off, and while those on the lower side of chamber were fixed, stained and observed using a Zeiss Photomicroscope. The results were analysed by counting at least five random fields.
2.9. Cytoplasmic and nuclear RNA purification
Cytoplasmic and nuclear RNA were purified from U87 cells using the Cytoplasmic & Nuclear RNA Purification Kit (Norgen, Belmont, CA, USA) following the instruction. After purification, the RNA was used for reverse transcription and qRT‐PCR as above described. U6 was employed as nuclear control, and while β‐actin was employed as cytoplasmic control. The primers used were as follows: for LINC00526, 5′‐TTCAGGCTTCTGGGTCTC‐3′ (sense) and 5′‐TTCACGGTTGGTATTTCGG‐3′ (antisense); for U6, 5′‐GCTTCGGCAGCACATATACTAAAAT‐3′ (sense) and 5′‐CGCTTCACGAATTTGCGTGTCAT‐3′ (antisense)35 and for β‐actin, 5′‐GGGAAATCGTGCGTGACATTAAG‐3′ (sense) and 5′‐TGTGTTGGCGTACAGGTCTTTG‐3′ (antisense).35
2.10. RNA immunoprecipitation (RIP) assay
RNA immunoprecipitation (RIP) assay was conducted using U87 cells with the Magna RIPTM RNA‐Binding Protein Immunoprecipitation Kit (Millipore) and EZH2 specific primary antibody (Millipore) in accordance with the protocol. The enriched RNA was used for reverse transcription and qRT‐PCR as above described. The primers used were as follows: for LINC00526, 5′‐TTCAGGCTTCTGGGTCTC‐3′ (sense) and 5′‐TTCACGGTTGGTATTTCGG‐3′ (antisense); for HOTAIR, 5′‐TTCCACAGACCAACACCC‐3′ (sense) and 5′‐CTAAATCCGTTCCATTCCA‐3′ (antisense) and for GAPDH, 5′‐GGTCTCCTCTGACTTCAACA‐3′ (sense) and 5′‐GTGAGGGTCTCTCTCTTCCT‐3′ (antisense).35
2.11. RNA pull‐down assay
LINC00526 full‐length sequences were in vitro transcribed and biotin‐labelled from pSPT19‐LINC00526 with the Biotin RNA Labeling Mix (Roche) and T7 RNA polymerase (Roche) following their protocols. LINC00526 antisense RNA was in vitro transcribed and biotin‐labelled from pSPT19‐LINC00526 with the Biotin RNA Labeling Mix (Roche) and Sp6 RNA polymerase (Roche) following their protocols. After treatment with DNase I (Takara), the in vitro transcribed RNA was purified by the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). Next, 3 µg of purified biotinylated RNA was incubated with 1 mg of U87 or U251 whole‐cell lysates at 25°C for 1 hour. Then, the complexes were isolated by streptavidin agarose beads (Invitrogen). The enriched proteins in the complexes were measured by Western blot.
2.12. Western blot
Total cell lysates were isolated from indicated glioma cells by the RIPA lysis buffer (Beyotime, Shanghai, China). Identical quantities of proteins were separated by sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and transferred into PVDF membrane (Millipore). After being blocked with 5% non‐fat milk, the blots were further incubated with primary antibodies against AXL (Cell Signaling Technology, Danvers, MA, USA), phosphorylated Akt (p‐Akt) (Cell Signaling Technology), Akt (Cell Signaling Technology), phosphorylated IκBα (p‐IκBα) (Cell Signaling Technology), EZH2 (Millipore) or GAPDH (Proteintech, Rosemont, IL, USA). After three washes, the blots were incubated with IRdye 700‐conjugated goat anti‐mouse IgG (Thermo Fisher Scientific) or IRdye 800‐conjugated goat anti‐rabbit IgG (Thermo Fisher Scientific). After three washes, the blots were detected by Odyssey Infrared Scanner (Li‐Cor, Lincoln, NE, USA).
2.13. Chromatin immunoprecipitation (ChIP) assay
ChIP assay was undertaken using indicated glioma cells with the EZ‐Magna ChIP™ A/G Chromatin Immunoprecipitation Kit (Millipore) and EZH2 specific primary antibody (Millipore), NFKB1 specific primary antibody (Sigma‐Aldrich, Saint Louis, MO, USA) or NFKB2 specific primary antibody (Sigma‐Aldrich) following the protocol. The enriched DNA was measured by qRT‐PCR as above described. The primers used were as follows: for LINC00526 promoter, 5′‐CCCGACTGTTTCCTACCC‐3′ (sense) and 5′‐GCCTCCCGACTTTTTGATG‐3′ (antisense); for AXL promoter, 5′‐GAGTGAGGGGGAATGAAGG‐3′ (sense) and 5′‐TCTGGGCTCTGTGTCTGGT‐3′ (antisense).
2.14. Statistical analysis
GraphPad Prism 5.0 was used to carry out statistical analyses. For comparisons, Kruskal‐Wallis test followed by Dunn's multiple comparison test, log‐rank test, Student's t test, one‐way ANOVA followed by Dunnett's multiple comparison test, or Pearson correlation analysis were carried out as indicated. P < 0.05 was considered as statistically significant.
3. RESULTS
3.1. LINC00526 was lowly expressed in glioma
We first measured LINC00526 expression level in 41 glioma tissues and 11 normal tissues. As shown in Figure 1A, LINC00526 was obviously lowly expressed in glioma tissues compared with normal tissues. Moreover, LINC00526 was further decreased in aggravated glioma tissues (Figure 1A). The Cancer Genome Atlas (TCGA) data revealed that lower expression of LINC00526 was associated with shorter survival time in brain low grade glioma patients (Figure 1B). Furthermore, LINC00526 expression level in normal glia cell line NHA and glioma cell lines LN18, U87, U251 and U138 was measured. The results showed that LINC00526 was lowly expressed in glioma cell lines compared with normal glia cell line (Figure 1C). Thus, these results indicated that LINC00526 was lowly expressed in glioma. Low expression of LINC00526 was associated with the aggravation and poor prognosis of glioma.
Figure 1.
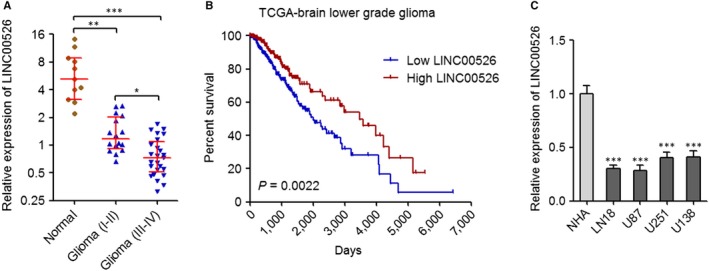
LINC00526 was lowly expressed in glioma. A, LINC00526 expression levels in 25 glioma tissues (III‐VI), 16 glioma tissues (I‐II) and 11 normal tissues were measured by qRT‐PCR. Results are shown as median with interquartile range. *P < 0.05, **P < 0.01, ***P < 0.001, Kruskal‐Wallis test followed by Dunn's multiple comparison test. B, Kaplan‐Meier survival analyses of the correlation between LINC00526 expression level and overall survival of brain lower grade glioma patients (n = 510) from TCGA data. LINC00526 median expression level was used as cut‐off. P = 0.0022, log‐rank test. C, LINC00526 expression levels in normal glia cell line NHA and glioma cell lines LN18, U87, U251 and U138 were measured by qRT‐PCR. Results are shown as mean ± SD of three independent experiments. ***P < 0.001, one‐way ANOVA followed by Dunnett's multiple comparison test
3.2. Ectopic expression of LINC00526 inhibited the proliferation, migration and invasion of glioma cells
To determine the influences of LINC00526 on glioma cells, we stably ectopically expressed LINC00526 in U87 and U251 cells (Figure 2A,2). CCK‐8 assays revealed that ectopic expression of LINC00526 obviously decreased the proliferation rate of U87 and U251 cells (Figure 2C,2). Moreover, EdU incorporation assays further supported the repressive roles of ectopic expression of LINC00526 on the proliferation of U87 and U251 cells (Figure 2E). Transwell migration assays revealed that ectopic expression of LINC00526 significantly decreased the migration of U87 and U251 cells (Figure 2F). Transwell invasion assays revealed that ectopic expression of LINC00526 significantly decreased the invasion of U87 and U251 cells (Figure 2G). Thus, these results indicated that ectopic expression of LINC00526 inhibited the proliferation, migration and invasion of glioma cells.
Figure 2.

Ectopic expression of LINC00526 repressed glioma cell proliferation, migration and invasion. A, LINC00526 expression levels in LINC00526 stably ectopically expressed and control U87 cells were measured by qRT‐PCR. B, LINC00526 expression levels in LINC00526 stably ectopically expressed and control U251 cells were measured by qRT‐PCR. C, Cell proliferation rates of LINC00526 stably ectopically expressed and control U87 cells were measured by CCK‐8 assays. OD values in 450 nm were detected to indicate cell proliferation. D, Cell proliferation rates of LINC00526 stably ectopically expressed and control U251 cells were measured by CCK‐8 assays. OD values in 450 nm were detected to indicate cell proliferation. E, Cell proliferation of LINC00526 stably ectopically expressed and control U87 and U251 cells were measured by EdU incorporation assays. Scale bars, 100 µm. F, Cell migration of LINC00526 stably ectopically expressed and control U87 and U251 cells were measured by transwell migration assays. Scale bars, 100 µm. G, Cell invasion of LINC00526 stably ectopically expressed and control U87 and U251 cells were measured by transwell invasion assays. Scale bars, 100 µm. Results are shown as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test
3.3. LINC00526 silencing promoted the proliferation, migration and invasion of glioma cells
To further confirm the tumour suppressive roles of LINC00526 in glioma, we stably silenced the expression of LINC00526 in U87 and U251 cells using two independent LINC00526 specific shRNAs (Figure 3A,3). CCK‐8 assays revealed that silencing of LINC00526 obviously increased the proliferation rate of U87 and U251 cells (Figure 3C,3). Moreover, EdU incorporation assays further supported the roles of LINC00526 silencing in promoting the proliferation of U87 and U251 cells (Figure 3E). Transwell migration assays revealed that LINC00526 silencing significantly promoted the migration of U87 and U251 cells (Figure 3F). Transwell invasion assays revealed that LINC00526 silencing significantly promoted the invasion of U87 and U251 cells (Figure 3G). Thus, these results indicated that LINC00526 silencing promoted the proliferation, migration and invasion of glioma cells.
Figure 3.
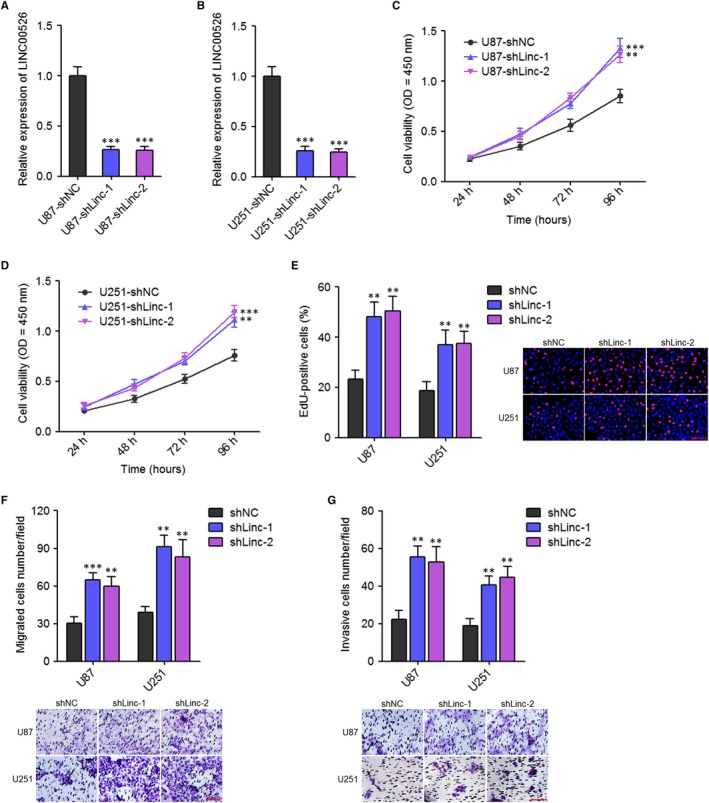
LINC00526 silencing promoted glioma cell proliferation, migration and invasion. A, LINC00526 expression levels in LINC00526 stably silenced and control U87 cells were measured by qRT‐PCR. B, LINC00526 expression levels in LINC00526 stably silenced and control U251 cells were measured by qRT‐PCR. C, Cell proliferation rates of LINC00526 stably silenced and control U87 cells were measured by CCK‐8 assays. OD values in 450 nm were detected to indicate cell proliferation. D, Cell proliferation rates of LINC00526 stably silenced and control U251 cells were measured by CCK‐8 assays. OD values in 450 nm were detected to indicate cell proliferation. E, Cell proliferation of LINC00526 stably silenced and control U87 and U251 cells were measured by EdU incorporation assays. Scale bars, 100 µm. F, Cell migration of LINC00526 stably silenced and control U87 and U251 cells were measured by transwell migration assays. Scale bars, 100 µm. G, Cell invasion of LINC00526 stably silenced and control U87 and U251 cells were measured by transwell invasion assays. Scale bars, 100 µm. Results are shown as mean ± SD of three independent experiments. **P < 0.01, ***P < 0.001, one‐way ANOVA followed by Dunnett's multiple comparison test
3.4. The expression of LINC00526 was inversely correlated with that of AXL in glioma tissues
To investigate the potential mechanisms mediating the tumour suppressive roles of LINC00526 in glioma, we analysed the expression correlations between LINC00526 and mRNAs in glioma tissues using TCGA data. Among the mRNAs whose expressions are correlated with LINC00526, we noted AXL, which is a receptor tyrosine kinase and has critical oncogenic roles in many cancers including glioma.36, 37 As shown in Figure 4A, AXL mRNA expression was significantly inversely correlated with that of LINC00526 in glioma tissues (r = −0.4249, P = 0.0022). In contrast to LINC00526, higher expression of AXL was associated with shorter survival time in glioma patients (Figure 4B).
Figure 4.
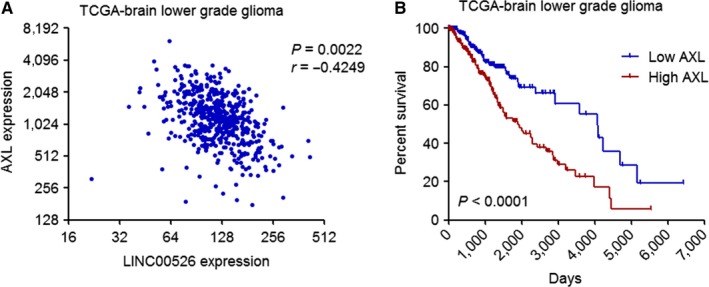
The expression levels of AXL were inversely correlated with that of LINC00526 in glioma tissues. A, The correlations between AXL expression level and LINC00526 expression level in brain lower grade glioma tissues (n = 510) from TCGA data. r = −0.4249, P = 0.0022, Pearson correlation analysis. B, Kaplan‐Meier survival analyses of the correlation between AXL expression level and overall survival of brain lower grade glioma patients (n = 510) from TCGA data. AXL median expression level was used as cut‐off. P < 0.0001, log‐rank test
3.5. LINC00526 repressed AXL transcription via interacting with EZH2
To determine the potential influences of LINC00526 on AXL, we first confirmed the subcellular location of LINC00526. The subcellular location of LINC00526 was predicted by lncLocator (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/).38 The results predicted that LINC00526 was located in the nucleus with a score of 0.63. Next, we performed cytoplasmic and nuclear RNA purification using U87 cells. The location of LINC00526 was measured by qRT‐PCR. The results showed that LINC00526 was mainly located in the nucleus (Figure 5A). Many nuclear lncRNAs were reported to interact with chromatin‐modifying complexes and modulate genes expression.17 About 20% nuclear lncRNAs were found to bind EZH2, a critical component of polycomb repressive complex 2 (PRC2).39 Interestingly, EZH2 was also reported to activate AXL transcription.34 Therefore, we further explore whether LINC00526 also binds EZH2 and whether LINC00526 influences AXL transcription via binding EZH2. RIP assays were performed in U87 cells. The results showed that LINC00526 was specifically enriched in EZH2 antibody group, which indicated the binding between LINC00526 and EZH2 (Figure 5B). Furthermore, RNA pulldown assays were performed using in vitro transcribed LINC00526 in U87 and U251 cells. As shown in Figure 5C, EZH2 was specifically enriched in LINC00526 group, which further supported the binding between LINC00526 and EZH2. Then, we investigated whether the binding between LINC00526 and EZH2 influences the binding of EZH2 on AXL promoter. ChIP assays using EZH2 specific antibody were performed in LINC00526 stably overexpressed and control U87 cells. As shown in Figure 5D, ectopic expression of LINC00526 decreased the binding of EZH2 on AXL promoter. ChIP assays using EZH2 specific antibody were also performed in LINC00526 stably silenced and control U251 cells. The results showed that LINC00526 silencing increased the binding of EZH2 on AXL promoter (Figure 5E). EZH2 was reported to activate AXL transcription in glioma cells. Next, the mRNA and protein levels of AXL in LINC00526 stably overexpressed and control U87 cells, and LINC00526 stably silenced and control U251 cells were measured by qRT‐PCR and Western blot. As shown in Figure 5F‐I, ectopic expression of LINC00526 decreased the mRNA and protein levels of AXL, and while LINC00526 silencing increased the mRNA and protein levels of AXL. As a receptor tyrosine kinase, AXL is well‐known to activate downstream PI3K/Akt/NF‐κB signalling. Next, we investigated the influences of LINC00526 on PI3K/Akt/NF‐κB signalling. As shown in Figure 5J, ectopic expression of LINC00526 decreased the phosphorylation levels of Akt and IκBα. LINC00526 silencing increased the phosphorylation levels of Akt and IκBα (Figure 5K). Taken together, these results suggested that LINC00526 bound to EZH2, decreased the binding of EZH2 to AXL promoter, repressed AXL transcription and repressed PI3K/Akt/NF‐κB signalling.
Figure 5.
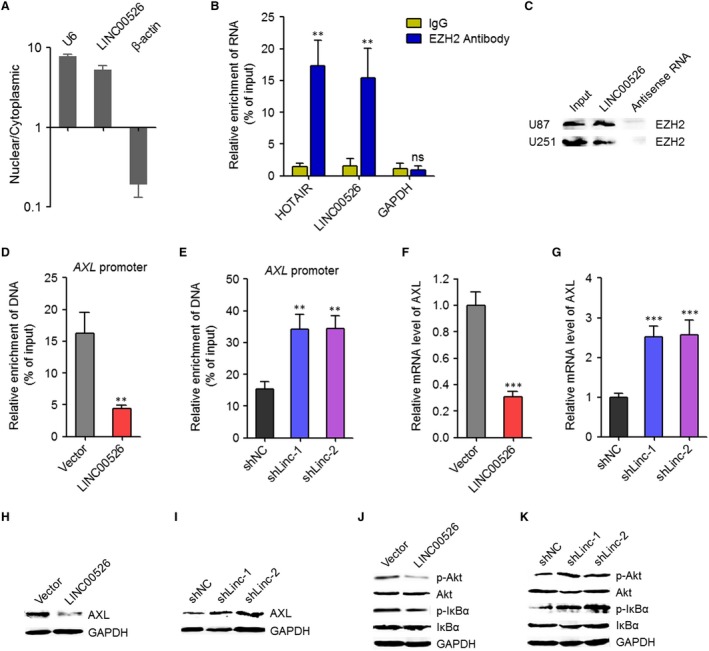
LINC00526 repressed AXL transcription via interacting with EZH2. A, LINC00526 subcellular location was detected by cytoplasmic and nuclear RNA purification, followed by qRT‐PCR. U6 and β‐actin were used as nuclear and cytoplasmic controls, respectively. B, RIP assays were performed in U87 cells using EZH2 specific antibody or nonspecific IgG. The enriched RNA was measured by qRT‐PCR. HOTAIR was used as positive control, and while GAPDH was used as negative control. C, RNA pull‐down assays were performed in U87 and U251 cells using in vitro transcribed biotin‐labelled LINC00526 or antisense RNA. The enriched proteins were detected by Western blot with EZH2 specific antibody. D, ChIP assays were performed in LINC00526 stably ectopically expressed and control U87 cells using EZH2 specific antibody. The enriched DNA was measured by qRT‐PCR and specific primers corresponding to AXL promoter. E, ChIP assays were performed in LINC00526 stably silenced and control U251 cells using EZH2 specific antibody. The enriched DNA was measured by qRT‐PCR and specific primers corresponding to AXL promoter. F, AXL mRNA expression levels in LINC00526 stably ectopically expressed and control U87 cells were measured by qRT‐PCR. G, AXL mRNA expression levels in LINC00526 stably silenced and control U251 cells were measured by qRT‐PCR. H, AXL protein expression levels in LINC00526 stably ectopically expressed and control U87 cells were measured by Western blot. I, AXL protein expression levels in LINC00526 stably silenced and control U251 cells were measured by Western blot. J, Phosphorylation levels of Akt and IκBα in LINC00526 stably ectopically expressed and control U87 cells were measured by Western blot. K, Phosphorylation levels of Akt and IκBα in LINC00526 stably silenced and control U251 cells were measured by Western blot. **P < 0.01, ***P < 0.001, ns, not significant, Student's t test (B, D, F) or one‐way ANOVA followed by Dunnett's multiple comparison test (E, G)
3.6. AXL repressed LINC00526 transcription via activating NF‐κB signalling
Due to the significant inverse correlation between the expression of AXL and LINC00526, we next explored whether AXL modulates the expression of LINC00526. Intriguingly, analysing the promoter region of LINC00526 using JASPAR (http://jaspar.genereg.net/),40 we found one NFKB1/NFKB2 binding site at −438‐−426 upstream of the transcription start site of LINC00526 (Figure 6A). After transient overexpressing AXL in U251 cells (Figure 6B), ChIP assays were performed using NFKB1 and NFKB2 specific antibodies. As shown in Figure 6C, NFKB1 and NFKB2 specifically bound to LINC00526 promoter. Furthermore, ectopic expression of AXL significantly promoted the binding of NFKB1 and NFKB2 to LINC00526 promoter (Figure 6C). After transient silencing AXL in U87 cells (Figure 6D), ChIP assays were performed using NFKB1 and NFKB2 specific antibodies. As shown in Figure 6E, NFKB1 and NFKB2 also specifically bound to LINC00526 promoter in U87 cells. Furthermore, AXL silencing significantly repressed the binding of NFKB1 and NFKB2 to LINC00526 promoter (Figure 6E). The influences of AXL on the transcription level of LINC00526 were investigated by qRT‐PCR. The results showed that ectopic expression of AXL significantly repressed the transcription of LINC00526, and while AXL silencing significantly promoted the transcription of LINC00526 (Figure 6F,G). To confirm whether the repression of LINC00526 transcription by AXL was mediated by the activation of NF‐κB signalling, the AXL overexpressed U251 cells were treated with NF‐κB inhibitor Andrographolide. As shown in Figure 6H, treatment with Andrographolide abolished the repressive roles of AXL on LINC00526 transcription. Thus, these results suggested that AXL repressed LINC00526 transcription via enhancing NF‐κB signalling. Next, we investigated whether LINC00526 regulates the effects of NF‐κB on LINC00526 via repressing AXL. ChIP assays were performed using NFKB1 and NFKB2 specific antibodies in LINC00526 stably overexpressed and control U87 cells, and LINC00526 stably silenced and control U251 cells. As shown in Figure 6I,J, in contrast to AXL, ectopic expression of LINC00526 significantly repressed the binding of NFKB1 and NFKB2 to LINC00526 promoter, and while LINC00526 silencing significantly promoted the binding of NFKB1 and NFKB2 to LINC00526 promoter. These results supported the auto‐regulatory roles of LINC00526 on itself.
Figure 6.
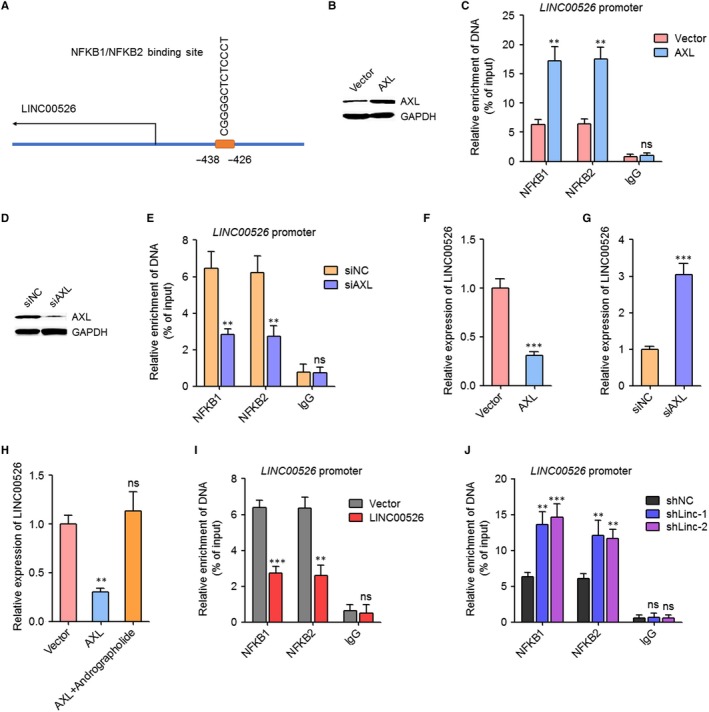
AXL repressed LINC00526 transcription via activating NF‐κB signalling. A, Schematic outlining the predicted binding sites of NFKB1/NFKB2 on LINC00526 promoter. B, AXL protein levels in U251 cells after transient overexpressing AXL were measured by Western blot. C, ChIP assays were performed in U251 cells after transient overexpressing AXL using NFKB1, NFKB2 specific antibody or non‐specific IgG. The enriched DNA was measured by qRT‐PCR and specific primers corresponding to LINC00526 promoter. D, AXL protein levels in U87 cells after transient silencing AXL were measured by Western blot. E, ChIP assays were performed in U87 cells after transient silencing AXL using NFKB1, NFKB2 specific antibody or non‐specific IgG. The enriched DNA was measured by qRT‐PCR and specific primers corresponding to LINC00526 promoter. F, LINC00526 expression levels in U251 cells after transient overexpressing AXL were measured by qRT‐PCR. G, LINC00526 expression levels in U87 cells after transient silencing AXL were measured by qRT‐PCR. H, LINC00526 expression levels in U251 cells after transient overexpressing AXL and treated with 10 μM Andrographolide for 2 days were measured by qRT‐PCR. I, ChIP assays were performed in LINC00526 stably overexpressed and control U87 cells using NFKB1, NFKB2 specific antibody or non‐specific IgG. The enriched DNA was measured by qRT‐PCR and specific primers corresponding to LINC00526 promoter. J, ChIP assays were performed in LINC00526 stably silenced and control U251 cells using NFKB1, NFKB2 specific antibody or non‐specific IgG. The enriched DNA was measured by qRT‐PCR and specific primers corresponding to LINC00526 promoter. **P < 0.01, ***P < 0.001, ns, not significant, Student's t test (C, E, F, G, I) or one‐way ANOVA followed by Dunnett's multiple comparison test (H, J)
3.7. The tumour suppressive roles of LINC00526 in glioma were dependent on AXL
The above results showed that LINC00526 interacted with EZH2, repressed AXL transcription, and repressed PI3K/Akt/NF‐κB signalling. AXL activated NF‐κB signalling and repressed LINC00526 transcription. Therefore, LINC00526 and AXL formed a double negative feedback loop. Next, we investigated whether the tumour suppressive roles of LINC00526 in glioma are dependent on AXL. We stably ectopically expressed AXL in LINC00526 stably overexpressed U87 cells (Figure 7A). CCK‐8 assays revealed that ectopic expression of AXL abolished the decrease in proliferation rate caused by LINC00526 overexpression (Figure 7B). Moreover, EdU incorporation assays further supported the roles of ectopic expression of AXL in the reverse of the proliferation decrease caused by LINC00526 overexpression (Figure 7C). Transwell migration and invasion assays revealed that ectopic expression of AXL abolished the decrease in migration and invasion caused by LINC00526 overexpression (Figure 7D,7). In addition, we treated LINC00526 stably silenced and control U87 cells with AXL inhibitor R428. CCK‐8 assays revealed that treatment with R428 blocked the increase in proliferation rate caused by LINC00526 silencing (Figure 7F). Moreover, EdU incorporation assays further showed that treatment with R428 blocked the pro‐proliferatory roles of LINC00526 silencing (Figure 7G). Transwell migration and invasion assays revealed that treatment with R428 blocked the increase in migration and invasion caused by LINC00526 silencing (Figure 7H,I). Thus, these results indicated that the roles of LINC00526 in the proliferation, migration and invasion of glioma cells are dependent on the regulation of AXL.
Figure 7.
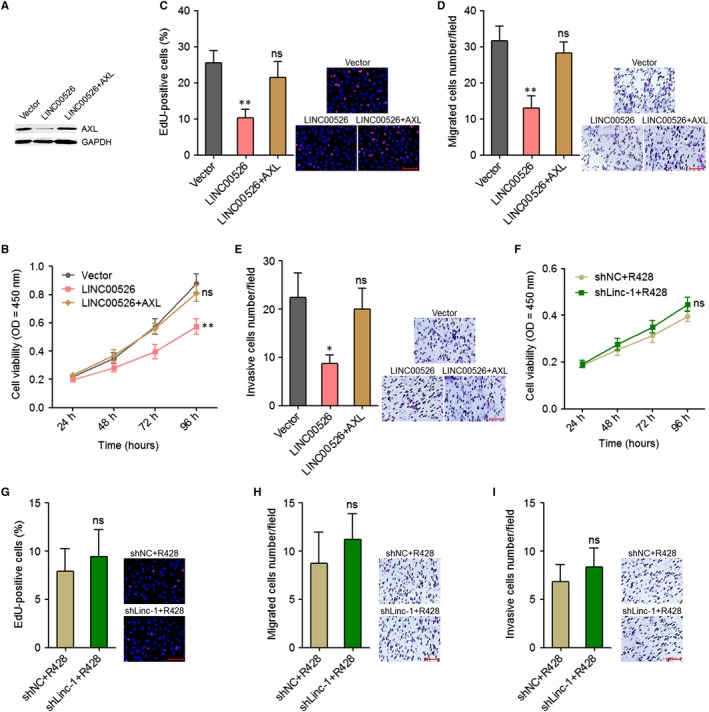
The tumour suppressive roles of LINC00526 in glioma were dependent on AXL. A, AXL protein levels in LINC00526 and AXL concurrently stably ectopically expressed and control U87 cells were measured by Western blot. B, Cell proliferation rates of LINC00526 and AXL concurrently stably ectopically expressed and control U87 cells were measured by CCK‐8 assays. OD values in 450 nm were detected to indicate cell proliferation. C, Cell proliferation of LINC00526 and AXL concurrently stably ectopically expressed and control U87 cells were measured by EdU incorporation assays. Scale bars, 100 µm. D, Cell migration of LINC00526 and AXL concurrently stably ectopically expressed and control U87 cells were measured by transwell migration assays. Scale bars, 100 µm. E, Cell invasion of LINC00526 and AXL concurrently stably ectopically expressed and control U87 cells were measured by transwell invasion assays. Scale bars, 100 µm. F, Cell proliferation rates of LINC00526 stably silenced and control U87 cells treated with 1 μM R428 were measured by CCK‐8 assays. OD values in 450 nm were detected to indicate cell proliferation. G, Cell proliferation of LINC00526 stably silenced and control U87 cells treated with 1 μM R428 were measured by EdU incorporation assays. Scale bars, 100 µm. D, Cell migration of LINC00526 stably silenced and control U87 cells treated with 1 μM R428 were measured by transwell migration assays. Scale bars, 100 µm. E, Cell invasion of LINC00526 stably silenced and control U87 cells treated with 1 μM R428 were measured by transwell invasion assays. Scale bars, 100 µm. Results are shown as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ns, not significant, one‐way ANOVA followed by Dunnett's multiple comparison test (B‐E) or Student's t test (F‐I)
4. DISCUSSION
In this study, we identified a novel lncRNA LINC00526, which is down‐regulated and functions as a tumour suppressor in glioma. We first confirmed the significant down‐regulation of LINC00526 in human glioma tissues. LINC00526 expression levels were inversely correlated with aggravation of glioma. Second, the correlation between LINC00526 expression level and prognosis of glioma patients was analysed using TCGA data. The TCGA data revealed that low expression of LINC00526 is associated with poor survival of glioma patients. Gain‐of‐function and loss‐of‐function assays demonstrated that ectopic expression of LINC00526 inhibited glioma cell proliferation, migration and invasion. Reciprocally, LINC00526 silencing promoted glioma cell proliferation, migration and invasion. Therefore, our data identified LINC00526 as a potential prognostic biomarker for glioma and suggested that ectopic expression of LINC00526 may be a potential therapeutic strategy for glioma. LncRNAs GAS5, NEAT1, GACAT3, CASC9, HMMR‐AS1, LINC00152, HOTAIRM1, AC003092.1, HOXD‐AS1, OIP5‐AS1, DANCR and so on have been reported to play oncogenic or tumour suppressive roles in glioma.32, 33, 41, 42, 43, 44, 45, 46, 47, 48, 49 Our study further proved the critical roles of lncRNAs in glioma and provided another candidate lncRNA for glioma prognosis and therapy. Multi‐centre analyses of the correlation between LINC00526 expression and prognosis of glioma patients would further detect the potential of LINC00526 as a prognostic biomarker, which needs further investigation.
Although the expression and roles of several lncRNAs in glioma have been sufficiently investigated, the molecular mechanisms mediating the roles of lncRNAs and the reasons contributing to the dysregulation of lncRNAs in glioma are relative less investigated. In this study, we further explored the molecular mechanisms mediating the down‐regulation and tumour suppressive roles of LINC00526 in glioma. Through searching TCGA data, we found that the expression of AXL was significantly inversely associated with that of LINC00526 in glioma tissues. AXL is a well‐known receptor tyrosine kinase.37 Several reports have revealed that AXL is overexpressed in human glioma and predicts poor prognosis of glioma patients.50, 51 AXL was also reported to promote glioma growth, migration, invasion, tumorigenesis and primary resistance to EGFR inhibition.37, 52, 53 The inverse expression patterns and inverse biological roles between LINC00526 and AXL implied that whether there is negative modulation between LINC00526 and AXL.
The molecular mechanisms mediating the roles of lncRNAs are complex and diverse.25, 28 As a class of regulatory RNAs, lncRNAs directly bind proteins, DNAs, mRNAs and/or microRNAs.35 Through interacting with other molecules, lncRNAs regulate the expression, location and/or functions of the interacting partners.35 In this study, we identified that LINC00526 directly interacted with EZH2, which is a histone lysine methyltransferase and epigenetically modulates target genes expression.54, 55 In a previous report, Ott et al showed that EZH2 induces AXL transcription.34 In this study, we further found that, the binding between LINC00526 and EZH2 decreased the binding of EZH2 to AXL promoter, and further decreased the transcription of AXL. Therefore, LINC00526 repressed AXL expression via occupying EZH2. Via repressing AXL, LINC00526 further repressed PI3K/Akt/NF‐κB signalling. Recuse assays further found that the tumour suppressive roles of LINC00526 were dependent on the repression of AXL.
Intriguingly, we further identified LINC00526 as a target of NF‐κB signalling. We identified a NFKB1/NFKB2 binding site at the promoter of LINC00526, and demonstrated that NF‐κB repressed LINC00526 transcription. Via activating NF‐κB signalling, AXL promoted the binding of NFKB1/NFKB2 to LINC00526 promoter and repressed LINC00526 transcription. Therefore, LINC00526 and AXL form a double negative feedback loop, which further promotes the down‐regulation of LINC00526 and the up‐regulation of AXL in glioma tissues. Several feedback regulatory loops have been identified in cancers.13, 56 The feedback regulatory loops could enlarge their modulatory roles in control gene expression and biological effects in cancers.13, 56 Because AXL/PI3K/Akt/NF‐κB signalling also regulates other genes, the effects of LINC00526 on these targets need further investigation.
In summary, this study identified a novel lncRNA LINC00526, which is lowly expressed in glioma. The down‐regulation of LINC00526 was correlated with aggravation and poor prognosis. LINC00526 inhibited glioma cell proliferation, migration and invasion via the LINC00526/EZH2/AXL/NF‐κB/LINC00526 feedback loop. Thus, this study identified LINC00526 as a potential prognostic biomarker and a candidate for therapy in glioma.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Yan J, Xu C, Li Y, et al. Long non‐coding RNA LINC00526 represses glioma progression via forming a double negative feedback loop with AXL. J Cell Mol Med. 2019;23:5518–5531. 10.1111/jcmm.14435
Jian Yan and Chunhua Xu contribute equally to this article.
Data Availability Statement: The data that support the findings of this study are available from the corresponding author upon reasonable request.
Contributor Information
Tao Hong, Email: ht2000@vip.sina.com.
Erming Zeng, Email: ermingzeng@aliyun.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Ricard D, Idbaih A, Ducray F, Lahutte M, Hoang‐Xuan K, Delattre JY. Primary brain tumours in adults. Lancet. 2012;379:1984‐1996. [DOI] [PubMed] [Google Scholar]
- 2. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492‐507. [DOI] [PubMed] [Google Scholar]
- 3. Knisely J, Schulder M. Radiation plus chemotherapy in Low‐grade glioma. N Engl J Med. 2016;375:490. [DOI] [PubMed] [Google Scholar]
- 4. Berger AC, Korkut A, Kanchi RS, et al. A comprehensive Pan‐cancer molecular study of gynecologic and breast cancers. Cancer Cell. 2018;33:690–705 e699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Du Z, Fei T, Verhaak R, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20:908–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. [DOI] [PubMed] [Google Scholar]
- 8. Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF‐beta promotes the invasion‐metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. [DOI] [PubMed] [Google Scholar]
- 9. Hu WL, Jin L, Xu AN, et al. GUARDIN is a p53‐responsive long non‐coding RNA that is essential for genomic stability. Nat Cell Biol. 2018;20:492–502. [DOI] [PubMed] [Google Scholar]
- 10. Zhang Z, Peng Z, Cao J, et al. Long noncoding RNA PXN‐AS1‐L promotes non‐small cell lung cancer progression via regulating PXN. Cancer Cell Int. 2019;19:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li J‐K, Chen C, Liu J‐Y, et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38‐MAPK signaling. Mol Cancer. 2017;16:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mondal T, Juvvuna PK, Kirkeby A, et al. Sense‐Antisense lncRNA pair encoded by locus 6p22.3 determines neuroblastoma susceptibility via the USP36‐CHD7‐SOX9 regulatory axis. Cancer Cell. 2018;33:417–434 e417. [DOI] [PubMed] [Google Scholar]
- 13. He S, Lin J, Xu Y, Lin L, Feng J. A positive feedback loop between ZNF205‐AS1 and EGR4 promotes non‐small cell lung cancer growth. J Cell Mol Med. 2019;23:1495–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang C, Yuan J, Hu H, et al. Long non‐coding RNA CHCHD4P4 promotes epithelial‐mesenchymal transition and inhibits cell proliferation in calcium oxalate‐induced kidney damage. Braz J Med Biol Res. 2017;51:e6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Michelini F, Pitchiaya S, Vitelli V, et al. Damage‐induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double‐strand breaks. Nat Cell Biol. 2017;19:1400–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iancu IV, Anton G, Botezatu A, et al. LINC01101 and LINC00277 expression levels as novel factors in HPV‐induced cervical neoplasia. J Cell Mol Med. 2017;21:3787–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu XT, Yuan JH, Zhu TT, Li YY, Cheng XY. Long noncoding RNA glypican 3 (GPC3) antisense transcript 1 promotes hepatocellular carcinoma progression via epigenetically activating GPC3. FEBS J. 2016;283:3739–3754. [DOI] [PubMed] [Google Scholar]
- 18. Wang Z, Yang B, Zhang M, et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell‐cycle progression in cancer. Cancer Cell. 2018;33:706–720 e709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin A, Hu Q, Li C, et al. The LINK‐A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat Cell Biol. 2017;19:238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grelet S, Link LA, Howley B, et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol. 2017;19:1105–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Salehi S, Taheri MN, Azarpira N, Zare A, Behzad‐Behbahani A. State of the art technologies to explore long non‐coding RNAs in cancer. J Cell Mol Med. 2017;21:3120–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang L, Yang FU, Yuan J‐H, et al. Epigenetic activation of the MiR‐200 family contributes to H19‐mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2013;34:577–586. [DOI] [PubMed] [Google Scholar]
- 23. Shao M, Yang Q, Zhu W, et al. LncHOXA10 drives liver TICs self‐renewal and tumorigenesis via HOXA10 transcription activation. Mol Cancer. 2018;17:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu H, Wang Y, Ding X, et al. Long non‐coding RNA XLOC_000647 suppresses progression of pancreatic cancer and decreases epithelial‐mesenchymal transition‐induced cell invasion by down‐regulating NLRP3. Mol Cancer. 2018;17:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu D, Yang F, Yuan JH, et al. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/beta‐catenin signaling. Hepatology. 2013;58:739–751. [DOI] [PubMed] [Google Scholar]
- 27. Luo X, Qiu Y, Jiang Y, et al. Long non‐coding RNA implicated in the invasion and metastasis of head and neck cancer: possible function and mechanisms. Mol Cancer. 2018;17:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peng Z, Liu C, Wu M. New insights into long noncoding RNAs and their roles in glioma. Mol Cancer. 2018;17:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fu C, Li D, Zhang X, Liu N, Chi G, Jin X. LncRNA PVT1 facilitates tumorigenesis and progression of glioma via regulation of MiR‐128‐3p/GREM1 axis and BMP signaling pathway. Neurotherapeutics. 2018;15:1139–1157. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30. Wang P, Peng X, Zhang J, et al. LncRNA‐135528 inhibits tumor progression by up‐regulating CXCL10 through the JAK/STAT pathway. Apoptosis. 2018;23:651–666. [DOI] [PubMed] [Google Scholar]
- 31. Liu C, Zhang Y, She X, et al. A cytoplasmic long noncoding RNA LINC00470 as a new AKT activator to mediate glioblastoma cell autophagy. J Hematol Oncol. 2018;11:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu N, Liu B, Lian C, et al. Long noncoding RNA AC003092.1 promotes temozolomide chemosensitivity through miR‐195/TFPI‐2 signaling modulation in glioblastoma. Cell Death Dis. 2018;9:1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou KE, Zhang C, Yao H, et al. Knockdown of long non‐coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR‐132. Mol Cancer. 2018;17:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ott M, Litzenburger UM, Sahm F, et al. Promotion of glioblastoma cell motility by enhancer of zeste homolog 2 (EZH2) is mediated by AXL receptor kinase. PLoS ONE. 2012;7:e47663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yuan J‐H, Liu X‐N, Wang T‐T, et al. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA‐PXN‐AS1. Nat Cell Biol. 2017;19:820–832. [DOI] [PubMed] [Google Scholar]
- 36. Wu G, Ma Z, Cheng Y, et al. Targeting Gas6/TAM in cancer cells and tumor microenvironment. Mol Cancer. 2018;17:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vouri M, An Q, Birt M, Pilkington GJ, Hafizi S. Small molecule inhibition of Axl receptor tyrosine kinase potently suppresses multiple malignant properties of glioma cells. Oncotarget. 2015;6:16183–16197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cao Z, Pan X, Yang Y, Huang Y, Shen HB. The lncLocator: a subcellular localization predictor for long non‐coding RNAs based on a stacked ensemble classifier. Bioinformatics. 2018;34:2185–2194. [DOI] [PubMed] [Google Scholar]
- 39. Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin‐modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Khan A, Fornes O, Stigliani A, et al. JASPAR 2018: update of the open‐access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D260–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu Q, Yu W, Zhu S, et al. Long noncoding RNA GAS5 regulates the proliferation, migration, and invasion of glioma cells by negatively regulating miR‐18a‐5p. J Cell Physiol. 2018;234:757–768. [DOI] [PubMed] [Google Scholar]
- 42. Wang J, Zhang M, Lu W. Long noncoding RNA GACAT3 promotes glioma progression by sponging miR‐135a. J Cell Physiol. 2018;234(7):10877–10887. [DOI] [PubMed] [Google Scholar]
- 43. Liu H, Li C, Yang J, et al. Long noncoding RNA CASC9/miR‐519d/STAT3 positive feedback loop facilitate the glioma tumourigenesis. J Cell Mol Med. 2018;22:6338–6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li J, Ji X, Wang H. Targeting long noncoding RNA HMMR‐AS1 suppresses and radiosensitizes glioblastoma. Neoplasia. 2018;20:456–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yu M, Xue Y, Zheng J, et al. Linc00152 promotes malignant progression of glioma stem cells by regulating miR‐103a‐3p/FEZF1/CDC25A pathway. Mol Cancer. 2017;16:110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46. Li Q, Dong C, Cui J, Wang Y, Hong X. Over‐expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up‐regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer Res. 2018;37:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen Y, Zhao F, Cui D, et al. HOXD‐AS1/miR‐130a sponge regulates glioma development by targeting E2F8. Int J Cancer. 2018;142(11):2313–2322. [DOI] [PubMed] [Google Scholar]
- 48. Liu X, Zheng J, Xue Y, et al. PIWIL3/OIP5‐AS1/miR‐367‐3p/CEBPA feedback loop regulates the biological behavior of glioma cells. Theranostics. 2018;8:1084–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ma Y, Zhou G, Li M, et al. Long noncoding RNA DANCR mediates cisplatin resistance in glioma cells via activating AXL/PI3K/Akt/NF‐kappaB signaling pathway. Neurochem Int. 2018;118:233–241. [DOI] [PubMed] [Google Scholar]
- 50. Hutterer M, Knyazev P, Abate A, et al. Axl and growth arrest‐specific gene 6 are frequently overexpressed in human gliomas and predict poor prognosis in patients with glioblastoma multiforme. Clin Cancer Res. 2008;14:130–138. [DOI] [PubMed] [Google Scholar]
- 51. Onken J, Vajkoczy P, Torka R, et al. Phospho‐AXL is widely expressed in glioblastoma and associated with significant shorter overall survival. Oncotarget. 2017;8:50403–50414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Onken J, Torka R, Korsing S, et al. Inhibiting receptor tyrosine kinase AXL with small molecule inhibitor BMS‐777607 reduces glioblastoma growth, migration, and invasion in vitro and in vivo. Oncotarget. 2016;7:9876–9889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vajkoczy P, Knyazev P, Kunkel A, et al. Dominant‐negative inhibition of the Axl receptor tyrosine kinase suppresses brain tumor cell growth and invasion and prolongs survival. Proc Natl Acad Sci USA. 2006;103:5799–5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Erkek S, Johann PD, Finetti MA, et al. Comprehensive analysis of chromatin states in atypical Teratoid/Rhabdoid tumor identifies diverging roles for SWI/SNF and Polycomb in gene regulation. Cancer Cell. 2019;35:95–110 e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zingg D, Debbache J, Peña‐Hernández R, et al. EZH2‐mediated primary cilium deconstruction drives metastatic melanoma formation. Cancer Cell. 2018;34:69–84 e14. [DOI] [PubMed] [Google Scholar]
- 56. Yuan J‐H, Yang FU, Chen B‐F, et al. The histone deacetylase 4/SP1/microrna‐200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology. 2011;54:2025–2035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


