Abstract
In South Africa, a progressive increase in listeriosis cases was noted from mid-June 2017, heralding what was to become the world's largest listeriosis outbreak. A total of 1060 cases were reported for the period January 1, 2017 to July 17, 2018. We describe laboratory activities, experiences, and results of whole-genome sequencing (WGS) analysis of Listeria monocytogenes isolates associated with this outbreak. Bacteria were identified using the VITEK-2 COMPACT 15 microbial identification system. WGS was performed using Illumina MiSeq technology. WGS data were analyzed using CLC Genomics Workbench Software and free-to-use on-line analysis tools/pipelines. Multilocus sequence typing (MLST) showed that 91% of clinical isolates were sequence type 6 (ST6), determining that the outbreak was largely associated with L. monocytogenes ST6. Epidemiological and laboratory findings led to investigation of a large ready-to-eat processed meat production facility in South Africa, named Enterprise Foods. L. monocytogenes ST6 was found in environmental sampling swabs of the production facility and in ready-to-eat processed meat products (including polony, a product similar to bologna sausage) manufactured at the facility. ST6 isolates, sourced at the Enterprise Foods production facility and from Enterprise food products, were shown by single nucleotide polymorphism (SNP) analysis to be highly related to clinical isolates; these nonclinical ST6 isolates showed <10 SNP differences when compared to clinical ST6 isolates. Core-genome MLST showed that clinical ST6 isolates and Enterprise-related ST6 isolates had no more than 4 allele differences between each other, suggestive of a high probability of epidemiological relatedness. WGS data interpreted together with epidemiological data concluded that the source of the listeriosis outbreak was ready-to-eat processed meat products manufactured by Enterprise Foods. Listeriosis has now been added to the South African list of mandatory notifiable medical conditions. Surveillance systems have been strengthened to facilitate prevention and early detection of listeriosis outbreaks.
Keywords: Listeria monocytogenes, whole-genome sequencing, sequence type 6, ST6, outbreak, listeriosis, South Africa
Introduction
Listeria monocytogenes is a Gram-positive bacterium that causes the disease named listeriosis. Acquisition of the L. monocytogenes pathogen occurs mainly by consumption of contaminated food. Infections with L. monocytogenes can result in mild febrile gastroenteritis in healthy individuals; however, invasive disease characterized by bacteremia, meningitis, pneumonia, endocarditis, and sepsis can occur, particularly in high risk groups (Ferreira et al., 2014). The pathogen most commonly affects immuno-compromised individuals, pregnant women, neonates, and the elderly. Listeriosis is associated with case fatality rates as high as 30% (Lomonaco et al., 2015).
Recently, there have been several international reports of L. monocytogenes associated foodborne disease outbreaks. These outbreaks have been linked to a variety of different food vehicles, including ice cream (Chen et al., 2017b; Li et al., 2017), ready-to-eat meat products and meat pâté (Kvistholm Jensen et al., 2016; Althaus et al., 2017; Gelbicova et al., 2018), ready-to-eat fish (Nakari et al., 2014; Schjorring et al., 2017), stone-fruits (Chen et al., 2016), cheese (Chen et al., 2017a), and caramel-apples (Angelo et al., 2017). Multilocus sequence typing (MLST) of isolates associated with outbreaks has shown that several sequence types (STs) of L. monocytogenes are capable of causing outbreaks. L. monocytogenes sequence type 6 (ST6) is one of the L. monocytogenes subtypes that have been associated with outbreaks. In 2013, a U.S. multistate cheese outbreak was associated with L. monocytogenes ST6 (www.cdc.gov/listeria/outbreaks/cheese-07-13). In 2016, meat pâté contaminated with L. monocytogenes ST6 caused an outbreak in Switzerland (Althaus et al., 2017). More recently, in early 2018, reports emerged of a L. monocytogenes ST6 outbreak (ongoing since 2015) in five European Union member states (Austria, Denmark, Finland, Sweden, and the United Kingdom); the outbreak was linked to frozen corn and possibly to other frozen vegetables (https://ecdc.europa.eu/en/news-events/listeria-monocytogenes-outbreak-47-cases-including-9-deaths).
In South Africa, comprehensive historic data for L. monocytogenes are lacking. This includes a lack of data related to prevalence, epidemiology, and description of clusters/outbreaks. The first documented report of a human listeriosis outbreak in South Africa, occurred over the period of August 1977 to April 1978; the outbreak occurred in the Gauteng Province of South Africa and involved 14 cases (Jacobs et al., 1978). In 1999, in the Mpumalanga Province of South Africa, an outbreak of listeriosis was described in cattle and sheep that were fed poor quality unmarketable potatoes (Van Vollenhoven, 1999). More recently, some reports from South Africa have described the presence of L. monocytogenes in a variety of human food items (Nel et al., 2004; van Nierop et al., 2005; Plessis et al., 2017). Notably, in 2015, a cluster of human cases caused by L. monocytogenes ST6 was described in the Western Cape Province of South Africa (Smith et al., 2016).
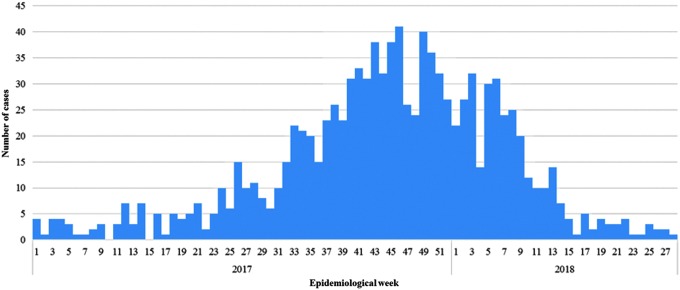
From mid-June 2017, a marked, progressive increase in listeriosis cases was noted in South Africa, which heralded the onset of an outbreak. A staggering total of 1060 cases were reported for the period January 1, 2017 to July 17, 2018 (Fig. 1). The World Health Organization (WHO) described this as the largest listeriosis outbreak that had ever been detected worldwide. The molecular epidemiology of the outbreak was investigated using whole-genome sequencing (WGS) analysis of the L. monocytogenes isolates; WGS was performed in as real time as possible. This was a massive undertaking for a relatively small reference public health laboratory in South Africa, the Centre for Enteric Diseases (CED), a laboratory that was still in its WGS “infancy” and still in the process of taking “small steps” toward the implementation of WGS for analysis of enteric pathogens. Herewith, we describe the CED laboratory activities, experiences and results of WGS analysis of L. monocytogenes isolates associated with the largest ever reported outbreak of listeriosis globally.
FIG. 1.
Epidemiological curve of laboratory-confirmed listeriosis showing number of cases by week (listed according to date of sample collection), South Africa, January 1, 2017 to July 17, 2018 (n = 1060).
Materials and Methods
Outbreak detection and public health response
During mid-June 2017, the National Institute for Communicable Diseases (NICD) was notified of an unusual increase in listeriosis cases at public sector hospitals in one province of South Africa. The NICD's CED and Outbreak Response Unit, launched an investigation into the reported increase in cases. A listeriosis outbreak was later declared. The CED focuses on surveillance and public health-focused research of pathogens associated with diarrhea and enteric fevers, and actively assists with the investigation and response to enteric disease outbreaks (including foodborne and waterborne disease outbreaks). The center provides specialized reference laboratory testing for enteric bacteria and viruses, including potential causes of outbreaks. It is a source of strategic data, technical support, and policy advice to the South African Department of Health and other major stakeholders, and contributes its expertise toward strengthening outbreak preparedness and response to public health emergencies.
Referral of isolates to the CED
Preceding this listeriosis outbreak, clinical microbiology laboratories across South Africa would forward L. monocytogenes isolates to the CED on an ad hoc basis. Upon declaration of the outbreak, all diagnostic microbiology laboratories across the country were formally requested to forward all isolates to the CED for further characterization. These included microbiology laboratories in all fields and disciplines, laboratories performing testing on every type of specimen, such as those sourced from clinical (humans), animals, food, food production facilities, environment, and others. The CED then performed phenotypic and genotypic characterization of all isolates.
Phenotypic identification of bacteria
Bacteria were received on Dorset-Egg transport media (Diagnostic Media Products [DMP]; National Health Laboratory Service, Johannesburg, South Africa) and sub-cultured onto 5% Blood Agar (DMP), to check viability and purity. The identification of bacterial cultures was confirmed using the VITEK-2 COMPACT 15 automated microbial identification system (bioMérieux, Marcy-l'Étoile, France).
Genomic DNA isolation from bacteria and WGS
Genomic DNA was isolated from bacteria using a Qiagen QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany). DNA was sequenced using Illumina MiSeq (Illumina, San Diego, CA) next generation sequencing technology; DNA libraries were prepared using a Nextera XT DNA Library Preparation Kit (Illumina), followed by 2 × 300 paired-end sequencing runs with ∼70 times coverage. Raw sequencing data (FastQ files for paired-end reads) were analyzed using tools available in the CLC Genomics Workbench Software, version 11 (CLC bio, Aarhus, Denmark); using the “trim sequences tool,” sequence reads were trimmed to include quality trimming and ambiguity trimming, and length trimming to discard reads below a length of 100 bases; trimmed reads were assembled using the “de novo assembly tool.”
MLST of bacteria
Assembled genome data were analyzed using the “MLST” on-line analysis pipeline available at the Center for Genomic Epidemiology (CGE) of the Technical University of Denmark (Larsen et al., 2012). MLST produces STs based on sequence analysis of seven housekeeping genes, as described at the Bacterial Isolate Genome Sequence Database (BIGSdb) for L. monocytogenes (BIGSdb-lm) database (http://bigsdb.pasteur.fr/listeria/listeria.html).
Single nucleotide polymorphism profiles and phylogenetic analysis of bacteria
Assembled genome data were analyzed using the “CSIPhylogeny 1.4” on-line analysis pipeline available at the CGE (Kaas et al., 2014). The CSIPhylogeny pipeline uses various publicly available programs; the analysis steps are briefly described as follows: assembled genome data are aligned against a reference genome and single nucleotide polymorphisms (SNPs) are called; SNPs are filtered and qualified; final qualified SNPs for each genome is concatenated to an alignment; phylogeny is then inferred based on a comparison of SNP alignments of strains. SNPs were called by alignment and referencing against a South African L. monocytogenes ST6 isolate (reference No. YA00053238). SNP alignments were analyzed with iTOL software (http://.itol.embl.de) to generate phylogenetic maximum-likelihood trees.
Core-genome multilocus sequence typing and phylogenetic analysis of bacteria
Raw sequencing data (FastQ files for paired-end reads) were further analyzed at the BIGSdb-lm database, to confirm MLST profiles of isolates and to determine the core-genome multilocus sequence typing (cgMLST) profiles of isolates. The cgMLST scheme is based on 1748 genes (Moura et al., 2016). Data (including cgMLST allele data) were exported from the BIGSdb-lm and then further captured into a BioNumerics Software version 7.6 database (Applied Maths, Sint-Martens-Latem, Belgium). Within this BioNumerics platform, phylogenetic cluster analysis of isolates was investigated using cgMLST data (categorical data values) analyzed using a single linkage algorithm; cluster analysis was depicted using a minimum spanning tree.
In silico analysis of sequencing data
Tools built into the BIGSdb-lm were used for further in silico analysis of sequencing data. These included tools for polymerase chain reaction (PCR) serogrouping.
Data availability
Genome sequences for 10 L. monocytogenes ST6 isolates associated with this South African outbreak have been deposited at National Center for Biotechnology Information (NCBI)/GenBank under the accession numbers QEXB00000000 to QEXK00000000 (BioProject No. PRJNA451422, BioSample Nos. SAMN08970424 to SAMN08970415, respectively) (Allam et al., 2018).
Results and Discussion
CED, PulseNet Africa and readiness for WGS analysis of L. monocytogenes
The CED is a member of the regional PulseNet Africa laboratory network, which forms part of the PulseNet International network (www.pulsenetinternational.org), a global molecular subtyping network for foodborne disease surveillance. In addition, the CED also coordinates the PulseNet Africa Network. PulseNet Africa was established in August 2010. Currently, formal membership includes 12 countries (South Africa, Kenya, Senegal, Cameroon, The Gambia, Malawi, Tanzania, Cote d'Ivoire, Ghana, Uganda, Mozambique, and Nigeria). Communication and discussion in the Network is achieved via a ListServ communication platform. The CED runs training courses in molecular subtyping methodologies, including pulsed-field gel electrophoresis (PFGE) analysis and multiple-locus variable-number tandem-repeats analysis (MLVA). The CED offers continuous assistance, advice, and guidance to African countries with regards to molecular subtyping of enteric pathogens; this includes assistance with PFGE analysis of isolates. The CED hosts/manages an “African” molecular subtyping database that includes hundreds of PFGE patterns from enteric pathogens isolated across Africa.
The CED has published extensively on the use of molecular subtyping methodologies for routine surveillance activities and for investigation of outbreaks involving enteric pathogens. This has included the use of PFGE analysis (Smith et al., 2011; Tau et al., 2012; Ismail et al., 2013), MLVA (Muvhali et al., 2017; Tau et al., 2017) and MLST (Smith et al., 2014, 2017). In late 2015, we were ready to take the first step toward the use of WGS for analysis of enteric pathogens. Our institution (the NICD) had just established a Sequencing Core Facility equipped with Illumina MiSeq next-generation sequencing equipment. Armed with CLC Genomics Workbench Software and various free-to-use on-line WGS analysis tools/pipelines, we were ready to commence with analysis of WGS data. In September 2015, we initiated our WGS activities by investigating a cluster of listeriosis cases reported from the Western Cape Province of South Africa (Smith et al., 2016). This analysis was timely, as the steering committee of the PulseNet International network was in discussions to start with implementation of WGS, of which the networks vision for implementation of WGS was recently published in 2017 (Nadon et al., 2017).
Listeriosis outbreak, description of clinical L. monocytogenes isolates and the ST6 outbreak strain
During mid-June 2017, several clinicians and microbiologists reported unusually high numbers of listeriosis cases at a number of sites in Gauteng Province of South Africa. A listeriosis outbreak was confirmed, with a total of 1060 cases eventually reported for the period January 1, 2017 to July 17, 2018 (Fig. 1). For the years 2013 to 2016, a range of 55–113 laboratory-confirmed cases of listeriosis occurred annually in South Africa. At the peak of the outbreak (mid-November 2017), 41 listeriosis cases were reported in a single week. Cases were reported from across the country; however, most cases were reported from Gauteng Province (58%), followed by Western Cape (13%) and KwaZulu-Natal (8%) provinces. Women accounted for 55% of the patients. The ages of patients ranged from birth to 93 years (median 19 years). Neonates (aged ≤28 d) were the most affected age group, accounting for 43% of cases. This was followed by adults of 15–49 years of age, accounting for 32% of cases. Final outcome was known for 806/1060 (76%) of cases; 27% (216/806) with known outcome died. These outcome data are in agreement with previously reported listeriosis case-fatality rates that can reach 30% (Lomonaco et al., 2015).
Laboratory analysis identified the causative species as L. monocytogenes. At the onset of the outbreak, we decided to investigate all bacterial isolates using WGS, with WGS data analysis conducted using various free-to-use on-line WGS analysis tools/pipelines. We certainly did not anticipate the mammoth task that awaited us. The weekly number of L. monocytogenes isolates received at our laboratory multiplied rapidly, increasing 40-fold at the height of the outbreak. The laboratory staff (two technicians, two technologists, and two scientists) were rapidly overwhelmed with the increased workload. WGS was performed in as real time as possible. From isolate reception at CED to WGS data availability, the timeline ranged from 7 to 10 working days. WGS was performed on 636 clinical isolates recovered over the period January 1, 2017 to July 17, 2018. MLST of WGS data showed that 91% (576/636) of isolates belonged to MLST ST6, while the remainder belonged to 14 different STs (ST1, ST101, ST155, ST2, ST204, ST219, ST224, ST3, ST5, ST54, ST7, ST8, ST87, and ST876). This determined that an ST6 strain was the most likely cause of the outbreak. The ST6 isolates were also found to belong to serogroup IVb, as determined by in silico PCR serogrouping. L. monocytogenes ST6 has previously been reported to be associated with outbreaks (Althaus et al., 2017). This ST6 subtype has also been described to be associated with an increased rate of unfavorable outcomes in patients with meningitis (Koopmans et al., 2013).
Source of the outbreak and analysis of non-human L. monocytogenes isolates
By early January 2018, food history interviews with patients suggested that “polony” was among the most commonly consumed foodstuff among persons with listeriosis. Polony is a ready-to-eat processed meat product, similar to bologna sausage. Epidemiological and laboratory findings led to the investigation of a large ready-to-eat processed meat production facility in South Africa, named Enterprise Foods. On February 2, 2018, the production facility was inspected and numerous environmental sampling swabs were collected throughout the facility. L. monocytogenes ST6 was isolated from the environment of numerous areas of the production facility, including postcooking areas. The same ST6 strain was also found in several food products (including polony) manufactured at the facility. On March 4, 2018, a recall of affected food products was initiated and Enterprise Foods' production facilities were shut down.
Phylogenetic analysis of L. monocytogenes isolates using SNP analysis and cgMLST
SNP analysis of clinical isolates showed that the ST6 isolates all clustered together on a phylogenetic tree (Fig. 2). Furthermore, almost all (99%) of the ST6 isolates were all highly related, with SNP differences ranging from zero to nine. ST6 isolates, sourced at the Enterprise Foods Polokwane production facility and from Enterprise Foods products, were shown by SNP analysis to be highly related to the clinical isolates; these nonclinical ST6 isolates showed <10 SNP differences when compared to clinical ST6 isolates. Similar SNP differences and clonal characteristics have previously been reported in studies describing L. monocytogenes foodborne outbreaks (Orsi et al., 2008; Kvistholm Jensen et al., 2016).
FIG. 2.
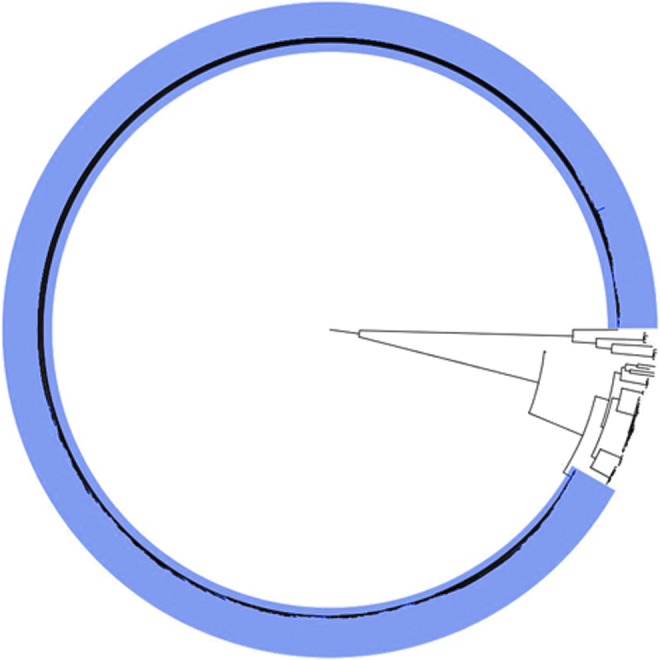
Maximum likelihood tree (circular tree) drawn using SNP alignments from WGS data of clinical Listeria monocytogenes isolates. The ST6 outbreak cluster representing 91% of the clinical isolates is highlighted with shading. SNP, single nucleotide polymorphism; ST6, sequence type 6; WGS, whole-genome sequencing.
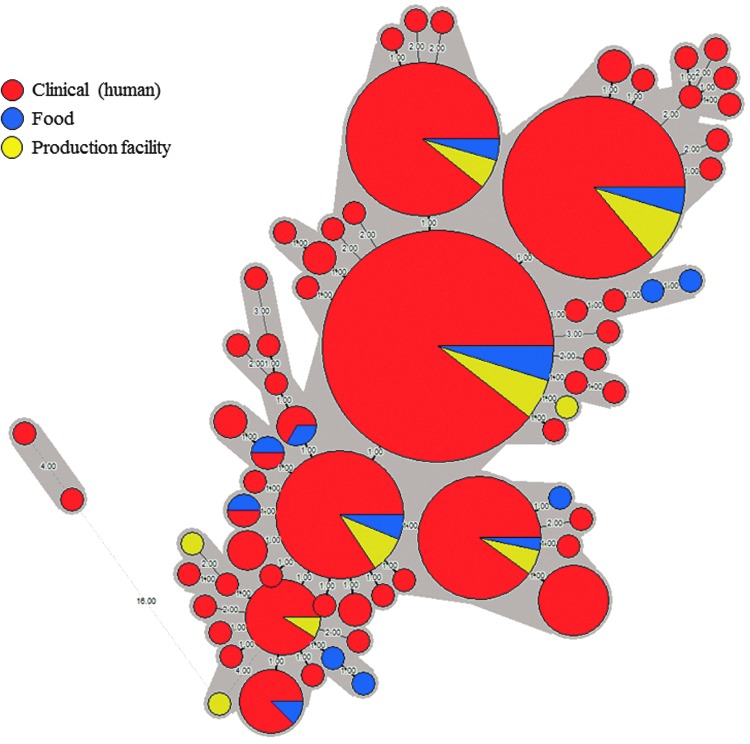
For cgMLST analysis of ST6 isolates from all sources, raw sequencing data were submitted to the BIGSdb-lm hosted by Institut Pasteur. BIGSdb-lm only accepts sequencing data that passes their strict quality control checks. To date, for ST6 isolates recovered over the years 2017–2018, sequencing data for 374 isolates have passed BIGSdb-lm quality control checks; these data were further investigated using cgMLST. With the exception of two clinical ST6 isolates recovered in 2017, all the remaining ST6 isolates (n = 372), had no more than four allele differences between each other, when investigated using cgMLST analysis (Fig. 3). This indicated that these 372 ST6 isolates were highly genetically related and suggested a high probability of epidemiological relatedness of the ST6 isolates, as concluded following analysis using this cgMLST scheme described by Moura et al. (2016, 2017). The BIGSdb-lm assigned these 372 ST6 isolates a cgMLST profile number 4148 (CT4148), a profile unique to our South African isolates and not reported from any other country worldwide. These 372 ST6 (CT4148) isolates included 326 clinical isolates, 22 Enterprise food isolates, and 24 environmental isolates recovered from Enterprise Foods Polokwane production facility (Fig. 3).
FIG. 3.
Minimum spanning tree drawn using cgMLST data from 374 Listeria monocytogenes ST6 isolates from several sources including clinical (human), food, and production facility. The circular nodes represent isolate(s) having the identical cgMLST profile; the larger the node, the more isolates are reflected. The number values between adjacent nodes indicate the number of allele differences between nodes. Gray shading encompasses all nodes (isolates) that have no more than four allele differences when compared to their adjacent neighboring nodes (isolates). The source of isolates is reflected in the coloring of the nodes. cgMLST, core-genome multilocus sequence typing.
Limitations of the Data Described in This Article
The data described in this article are preliminary and so has limitations. The data described are a prelude to more detailed articles that will soon follow. All epidemiological aspects of this outbreak investigation have not been presented and discussed—these aspects will be the subject of a larger “epidemiology article” that will soon follow and be published separately. Likewise, all molecular and genomic aspects of the L. monocytogenes isolates have not been presented and discussed—these aspects will be the subject of a larger “molecular article” that will soon follow and be published separately.
Conclusions
WGS data interpreted together with epidemiological data have determined that the source of the listeriosis outbreak in South Africa was ready-to-eat processed meat products manufactured by Enterprise Foods. This was the largest ever reported outbreak of listeriosis globally. Outbreak milestones included an alert in mid-June 2017, a peak in mid-November 2017, and finally the identification of the outbreak source in mid-February 2018. This 8-month timeline was rather remarkable, considering the large number of cases involved and the limited capacity and resources available for foodborne disease outbreak investigations in South Africa. As a direct result of this outbreak, listeriosis has now been added to the South African list of mandatory notifiable medical conditions. Surveillance systems have been strengthened to facilitate prevention and early detection of listeriosis outbreaks.
Acknowledgments
For their assistance with the outbreak investigation, we thank the Outbreak Response Unit of the NICD, South Africa; and the National Department of Health, South Africa, including all provincial, metropolitan, and district environmental health departments. For their assistance with WGS, we thank the Sequencing Core Facility, NICD, South Africa. For their assistance with cgMLST analysis, we thank the National Reference Centre and WHO Collaborating Center for Listeria, Biology of Infection Unit, Institut Pasteur, France; and the Enteric Diseases Laboratory Branch, Centers for Disease Control and Prevention, Atlanta, Georgia.
Disclosure Statement
No competing financial interests exist.
References
- Allam M, Tau N, Smouse SL, Mtshali PS, Mnyameni F, Khumalo ZTH, Ismail A, Govender N, Thomas J, Smith AM. Whole-genome sequences of Listeria monocytogenes sequence type 6 isolates associated with a large foodborne outbreak in South Africa, 2017 to 2018. Genome Announc 2018;6:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althaus D, Jermini M, Giannini P, Martinetti G, Reinholz D, Nuesch-Inderbinen M, Lehner A, Stephan R. Local outbreak of Listeria monocytogenes serotype 4b sequence type 6 due to contaminated meat pate. Foodborne Pathog Dis 2017;14:219–222 [DOI] [PubMed] [Google Scholar]
- Angelo KM, Conrad AR, Saupe A, Dragoo H, West N, Sorenson A, Barnes A, Doyle M, Beal J, Jackson KA, Stroika S, Tarr C, Kucerova Z, Lance S, Gould LH, Wise M, Jackson BR. Multistate outbreak of Listeria monocytogenes infections linked to whole apples used in commercially produced, prepackaged caramel apples: United States, 2014–2015. Epidemiol Infect 2017;145:848–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Burall LS, Luo Y, Timme R, Melka D, Muruvanda T, Payne J, Wang C, Kastanis G, Maounounen-Laasri A, De Jesus AJ, Curry PE, Stones R, K'Aluoch O, Liu E, Salter M, Hammack TS, Evans PS, Parish M, Allard MW, Datta A, Strain EA, Brown EW. Listeria monocytogenes in stone fruits linked to a multistate outbreak: enumeration of cells and whole-genome sequencing. Appl Environ Microbiol 2016;82:7030–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Luo Y, Carleton H, Timme R, Melka D, Muruvanda T, Wang C, Kastanis G, Katz LS, Turner L, Fritzinger A, Moore T, Stones R, Blankenship J, Salter M, Parish M, Hammack TS, Evans PS, Tarr CL, Allard MW, Strain EA, Brown EW. Whole genome and core genome multilocus sequence typing and single nucleotide polymorphism analyses of Listeria monocytogenes associated with an outbreak linked to cheese, United States, 2013. Appl Environ Microbiol 2017a;83:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Luo Y, Curry P, Timme R, Melka D, Doyle M, Parish M, Hammack TS, Allard MW, Brown EW, Strain EA. Assessing the genome level diversity of Listeria monocytogenes from contaminated ice cream and environmental samples linked to a listeriosis outbreak in the United States. PLoS One 2017b;12:e0171389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira V, Wiedmann M, Teixeira P, Stasiewicz MJ. Listeria monocytogenes persistence in food-associated environments: Epidemiology, strain characteristics, and implications for public health. J Food Prot 2014;77:150–170 [DOI] [PubMed] [Google Scholar]
- Gelbicova T, Zobanikova M, Tomastikova Z, Van Walle I, Ruppitsch W, Karpiskova R. An outbreak of listeriosis linked to turkey meat products in the Czech Republic, 2012–2016. Epidemiol Infect 2018;146:1407–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail H, Smith AM, Tau NP, Sooka A, Keddy KH. Cholera outbreak in South Africa, 2008–2009: Laboratory analysis of Vibrio cholerae O1 strains. J Infect Dis 2013;208(Suppl 1):39–45 [DOI] [PubMed] [Google Scholar]
- Jacobs MR, Stein H, Buqwane A, Dubb A, Segal F, Rabinowitz L, Ellis U, Freiman I, Witcomb M, Vallabh V. Epidemic listeriosis. Report of 14 cases detected in 9 months. S Afr Med J 1978;54:389–392 [PubMed] [Google Scholar]
- Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 2014;9:e104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans MM, Brouwer MC, Bijlsma MW, Bovenkerk S, Keijzers W, van der Ende A, van de Beek D. Listeria monocytogenes sequence type 6 and increased rate of unfavorable outcome in meningitis: Epidemiologic cohort study. Clin Infect Dis 2013;57:247–253 [DOI] [PubMed] [Google Scholar]
- Kvistholm Jensen A, Nielsen EM, Bjorkman JT, Jensen T, Muller L, Persson S, Bjerager G, Perge A, Krause TG, Kiil K, Sorensen G, Andersen JK, Molbak K, Ethelberg S. Whole-genome sequencing used to investigate a nationwide outbreak of listeriosis caused by ready-to-eat delicatessen meat, Denmark, 2014. Clin Infect Dis 2016;63:64–70 [DOI] [PubMed] [Google Scholar]
- Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 2012;50:1355–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Perez-Osorio A, Wang Y, Eckmann K, Glover WA, Allard MW, Brown EW, Chen Y. Whole genome sequencing analyses of Listeria monocytogenes that persisted in a milkshake machine for a year and caused illnesses in Washington State. BMC Microbiol 2017;17:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonaco S, Nucera D, Filipello V. The evolution and epidemiology of Listeria monocytogenes in Europe and the United States. Infect Genet Evol 2015;35:172–183 [DOI] [PubMed] [Google Scholar]
- Moura A, Criscuolo A, Pouseele H, Maury MM, Leclercq A, Tarr C, Bjorkman JT, Dallman T, Reimer A, Enouf V, Larsonneur E, Carleton H, Bracq-Dieye H, Katz LS, Jones L, Touchon M, Tourdjman M, Walker M, Stroika S, Cantinelli T, Chenal-Francisque V, Kucerova Z, Rocha EP, Nadon C, Grant K, Nielsen EM, Pot B, Gerner-Smidt P, Lecuit M, Brisse S. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat Microbiol 2016;2:16185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura A, Tourdjman M, Leclercq A, Hamelin E, Laurent E, Fredriksen N, Van Cauteren D, Bracq-Dieye H, Thouvenot P, Vales G, Tessaud-Rita N, Maury MM, Alexandru A, Criscuolo A, Quevillon E, Donguy MP, Enouf V, de Valk H, Brisse S, Lecuit M. Real-Time Whole-genome sequencing for surveillance of Listeria monocytogenes, France. Emerg Infect Dis 2017;23:1462–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muvhali M, Smith AM, Rakgantso AM, Keddy KH. Investigation of Salmonella Enteritidis outbreaks in South Africa using multi-locus variable-number tandem-repeats analysis, 2013–2015. BMC Infect Dis 2017;17:661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadon C, Van Walle I, Gerner-Smidt P, Campos J, Chinen I, Concepcion-Acevedo J, Gilpin B, Smith AM, Man Kam K, Perez E, Trees E, Kubota K, Takkinen J, Nielsen EM, Carleton H. PulseNet International: Vision for the implementation of whole genome sequencing (WGS) for global food-borne disease surveillance. Euro Surveill 2017;22:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakari UM, Rantala L, Pihlajasaari A, Toikkanen S, Johansson T, Hellsten C, Raulo SM, Kuusi M, Siitonen A, Rimhanen-Finne R. Investigation of increased listeriosis revealed two fishery production plants with persistent Listeria contamination in Finland in 2010. Epidemiol Infect 2014;142:2261–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nel S, Lues JF, Buys EM, Venter P. Bacterial populations associated with meat from the deboning room of a high throughput red meat abattoir. Meat Sci 2004;66:667–674 [DOI] [PubMed] [Google Scholar]
- Orsi RH, Borowsky ML, Lauer P, Young SK, Nusbaum C, Galagan JE, Birren BW, Ivy RA, Sun Q, Graves LM, Swaminathan B, Wiedmann M. Short-term genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genomics 2008;9:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessis EMD, Govender S, Pillay B, Korsten L. Exploratory study into the microbiological quality of spinach and cabbage purchased from street vendors and retailers in Johannesburg, South Africa. J Food Prot 2017;80:1726–1733 [DOI] [PubMed] [Google Scholar]
- Schjorring S, Gillesberg Lassen S, Jensen T, Moura A, Kjeldgaard JS, Muller L, Thielke S, Leclercq A, Maury MM, Tourdjman M, Donguy MP, Lecuit M, Ethelberg S, Nielsen EM. Cross-border outbreak of listeriosis caused by cold-smoked salmon, revealed by integrated surveillance and whole genome sequencing (WGS), Denmark and France, 2015 to 2017. Euro Surveill 2017;22:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Keddy KH, Ismail H, Thomas J, van der Gryp R, Manamela MJ, Huma M, Sooka A, Theobald LK, Mennen MA, O'Reilly LC. International collaboration tracks typhoid fever cases over two continents from South Africa to Australia. J Med Microbiol 2011;60:1405–1407 [DOI] [PubMed] [Google Scholar]
- Smith AM, Mthanti MA, Haumann C, Tyalisi N, Boon GP, Sooka A, Keddy KH. Nosocomial outbreak of Salmonella enterica serovar Typhimurium primarily affecting a pediatric ward in South Africa in 2012. J Clin Microbiol 2014;52:627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Naicker P, Bamford C, Shuping L, McCarthy KM, Sooka A, Smouse SL, Tau N, Keddy KH. Genome sequences for a cluster of human isolates of Listeria monocytogenes identified in South Africa in 2015. Genome Announc 2016;4:1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Smouse SL, Tau NP, Bamford C, Moodley VM, Jacobs C, McCarthy KM, Lourens A, Keddy KH. Laboratory-acquired infections of Salmonella enterica serotype Typhi in South Africa: Phenotypic and genotypic analysis of isolates. BMC Infect Dis 2017;17:656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tau NP, Meidany P, Smith AM, Sooka A, Keddy KH. Escherichia coli O104 associated with human diarrhea, South Africa, 2004–2011. Emerg Infect Dis 2012;18:1314–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tau NP, Smith AM, Wain JR, Tarupiwa A, Coulibaly KJ, Keddy KH, Germs S. Development and evaluation of a multiple-locus variable-number tandem-repeats analysis assay for subtyping Salmonella Typhi strains from sub-Saharan Africa. J Med Microbiol 2017;66:937–945 [DOI] [PubMed] [Google Scholar]
- van Nierop W, Duse AG, Marais E, Aithma N, Thothobolo N, Kassel M, Stewart R, Potgieter A, Fernandes B, Galpin JS, Bloomfield SF. Contamination of chicken carcasses in Gauteng, South Africa, by Salmonella, Listeria monocytogenes and Campylobacter. Int J Food Microbiol 2005;99:1–6 [DOI] [PubMed] [Google Scholar]
- van Vollenhoven E. An outbreak of listeriosis in cattle and sheep ingesting potato offcuts. Tydskr S Afr Vet Ver 1999;70:50–57 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genome sequences for 10 L. monocytogenes ST6 isolates associated with this South African outbreak have been deposited at National Center for Biotechnology Information (NCBI)/GenBank under the accession numbers QEXB00000000 to QEXK00000000 (BioProject No. PRJNA451422, BioSample Nos. SAMN08970424 to SAMN08970415, respectively) (Allam et al., 2018).