Abstract
PulseNet USA is the molecular surveillance network for foodborne disease in the United States. The network consists of state and local public health laboratories, as well as food regulatory agencies, that follow PulseNet's standardized protocols to perform pulsed-field gel electrophoresis (PFGE) and whole genome sequencing (WGS) and analyze the results using standardized software. The raw sequences are uploaded to the GenomeTrakr or PulseNet bioprojects at the National Center for Biotechnology Information. The PFGE patterns and analyzed sequence data are uploaded in real time with associated demographic data to the PulseNet national databases managed at the Centers for Disease Control and Prevention. The PulseNet databases are organism specific and provide a central storage location for molecular and demographic data related to an isolate. Sequences are compared in the databases, thereby facilitating the rapid detection of clusters of foodborne diseases that may represent widespread outbreaks. WGS genotyping data, for example, antibiotic resistance and virulence profiles, are also uploaded in real time to the PulseNet databases to improve food safety surveillance activities.
Keywords: PulseNet, whole genome sequencing, data analysis, foodborne outbreak detection
Introduction
PulseNet USA is the national molecular subtyping network for foodborne disease surveillance (Swaminathan et al., 2001; Gerner-Smidt et al., 2006; Ribot et al., in press). The network enables the rapid detection of local and national clusters of foodborne illness, reducing the likelihood of foodborne outbreaks becoming large and widespread, thus preventing illness and reducing health care costs (Scharff et al., 2016). Before PulseNet, these multistate outbreaks often went undetected because an individual state might only have had one case and without PulseNet the state may have been unaware that its case was part of a larger multistate outbreak.
The network consists of 82 state and local public health laboratories and food regulatory federal agencies coordinated by Centers for Disease Control and Prevention (CDC) and the Association of Public Health Laboratories. Individuals in each laboratory are trained and certified in pulsed-field gel electrophoresis (PFGE) and whole genome sequencing (WGS) laboratory methods and data analysis. Laboratories analyze PFGE and WGS data using a PulseNet-customized version of BioNumerics (Applied Maths, Sint-Martens-Latem, Belgium), a commercial off-the-shelf data analysis and management software. Analyzed data are uploaded to national databases that contain both PFGE and WGS data and are housed at CDC. This article provides an update on the submission of data to the databases as well as their management, including the types of analyses performed.
PFGE Historical Databases
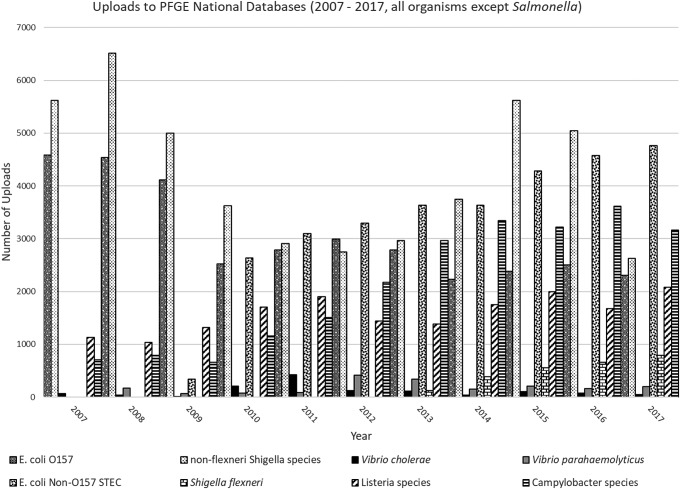
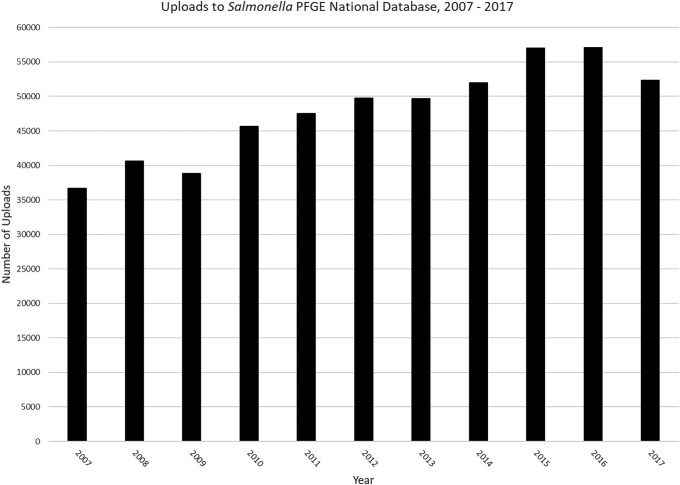
Before the combined PFGE and WGS databases, CDC maintained nine separate PFGE-only national databases. Each database contained PFGE pattern images and associated demographic information separated by organism: Escherichia coli O157, non-O157 Shiga toxin–producing E. coli (STEC), non-flexneri Shigella species, Shigella flexneri, Vibrio cholerae, Vibrio parahaemolyticus, Salmonella, Listeria, or Campylobacter. By the end of 2017, the historical PFGE databases had a combined total of >800,000 entries representing bacterial isolates from human, food, animal, and environmental sources (Fig. 1). Salmonella is the most common foodborne bacteria monitored by PulseNet, causing an estimated one million illnesses each year; therefore, the Salmonella PFGE national database contains the majority of PFGE patterns uploaded to PulseNet (Scallan et al., 2011) (Fig. 2).
FIG. 1.
Graph of the number of PFGE pattern uploads to the Escherichia coli O157, E. coli non-O157 STEC, non-flexneri Shigella species, Shigella flexneri, Vibrio cholerae, Vibrio parahaemolyticus, Listeria species, and Campylobacter species PFGE National Databases from 2007 through 2017. PFGE, pulsed-field gel electrophoresis; STEC, Shiga toxin–producing E. coli.
FIG. 2.
Graph of the number of PFGE pattern uploads to the Salmonella PFGE National Database from 2007 through 2017.
PulseNet participating laboratories maintained their own local PFGE databases containing tagged image file format (TIFF) images of PFGE patterns. The TIFF images were analyzed using BioNumerics software with PulseNet MasterScripts installed. MasterScripts were customizations in BioNumerics developed by Applied Maths for PulseNet. The scripts ensured that data were standardized and comparable within each database and provided reference standards, information fields, and PFGE experiment types for the organism-specific entries (Gerner-Smidt et al., 2006).
PFGE TIFF images were analyzed in the local databases by marking bands that corresponded to their molecular weights. Each lane on the TIFF image represented a single bacterial fingerprint, or PFGE pattern, digested with a restriction enzyme.
Demographic data associated with each pattern were entered into the database. The demographic data included information such as isolate source (human, food, environmental, and animal), serotype or species data, and isolate collection date. Personal information such as name or street address was not uploaded for isolates collected from patients, but certain data needed to characterize foodborne disease clusters, such as patient location, gender, and age, were included.
Within 4 working days of receiving isolates in the laboratory, the analyzed TIFF image and the demographic data were uploaded to the organism-specific national databases located at CDC. Once patterns were uploaded, database managers assigned alphanumeric pattern names to the uploads within 24 h. Each pattern name had three components. The first three characters represented the organism, the next three characters represented the enzyme used to generate the pattern, and the last four characters represented the pattern number, for example, EXHX01.0001, PFGE XbaI pattern one for E. coli O157. Indistinguishable patterns were given the same pattern name (Gerner-Smidt et al., 2006). Pattern names allowed database managers to quickly graph indistinguishable patterns over time to detect clusters that may become widespread outbreaks.
PFGE clusters were defined as two or more clinical isolates that shared the same pattern and whose frequency exceeded that pattern's historical occurrence when compared with the previous 4 years. When a cluster was detected, a unique cluster code was assigned to database entries included in the cluster. The cluster code identified the time (month, year) when the cluster was detected, the geographic distribution (state or multistate), and the organism involved, for example, 1702MLEXH-1, the first multistate cluster of E. coli O157 detected in February 2017 (Gerner-Smidt et al., 2006).
Although PFGE had long been the gold standard for PulseNet, it did have limitations. Not all bacterial strains produce PFGE patterns when cut with standard restriction enzymes, and PFGE may not fully distinguish background cases from outbreak cases for clonal organisms such as Salmonella serotype Enteritidis (Deng et al., 2014). Isolates with different PFGE patterns can also be highly related. If PFGE data are the only method used for cluster detection, those cases could be excluded from the cluster (Besser et al., in press). To further characterize the isolates, laboratories also determined the serotype, antibiotic resistance, and/or virulence using conventional methods. Marking pattern bands is subjective and at times bands were not marked the same way across laboratories.
To enhance cluster detection and surveillance, PFGE has been phased out and replaced with WGS as PulseNet's primary surveillance tool. WGS provides superior discrimination compared with PFGE and allows multiple characterizations of isolates with a single workflow (Besser et al., 2018). With the introduction of WGS, the nine PFGE databases were merged and migrated into five databases (see Combined PFGE and WGS Databases section) in 2019. This configuration enables the users to still access historical PFGE data as well as WGS data in the same database.
Combined PFGE and WGS Databases
The first organism to become routinely subtyped by WGS in PulseNet was Listeria monocytogenes. Subtyping L. monocytogenes using WGS began as a pilot project in 2013 (Jackson et al., 2016). The Listeria Pilot Project was one of the U.S. Department of Health and Human Services (HHS) secretary's picks in 2014 in the HHS innovates competition, and also received the CDC Director's Award for Innovation the same year. For a number of years, WGS and PFGE were performed in parallel in PulseNet, but since 2018, WGS has been the only method PulseNet participants have been required to use for Listeria subtyping.
The combined WGS and PFGE databases are being managed at CDC using the most recent version of BioNumerics (v 7.6), which has added functionality for analyzing nucleic acid sequence data. Whereas there were nine separate databases for PFGE, there are only five databases for WGS: Escherichia, which combines E. coli O157, non-O157 STEC, non-flexneri Shigella species, and Shigella flexneri into one database; Salmonella; Listeria; Campylobacter; and Vibrio, which combines V. cholerae and V. parahaemolyticus into one database and will also include V. vulnificus. The four former databases have been validated and are implemented, but Vibrio is still under validation. Organisms historically separated in PFGE databases were combined by genus/species in WGS databases due to the genetic similarity of the organisms. As of the writing of this article, the databases had a combined total of >125,000 sequences.
Database plugins are used to add PulseNet data fields, experiment types, and WGS-related features. If both PFGE and WGS profiles are available, the isolate accession number and demographic data are linked to both PFGE and WGS data in the combined PFGE and WGS national databases. This allows both data types to be used simultaneously in cluster investigations.
PulseNet participants maintain their own local reference identification database and their own local combined PFGE and WGS databases, giving them access to the same functionalities as the national databases. After the raw sequence data are linked to an entry in the local reference identification database, the sequence is submitted to a high capacity computing cluster at CDC, the “calculation engine,” for de novo assembly and average nucleotide identity to determine the taxonomic identification (genus and species) of the isolate (Thompson et al., 2013).
The sequence is checked for contamination, and basic quality metrics (Q-scores of reads 1 and 2, average coverage, and sequence length) are assessed on the assembled sequences. Assemblies, quality metrics, and the taxa identification for sequences that pass the initial quality check are imported into a local organism-specific database that contains both PFGE and WGS data generated by the local laboratory. The organism-specific database contains demographic data associated with the isolate, including, but not limited to, source information (human, food, environmental, and animal), serotype or species data, and isolate collection date.
After the data have been imported into the organism-specific database, each laboratory again submits its assemblies to the calculation engine, and allele calls, as well as genotyping data such as serotype, antibiotic resistance, virulence, and plasmid identification, are retrieved. Additional quality data determining percentage core loci identified are also obtained.
The raw sequence data of good quality sequences (those with percentage core ≥95%) are uploaded to the GenomeTrakr or PulseNet bioprojects at National Center for Biotechnology Information (NCBI) (Timme et al., 2017). Sequence files are large and require a large amount of disk space. Uploading the raw data to NCBI allows the public health laboratories to store their data safely without purchasing extra disk space for storage. Since NCBI is a publically available website, the sequence is given an anonymized unique identifier before uploading to protect the identity of the case patient. To further anonymize the data, minimal demographic information is included (Ribot et al., in press). After the sequence has been submitted to NCBI, NCBI-assigned accession numbers are downloaded into the local database so isolate information can be easily linked back to NCBI.
All laboratories are responsible for uploading their own sequence data and demographic data to NCBI. Making data publically available on NCBI allows comparison of sequence data with non-PulseNet laboratories. This is especially valuable when contaminated food is imported or exported globally. The raw sequence data from non-PulseNet laboratories on NCBI can be linked to an entry in BioNumerics and processed through the calculation engine at CDC to compare with sequences in the PulseNet network.
Once data are analyzed, laboratories upload allele calls, genotyping data, demographic data, and NCBI information to the national organism-specific databases managed at CDC. Laboratories are expected to upload analyzed data within 7 working days of receiving a pure culture for DNA extraction in the laboratory. The uploaded sequence data receive an automatically assigned allele code similar to the single nucleotide polymorphism (SNP) address concept developed and used by Public Heath England for foodborne disease surveillance in Europe (Inns et al., 2017). An allele code is a unique name given to all sequences based on the core genome multilocus sequence typing (cgMLST) profile. Like a PFGE pattern name, an allele code provides a way of communicating strain information; however, allele codes demonstrate phylogenetic relatedness between isolates.
The code comprises the organism identifier and version of the allele code naming script used, followed by a set of up to six numbers representing the genetic similarity of the isolate at different allelic thresholds compared with the rest of the database, for example, at 5, 10, 25, 50, 100, and 250 allelic differences. The codes are based on the allelic thresholds that were shown to be most stable for each organism during the validations of the databases. Ninety-five percent of the core genome must be present in order for an allele code to be assigned (Moura et al., 2016). If a sequence fails quality, the reason for failing is displayed in place of an allele code. This provides the generating laboratory information as to why the isolate should have sequencing repeated.
Clusters are detected based on cgMLST. The allele range for cases to be considered closely related and coded into a cluster without other information varies by organism. In general, a cluster is coded if there are three or more clinical isolates within 10 alleles in the past 120 d (for Listeria) or 60 d (for all other organisms), and 2 of those cases are within 5 alleles of each other. Any historical sequences of interest that are closely related to a newly identified cluster are included in a report to epidemiologists for follow-up (Besser et al., in press). Depending on the discriminatory ability of cgMLST, additional typing schema may be used such as whole genome multilocus sequence typing (wgMLST) (www.applied-maths.com/applications/wgmlst) and SNP analysis (Ribot et al., in press). Clusters are communicated and named using the same nomenclature for WGS as PFGE by the database team at CDC.
After a cluster is detected and throughout the cluster investigation, information pertaining to the cluster such as demographic data and allele code is communicated to CDC epidemiologists and to the laboratories in the PulseNet network through e-mail, the PulseNet SharePoint site, and at meetings. New cases are added to the cluster as uploads are received and evaluated (Gerner-Smidt et al., 2006).
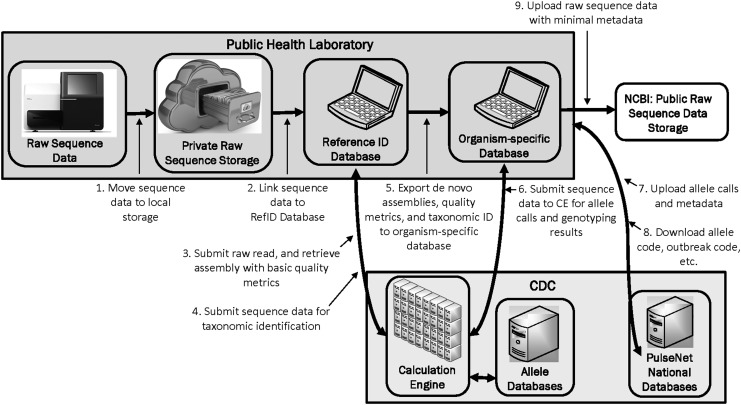
PulseNet participants can download cluster codes, allele codes, and PFGE pattern names back into their laboratory's local databases, enabling them to manage their isolates involved in clusters locally and to search for clusters at the local level. In addition, laboratories can connect to the national databases to view PFGE, WGS, and demographic data uploaded by all PulseNet participating laboratories, which allows laboratories to see whether sequences or patterns of interest are being seen in other areas of the country (Fig. 3).
FIG. 3.
Diagram illustrating the PulseNet USA WGS data analysis workflow. WGS, whole genome sequencing.
Discussion
PulseNet's transition to WGS as the primary subtyping tool for surveillance was completed for Listeria, Salmonella, E. coli, Shigella, and Campylobacter in 2019. Vibrio, Yersinia, and Cronobacter will follow.
Having a database containing both PFGE and WGS data allows laboratories to analyze all data in one location while maintaining the historical context of the data. With the increased resolution of WGS, it has become possible to investigate outbreaks stretching over many years, such as the L. monocytogenes outbreak linked to ice cream (https://www.cdc.gov/listeria/outbreaks/ice-cream-03-15/index.html). However, only a fraction of historical isolates, mainly from past outbreaks, have been sequenced. If a new cluster is detected using WGS, PFGE may be run on representative isolates in the cluster. The PFGE patterns of representative isolates from the newly detected cluster can then be compared with historical PFGE patterns. Historical patterns found to be indistinguishable from the new cluster can then be sequenced. By maintaining the PFGE databases, determining which historical isolates should be sequenced can be prioritized, providing the possibility of finding historical isolates closely related to current clusters.
The WGS workflow generates a wealth of data that, although costly compared with PFGE (100–200 USD per isolate at CDC for WGS vs. 25–30 USD per isolate at CDC for PFGE), will make its use increasingly cost efficient. Because the serotype, virulence profile, and complete antimicrobial resistance profile can be predicted from the sequence, the public health laboratories may largely replace the traditional methods previously used to generate this kind of data.
Using WGS to detect clusters reduces the number of background cases erroneously included in outbreaks compared with PFGE (Jackson et al., 2016). The availability of resistance and virulence data for all isolates in real time has also proven useful when interpreting outbreak data and understanding the virulence of the organism (https://www.cdc.gov/salmonella/reading-07-18/index.html; https://www.cdc.gov/campylobacter/outbreaks/puppies-9-17/index.html).
Throughout the transition to WGS-based surveillance, it has been critical to create a functionality similar to PFGE. Continuing to use the same BioNumerics software has made training and implementation of the method easier and ensures that microbiologists with limited bioinformatics skills can perform the data analysis. As with PFGE, laboratories can connect to the national databases and view other laboratory's WGS and PFGE data. Laboratorians can analyze their own data and take full advantage of WGS for local investigations. Also, as with PFGE, nomenclature information and cluster codes can be downloaded back into the local database. It is critical to communicate this information efficiently with all federal and state partners in outbreak investigations.
Although PFGE has proved an invaluable tool for outbreak surveillance for >20 years (Ribot and Hise, 2016), saving millions of dollars in health care and productivity-related costs (Scharff et al., 2016), WGS provides better resolution in a likely more cost-effective approach, which will result in more solved outbreaks with fewer cases (Jackson et al., 2016) and improved food safety in the United States and in other countries.
Conclusions
With WGS, PulseNet has moved beyond subtyping for outbreak detection and investigation, although this remains the core function of the network. The participating laboratories are now able to perform most strain characterization, for example, species identification, serotyping, virulence, and resistance profiling, using one semiautomatic workflow that before WGS was done using a multitude of different phenotypic and molecular methods. With these enhancements, as well as improved cluster detection, public health laboratory surveillance in the United States has become more efficient and precise than ever before, leading to better protection of the American people from foodborne infections.
Disclosure Statement
No competing financial interests exist.
References
- Besser J, Carleton HA, Gerner-Smidt P, Lindsey RL, Trees E. Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin Microbiol Infect 2018;24:335–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Desai PT, den Bakker HC, Mikoleit M, Tolar B, Trees E, Hendriksen RS, Frye JG, Porwollik S, Weimer BC, Wiedmann M, Weinstock GM, Fields PI, McClelland M. Genomic epidemiology of Salmonella enterica serotype Enteritidis based on population structure of prevalent lineages. Emerg Infect Dis 2014;20:1481–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner-Smidt P, Hise K, Kincaid J, Hunter S, Rolando S, Hyytiä-Trees E, Ribot EM, Swaminathan B. PulseNet USA: A five-year update. Foodborne Pathog Dis 2006;3:9–19 [DOI] [PubMed] [Google Scholar]
- Inns T, Ashton PM, Herrera-Leon S, Lighthill J, Foulkes S, Jombart T, Rehman Y, Fox A, Dallman T DE.Pinna E, Browning L, Coia JE, Edeghere O, Vivancos R. Prospective use of whole genome sequencing (WGS) detected a multi-country outbreak of Salmonella Enteritidis. Epidemiol Infect 2017;145:289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson BR, Tarr C, Strain E, Jackson KA, Conrad A, Carleton H, Katz LS, Stroika S, Gould LH, Mody RK, Silk BJ, Beal J, Chen Y, Timme R, Doyle M, Fields A, Wise M, Tillman G, Defibaugh-Chavez S, Kucerova Z, Sabol A, Roache K, Trees E, Simmons M, Wasilenko J, Kubota K, Pouseele H, Klimke W, Besser J, Brown E, Allard M, Gerner-Smidt P. Implementation of nationwide real-time whole-genome sequencing to enhance listeriosis outbreak detection and investigation. Clin Infect Dis 2016;63:380–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura A, Criscuolo A, Pouseele H, Maury MM, Leclercq A, Tarr C, Björkman JT, Dallman T, Reimer A, Enouf V, Larsonneur E, Carleton H, Bracq-Dieye H, Katz LS, Jones L, Touchon M, Tourdjman M, Walker M, Stroika S, Cantinelli T, Chenal-Francisque V, Kucerova Z, Rocha EP, Nadon C, Grant K, Nielsen EM, Pot B, Gerner-Smidt P, Lecuit M, Brisse S. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat Microbiol 2016;2:16185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot EM, Hise KB. Future challenges for tracking foodborne diseases: PulseNet, a 20-year-old US surveillance system for foodborne diseases, is expanding both globally and technologically. EMBO Rep 2016;17:1499–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M, Roy SL, Jones JL, Griffin PM. Foodborne illness acquired in the United States—Major pathogens. Emerg Infect Dis 2011;17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff RL, Besser J, Sharp DJ, Jones TF, Gerner-Smidt P, Hedberg CW. An economic evaluation of PulseNet: A network for foodborne disease surveillance. Am J Prev Med 2016;50(5 Suppl 1):S66–S73 [DOI] [PubMed] [Google Scholar]
- Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV. PulseNet: The molecular subtyping network for foodborne bacterial disease surveillance—United States. Emerg Infect Dis 2001;7:382–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CC, Chimetto L, Edwards RA, Swings J, Stackebrandt E, Thompson FL. Microbial genomic taxonomy. BMC Genomics 2013;14:913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme RE, Rand H, Shumway M, Trees EK, Simmons M, Agarwala R, Davis S, Tillman G, Defibaugh-Chavez S, Carleton HA, Klimke WA, Katz LS. Benchmark datasets for phylogenomic pipeline validation, applications for foodborne pathogen surveillance. PeerJ 2017;5:e3893. [DOI] [PMC free article] [PubMed] [Google Scholar]