Abstract
Significance: p62/SQSTM1 is a multifunctional scaffolding protein involved in the regulation of various signaling pathways as well as autophagy. In particular, p62/SQSTM1 serves as an essential adaptor to identify and deliver specific organelles and protein aggregates to autophagosomes for degradation, a process known as selective autophagy.
Critical Issues: With the emergence of autophagy as a critical process in cellular metabolism and the development of cardiometabolic diseases, it is increasingly important to understand p62's role in the integration of signaling and autophagic pathways.
Recent Advances: This review first discusses the features that make p62/SQSTM1 an ideal chaperone in integrating signaling pathways with autophagy and details the current understanding of its diverse roles in selective autophagy processes. Distinct and overlapping roles of other chaperones with similar functions are then discussed in the context of p62/SQSTM1. Finally, the recent literature focusing on p62 and selective autophagy in metabolism and the spectrum of cardiometabolic diseases including atherosclerosis, fatty liver disease, and obesity is evaluated.
Future Directions: A comprehensive understanding of the nuanced roles p62/SQSTM1 plays in mediating distinct autophagy pathways would provide new insights into the mechanisms of this critical degradative pathway. This will, in turn, facilitate our understanding of cardiovascular and cardiometabolic disease pathology and the development of novel autophagy-modulating therapeutic strategies.
Keywords: p62/SQSTM1, selective autophagy, cardiometabolic disease, cardiovascular disease, atherosclerosis, fatty liver disease
The Structure of p62/SQSTM1
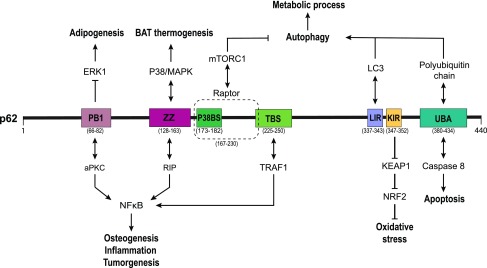
P62/SQSTM1 (hereafter called p62) contains a series of distinct domains that act in concert with binding partners to integrate and modulate a variety of cellular processes. As we describe hereunder, prominent p62 domains include the N-terminus PB1 (Phox/Bem1p), followed by the ZZ-type zinc finger (ZZ) domain, the tumor necrosis factor receptor-associated factor 6 (TRAF6)-binding sequence (TBS), the microtubule-associated protein light chain 3 (LC3)-interacting region (LIR), the Kelch-like ECH-associated protein 1 (Keap1)-interacting region (KIR), and the C-terminal ubiquitin-associated (UBA) domain (Fig. 1).
FIG. 1.
The structural domain organization, interacting partners, and functions of p62. p62 is a multidomain protein with roles in the regulation of numerous signaling pathways through distinct binding partners. Depicted are the N-terminal PB1 domain, followed by the ZZ domain, the p38-binding sequence, the TRAF6 binding sequence, LIR, KIR, and a C-terminal UBA domain. aPKC, atypical protein kinase C; BAT, brown adipose tissue; ERK1, extracellular signal-regulated kinase 1; Keap1, Kelch-like ECH-associated protein 1; KIR, Keap1-interacting region; LC3, microtubule-associated protein light chain 3; LIR, LC3-interacting region; MAPK, mitogen-activated protein kinase; mTORC1, mammalian target of rapamycin complex 1; NF-κB, nuclear factor kappa B; NRF2, nuclear factor erythroid 2-related factor 2; p62, p62/SQSTM1; PB1, Phox/Bem1p; RIP, receptor-interacting protein; TBS, TRAF6-binding sequence; TRAF6, tumor necrosis factor receptor-associated factor 6; UBA, ubiquitin associated; ZZ, ZZ-type zinc finger. Color images are available online.
The PB1 domain of p62 can bind atypical protein kinase C (aPKC) and activate aPKC-mediated nuclear factor kappa B (NF-κB) signaling (89, 90) to affect inflammatory responses. The PB1 domain also interacts and negatively regulates extracellular signal-regulated kinase 1 (ERK1), with crucial roles during adipogenesis and obesity (58). Furthermore, PB1 domain is required for p62 oligomerization and for binding with other PB1-containing protein, contributing to its role as a scaffolding protein (72).
The ZZ domain of p62 interacts with receptor-interacting protein 1 (RIP1) and serves to recruit aPKC to the tumor necrosis factor α (TNFα) signaling cascade, stimulating NF-κB and its downstream activation of the inflammatory response (90). In addition, a p38-binding sequence next to the ZZ domain can activate p38 mitogen-activated protein kinase (MAPK) and its targets, PPARγ coactivator-1α (PGC1α) and uncoupling protein 1 (UCP1), affecting mitochondrial function and brown adipose tissue (BAT) thermogenesis (74).
The TBS has been identified as TRAF6 binding motif. The p62–TRAF6 interaction in conjunction with the binding of aPKC through the PB1 domain fosters nerve growth factor (NGF) signaling (111), receptor activator of NF-κB (RANK)-induced NF-κB activation in osteoclastogenesis and bone remodeling (19), and Ras-induced tumorigenesis (18, 70).
The region between the ZZ and TBS domains is important for the interaction of p62 and raptor, a core subunit of mammalian target of rapamycin (mTOR) complex 1 (mTORC1) and crucial for mTOR activation (17) and its inhibitory effects on autophagy (71).
Although the p62–Raptor interaction is clearly important for p62's effects in metabolism, a more direct role for p62 in autophagy is its direct binding to the autophagosome coat protein LC3/Atg8 and other mammalian Atg8 homologues through the LIR motif (82). Not only is this interaction critical for delivery of cargo to autophagosomes, but it also potentially serves an important role in the formation of autophagosomes themselves (27, 42).
Another well-characterized p62 domain is the KIR motif wherein direct binding of p62 with Keap1 induces its autophagic degradation (56), thereby neutralizing Keap1's inhibitory effects on the nuclear factor erythroid 2-related factor 2 (NRF2), a transcription factor responsive to oxidative stress (36). Interestingly, the p62–Keap1 interaction is dependent on p62 oligomerization via its PB1 domain (36), demonstrating the importance of multiple p62 domains in its modulation of signaling pathways.
Finally, The C-terminal UBA domain of p62 is known to bind and sequester ubiquitinated proteins, primarily targeting them for autophagic degradation via the LC3-interacting motif already mentioned (34, 63, 73). More nuanced regulation also exists at the UBA domain. For example, the phosphorylation of serine 403 (S403) by TANK binding kinase 1 (TBK1) and casein kinase 2 (CK2) or the ubiquitination of lysine 420 (K420) by Keap1/cullin3, both of which are located in the UBA, modulates its binding affinity for polyubiquitinated chains and subsequent autophagic degradation (59, 68, 85). Furthermore, the UBA has also been shown to bind cullin3-mediated caspase-8 polyubiquitination, thus controlling apoptotic signaling (39).
p62 As a Chaperone for Selective Autophagy
Autophagy is the predominant mechanism for delivering protein aggregates and long-lived/damaged organelles to lysosomes for degradation. In addition to playing the critical role of eliminating unwanted or damaged cellular cargo and thus protecting the cell from intracellular stressors, autophagy also provides essential nutrients for cellular survival in states of starvation. These classical functions of autophagy have long been considered to constitute a nonselective form of degradation, where adjacent cargo is handled in bulk. However, there is increasing evidence that some forms of autophagy are highly selective and well-organized processes involving specifically marked cargo recognized by the autophagic machinery (22). This selective pathway requires adaptor proteins that can specifically bind to cytosolic cargo and deliver them to autophagosomes.
The prototypical and most studied chaperone is p62 but others include neighbor of BRCA1 gene (NBR1), optineurin (OPTN), NDP52/CALCOCO2 (hereafter called NDP52, nuclear dot protein 52 kDa/calcium-binding and coiled-coil domain 2), toll-interacting protein (TOLLIP), BNIP3L/Nix, FUN14 domain containing 1 (FUNDC1), and starch binding domain 1 (STBD1). These receptors can recognize and sort cytosolic cargo usually via an ubiquitin binding domain and interact with the LC3/Atg8 family of autophagy proteins for targeting to autophagosomes. Detailed understanding of this selective autophagy requires uncovering how a particular organelle or protein is labeled, which specific adaptors recognize the target, and what autophagy proteins act as their binding partners. In the following discussion, we describe various forms of selective autophagy and detail the known roles for p62 in mediating their degradation. Furthermore, we correlate how each of these p62-dependent processes contributes to the progression or protection of metabolic diseases.
Aggrephagy
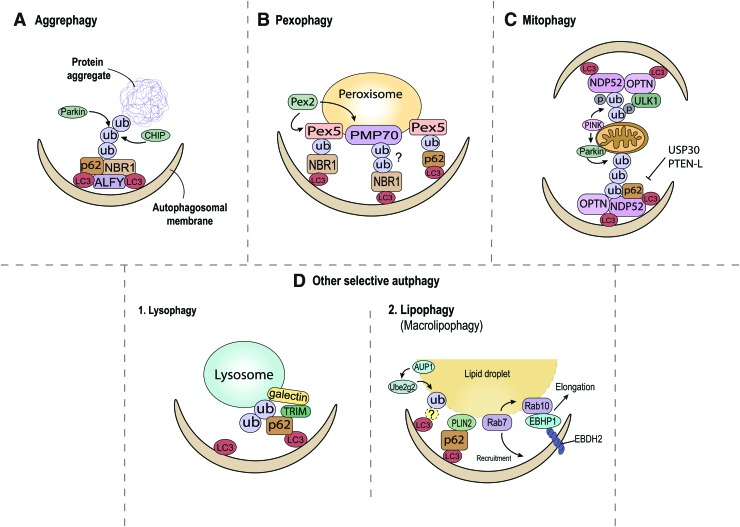
Aggrephagy, autophagic degradation of intracellular protein aggregates, is one of the earliest recognized forms of selective autophagy, first described in neurodegenerative diseases (113). Misfolded proteins originate from mistranslation of defective ribosomal products, misfolding after translation, abnormal protein modification, or mutations in transcripts incurred by genomic damage. Under steady state conditions, cells can defend against the toxicity of accumulated misfolded proteins through chaperones involved in monitoring protein folding, trafficking, and assisting in the protein refolding, and preventing aggregation (41). Alternatively, chaperones can also aid in the degradation of misfolded proteins through the ubiquitin–proteasome system (UPS) or the autophagy pathway by conjugating with E3 ubiquitin ligase, such as c-terminus of heat shock chaperone 70-interacting protein (CHIP) or Parkin (11, 75).
Ubiquitination is a post-translational modification that plays an essential role in substrate recognition and degradation of proteins both by the UPS and selective autophagy (52, 84). The central feature of this ubiquitin tagging system uses a series of enzymatic reactions involving the E1 ubiquitin activating enzyme, E2 ubiquitin conjugating enzyme, and E3 ubiquitin protein ligase. Ubiquitin has seven internal lysine residues (K6, K11, K27, K29, K33, K48, and K63) that are used to conjugate to proteins as monomers or as polyubiquitin chains. K48-linked polyubiquitin chains have been identified as potent signals for protein targeting to protoesomes, whereas K27- and K63-linked mono- or polyubiquitin chains have been shown to target proteins for autophagic degradation. These different patterns of ubiquitination offer the specificity needed to initiate the next steps of autophagic degradation.
Recognition and delivery of ubiquitinated protein aggregates to autophagosomes then require proteins containing ubiquitin-binding domains. p62 is one of the most well studied of these adaptors with the ability to recognize and bind to ubiquitinated proteins through its UBA domain followed by interaction with the core autophagic protein LC3 through the LIR domain (8, 50). Autophagy-linked FYVE protein (ALFY, also known as WDFY3) has also been shown to be a central scaffold protein in aggrephagy (12). ALFY is essential for p62-dependent aggrephagy by interacting with p62 as well as components of the autophagy initiation complex (Atg5, Atg12) and phosphatidylinositol-3-phosphate, an important lipid species regulating autophagosome membrane formation (23) (Fig. 2A). Other adaptor proteins such as histone deacetylases 6, a microtubule-associated deacetylase with a ubiquitin-binding zinc-finger domain mediates ubiquitin binding, have also been shown to enhance transport of ubiquitinated proteins into aggresomes, and promote fusion of autophagosome with lysosome (44, 54); however, more studies are needed to understand the relative importance of p62 and these adaptors in aggrephagy.
FIG. 2.
Selective autophagy related to p62. (A) Aggrephagy: CHIP and Parin are the major E3 ubiquitin ligase for protein aggregates. p62, NBR1, and ALFY enhance transport of polyubiquitinated proteins into autophagosome, and promote aggrephagy. (B) Pexophagy: The ubiquitination of PEX5 is mainly induced by the PEX2 E3 ubiquitin ligase. P62 and NBR1 mediate interaction of ubiquitinated peroxisomes and autophagosomes. (C) Mitophagy: PINK1 affects both of ubiquitin phosphorylation and Parkin E3 ubiquitin ligase activation. The mitochondrial deubiquitinase USP30 and phosphatase PTEN-L antagonize PINK1/Parkin-mediated ubiqutination. p62, NDP52, and OPTN are adaptors implicated in mitophagy. (D) (Left) Lysophagy: Galectins bind to exposed innermembrane glycosidase and recruit TRIM proteins. Unknown ubiqutinated lysosomal proteins and TRIM interact with p62 directly to promote lysophagy. (Right) Lipopagy: AUP1 recruits the Ube2g2 E2 ubiquitin conjugase. The LDs coated proteins PLIN2 and unknown ubiqutinated surface proteins can bind with p62 for the progression of lipophagy. Also, Rab7 small GTPase directly recruits LDs and Rab10 to the autophagosomal membrane by promoting interaction with the EHBP1 and EBDH2 complex on the autophagosome. ALFY, autophagy-linked FYVE protein; CHIP, c-terminus of heat shock chaperone 70-interacting protein; LDs, lipid droplets; NBR1, neighbor of BRCA gene 1; NDP52, nuclear dot protein 52kDa/calcium-binding and coiled-coil domain 2; OPTN, optineurin; PEX, peroxin; PINK1, PTEN-induced putative kinase 1; PLIN2, Perilipin 2; PTEN, phosphatase and tensin homolog; PTEN-L, PTEN-long.; USP, ubiquitin carboxyl-terminal hydrolase. Color images are available online.
Pexophagy
Peroxisomes are important metabolic organelles involved in lipid metabolism by catabolizing long chain fatty acid β-oxidation as well as reduction of reactive oxygen species (ROS), specifically hydrogen peroxide. ROS produced as a by-product of aerobic respiration is removed via peroxisomal enzymes, a process essential for ameliorating the damaging effects of ROS. Since impaired peroxisomes lead to accumulation of ROS within the cell, dysfunctional peroxisomes are cleared by the autophagy process called pexophagy. Work thus far has again implicated the importance of ubiquitination followed by recognition by the adaptor p62 and targeting to autophagosomes (45). However, despite the importance of this clearance mechanism in peroxisomal homeostasis, we only have a rudimentary understanding of the components and overall regulation of pexophagy in mammalian cells.
Recent studies have focused on peroxisomal matrix protein receptor peroxin 5 (PEX5) as the ubiquitinated target in pexophagy (45, 78). PEX5 can be shuttled between the peroxisomal membrane and the cytosol in a ubiquitin-dependent manner (14). In response to ROS, ataxia-telangiectasia-mutated signaling activates phosphorylation and monoubiquitination of PEX5 at the peroxisomal membrane, upon which it can be recognized by p62, leading to autophagosome targeting and pexophagy (114). Furthermore, PEX1/PEX6 complexes, which form a single and heterohexameric Type 2 AAA-ATPase motor, appear to also play a role in pexophagy by regulating shuttling of ubiquitinated PEX5 (25). The ubiquitination of PEX5 is induced by the peroxisomal E3 ubiquitin ligase PEX2, particularly during amino acid starvation (92). There is also recent evidence that PEX5 might not be the only target for initiating pexophagy. The ATP-binding cassette transporter 70 kDa peroxisomal membrane protein 70 (PMP70) is also ubiquitinated by PEX2 and might be another key substrate for adaptor binding and autophagosome targeting (92). Lastly, NBR1 has also been implicated as an essential adaptor protein that can promote pexophagy (16). Although independent of p62, the efficiency of NBR1 interaction with the peroxisome might also be enhanced by p62.
The series of proteins implicated thus far in pexophagy are summarized in Figure 2B. Overall, many unanswered questions remain in the process of pexophagy, including identification of the critical ubiquitination targets of PEX2, uncovering the exact role of PMP70 ubiquitination, and deciphering the relative importance of p62 and other adaptor proteins in autophagosome targeting.
Mitophagy
Mitochondria play a central role in various metabolic process, including production of the majority of cellular ATP through oxidative phosphorylation and regulation of the intrinsic pathway of apoptosis. In turn, mitochondrial dysfunction is associated with excessive ROS production, loss of control over apoptotic signals, and adverse effects on metabolic homeostasis. Mitophagy is a critical mechanism by which cells maintain metabolic homeostasis by eliminating damaged or long-lived mitochondria via autophagic degradation. Important proteins in mitophagy including the adaptor p62 have been identified, which we describe hereunder.
The phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1) and the E3 ubiquitin ligase Parkin constitute the most studied mitophagy pathway responsible for mitochondrial quality control (76, 66, 105). Although they likely play important roles in a variety of diseases, both PINK1 and Parkin initially came to attention as Parkinson's disease-related genes (48, 98, 104). PINK1 requires depolarization of mitochondrial membrane potential for its inner mitochondrial inner membrane import. In healthy conditions, PINK1 is cleaved by mitochondrial intramembrane protease presenilin-associated rhomboid like in the transmembrane segment and subsequently released to the cytosol for proteasomal degradation (38). However, mitochondrial damage and resultant loss of mitochondrial membrane potential prevent PINK1 cytosolic export, leading to its accumulation on depolarized mitochondria and phosphorylation of downstream targets such as ubiquitin, mitofusin 2 (MFN2), and Parkin (51, 108). Parkin not only assembles ubiquitin chains on the mitochondrial outer membrane (MOM) but also acts to amplify the “phospho-ubiquitin” mitophagy signal produced by PINK1 by recruiting more Parkin to MOM (43). Phosphorylated MFN2 by PINK1 can also promote Parkin recruitment for generating ubiquitin chains on MOM priming the mitochondria for mitophagy. It should also be noted that this ubiquitination process can also be reversed providing an early break on this process. Both mitochondrial deubiquitinase ubiquitin carboxyl-terminal hydrolase 30 (USP30) (7) and protein phosphatase PTEN long (106) have been shown to antagonize PINK1/Parkin-mediated ubiqutination through removal of phosphoubiquitin or ubiquitin chains from MOM.
Upon priming of mitochondria by ubiquitination, a large body of evidence has implicated p62 as an essential chaperone mediating Parkin-dependent mitophagy (26, 79, 112). p62 can bind the polyubiquitin chains of MOM through its UBA domain and by interacting with Atg8/LC3 on autophagosome through its LIR domain, and can target mitochondria for autophagic clearance. It should be noted that this traditional model has been challenged recently, where, in lieu of p62, PINK1 recruits two other adaptors OPTN and UDP52, leading to recruitment of the critical autophagy Ser/Thr kinase unc-51-like autophagy activating kinase (ULK1), and activation of mitophagy. Importantly, this mechanism is also independent of Parkin (57). These two competing mechanisms are shown in Figure 2C. The relative importance of each in various cell types and their relevance to metabolic diseases remain unknown.
Lysophagy
P62-tagged cargoes are ultimately routed to the lysosome, wherein an acidic environment and a diverse array of hydrolases mediate catabolism into basic metabolites. However, lysosomes themselves are also vulnerable to damage, rupture, and increases in membrane permeability often caused by indigestible crystalline material (83). These changes both disrupt the normal degradative functions of the lysosome and release hydrolases, which can damage other cellular components and initiate cell death. Lysophagy is the selective autophagic degradation of damaged lysosomes and acts to limit these adverse consequences (83). The proximal events mediating lysophagy are thought to first include recognition of exposed inner-membrane glycans by galectins and recruitment of TRIM proteins, which serve as scaffolds to initiate a variety of autophagy signaling cascades (46, 83). A number of TRIM proteins are known to interact with p62 directly (46), and also recruit E3 ubiquitin ligases to target sites of membrane damage, (4) which may favor further ubiquitination, p62 binding, and lysophagy. Indeed, the seminal reports characterizing lysophagy observed ubiquitin and p62 colocalized at sites of lysosomal membrane damage (31, 64). In addition, p62 is required for full recruitment of LC3 to damaged endolysosomes (10). p62 has also been observed to be strongly localized to the lysosome in contexts related to its signaling actions (discussed in depth in subsequent sections). For example, p62 interacts with mTOR and atypical PKCs at the lysosome and is a crucial part of these signaling cascades (1, 65). The functional differences in p62 action at the lysosome to either favor lysophagy under conditions of lysosomal damage or serve these types of signaling roles are likely determined by its specific binding partners under either condition.
Lipophagy
Lipophagy, the selective degradation of lipid droplets (LDs) via autophagy (115), was initially observed in hepatocytes during starvation. Although starvation is thought to induce global autophagy not overtly specific to any cargo, a particular enrichment in autophagosomes containing only LDs was observed, suggesting both the presence of a selective mechanism and that macroautophagy is a primary mechanism of LDs delivery to the lysosome (99). Regardless of the mode of lipid delivery, lysosomal hydrolysis of LD cholesterol esters and triglycerides is mediated by lysosomal acid lipase. Other major conceptual advances in the field include recognition of the many interactions between autophagy and cytosolic lipases such as adipose triglyceride lipase, and the appreciation of microlipophagy as an important mechanism of LDs delivery to the lysosome in some contexts (93). In general, the characterization of the molecular mechanisms initiating selective recruitment of autophagosomes to LDs remains undercharacterized, and many genetic or pharmacological manipulations reported to alter lipophagy seem to act downstream of these putative initial events. p62 is one of many potential molecular mediators conferring selectivity for lipophagy and has recently been observed at the LD in a novel screen for bona fide LD proteins (5). One mechanism that could target LDs for lipophagy is ubiquitination that is observed on many resident LD proteins (20, 77, 100), although direct binding of these ubiquitin moieties by p62 or other chaperones and relevance to lipophagy are unconfirmed. Several earlier articles have assessed the relevance of p62 to lipophagy. Wang et al. demonstrated that p62 is required for full LC3 recruitment to LDs upon ethanol-induced lipophagy, and that p62 silencing accordingly resulted in triglyceride and cholesterol accumulation (107). In another study, p62 was found to colocalize with LDs and coimmunoprecipitate with the LD protein PLIN2 (Perilipin 2) during lipophagy (55). It remains uncertain whether p62 truly serves as a lipophagy chaperone in these settings as p62 deficiency could impact lipolysis through many connected signaling cascades. To test whether p62 at the LDs was sufficient to induce lipophagy, Tatsumi et al. (101) created a forced lipophagy system, wherein p62 was fused to a LD localization domain from TIP47. Overexpression of this fusion protein was found to reduce lipid content in an autophagy-dependent manner and similar phenotypes were not observed with simple p62 overexpression. The construct also strongly colocalized with LC3/Lamp1, further suggesting that p62 is capable of inducing lipophagy in this setting. Additional studies might investigate whether p62 LIR domain deletion abrogates these phenotypes, and a great deal of work is still required to discover the proximal events initiating lipophagy, including how p62 or other chaperones are recruited to the LD.
Noncanonical Domains of p62 Involved in Selective Autophagy
Beside the prototypical p62 domains mediating selective autophagy (i.e., the UBA's association with ubiquitin-tagged proteins and the LIR's binding with the autophagy coat protein LC3), other domains of p62 appear to also be involved in various aspects of the initiation and regulation of selective autophagy. These are depicted in Figure 3 and described hereunder.
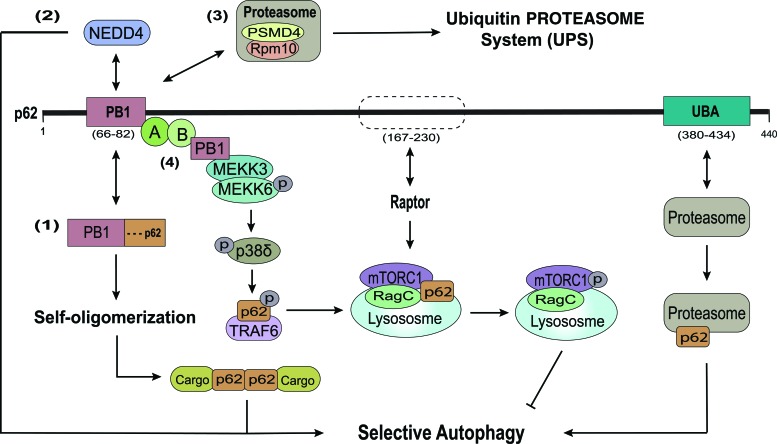
FIG. 3.
Noncanonical domains of p62 involved in selective autophagy. Beside classical LIR and UBA domains mediating selective autophagy, other domains of p62 also play important roles in autophagy as receptors or regulators. (1) The N-terminal PB1 domain is a key component by functioning as an adaptor for the oligomerization of p62 and mediating autophagy initiation in an LIR/UBA-independent manner. (2) The PB1 domain is also recognized and ubiquitinated by the HECT E3 ubiquitin ligase NEDD4, thus enhancing selective autophagic degradation of inclusion bodies. Additional interaction with the 19S proteasomal subunit PSMD4/Rpn10 accelerates delivery and clearance of ubiquitinated cargo to the UPS. (3) The p62 UBA domain also recognizes the proteasome for autophagic degradation. (4) Interaction of PB1 and MEKK3 activates a phosphorylation cascade leading to recruitment of TRAF-6 to p62 TB domain. TRAF-6 promotes translocation of mTOR to lysosomes, its K63 ubiquitination, activation of mTOR signaling, and inhibition of autophagy. A region between the p62 ZZ and TB domains (amino acids 167–230) also participates in lysosomal translocation and activation of mTOR by binding with Raptor and RagC. mTOR, mammalian target of rapamycin; NEDD, neural precursor cell expressed developmentally downregulated protein; UPS, ubiqitin proteasome system. Color images are available online.
Role of the PB1 domain in clearance of nonubiquitinated protein aggregates
The N-terminal PB1 domain plays a critical role in oligomerization of p62 by mediating interactions between basic and acidic charge clusters through electrostatic forces (72). In one study, oligomerization of p62 through PB1 promoted its recruitment to endoplasmic reticulum-located autophagosome formation sites in fibroblasts and was independent of the interaction with the phagophore mediated by the LIR domain, implying a different mechanism for autophagy initiation (35). Another study also showed the clearance of an aggregation-prone isoform of signal transducer and activator of transcription (STAT)5A to be dependent on interaction with the p62 PB1 domain (109). Although aggregate formation of this isoform is ubiquitination independent, the PB1-mediated oligomerization of p62 may improve its interaction with STAT5A aggregates and trigger their autophagic clearance. Moreover, the HECT E3 ubiquitin ligase, neural precursor cell expressed developmentally downregulated protein (NEDD)4, also interacts and ubiquitinates the PB1 domain of p62, in turn, facilitating selective autophagy and the degradation of inclusion bodies (60).
Involvement of the PB1 and UBA domains in the UPS
Autophagy and the UPS constitute two prominent and distinct processes for the degradation of ubiquitin-labeled intracellular cargo. Akin to its critical role in selective autophagy, p62 also contributes to shuttling of ubiquitinated proteins to proteasomes. However, instead of the LIR domain of p62 for cargo delivery, ubiquitinated substrates are delivered via interaction of the PB1 domain with the 19S proteasomal subunit PSMD4/Rpn10 (94).
p62 has also recently been implicated in targeting and removal of proteasomes themselves, a mechanism that has long been enigmatic. Recent study by Cohen-Kaplan et al. shows that p62-mediated autophagy is likely the main degradation pathway for the mammalian proteasome (13). Specifically, amino acids starvation induces ubiquitination of select residues on the 19S proteasomal subunits, which are then recognized and interact with the p62 UBA domain for delivery to the autophagosome. These data demonstrate p62's unique role as an adaptor linking these two degradation systems.
p62-mediated negative regulation of autophagy by activation of mTOR signaling
Known as a critical regulator of intracellular metabolism and cell growth, the mTOR predominantly inhibits autophagy through the ULK1-FIP200-Atg13 complex. Interestingly, unlike its classical role as an autophagy chaperone, p62 has also been paradoxically shown to negatively regulate autophagy by activating mTOR. There are several proposed mechanisms by which this occurs.
First, a region between the p62 ZZ and TB domains (amino acids 167–230) is necessary for its interaction with raptor, a main component of the mTORC1 complex. This interaction appears to foster additional association of p62 with RagC, activation of the regulator complex, docking of mTOR on the lysosomal surface, and activation of downstream mTORC1 signaling (17). Aside from the proposed p62–raptor–RagC interaction, Linares et al. have demonstrated another novel mechanism of mTOR activation where the p62 TB and PB1 domains are involved (61). Interaction of MEKK3 with the PB1 domain creates a molecular platform where amino acid stimulation leads to successive activation of MEKK3, p38δ, and phosphorylation of T269 and S272 residues of p62. This phosphorylation then recruits TRAF-6, a lysine 63 (K63) E3 ligase, to the p62 TB domain, which, in turn, promotes its K63 ubiquitination as well as mTOR translocation to the lysosomal surface and mTORC1 activation. The relationship between these two mechanisms of p62-mediated mTORC1 activation remains unclear.
Although the UBA and LIR domains of p62 are the most studied and well-characterized contributors to selective autophagy, the other p62 domains appear to also impact this process in noncanonical ways including the potential of even inhibiting autophagy. Such opposing effects of p62 in autophagic degradation deserve continued investigation as they reflect intricate feedback mechanisms that can play unique roles in different cell types and during exposure to cellular stress.
Crosstalk Between p62 and Other Chaperones in Selective Autophagy
p62 is not the only adaptor/chaperone protein involved in selective autophagy. As briefly listed previously, other p62-like proteins serving this role include NBR1, NDP52, OPTN, and STBD1. In fact, degradation of different cargos appears to be preferentially mediated by distinct chaperones. In addition, interaction and cooperation between p62 and these chaperones are not only important for the initiation and processing of select autophagic substrates, but also provide intricate regulatory mechanisms in the clearance of protein aggregates and damaged organelles. We describe some of these hereunder.
Neighbor of BRCA gene 1
NBR1 is a 966-amino acid protein that has domain organization similar to p62, including N-terminal PB1, ZZ zinc finger, LIR, and C-terminal UBA domains (47). NBR1 has been identified in intracellular inclusion bodies composed of aggregated polyubiqitinated proteins. During aggrephagy, NBR1 can bind p62 through its PB1 domain and promote formation of the autophagic complex with other ATG family members. Although NBR1 cannot dimerize by itself, it appears to still be able to mediate autophagic clearance of aggregates in p62-deficient cells. For example, NBR1 can also act as an autophagy adaptor for peroxisomes in a process similar to that of p62. Deosaran et al. demonstrated that several domains of NBR1 including an amphipathic a-helical J domain along with UBA, LIR, coiled-coil domains are necessary for NBR1-mediated pexophagy (16). Despite the ability of NBR1 to promote pexophagy independent of p62, its binding to p62 increases the efficiency of peroxisome targeting to autophagosomes (47).
Nuclear dot protein 52 kDa and optineurin
NDP52 is another autophagy adaptor with reported roles in mitophagy, aggrephagy, and xenophagy (102). In some cases, it functions independently of p62 (e.g., NDP52 can cooperate with OPTN in Pink1–Parkin-mediated mitophagy) (30). However, p62 can exert its influence on NDP52 transcript levels. NDP52 transcription is dependent on NRF2, whose nuclear translocation is regulated by p62 via its competitive binding with Keap1 (40). Thus, p62 can also regulate selective autophagy indirectly by controlling expression of other chaperones.
Although less investigated, OPTN is another autophagy adaptor with proposed p62-independent roles in mitophagy (as already described) and xenophagy (particularly in clearance of invading Salmonella) (110). In the context of p62, OPTN ubiquitination by the tumor suppressor HACE1, an ubiquitin ligase, promotes its interaction with p62, formation of an autophagy adaptor complex, and acceleration of autophagy flux (62).
TANK binding kinase 1
A further level of regulation worthy of mention is the effect of phosphorylation on the efficiency of p62-mediated autophagic clearance. The ability of p62 to associate with LC3/ATG8 is increased upon phosphorylation of residues adjacent to the LIR (68). Phosphorylation of the p62 UBA domain at Ser-403 by the TBK1, a known p62-binding partner, improves affinity of p62 for ubiquitin chains and enhances aggrephagy (85). Furthermore, during Parkin-dependent mitophagy, phosphorylated p62 by TBK1 during mitochondrial depolarization synergizes mitochondrial engulfment by autophagosomes (67).
Regulation of p62
Both transcriptional and post-translational regulations of p62 constitute important mechanisms by which autophagic processing can be modulated. Several upstream signaling pathways and their transcription factor effectors have been implicated in upregulation of p62 transcripts, thus providing enough of this chaperone to facilitate ongoing autophagy. Transcription factor EB, known as the master regulator of the autophagy-lysosomal biogenesis, is responsible for broad transcriptional upregulation of the autophagic machinery including p62 (21, 96, 96a). Under conditions of oxidative stress where autophagy is concomitantly induced, the transcription factor NRF2 directly induces p62 expression by binding to conserved antioxidant-responsive elements in the p62 promoter (15, 49). In a positive feedback cycle, p62 can then stimulate NRF2 activity by interacting with and neutralizing KEAP1, an inhibitor of NRF2 (36). In response to other cellular stressors such as amino acid deficiency and endoplasmic reticulum stress, activating transcription factor 4 plays an important role in transcriptional regulation of autophagy genes including p62 by direct binding of their promoters in conjunction with CCAAT/enhancer-binding protein homologous protein (1, 24).
Besides modulation of transcript levels, p62 can also be regulated via post-translational modification such as phosphorylation and ubiqutination. Phosphorylation of serine 403 (S403) located in the UBA domain of p62 by TBK1 and CK2 promotes its binding affinity for ubiquitinated cargos destined for autophagic degradation (67, 68, 85). Phosphorylation of serine 351 (S351) at the p62 KIR domain enhances its affinity for KEAP1, which not only promotes the autophagic removal of ubiquitinated cargo but also liberates NRF2 to transcriptionally upregulate p62 as already discussed (32). Ubiquitination of lysine 420 (K420) located in the p62 UBA domain by Keap1/Cullin3 also enhances autophagic degradation (59), whereas ubiquitination of lysine 7 (K7) in the p62 PB domain by the E3 ubiquitin ligase TRIM21 interferes with p62 oligomerization and cargo sequestration (81). Although these findings suggest the importance of nuanced p62 regulation in autophagy, further study in needed to delineate whether this regulation differs in the various types of selective autophagy.
Roles for p62 in Cardiometabolic Disease
Mutations in a variety of p62 domains have long been known to underlie autosomal dominant diseases, such as frontotemporal dementia, amyotrophic lateral sclerosis, Paget's disease, and distal myopathy. Although variants in p62 have not yet been uncovered in genome-wide association studies of common cardiometabolic diseases, experimental evidence clearly implicates p62 as a major player in atherosclerosis, cardiac proteinopathies, nonalcoholic fatty liver disease, obesity, and type 2 diabetes (Fig. 4).
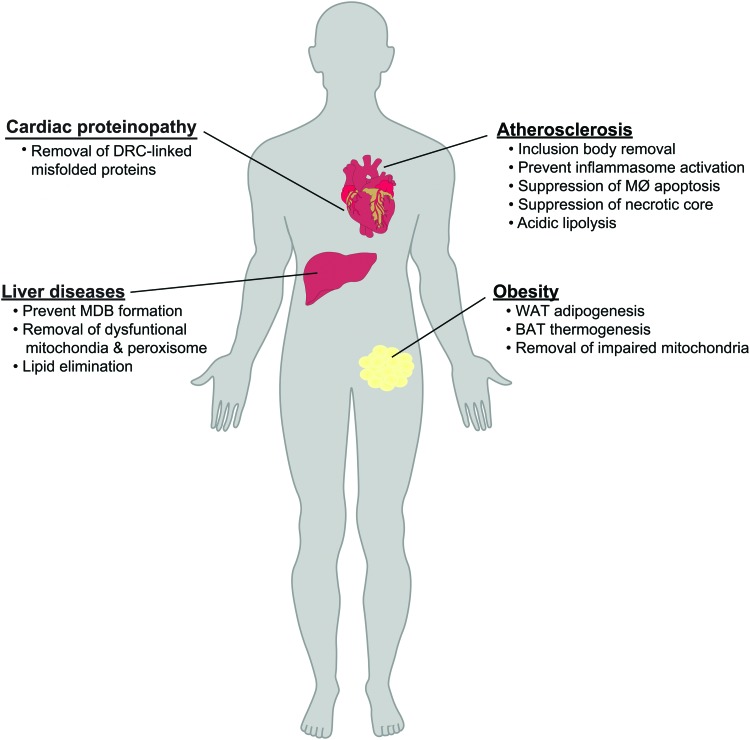
FIG. 4.
Roles for p62 in cardiometabolic diseases. The ability of p62 to mediate distinct autophagic processes implicates it in a variety of cardiometabolic diseases. In atherosclerosis, p62-dependent aggrephagy and removal of inclusion bodies ameliorates atherosclerosis by suppressing macrophage inflammasome activation, apoptosis, and necrotic core formation. In the cardiac proteinopathy, p62-linked autophagy can mediate aggresome formation for removing the DRC-linked proteins. In the liver, p62-dependent aggrephagy prevents accumulation of cytotoxic MDBs) and p62-dependent mitophagy/pexophagy prevents accumulation of dysfunctional mitochondria and peroxisomes, which together ameliorate the progression of fatty liver disease. In obesity, p62-dependent mitophagy modulates WAT adipogenesis, BAT thermogenesis, and the progression of obesity. DRC, desmin-related cardiomyopathy; MDB, Mallory–Denk body; WAT, white adipose tissue. Color images are available online.
In the atherosclerotic plaque, appropriate function of the autophagy–lysosome pathway in crucial cell types such as macrophages is essential for limiting inflammation, promoting cell survival, and appropriately handling lipids. Several studies provide strong evidence that p62-linked selective autophagy is particularly relevant to atherosclerosis in this regard. As discussed, p62 is known to tag ubiquitinated protein aggregates, serve as a link between ubiquitinated targets and autophagic machinery, and, therefore, accumulate as a consequence of dysfunctional autophagy (9). Atherosclerotic plaques are no exception and both human and murine samples feature marked p62 accumulation (21, 95, 96). Furthermore, macrophage-specific autophagy deficiency worsens atherosclerosis through accumulation of a variety of selective autophagy cargo related to p62. For example, lipophagy is essential for targeting macrophage LDs for lysosomal hydrolysis and cholesterol efflux (37, 80), and it would be worthwhile to assess the role of p62 in this process. Another key phenotype of macrophage-specific autophagy deficiency is inflammasome activation, particularly in the setting of cholesterol crystal accumulation (86). Mitochondrial dysfunction and inflammation in this setting may be downstream of defective aggrephagy, mitophagy, and lysophagy.
Evidence from our laboratory characterizing p62-deficient mouse models further supports the important roles of p62 in atherosclerosis. Mice deficient in p62 have increased atherosclerotic plaque burden with elevated macrophage inflammation, macrophage apoptosis, necrotic core formation, and plaque complexity (95). Mechanistically, p62-linked protein aggregation and aggrephagy limit inflammasome activation in a manner that is dependent on ubiquitin binding and that is not impacted by NRF2, ERK, p38, or NF-κB inhibition (83, 101). Overall, p62's role in selective autophagy seems to be a primary mechanism of atheroprotection, but more specific investigation of the impacts of deficiency on specific cargoes such as mitochondria and LDs is warranted.
There is also emerging evidence for the role of p62 and selective autophagy pathways in the spectrum of cardiac diseases known as cardiac proteinopathies. Desmin-related cardiomyopathy (DRC), which represents a prototypical cardiac proteinopathy, is characterized by an accumulation of misfolded proteins and protein aggregates in cardiomyocytes resulting in cytotoxicity (88). Recent studies have shown that p62 expression is significantly increased in the myocardium of a DRC mouse model in conjunction with accumulation of protein aggregates (6). Cardiomyocyte p62 mediates aggresome formation as well as activation of autophagy of the DRC-linked misfolded proteins (116). Although the precise molecular mechanisms of the disposal of these aggregates in cardiomyocytes remain undefined, such initial observations suggest that impairment of p62-dependent aggrephagy plays a prominent role in the pathogenesis of cardiac proteinopathies.
In the context of liver disease, Singh et al. were the first to suggest that autophagy plays a pivotal role in hepatocytes to deliver LDs to lysosomes for hydrolysis and provided some of the earliest experimental evidence for lipophagy in vivo. Inhibition of lipophagy led to decreased triglyceride hydrolysis and fatty acid β-oxidation, which resulted in LD accumulation in cultured hepatocytes and lipid-laden livers in mice (99). This study also highlighted that dysfunctional lipophagy in hepatocytes could contribute to human liver disease caused by excessive accumulation of lipid, such as nonalcoholic fatty liver disease (NAFLD). Also, recent studies have found that progressive insulin resistance and oxidative stress in fatty livers can instigate autophagy dysfunction, increased p62 levels, and massive accumulation of mitochondria and peroxisome in liver (28). Reductions in this p62-dependent selective autophagy lead to deleterious accumulation of cytotoxic organelles and exacerbate liver injury.
Accumulation of protein inclusion called Mallory–Denk bodies (MDBs) or Mallory's hyaline has also been described in livers of patients with alcoholic liver disease and NAFLD (53). Dysfunction in p62-dependent aggrephagy in hepatocytes appears to be a major cause of these accumulated inclusion bodies. Interestingly, p62-laden MDBs are not a phenomenon exclusive to hepatosteatosis as it has also been described in hepatocellular carcinoma among other cancers (65, 103). A related consequence of this p62 buildup is increased interactions with Keap1, persistent activation of NRF2, and progression of hepatocellular adenomas (33). Thus, impaired hepatic autophagy with resulting accumulation of cytotoxic organelles and protein aggregates creates multiple independent insults to the liver.
Among the most striking phenotypes of whole-body murine p62 deficiency is the clear development of mature-onset obesity and associated insulin resistance (87). Enhanced ERK1 activity in this model has been proposed as key mediator of these phenotypes. Impressively, ERK1 deficiency alone has minimal impact on energy expenditure, obesity, adipogenesis, or insulin resistance, but can almost entirely normalize the abnormalities in these phenotypes observed with p62 deficiency (58). Another aspect of p62 function relevant to obesity is linked to signaling through the p38-MAPK pathway, an important input to transcriptional control of brown/beige adipose tissue thermogenesis. p62 deficiency is associated with decreased UCP-1 and PGC-1α levels in BAT and deficiency impairs mitochondrial gene expression and functional thermogenesis in a cell-autonomous manner. The impacts of p62 deficiency on impaired mitophagy and lipophagy in this context deserves future attention, as defects in either pathway could also contribute to this phenotype. Whether these phenotypes of p62-deficient mice are entirely related to action at the level of adipocytes is also unclear. On one hand, aP2-Cre mediated tissue-specific knockout of p62 recapitulated key phenotypes of whole body deficiency including obesity, glucose intolerance, and insulin resistance, same with p62 whole body deficient mice (74). However, aP2-Cre has now been shown multiple times to elicit recombination in several nonadipocyte tissues including neurons, and neuron-specific p62-deficient mice (Nestin-cre/p62-floxed mice) also display an obese phenotype (29). This model displayed leptin resistance linked to defective STAT3 activity, hyperphagia, and mature-onset obesity in brain-specific p62 deficiency.
Concluding Remarks/Future Directions
There is increasing evidence that p62 is a major adaptor protein linking aggregated proteins and damaged cellular organelles to autophagic degradation. A number of selective autophagy processes including aggrephagy, mitophagy, pexophagy, lysophagy, and lipophagy use p62 as the critical chaperone for cargo selection. Disruptions in the ability of p62 to deliver specific cargo for degradation, it can lead disruptions in the homeostasis of cellular signaling and metabolic pathways with important ramifications on a myriad of cardiovascular and cardiometabolic diseases. Although the intricacies of these pathways have not been fully elucidated in relevant organ systems, our progressive understanding of the involved processes will clarify not only disease pathogenesis but also provide a basis for the development of new therapies that harness autophagy and cellular degradation pathways.
Acknowledgments
This study was supported by grants from National Institutes of Health (R01 HL125838 and P30 DK020579), the U.S. Department of Veterans Affairs (VA MERIT I01 BX003415), and the American Diabetes Association (Innovative Basic Science 1-18-IBS-029) to B.R., and National Institutes of Health (F31 HL132434) to T.D.E.
Abbreviations Used
- ALFY
autophagy-linked FYVE protein
- aPKC
atypical protein kinase C
- BAT
brown adipose tissue
- CHIP
c-terminus of heat shock chaperone 70-interacting protein
- CK2
casein kinase 2
- DRC
desmin-related cardiomyopathy
- ERK1
extracellular signal-regulated kinase 1
- Keap1
Kelch-like ECH-associated protein 1
- KIR
Keap1-interacting region
- LC3
microtubule-associated protein light chain 3
- LDs
lipid droplets
- LIR
LC3-interacting region
- MAPK
mitogen-activated protein kinase
- MDB
Mallory–Denk body
- MFN
mitofusin
- MIM
mitochondrial inner membrane
- MOM
mitochondrial outer membrane
- mTOR
mammalian target of rapamycin
- mTORC1
mammalian target of rapamycin complex 1
- NAFLD
nonalcoholic fatty liver disease
- NBR1
neighbor of BRCA gene 1
- NDP52/CALCOCO2
nuclear dot protein 52 kDa/calcium-binding and coiled-coil domain 2
- NEDD
neural precursor cell expressed developmentally downregulated protein
- NF-κB
nuclear factor kappa B
- NRF2
nuclear factor erythroid 2-related factor 2
- OPTN
optineurin
- p62
p62/SQSTM1
- PB1
Phox/Bem1p
- PEX
peroxin
- PGC-1α
PPARγ coactivator-1α
- PINK1
PTEN-induced putative kinase 1
- PLIN2
Perilipin 2
- PMP70
peroxisomal membrane protein 70
- PTEN
phosphatase and tensin homolog
- RIP1
receptor-interacting protein 1
- ROS
reactive oxygen species
- STAT
signal transducer and activator of transcription
- STBD1
starch binding domain 1
- TBK1
TANK binding kinase 1
- TBS
TRAF6-binding sequence
- TRAF6
tumor necrosis factor receptor-associated factor 6
- UBA
ubiquitin associated
- UCP1
uncoupling protein 1
- ULK
Unc-51 like autophagy activating kinase
- UPS
ubiquitin-proteasome system
- USP
ubiquitin carboxyl-terminal hydrolase
- ZZ
ZZ-type zinc finger
References
- 1. B'Chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, Parry L, Stepien G, Fafournoux P, and Bruhat A. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res 41: 7683–7699, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. This reference has been deleted.
- 3. This reference has been deleted.
- 4. Bell JL, Malyukova A, Holien JK, Koach J, Parker MW, Kavallaris M, Marshall GM, and Cheung BB. TRIM16 acts as an E3 ubiquitin ligase and can heterodimerize with other TRIM family members. PLoS One 7: e37470, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bersuker K, Peterson CWH, To M, Sahl SJ, Savikhin V, Grossman EA, Nomura DK, and Olzmann JA. A proximity labeling strategy provides insights into the composition and dynamics of lipid droplet proteomes. Dev Cell 44: 97–112.e7, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhuiyan MS, Pattison JS, Osinska H, James J, Gulick J, McLendon PM, Hill JA, Sadoshima J, and Robbins J. Enhanced autophagy ameliorates cardiac proteinopathy. J Clin Invest 123: 5284–5297, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, Foreman O, Kirkpatrick DS, and Sheng M. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 510: 370–375, 2014 [DOI] [PubMed] [Google Scholar]
- 8. Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, Stenmark H, and Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171: 603–614, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bjørkøy G, Lamark T, Pankiv S, Øvervatn A, Brech A, and Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol 452: 181–197, 2009 [DOI] [PubMed] [Google Scholar]
- 10. Chen X, Khambu B, Zhang H, Gao W, Li M, Chen X, Yoshimori T, and Yin XM. Autophagy induced by calcium phosphate precipitates targets damaged endosomes. J Biol Chem 289: 11162–11174, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chin LS, Olzmann JA, and Li L. Parkin-mediated ubiquitin signalling in aggresome formation and autophagy. Biochem Soc Trans 38: 144–149, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, Øvervatn A, Stenmark H, Bjørkøy G, Simonsen A, and Johansen T. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 6: 330–344, 2010 [DOI] [PubMed] [Google Scholar]
- 13. Cohen-Kaplan V, Livneh I, Avni N, Fabre B, Ziv T, Kwon YT, and Ciechanover A. p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc Natl Acad Sci U S A 113: E7490–E7499, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dammai V. and Subramani S. The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell 105: 187–196, 2001 [DOI] [PubMed] [Google Scholar]
- 15. Darvekar SR, Elvenes J, Brenne HB, Johansen T, and Sjøttem E. SPBP is a sulforaphane induced transcriptional coactivator of NRF2 regulating expression of the autophagy receptor p62/SQSTM1. PLoS One 9: e85262, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deosaran E, Larsen KB, Hua R, Sargent G, Wang Y, Kim S, Lamark T, Jauregui M, Law K, Lippincott-Schwartz J, Brech A, Johansen T, and Kim PK. NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci 126: 939–952, 2013 [DOI] [PubMed] [Google Scholar]
- 17. Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, and Diaz-Meco M. P62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell 44: 134–146, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, and Moscat J. The signaling adaptor p62 is an important NF-κB mediator in tumorigenesis. Cancer Cell 13: 343–354, 2008 [DOI] [PubMed] [Google Scholar]
- 19. Durán A, Serrano M, Leitges M, Flores JM, Picard S, Brown JP, Moscat J, and Diaz-Meco MT. The atypical PKC-interacting protein p62 is an important mediator of RANK-activated osteoclastogenesis. Dev Cell 6: 303–309, 2004 [DOI] [PubMed] [Google Scholar]
- 20. Eastman SW, Yassaee M, and Bieniasz PD. A role for ubiquitin ligases and Spartin/SPG20 in lipid droplet turnover. J Cell Biol 184: 881–894, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Emanuel R, Sergin I, Bhattacharya S, Turner J, Epelman S, Settembre C, Diwan A, Ballabio A, and Razani B. Induction of lysosomal biogenesis in atherosclerotic macrophages can rescue lipid-induced lysosomal dysfunction and downstream sequelae. Arterioscler Thromb Vasc Biol 34: 1942–1952, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Evans TD, Sergin I, Zhang X, and Razani B. Target acquired: selective autophagy in cardiometabolic disease. Sci Signal 10: 1–18, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, Krainc D, Brech A, Stenmark H, Simonsen A, and Yamamoto A. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein alfy. Mol Cell 38: 265–279, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fitz-Gibbon S, Tomida S, Chiu BH, Nguyen L, Du C, Liu M, Elashoff D, Erfe MC, Loncaric A, Kim J, Modlin RL, Miller JF, Sodergren E, Craft N, Weinstock GM, and Li H. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. Cell Metab 133: 2152–2160, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gardner BM, Castanzo DT, Chowdhury S, Stjepanovic G, Stefely MS, Hurley JH, Lander GC, and Martin A. The peroxisomal AAA-ATPase Pex1/Pex6 unfolds substrates by processive threading. Nat Commun 9: 135, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, and Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12: 119–131, 2010 [DOI] [PubMed] [Google Scholar]
- 27. Geng J. and Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. “Protein modifications: beyond the usual suspects” review series. EMBO Rep 9: 859–864, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haga S, Ozawa T, Yamada Y, Morita N, Nagashima I, Inoue H, Inaba Y, Noda N, Abe R, Umezawa K, and Ozaki M. p62/SQSTM1 plays a protective role in oxidative injury of steatotic liver in a mouse hepatectomy model. Antioxid Redox Signal 21: 2515–2530, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harada H, Warabi E, Matsuki T, Yanagawa T, Okada K, Uwayama J, Ikeda A, Nakaso K, Kirii K, Noguchi N, Bukawa H, Siow RC, Mann GE, Shoda J, Ishii T, and Sakurai T. Deficiency of p62/sequestosome 1 causes hyperphagia due to leptin resistance in the brain. J Neurosci 33: 14767–14777, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heo JM, Ordureau A, Paulo JA, Rinehart J, and Harper JW. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol Cell 60: 7–20, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hung YH, Chen LMW, Yang JY, and Yuan Yang W. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat Commun 4: 1–7, 2013 [DOI] [PubMed] [Google Scholar]
- 32. Ichimura Y, Waguri S, Sou Y, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, Hoshii T, Hirao A, Takagi K, Mizushima T, Motohashi H, Lee M, Yoshimori T, Tanaka K, Yamamoto M, and Komatsu M. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell 51: 618–631, 2013 [DOI] [PubMed] [Google Scholar]
- 33. Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, Lee MS, Tanaka K, and Komatsu M. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol 193: 275–284, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Isogai S, Morimoto D, Arita K, Unzai S, Tenno T, Hasegawa J, Sou YS, Komatsu M, Tanaka K, Shirakawa M, and Tochio H. Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin. J Biol Chem 286: 31864–31874, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Itakura E. and Mizushima N. p62 targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol 192: 17–27, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jain A, Lamark T, Sjøttem E, Larsen KB, Awuh JA, Øvervatn A, McMahon M, Hayes JD, and Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem 285: 22576–22591, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jeong SJ, Kim S, Park JG, Jung I, Lee M, Jeon S, Kweon HY, Yu D, Lee S, Jang Y, Kang SW, Han K, Miller YI, Park YM, Cheong C, Choi J, and Oh GT. Prdx1 (peroxiredoxin 1) deficiency reduces cholesterol efflux via impaired macrophage lipophagic flux. Autophagy 14: 120–133, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, and Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol 191: 933–942, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, and Ashkenazi A. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 137: 721–735, 2009 [DOI] [PubMed] [Google Scholar]
- 40. Jo C, Gundemir S, Pritchard S, Jin YN, Rahman I, and Johnson GVW. Nrf2 reduces levels of phosphorylated tau protein by inducing autophagy adaptor protein NDP52. Nat Commun 5: 3496, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jolly C. and Morimoto RI. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J Natl Cancer Inst 92: 1564–1572, 2000 [DOI] [PubMed] [Google Scholar]
- 42. Kabeya Y. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19: 5720–5728, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, and Youle RJ. PINK1 phosphorylates ubiquitin to activate parkin E3 ubiquitin ligase activity. J Cell Biol 205: 143–153, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, and Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115: 727–738, 2003 [DOI] [PubMed] [Google Scholar]
- 45. Kim PK, Hailey DW, Mullen RT, and Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A 105: 20567–20574, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kimura T, Mandell M, and Deretic V. Precision autophagy directed by receptor regulators—emerging examples within the TRIM family. J Cell Sci 129: 881–891, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, Bilusic I, Theurillat JP, Øvervatn A, Ishii T, Elazar Z, Komatsu M, Dikic I, and Johansen T. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 33: 505–516, 2009 [DOI] [PubMed] [Google Scholar]
- 48. Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, and Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392: 605–608, 1998 [DOI] [PubMed] [Google Scholar]
- 49. Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, and Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12: 213–223, 2010 [DOI] [PubMed] [Google Scholar]
- 50. Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, and Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131: 1149–1163, 2007 [DOI] [PubMed] [Google Scholar]
- 51. Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, Endo T, Fon EA, Trempe JF, Saeki Y, Tanaka K, and Matsuda N. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510: 162–166, 2014 [DOI] [PubMed] [Google Scholar]
- 52. Kraft C, Peter M, and Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol 12: 836–841, 2010 [DOI] [PubMed] [Google Scholar]
- 53. Lahiri P, Schmidt V, Smole C, Kufferath I, Denk H, Strnad P, Rülicke T, Fröhlich LF, and Zatloukal K. p62/Sequestosome-1 is indispensable for maturation and stabilization of Mallory-Denk bodies. PLoS One 11: e0161083, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lam HC, Cloonan SM, Bhashyam AR, Haspel JA, Singh A, Sathirapongsasuti JF, Cervo M, Yao H, Chung AL, Mizumura K, An CH, Shan B, Franks JM, Haley KJ, Owen CA, Tesfaigzi Y, Washko GR, Quackenbush J, Silverman EK, Rahman I, Kim HP, Mahmood A, Biswal SS, Ryter SW, and Choi AM. Histone deacetylase 6-mediated selective autophagy regulates copd-associated cilia dysfunction. J Clin Invest 123: 5212–5230, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lam T, Harmancey R, Vasquez H, Gilbert B, Patel N, Hariharan V, Lee A, Covey M, and Taegtmeyer H. Reversal of intramyocellular lipid accumulation by lipophagy and a p62-mediated pathway. Cell Death Discov 2: 16061, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lau A, Zheng Y, Tao S, Wang H, Whitman SA, White E, and Zhang DD. Arsenic inhibits autophagic flux, activating the Nrf2-Keap1 pathway in a p62-dependent manner. Mol Cell Biol 33: 2436–2446, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, and Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524: 309–314, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee SJ, Pfluger PT, Kim JY, Nogueiras R, Duran A, Pagès G, Pouysségur J, Tschöp MH, Diaz-Meco MT, and Moscat J. A functional role for the p62-ERK1 axis in the control of energy homeostasis and adipogenesis. EMBO Rep 11: 226–232, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lee YJ, Chou TF, Pittman SK, Keith AL, Razani B, and Weihl CC. Keap1/Cullin3 modulates p62/SQSTM1 activity via UBA domain ubiquitination. Cell Rep 20: 188–202, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lin Q, Dai Q, Meng H, Sun A, Wei J, Peng K, Childress C, Chen M, Shao G, and Yang W. The HECT E3 ubiquitin ligase NEDD4 interacts with and ubiquitylates SQSTM1 for inclusion body autophagy. J Cell Sci 130: 3839–3850, 2017 [DOI] [PubMed] [Google Scholar]
- 61. Linares JF, Duran A, Yajima T, Pasparakis M, Moscat J, and Diaz-Meco MT. K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol Cell 51: 283–296, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu Z, Chen P, Gao H, Gu Y, Yang J, Peng H, Xu X, Wang H, Yang M, Liu X, Fan L, Chen S, Zhou J, Sun Y, Ruan K, Cheng S, Komatsu M, White E, Li L, Ji H, Finley D, and Hu R. Ubiquitylation of autophagy receptor optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell 26: 106–120, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Long J, Gallagher TRA, Cavey JR, Sheppard PW, Ralston SH, Layfield R, and Searle MS. Ubiquitin recognition by the ubiquitin-associated domain of p62 involves a novel conformational switch. J Biol Chem 283: 5427–5440, 2008 [DOI] [PubMed] [Google Scholar]
- 64. Maejima I, Takahashi A, Omori H, Kimura T, Takabatake Y, Saitoh T, Yamamoto A, Hamasaki M, Noda T, Isaka Y, and Yoshimori T. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J 32: 2336–2347, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, Dipaola RS, Karantza-Wadsworth V, and White E. Autophagy suppresses tumorigenesis through elimination of p62. Cell 137: 1062–1075, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, Kimura M, Komatsu M, Hattori N, and Tanaka K. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 189: 211–221, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Matsumoto G, Shimogori T, Hattori N, and Nukina N. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum Mol Genet 24: 4429–4442, 2015 [DOI] [PubMed] [Google Scholar]
- 68. Matsumoto G, Wada K, Okuno M, Kurosawa M, and Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell 44: 279–289, 2011 [DOI] [PubMed] [Google Scholar]
- 69. This reference has been deleted.
- 70. Moscat J. and Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 137: 1001–1004, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Moscat J. and Diaz-Meco MT. Feedback on fat: P62-mTORC1-autophagy connections. Cell 147: 724–727, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Moscat J, Diaz-Meco MT, Albert A, and Campuzano S. Cell signaling and function organized by PB1 domain interactions. Mol Cell 23: 631–640, 2006 [DOI] [PubMed] [Google Scholar]
- 73. Moscat J, Diaz-Meco MT, and Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci 32: 95–100, 2007 [DOI] [PubMed] [Google Scholar]
- 74. Müller TD, Lee SJ, Jastroch M, Kabra D, Stemmer K, Aichler M, Abplanalp B, Ananthakrishnan G, Bhardwaj N, Collins S, Divanovic S, Endele M, Finan B, Gao Y, Habegger KM, Hembree J, Heppner KM, Hofmann S, Holland J, Küchler D, Kutschke M, Krishna R, Lehti M, Oelkrug R, Ottaway N, Perez-Tilve D, Raver C, Walch AK, Schriever SC, Speakman J, Tseng YH, Diaz-Meco M, Pfluger PT, Moscat J, and Tschöp MH. p62 links β-adrenergic input to mitochondrial function and thermogenesis. J Clin Invest 123: 469–478, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Murata S, Minami Y, Minami M, Chiba T, and Tanaka K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep 2: 1133–1138, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, and Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8: e1000298, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nian Z, Sun Z, Yu L, Toh SY, Sang J, and Li P. Fat-specific protein 27 undergoes ubiquitin-dependent degradation regulated by triacylglycerol synthesis and lipid droplet formation. J Biol Chem 285: 9604–9615, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Nordgren M, Francisco T, Lismont C, Hennebel L, Brees C, Wang B, Van Veldhoven PP, Azevedo JE, and Fransen M. Export-deficient monoubiquitinated PEX5 triggers peroxisome removal in SV40 large T antigen-transformed mouse embryonic fibroblasts. Autophagy 11: 1326–1340, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Okatsu K, Saisho K, Shimanuki M, Nakada K, Shitara H, Sou YS, Kimura M, Sato S, Hattori N, Komatsu M, Tanaka K, and Matsuda N. P62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells 15: 887–900, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ouimet M, Franklin V, Mak E, Liao X, Tabas I, and Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab 13: 655–667, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Pan JA, Sun Y, Jiang YP, Bott AJ, Jaber N, Dou Z, Yang B, Chen J, Catanzaro JM, Du C, Ding W, Diaz-Meco MT, Moscat J, Ozato K, Lin RZ, and Zong W. TRIM21 ubiquitylates SQSTM1/p62 and suppresses protein sequestration to regulate redox homeostasis. Mol Cell 61: 720–733, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, and Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145, 2007 [DOI] [PubMed] [Google Scholar]
- 83. Papadopoulos C. and Meyer H. Detection and clearance of damaged lysosomes by the endo-lysosomal damage response and lysophagy. Curr Biol 27: R1330–R1341, 2017 [DOI] [PubMed] [Google Scholar]
- 84. Pickart CM. and Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta 1695: 55–72, 2004 [DOI] [PubMed] [Google Scholar]
- 85. Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, Dupont N, Ornatowski W, Jiang S, Bradfute SB, Bruun JA, Hansen TE, Johansen T, and Deretic V. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 37: 223–234, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Razani B, Feng C, Coleman T, Emanuel R, Wen H, Hwang S, Ting JP, Virgin HW, Kastan MB, and Semenkovich CF. Autophagy links inflammasomes to atherosclerotic progression. Cell Metab 15: 534–544, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Rodriguez A, Durán A, Selloum M, Champy MF, Diez-Guerra FJ, Flores JM, Serrano M, Auwerx J, Diaz-Meco MT, and Moscat J. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab 3: 211–222, 2006 [DOI] [PubMed] [Google Scholar]
- 88. Sanbe A, Osinska H, Saffitz JE, Glabe CG, Kayed R, Maloyan A, and Robbins J. Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis. Proc Natl Acad Sci U S A 101: 10132–10136, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sanchez P, De Carcer G, Sandoval IV, Moscat J, and Diaz-Meco MT. Localization of atypical protein kinase C isoforms into lysosome-targeted endosomes through interaction with p62. Mol Cell Biol 18: 3069–3080, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sanz L, Sanchez P, Lallena MJ, Diaz-Meco MT, and Moscat J. The interaction of p62 with RIP links the atypical PKCs to NF-kappaB activation. EMBO J 18: 3044–3053, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. This reference has been deleted.
- 92. Sargent G, van Zutphen T, Shatseva T, Zhang L, Di Giovanni V, Bandsma R, and Kim PK. PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. J Cell Biol 214: 677–690, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sathyanarayan A, Mashek MT, and Mashek DG. ATGL promotes autophagy/lipophagy via SIRT1 to control hepatic lipid droplet catabolism. Cell Rep 19: 1–9, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR. and Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol 24: 8055–8068, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sergin I, Bhattacharya S, Emanuel R, Esen E, Stokes CJ, Evans TD, Arif B, Curci JA, and Razani B. P62-enriched inclusion bodies in macrophages protect against atherosclerosis. Sci Signal 9: ra2, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sergin I, Evans TD, Zhang X, Bhattacharya S, Stokes CJ, Song E, Ali S, Dehestani B, Holloway KB, Micevych PS, Javaheri A, Crowley JR, Ballabio A, Schilling JD, Epelman S, Weihl CC, Diwan A, Fan D, Zayed MA, and Razani B. Exploiting macrophage autophagy-lysosomal biogenesis as a therapy for atherosclerosis. Nat Commun 8: 15750, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96a. Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, and Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science 332: 1429–1433, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, and Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol 13: 255–263, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Shimura H, Hattori N, Kubo Si, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K, and Suzuki T. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet 25: 302–305, 2000 [DOI] [PubMed] [Google Scholar]
- 99. Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, and Czaja MJ. Autophagy regulates lipid metabolism. Nature 458: 1131–1135, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Spandl J, Lohmann D, Kuerschner L, Moessinger C, and Thiele C. Ancient ubiquitous protein 1 (AUP1) localizes to lipid droplets and binds the E2 ubiquitin conjugase G2 (Ube2g2) via its G2 binding region. J Biol Chem 286: 5599–5606, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Tatsumi T, Takayama K, Ishii S, Yamamoto A, Hara T, Minami N, Miyasaka N, Kubota T, Matsuura A, Itakura E, and Tsukamoto S. Forced lipophagy reveals that lipid droplets are required for early embryonic development in mouse. Development 145: pii:, 2018 [DOI] [PubMed] [Google Scholar]
- 102. Thurston TLM, Ryzhakov G, Bloor S, von Muhlinen N, and Randow F. The tbk1 adaptor and autophagy receptor ndp52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 10: 1215–1221, 2009 [DOI] [PubMed] [Google Scholar]
- 103. Umemura A, He F, Taniguchi K, Nakagawa H, Yamachika S, Font-Burgada J, Zhong Z, Subramaniam S, Raghunandan S, Duran A, Linares JF, Reina-Campos M, Umemura S, Valasek MA, Seki E, Yamaguchi K, Koike K, Itoh Y, Diaz-Meco MT, Moscat J, and Karin M. p62, upregulated during preneoplasia, induces hepatocellular carcinogenesis by maintaining survival of stressed HCC-initiating cells. Cancer Cell 29: 935–948, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Valente EM, Abou-sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, González-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, and Wood NW. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304: 1158–1161, 2004 [DOI] [PubMed] [Google Scholar]
- 105. Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, Magrané J, Moore DJ, Dawson VL, Grailhe R, Dawson TM, Li C, Tieu K, and Przedborski S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A 107: 378–383, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wang L, Cho Y, Tang Y, Wang J, Park JE, Wu Y, Wang C, Tong Y, Chawla R, Zhang J, Shi Y, Deng S, Lu G, Wu Y, Tan HW, Pawijit P, Lim GG, Chan HY, Zhang J, Fang L, Yu H, Liou YC, Karthik M, Bay BH, Lim KL, Sze SK, Yap CT, and Shen HM. PTEN-L is a novel protein phosphatase for ubiquitin dephosphorylation to inhibit PINK1–Parkin-mediated mitophagy. Cell Res 28:787–802, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wang L, Zhou J, Yan S, Lei G, Lee CH, and Yin XM. Ethanol-triggered lipophagy requires SQSTM1 in AML12 hepatic cells. Sci Rep 7: 12307, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, and Schwarz TL. PINK1 and Parkin target miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147: 893–906, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Watanabe Y. and Tanaka M. p62/SQSTM1 in autophagic clearance of a non-ubiquitylated substrate. J Cell Sci 124: 2692–2701, 2011 [DOI] [PubMed] [Google Scholar]
- 110. Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dötsch V, Bumann D, and Dikic I. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333: 228–233, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wooten MW, Seibenhener ML, Mamidipudi V, Diaz-Meco MT, Barker PA, and Moscat J. The atypical protein kinase C-interacting protein p62 is a scaffold for NF-κB activation by nerve growth factor. J Biol Chem 276: 7709–7712, 2001 [DOI] [PubMed] [Google Scholar]
- 112. Yamada T, Murata D, Adachi Y, Itoh K, Kameoka S, Igarashi A, Kato T, Araki Y, Huganir RL, Dawson TM, Yanagawa T, Okamoto K, Iijima M, and Sesaki H. Mitochondrial stasis reveals p62-mediated ubiquitination in Parkin-independent mitophagy and mitigates nonalcoholic fatty liver disease. Cell Metab 28: 588–604.e5, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M, Aguzzi A, and Denk H. P62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol 160: 255–263, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zhang J, Tripathi DN, Jing J, Alexander A, Kim J, Powell RT, Dere R, Tait-Mulder J, Lee JH, Paull TT, Pandita RK, Charaka VK, Pandita TK, Kastan MB, and Walker CL. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol 17: 1259–1269, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zhang X, Evans TD, Jeong SJ, and Razani B. Classical and alternative roles for autophagy in lipid metabolism. Curr Opin Lipidol 29: 203–211, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zheng Q, Su H, Ranek MJ, and Wang X. Autophagy and p62 in cardiac proteinopathy. Circ Res 109: 296–308, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. This reference has been deleted.