Abstract
Significance: Alterations in adipose tissue function have profound consequences on whole body energy homeostasis because this tissue is central for fat accumulation, energy expenditure, glucose and insulin metabolism, and hormonal regulation. With the obesity reaching epidemic proportions globally, it is important to understand the mechanisms leading to adipose tissue malfunction.
Recent Advances: Autophagy has originally been viewed as an adaptive response to cellular stress, but in recent years this process was shown to regulate important cellular processes. In adipose tissue, autophagy is a key regulator of white adipose tissue (WAT) and brown adipose tissue (BAT) adipogenesis, and dysregulated autophagy impairs fat accumulation both in vitro and in vivo. Animal studies have also suggested an important role for autophagy and mitophagy during the transition from beige to white fat. Human studies have provided evidence for altered autophagy in WAT, and these alterations correlated with the degree of insulin resistance.
Critical Issues: Despite these important advances in the study of autophagy in adipose tissue, we still do not understand the physiological role of autophagy in mature white and brown adipocytes. Furthermore, several human studies involving autophagy assessment were performed on whole adipose tissue, which complicates the interpretation of the results considering the cellular heterogeneity of this tissue.
Future Directions: Future studies will undoubtedly expand our understanding of the role of autophagy in fully differentiated adipocytes, and uncover novel cross-talks between this tissue and other organs in regulating lipid metabolism, redox signaling, energy homeostasis, and insulin sensitivity.
Keywords: obesity, autophagy, adipose tissue, adipogenesis, thermogenesis, insulin resistance
Introduction
Adipose tissue is vital for the body, and it is now recognized as an endocrine organ that secretes key adipokines and hormones to maintain metabolic homeostasis. There are three types of adipose tissues with distinct cellular origin: white adipose tissue (WAT), brown adipose tissue (BAT), and bright or beige adipose tissue. The function of WAT is fat storage when calories are in excess and fat mobilization when calories are sparse. In contrast, BAT is involved in thermogenesis owing to its capacity to dissipate energy as heat through mitochondrial uncoupling. The beige adipose tissue is an intermediary phenotype between WAT and BAT, in which energy dissipation can be induced in response to β-adrenergic stimulation.
Adipose tissue is an active metabolic organ that requires intrinsic systems to maintain its function and health. One of these systems is autophagy, which is a homeostatic process by which most mammalian cells recycle cellular components and eliminate damaged proteins and organelles. In the last decade, several studies have highlighted the role of autophagy in adipose tissue function such as its role in adipogenesis and thermogenesis. Furthermore, the autophagic process is impaired in adipose tissue of obese and diabetic humans and animals, thus providing evidence for the importance of this degradation pathway in the maintenance of metabolic health.
In this review, we briefly summarize the molecular mechanisms of autophagy and its regulation, and describe its role in adipogenesis and thermogenesis in WAT and BAT. We also review key aspects of autophagy impairments in WAT and BAT during obesity and diabetes, and discuss its involvement in adipose tissue inflammation and insulin resistance.
Autophagy and Its Regulation
The term “autophagy” comes from the Greek world “self-eating” and was first used by the Nobel Prize laureate Christian de Duve in 1974 (14). However, the molecular pathways involved in the autophagic process and its regulation were only recently discovered, owing to the advancement in genetic approaches that were used to target this process (95, 131). Most of what we know about autophagy has been discovered in the unicellular eukaryote Saccharomyces cerevisiae, and this knowledge yielded another Nobel Prize for Dr. Yoshinori Ohsumi in 2016.
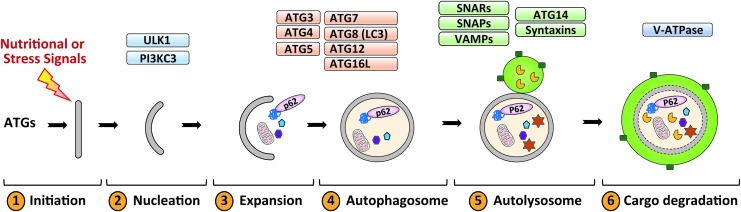
The autophagic process has been extensively reviewed elsewhere (17, 21, 36), and we briefly describe the main steps and the key regulators of this process in mammalian cells. The autophagic process consists of six steps: (i) initiation, (ii) nucleation, (iii) expansion, (iv) autophagosome formation, (v) autophagosome–lysosome fusion, and (vi) lysosomal degradation of the cargo (Fig. 1). The initiation of autophagy involves two main players: Unc-51-like kinase 1 (ULK1) and the class III phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K3C). The activation of these kinases is accompanied by the recruitment of autophagy related genes (ATGs)-containing vesicles, which deliver lipids and proteins necessary for membrane expansion (46, 92). The ATGs involved are mostly the ubiquitin-like ATG8 family members, including ATG3, 4, 5, 7, 12, and 16L1 (120). This conjugation process results in the conversion of the freely diffused form of ATG8 (known as LC3I) to the membrane-tethered and lipidated form (known as LC3II) (20, 34, 56). Once the phagophore is sealed, the autophagosome undergoes maturation, which involves the gradual clearance of ATGs from the outer membrane and recruitment of the machinery responsible for lysosomal delivery and fusion comprising the SNAREs, syntaxins, SNAPs, and VAMPs (15, 40). Once fused with lysosomes, the autophagic cargo is degraded in the acidic lumen of the lysosomes (97).
FIG. 1.
The autophagic steps in mammalian cells. Autophagy is initiated upon activation of upstream signals, and results in activation of ULK1 and class III PI3KC3. Elongation of the phagophore and maturation of autophagosome requires a conjugation system consisting of ATG3, ATG4, ATG5-ATG12, ATG7, ATG8 (or LC3), ATG10, and ATG16L. Cellular material targeted for degradation is marked by adaptor proteins such as p62 (or sequestosome 1), and then engulfed in autophagosomes. The mature autophagosome then fuses with lysosomes, and the cargo is degraded by lysosomal hydrolases. ATGs, autophagy related genes; ATP, adenosine triphosphate; PI3K3C, phosphatidylinositol-4,5-bisphosphate 3-kinase; ULK1, Unc-51-like kinase 1. Color images are available online.
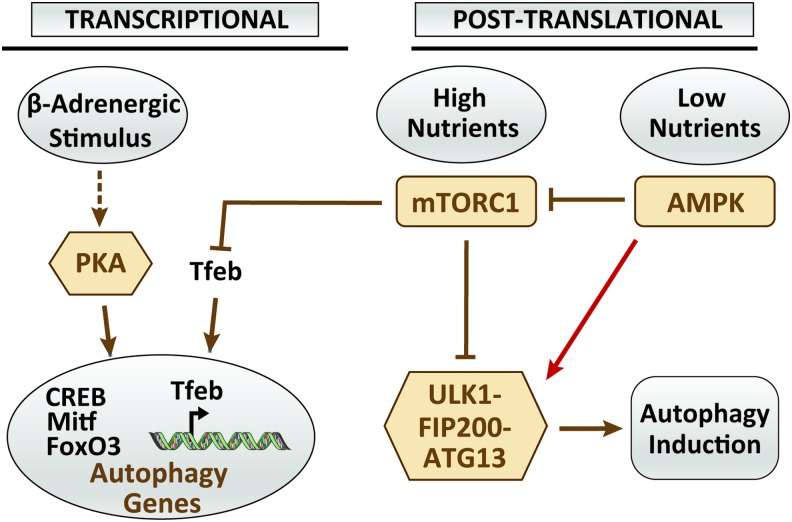
Because autophagy is an adaptive response to mainly nutrient stress, it is regulated predominantly by energy stress sensors such as the mammalian target of rapamycin (mTOR) and the adenosine monophosphate-activated protein kinase (AMPK) (Fig. 2). There are two distinct mTOR complexes: mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2); however, only mTORC1 was shown to directly regulate autophagy (43). The mechanisms by which mTORC1 inhibit autophagy involve ULK1, which is phosphorylated and inactivated by mTORC1 in high nutrient conditions (38). In contrast, ULK1 is dephosphorylated and dissociated from mTORC1 under starvation (38). During nutritional stress (drop in adenosine triphosphate [ATP]) AMPK is activated, which leads to mTORC1 inactivation through phosphorylation and activation of the mTORC1 inhibitor tuberous sclerosis 2 (TSC2) (39). In addition, AMPK can activate ULK1 upon phosphorylation at various serine residues, thus activating autophagy (18). Aside from mTOR and AMPK, work on yeast identified protein kinase A (PKA) as an inhibitor of autophagy (124), an observation recently extended to mammalian cells (91, 111). The mechanisms by which PKA and its downstream transcription factor, cAMP response element binding (CREB) protein, negatively regulate autophagy in mammalian cells involved the transcription of Itm2a, a membrane protein that interferes with the vacuolar ATPase function and thus the autophagic flux (91). Relevant to adipose tissue, PKA was also shown to inhibit autophagy and mitophagy through the modulation of gene expression of the transcription factors Mitf and FoxO3 and their autophagic/lysosomal targets (1, 2, 10). This will be further discussed under the Autophagy/Mitophagy and Thermogenesis section. Another key transcriptional factor involved in autophagy regulation is the transcription factor EB (Tfeb). Tfeb is negatively regulated by mTORC1 and released upon starvation to induce the expression of genes involved in lysosomal biogenesis and lipid catabolism (110).
FIG. 2.
The main regulators of autophagy in adipose tissue. Autophagy is regulated both transcriptionally and post-translationally. Nutrient stress such as overfeeding or starvation modulates autophagy through two main arms: mTORC1 activation (overfeeding) and AMPK (starvation). Phosphorylation of ULK1-FIP200-ATG13 complex by mTORC1 leads to its dissociation and inactivation. In contrast, during starvation, AMPK is activated, leading to ULK1/FIP200-ATG13 activation and autophagy initiation. AMPK also phosphorylates the mTORC1 inhibitor TSC2, thus activating autophagy. PKA activates autophagy in beige adipose tissue in response to β-adrenergic stimulation. The effect of PKA on autophagy may involve the transcription factors CREB, Mitf, and FoxO3. Similarly, mTORC1 activation inhibits the nuclear translocation of Tfeb, a transcription factor important for autophagy/lysosomal function. AMPK, adenosine monophosphate-activated protein kinase; CREB, cAMP response element binding; mTOR, mammalian target of rapamycin; mTORC1, mTOR complex 1; PKA, protein kinase A; Tfeb, transcription factor EB; TSC2, tuberous sclerosis 2. Color images are available online.
Selective Autophagy in Adipose Tissue
Although initially described as a bulk nonselective degradation process, autophagy can be selective. Thus, chaperone-mediated autophagy (CMA) is responsible for degrading certain proteins containing a specific amino acid sequence (16, 47). In addition to selectivity toward cellular proteins (aggrephagy), autophagy can selectively target organelles, including lipid droplets (LDs; lipophagy), mitochondria (mitophagy), peroxisomes (pexophagy), ribosomes (ribophagy), and endoplasmic reticulum (ER; reticulophagy) (23). The mechanisms involved in selective autophagy are partially understood but seem to involve a cargo recognition step involving specific receptors/adaptors [a topic extensively reviewed elsewhere in Anding and Baehrecke (3), Sica et al. (117), Stolz et al. (125), and Zaffagnini and Martens (141)]. For the purpose of this review, we discuss the importance of lipophagy and mitophagy in WAT and BAT function and maintenance.
Lipophagy
LDs are composed of hydrophobic triglyceride (TG) core and sterol esters enwrapped by a polar lipid monolayer associated with various proteins (28, 30). Although found in many cell types, LDs are the core cytosolic components of white adipocytes and serve as a reservoir for TGs to be mobilized in times of energy demand (102). The mobilization of lipids from LDs in adipocytes occurs mainly through hydrolysis by cytosolic lipases such as LD-associated adipose triglyceride lipase (ATGL); the cytosolic hormone-sensitive lipase (HSL); and monoacylglycerol lipase (MGL), which are all regulated by nutrients and hormonal signals (28). Another form of mobilization of LDs uses autophagy and is termed lipophagy. Lipophagy was first discovered in the liver (118), and then subsequently found in other cell types such as neurons, macrophages, and tumor cells. Since then, the question was raised to whether lipophagy also regulates lipid mobilization in adipocytes, a cell dedicated to lipid storage and degradation. Lipophagy in the liver was shown to depend on an intact autophagy machinery (118), and lack of this process in adipose tissue was rather associated with a decrease in adipocyte LDs content (119, 142). This contradicting finding may be explained by the impaired adipogenesis and TGs accumulation in adipose-specific Atg7-deficient mice (119, 142). In fact, reduced fat accumulation in mice lacking Atg7 in adipose tissue is rather consistent with a role of autophagy in LDs biogenesis rather than degradation, which was recently reported in hepatocytes (65, 115). The mechanisms regulating the recruitment of the autophagic machinery to LDs are still not fully characterized. A recent study showed that autophagy is necessary for LDs degradation upon β2-adrenergic-stimulated lipolysis in 3T3-L1 adipocytes in vitro, which involved the Ras-related protein Rab-7 (72). Rab-7 is associated with LDs, and helped recruit the autophagic machinery and lysosomal fusion (72). Aside from Rab-7, the autophagy adaptor p62 (SQSTM1) was recently shown to associate with LD proteins both in hepatocytes and in myotubes (7, 58). Indeed, p62 is necessary for LC3 recruitment to LDs, and its silencing resulted in TG accumulation in hepatocytes (134). Whether p62 has similar roles in lipophagy in adipocytes is not clear as lack of p62 in brown preadipocytes had no effect on lipid accumulation (88).
Lipophagy and lipolysis are often connected, and more evidence now suggests that these two processes work together to optimize lipolysis in BAT in response to β-adrenergic stimulation (80). Lipophagy induction in BAT and liver is associated with lipolysis activation through the interaction of ATGL with LC3 on autophagosomes to maximize lipolysis. Finally, altered autophagy through downregulation of Bif-1 (a membrane-curvature-inducing protein) increased obesity with high-fat feeding through impairment of basal but not hormone-stimulated lipolysis (71). Thus, to better clarify the role of autophagy and adipose tissue lipophagy and to define its interaction with lipolysis, a specific targeting strategy of autophagy in mature adipocytes is needed.
Mitophagy
While WAT mitochondrial content is low, brown and beige adipose tissues are characterized by the abundance of mitochondria. Mitochondrial content in brown and beige adipose tissues is dynamic, and varies in response to nutritional and hormonal stimuli. Mitophagy, which is the selective degradation of mitochondria, is necessary to segregate and eliminate damaged/dysfunctional organelles in cells (64). This process is usually coupled with mitochondrial biogenesis and mitochondrial dynamics (change in mitochondrial shape) to assure an adequate number that matches the energetic demand. The molecular mechanisms of mitophagy and key mitophagy receptors have been extensively studied and characterized in cells after mitochondrial depolarization by synthetic ionophores such as carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP). Most mitophagy receptors contain a region allowing them to interact with mitochondria and an LC3-interacting region (LIR) allowing them to recruit the autophagic machinery. These receptors are tissue specific, and their elimination of mitochondria can be dependent or independent of autophagy. For example, Nix, also called Bcl2/adenovirus E1B-interacting protein 3-like (Bnip3l), is the mitophagy receptor for the elimination of mitochondria in reticulocytes (105, 108). A homolog of Nix called Bnip3 has also been shown to participate along with Nix to hypoxia-mediated mitochondrial clearance in the heart (33). The most characterized mitophagy pathway involves the PTEN-induced putative kinase 1 (Pink1) and the cytosolic E3 ubiquitin ligase Parkin [reviewed elsewhere in Pickles et al. (98)]. In brief, upon depolarization of mitochondria, Pink1 is translocated to the mitochondrial outer membrane, where it undergoes cleavage and autophosphorylation. Pink1 then phosphorylates Parkin and ubiquitin at Ser65, increasing its E3 ligase activity (51, 114). It appears that phosphorylation of ubiquitin is sufficient to induce basal mitophagy even in the absence of Parkin, suggesting the existence of redundant pathways (44). The synthesis of ubiquitin chain linkages on the mitochondrial outer membrane by the concerted action of Pink1 and Parkin allows mitophagy receptors to bind to mitochondria through their ubiquitin-binding domains (125). There are five mitophagy receptors identified in mammalian cells, including sequestosome 1 or p62, Nbr1, Ndp52, optineurin (Optn), and Tax1bp1. Lack of all these receptors in a single cell line (also called PentaKO cells) abolished Pink1-Parkin-mediated mitophagy (60).
Among the earliest studies that suggested a role for mitophagy in adipose tissue function is the work from Jorge Moscat's laboratory (88) using aP2-driven deletion of p62 in adipose tissue. Lack of p62 impaired thermogenesis through a reduction in β-adrenergic-stimulated mitochondrial biogenesis and mitochondrial respiration (88). Whether these effects of p62 deletion in adipose tissue are a consequence of impaired mitophagy is still not clear as mitochondrial content was rather decreased in BAT of these mice. More recent studies associated whitening of BAT with enhanced expression of mitophagy markers after high-fat diet (HFD), whereas activation of BAT after cold exposure or β-adrenergic stimulation is characterized by a reduction in mitophagy (10, 116). These results imply that to maintain higher mitochondrial content, BAT mitophagy should be inhibited. This is technically challenging to test as there are currently no tools to target mitophagy specifically without affecting the autophagy process. More recently, three mouse models were developed to study mitophagy in vivo such as the mito-QC, mito-timer, or mt-keima mice (83, 126, 127). These models all use pH-sensitive mitochondria-targeted signal, and could be instrumental to study mitophagy in WAT and BAT.
Autophagy and Adipogenesis
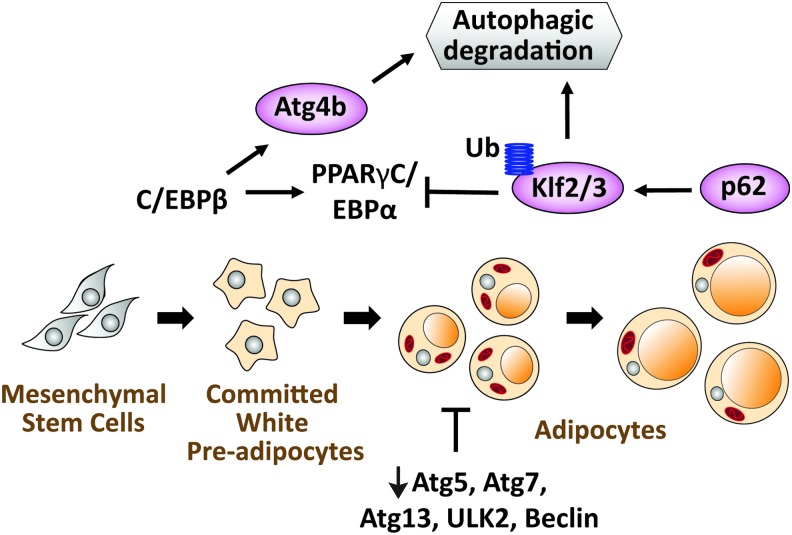
The first evidence for an inhibitory effect of autophagy on adipogenesis came from a study using mouse embryonic fibroblasts (MEFs) from whole body Atg5-deficient mice (5). This study demonstrated that adipogenesis is associated with an increase in autophagy in wild-type cells and is impaired in Atg5-deficient cells (5). Furthermore, impaired autophagy through deletion of Atg7 in aP2+ cells or 3T3-L1 preadipocytes similarly impaired adipogenesis and reduced fat accumulation in mice (119, 142). Mechanistically, it was first demonstrated that lack of autophagy does not interfere with early events of adipogenesis such as the induction of adipogenic genes but rather impeded the later stages associated with fat accumulation (5). However, this idea has been challenged by a recent study showing that autophagy is rather necessary for the degradation of adipogenic inhibitors such as Klf2/3 through a C/EBPβ-mediated induction of Atg4 expression (29) (Fig. 3). The importance of autophagy in adipogenesis is not unique to WAT as this process is also impaired in BAT of mice lacking Atg7 in Myf5+ BAT precursors (79). Aside from degrading adipogenic inhibitors, one possible role of autophagy in white adipocytes could be the clearance of cytosolic components such as organelles to accommodate the growing LDs. In support of this idea, electron microscopy examination revealed a higher number of mitochondria of Atg7-deficient adipocytes (142). However, this explanation will be inconsistent with the maintenance of a higher mitochondrial number in BAT adipocytes. One may argue that the regulation of mitochondrial number in WAT and BAT is very different. Thus, WAT uses less mitochondrial biogenesis and more autophagy to maintain a lower number of mitochondria, whereas higher mitochondrial biogenesis and lower mitophagy are characteristic of BAT. Indeed, white differentiation of 3T3-L1 preadipocyte is associated with an increase in the number of autophagosomes containing mitochondria (26, 93), while β-adrenergic stimulation of BAT led to the inhibition of autophagy/mitophagy (10).
FIG. 3.
The role of autophagy in adipogenesis. Autophagy is upregulated during the conversion of preadipocytes to adipocytes. Deletion of autophagy proteins (Atg5, Atg7, Atg13, ULK2, or beclin) reduced adipogenesis in vitro and in vivo. One of the mechanisms involved in autophagy induction during adipogenesis is the transcriptional induction of Atg4b by C/EBPβ. The induction of Atg4b expression and the ubiquitination of Klf2 and 3 by p62 help eliminate these two negative regulators of adipogenesis. Color images are available online.
The Role of mTOR in Adipose Tissue Autophagy
Activation of mTOR through growth factors signaling (insulin in the case of adipogenesis) is known to suppress autophagy (36, 139). However, treatment of 3T3-L1 with the mTOR inhibitor rapamycin blocked adipogenesis through a reduction in clonal expansion, a step required for terminal differentiation (6, 138, 139). This is in line with a recent report showing that rapamycin suppresses adipogenesis in 3T3-L1 preadipocytes in an autophagy-independent manner (103). Consistent with impaired adipogenesis, lack of the mTORC1 regulatory subunit raptor in aP2+ cells produced a similar phenotype to the Atg7-deficient mice with reduced fat mass and resistance to diet-induced obesity (DIO) (100). Contrary to this study, targeting raptor to mature white and brown adipocytes using the adiponectin-Cre promoter produced a lipodystrophic phenotype associated with metabolic disease (62). This discrepancy highlights again the inefficient and the off-target effects of the aP2-Cre system. The lipodystrophic phenotype of adipocytes-specific raptor knock-out mice cannot be solely explained by an increase in autophagy because mTORC1 has several other downstream targets other than autophagy. However, enhanced autophagy and lipophagy may have played a role in the clearance of LDs in these mice, resulting in heterogeneity in LD size. Deletion of mTOR has also profound consequences on BAT as evidenced by lack of adaptation to prolonged cold exposure in raptor-adiponectin-Cre mice (57). Furthermore, inhibition of mTOR by rapamycin was shown to inhibit WAT browning in response to cold exposure or to β-adrenergic stimulation (69, 130). It remains to be determined whether these effects of mTORC1 deletion in white and brown adipocytes are mediated by the induction of autophagy or by another arm of mTOR signaling. Moreover, as mTORC1 was recently shown to be phosphorylated by PKA in adipocytes (68), future studies may unveil autophagy-dependent versus autophagy-independent processes regulated by mTOR in WAT and BAT.
Autophagy/Mitophagy and Thermogenesis
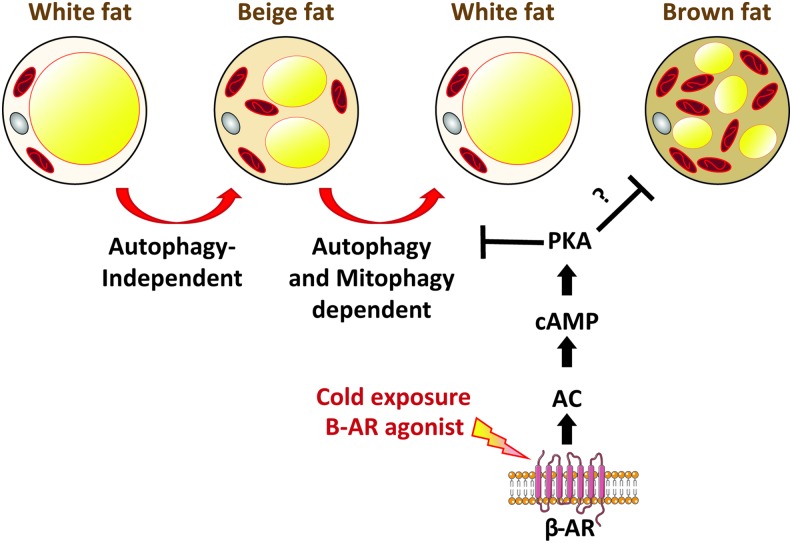
Brown fat has the capacity to dissipate energy as heat due to the presence of the uncoupling protein 1 (UCP1) in the inner mitochondrial membrane. UCP1 uncouples electron transfer from ATP synthesis allowing more oxidation of substrates to maintain a membrane potential. This uncoupling is an exciting strategy to dissipate excess energy and to burn extra calories. Therefore, agents that can uncouple mitochondrial respiration from ATP synthesis were viewed as a potential therapeutic avenue to fight obesity. However, due to the presence of other uncoupling proteins in other organs (mainly UCP2 and UCP3 in heart, muscle, and β cells), it became problematic to use uncoupling agents without the undesirable cardiac phenotypes associated. Another type of fat that has recently gained much attention is beige fat, which shares functional similarities and thermogenic capacity with brown fat despite having a distinct developmental origin (27, 63, 107, 109). One main difference between beige and brown fat is that beige fat is inducible and can be recruited in response to cold or to β-adrenergic stimulation, whereas brown fat is constitutively active. The presence of beige fat in humans and its ability to be recruited and activated in response to cold spared interest in studying the functionality and the recruitment of this fat. It has been recently reported that autophagy is differentially regulated in brown versus beige fat (Fig. 4).
FIG. 4.
Repression of autophagy/mitophagy is necessary for beige fat maintenance. Upon β-adrenergic stimulation, beige fat is recruited into white adipose tissue through a process that does not require autophagy. Upon removal of the β-adrenergic stimulation, beige fat converts back to white fat through the autophagic/mitophagic clearance of mitochondria. Adenylate cyclase (AC)/cAMP/PKA represses autophagy to maintain beige fat upon β-adrenergic stimulation. In brown fat, some studies showed that the same cAMP/PKA pathway can repress autophagy during β-adrenergic stimulation. ? represents an inhibition that is not yet confirmed. Color images are available online.
Brown fat
Brown fat is the main site for nonshivering thermogenesis in mammals. During cold exposure, brown fat undergoes tremendous remodeling including the increase in mitochondrial biogenesis and mitochondrial turnover to allow substrate oxidation and mitochondrial uncoupling (27, 135). Indeed, work by Wikstrom et al. (135) elegantly demonstrated that mitochondrial fragmentation upon norepinephrine (NE) stimulation of brown adipocytes was required for mitochondrial uncoupling in vitro. Perhaps contradicting these results, in vivo deletion of the profusion mitofusin 2 (Mfn2) in white and brown adipocytes or in brown and beige adipocytes led to whitening of BAT and a reduction in cold-induced thermogenesis in mice (9, 76). These results suggest that fragmentation of mitochondrial network may be necessary for enhancing the organelles' contact with LDs to liberate and utilize fatty acids, but this fragmentation cannot be sustained once the stimulus is removed. Thus, a balance between fusion and fission is key to maintain the metabolic health of adipose tissue.
Initial studies examining the role of autophagy in adipose tissue reported an increase in BAT mass in Atg7fl/fl-aP2-Cre mice (119). The reasons for increased BAT mass were not fully examined but enhanced mitochondrial content was observed, suggesting a failure to clear mitochondria or impaired mitophagy (142). Consistent with this idea, short-term (24 h) exposure of mice to cold (4°C) resulted in a significant reduction in the expression of autophagy genes in BAT, whereas thermogenic gene expression was elevated (10). Similar to cold exposure, treatment of brown adipocytes with NE also suppressed autophagy, indicating that the thermogenic program inversely correlates with the autophagy program (10). Cold or NE-mediated suppression of autophagy in BAT occurred both at the transcriptional and at the post-translational level. The transcriptional regulation involved cAMP/PKA/CREB and p38MAPK because inhibitors of these pathways partially restored autophagy in activated BAT (2, 10). Furthermore, the post-translational regulation may occur in part through the reduction in LC3 association with LDs in activated BAT (10). However, these findings are in contrast with recent reports showing that activation of BAT by cold or by β-adrenergic stimulation is associated with rather an increase in autophagy and mitophagy (74, 80). Surprisingly, one of the studies that showed that mitophagy was increased in BAT upon activation reported an accumulation of the autophagy adaptor protein p62, which is a reflection of autophagy inhibition and not activation (74). The reasons for the discrepancy in BAT mitophagy are not fully understood but may be related to the duration of cold exposure, which was quite different between these studies (1 h, 24 h, and 7 days, respectively). However, the fact that PKA can phosphorylate and activate mTOR upon β-adrenergic stimulation in BAT (68) is consistent with an inhibition of autophagy and perhaps mitophagy under this condition. Additional studies are needed to define the homeostatic role of autophagy and mitophagy and their contribution to BAT remodeling in response to a β-adrenergic stimulation. It will also be important to clearly define the role of PKA/mTOR in autophagy in BAT.
Beige fat
While the involvement of autophagy/mitophagy in classical brown fat maintenance and remodeling upon activation is still debated, inhibition of mitophagy is important for the maintenance of beige fat. Thus, WAT of Atg7fl/fl-aP2-Cre mice acquired characteristic of the BAT, with the abundance of small multilocular LDs and mitochondria (119, 142). These changes that caused increased fatty acid oxidation and energy expenditure protected the mice from DIO. However, the use of aP2-Cre in these studies raised the question whether the recruitment of beige fat was mediated by nonadipose tissues, including skeletal muscle, since Atg7 deletion in this tissue elicited beige fat recruitment (48, 89). Furthermore, the expression of aP2 in the central nervous system (CNS) and the deletion of autophagy in this organ may have contributed to beige fat recruitment (35). In fact, the short life span observed in Atg7fl/fl-aP2-Cre mice resembles the phenotype of Atg7 or Atg5 deletion in the CNS (50, 55). Therefore, it was important to address the role of autophagy/mitophagy in beige fat specifically. An elegant study from the Kajimura group (2) used Ucp1-Cre mice to target autophagy through Atg12 or Atg5 deletion in classical brown and beige fat. Findings from this study clearly demonstrated that autophagy is not necessary for beige fat recruitment in WAT after cold exposure or in response to β-adrenergic stimulation. In contrast, autophagy was necessary for the clearance of mitochondria during the transition from beige-to-white fat after withdrawal of the β-adrenergic stimulus (2). The mechanisms involved in autophagy induction during beige-to-white transition were mediated by a reduction in cAMP/PKA signaling and the transcriptional repression of Mitf and FoxO3 (2). The same group further demonstrated that Parkin is required for mitochondrial clearance by autophagy during the transition from beige-to-white fat, and that NE through the PKA pathway inhibited the recruitment of Parkin to mitochondria (73). In the same study, it was also reported that mitochondrial uncoupling and thus membrane depolarization are not required for Parkin translocation to mitochondria during beige-to-white fat transition (73), suggesting the existence of distinct signal (106). Other groups also found an inverse relationship between Parkin and UCP1 expression in inguinal white adipose tissue (iWAT) of mice treated with the β-adrenergic receptor agonist CL316,243. Treatment of mice with CL316,243 significantly reduced Parkin and increased UCP1 expression in iWAT in mice (129). Furthermore, the increase in Parkin expression in epididymal WAT of obese mice may explain the resistance of this tissue to browning (12, 13, 81). The same increase in Parkin and Pink1 was also observed in BAT of obese mice, which may be involved in BAT whitening (116). Altogether, these findings suggest that autophagy/mitophagy antagonizes WAT browning and is under the direct regulation of cAMP/PKA signaling in adipose tissue (Fig. 2). It remains to be determined, however, why is brown and beige fat different in their response to autophagy inhibition if the main regulator of this process is the cAMP/PKA signaling? Why is autophagy indispensable for beige fat recruitment and BAT activation?
Alterations in Adipose Tissue Autophagy in Obesity and Insulin Resistance
Since the initial studies show an improved metabolic profile in Atg7fl/fl-aP2-Cre mice, several investigations started examining the level of autophagy in adipose tissue of obese humans and animals (Fig. 5). Thus, it was proposed that autophagy could be targeted pharmacologically to alleviate metabolic derangements in the obese state.
FIG. 5.
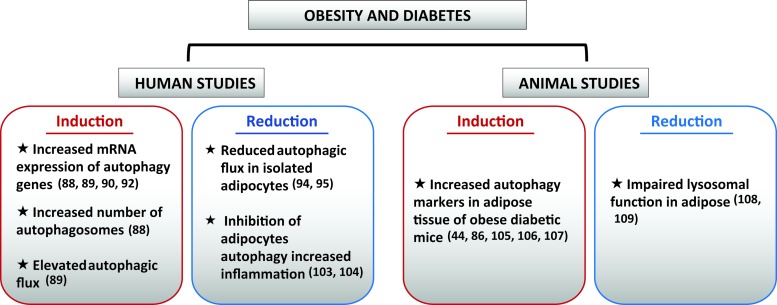
The state of adipose autophagy in obese diabetic humans and animals. The state of autophagy in adipose tissue of obese diabetic humans and animals is controversial. We summarized in this schematic studies showing enhanced or reduced autophagy in obese diabetic adipose tissue of humans and animals (mostly rodents). References are provided in parentheses and are cited in the main text. mRNA, messenger RNA. Color images are available online.
Adipose Autophagy/Mitophagy Modulation by Phospholipids
Different roles that autophagy plays in BAT and WAT may arise from differences in cellular lipid composition in these tissues. BAT contains greater amounts of cardiolipin (CL) and phosphoethanolamine (PE) compared with WAT (82, 128). These findings likely reflect the greater mitochondrial content in BAT compared with WAT, as CL and PE are conically shaped phospholipids that are highly concentrated in cristae where the enzymes of electron transport chain reside (37). Cold stimulus induces CL and PE biosynthesis (75, 78, 128), and the lack of CL impairs the thermogenic capacity of BAT (128). CL and PE are both essential for autophagy/mitophagy. In particular, mitochondrial PE appears to be the main source of LC3 lipidation (104) and externalization of CL to outer mitochondrial membrane signals mitophagy by LC3 binding (11). As cold induction reduces mitophagic flux, we speculate that there must be a BAT-specific quality control mechanism by which mitochondrial CL or PE does not initiate autophagy. It is unclear whether greater PE or CL in BAT compared with WAT plays a role in how these tissues differentially respond to autophagy. Phospholipids from BAT also contain higher proportion of polyunsaturated fatty acids (PUFAs) compared with WAT (82), also likely contributed by greater mitochondrial content. In contrast, WAT contains greater amounts of phosphatidic acid (PA), diacylglycerol (DAG), TGs, cholesterol-ester, sphingomyelin, and ceramides compared with BAT (82). It is curious that PA and DAG contents are higher in WAT, as these two lipids represent the precursors for all phospholipid molecules (132). Phosphatidylglycerol, phosphatidylinositol (PI), and CL are generated from PA, whereas PE, phosphatidylcholine, and phosphatidylserine are generated from DAG. Greater PA and DAG content in WAT may suggest greater need for membrane recycling compared with BAT, consistent with autophagy-dependent mitochondrial clearance in WAT (2). PA is a known activator for mTOR (19, 85), and PI is a mediator of PI3K3C that is essential for nucleation of phagaphore (112). Thus, these signals may partly mediate accelerated mitophagy in WAT compared with BAT.
Human studies
Human studies involving adipose tissue rely solely on frozen subcutaneous and omental biopsies taken during elective general abdominal surgery or during bariatric surgery. The first study that examined the level of autophagy in human adipose tissue used subcutaneous adipose tissue from type 2 diabetes (T2D) patients who were overweight (body mass index [BMI] > 27) and compared them with nondiabetic controls who were not matched for age, gender, or BMI (96). The study showed that autophagy was increased in the subcutaneous adipose tissue of T2D patients as evidenced by the abundance of LC3 puncta and autophagosomes, and this increase was associated with reduced mTOR signaling (96). A subsequent study using subcutaneous and omental WAT from two different cohorts of obese and nonobese patients reported an elevated expression of autophagy genes and LC3II in the omental fat of obese individuals (54). Furthermore, this study examined the autophagic flux by incubating adipose tissue explants of the same subjects with the lysosomal inhibitors bafilomycin A1 and leupeptin, and showed that p62, which is normally degraded by autophagy, accumulated more in WAT of obese individuals (54). Finally, to determine the contribution of adipocyte and nonadipocyte cells to the increase in autophagy in WAT of obese patients, the same group examined the expression of autophagy genes and LC3II protein in collagenase-digested WAT, and demonstrated that the increase in autophagy markers occurred only in adipocytes (54). In subsequent studies from the same group, there was an inverse correlation between omental adipose tissue ATG5 and LC3A messenger RNA (mRNA) and circulating adiponectin levels and insulin sensitivity in obese humans (121). They further demonstrate that inhibition of autophagy in adipose tissue explants from the same patients recovered adiponectin levels (121), suggesting that autophagy induction in adipose tissue of obese patients may be a cause for metabolic dysfunction. The transcriptional regulation of autophagy genes in adipose tissue of obese humans involved the E2F family of transcriptional regulators (77). E2F1 protein and mRNA expression were elevated in the omental fat of obese patients, and correlated with the mRNA expression of ATG5 and LC3B (31). Indeed, increased binding of E2F1 to putative binding sites in the LC3B promoter was observed in the omental fat of obese patients (32). Finally, autophagic flux and expression of autophagy genes were suppressed in MEFs from E2F1-deficient mice that were differentiated into adipocytes (31). Surprisingly, adipogenesis was not affected in these cells despite the known regulation of the cell cycle by E2F1 and the importance of clonal expansion for adipogenic differentiation. In contrast to these studies, reduced autophagic flux was seen in isolated subcutaneous adipocytes from obese patients compared with nonobese controls (123). Interestingly, weight loss after bariatric surgery partially ameliorated adipocytes autophagy, suggesting that autophagy inhibition may be caused by lipid overload in adipocytes (122, 123). This observation is consistent with the known suppressive effect of lipids on autophagic/lysosomal degradation previously reported in liver (49, 69, 137), β cells (59), heart (41, 99), and hypothalamus (84). The use of adipose tissue explants to examine autophagic flux is complicated by the fact that adipose tissue cellular composition is complex, making it difficult to ascertain that autophagy is equivalently affected across cell types. In addition, adipose tissue from obese individuals contains many immune cells when compared with adipose tissue of lean individuals. Because of the known suppressive effect of autophagy on inflammation, we can speculate that autophagy induction in immune cells or other cell types within adipose tissue plays a protective role to counteract inflammation. Indeed, pharmacological inhibition of autophagy in adipose tissue explants or adipocytes significantly increases proinflammatory cytokine secretion (42, 140). The potential role of autophagy in adipose tissue inflammation will be discussed later.
Animal studies
Similar to human studies, animal studies examining the level of autophagy and its role in metabolic homeostasis have been inconsistent. Associative studies in rodents have reported an increase in autophagy markers in WAT of genetically obese db/db and ob/ob mice and obese WOKW rats with the metabolic syndrome (53, 66, 143). Similarly, p62 and LC3I levels were reduced, and LC3II/LC3I ratios were increased in WAT of DIO mice (13, 116). Contrary to these studies that examined the static level of autophagic markers in whole WAT, autophagic flux measurement in WAT of DIO mice revealed a defect in lysosomal clearance and an increase in autophagosomes formation (86, 87), which may explain in part the discordance in the results obtained with static autophagic measurements.
The use of genetic approaches to study autophagy in adipose tissue has further confirmed a role for this process in adipose tissue development and homeostasis. Thus, deletion of Atg7 using the fatty acid binding protein 4 (Fabp4 or aP2) resulted in a lean mouse with improved resistance to obesity and insulin resistance (119, 142). One important finding in these studies is the importance of autophagy in the process of adipogenesis, which was concomitantly observed in MEFs of Atg5-deficient mice (5). Due to the defect in adipocytes development in aP2-Cre-driven autophagy deletion, it is not possible to conclude whether the role of autophagy in mature adipocytes is beneficial or detrimental for metabolic health. Furthermore, reduced survival of otherwise healthy Atg7fl/fl-aP2-Cre mice suggests the existence of central defects that may have affected energy expenditure independently of adipose autophagy (119). Indeed, targeted deletion of autophagy in hypothalamic pro-opiomelanocortin (POMC) neurons is sufficient to increase energy expenditure in mice (80, 136). Thus, the initial phenotype of Atg7fl/fl-aP2-Cre mice could be due to lack of autophagy in POMC or other neuronal populations since aP2 is also expressed in the brain (35). One other intriguing observation in adipose tissue of Atg7fl/fl-aP2-Cre mice is a marked increase in macrophage recruitment in epididymal WAT, which was not consistent with their improved metabolic profile of these mice and may suggest that autophagy is necessary to counteract inflammation in adipose tissue (119). Consistent with this idea, heterozygous deletion of Atg7 in ob/ob mice enhanced systemic and adipose tissue inflammation and exacerbated insulin resistance (66).
Autophagy and Adipose Tissue Inflammation
Autophagy is a lysosomal degradation process that not only participates in the control of intracellular homeostasis by regulating organelle and protein turnover but also contributes to the regulation of innate and adaptive immune responses. Both inflammation and autophagy constitute a natural response to stress, and the relationship between them is relatively complex. During infections, defects in autophagy lead to excessive proliferation of the pathogens and/or exacerbated immune response, both of which are deleterious. Through autophagy, cells can eliminate harmful components such as pathogens, allowing them to survive in response to environmental stress (101, 133). Like in infectious diseases induced by micro-organisms, chronic inflammatory diseases, such as metabolic disorders, induced by endogenous compounds, are generally associated with inappropriate modulation of autophagy, which can play a pivotal role in inflammation by influencing the development, homeostasis, and survival of inflammatory cells, including lymphocytes, macrophages, and neutrophils as well as regulating cytokines secretion (101).
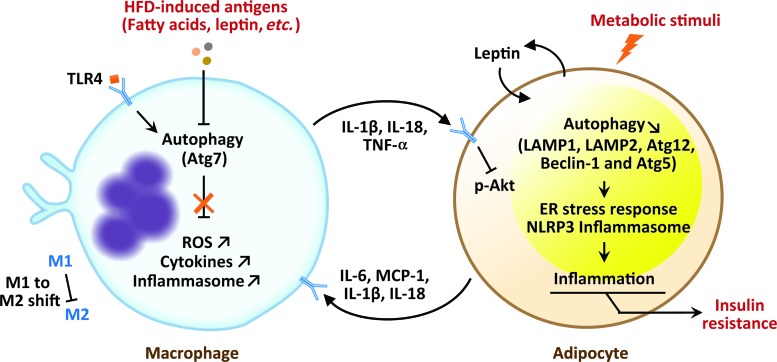
Autophagic proteins have important roles in the regulation of several immune system functions such as lymphocyte development, antigen presentation, and cytokines production by immune cells and nonimmune cells (133). In vitro and in vivo studies revealed that autophagy is an important regulator of adipocyte inflammation during metabolic diseases (Fig. 6). Adipocytes autophagy impairment, attested by reduced autophagy-related gene expression, such as LAMP1, LAMP2, and Atg5, in insulin-resistant and in both DIO (42) and age-associated obesity (24) models is closely associated with aberrant ER stress response (22, 42, 140). Adipocyte-derived proteins such as leptin modulate adipose tissue inflammation by reduction in adipose autophagy. As shown in ob/ob mice, leptin inhibits autophagy through the downregulation of Atg12, Beclin 1 as well as the Atf4/Atg5 complex formation and reduces degradation of IκB with IL-18 and IL-1β reduction in adipocytes, thus impacting ER stress response and inflammation (22). Moreover, fatty acids upregulated during obesity (4) may act as a harmful signal, which activates innate immune receptors TLR (toll-like receptor) to initiate an immune response, which participates in the regulation of energy balance and insulin resistance (113). Evidence using old TLR4-deficient mice suggests that TLR4 could initiate autophagy and promote adipose tissue inflammation during age-associated obesity (25). Experiments based on models of genetic deficiencies in autophagy have demonstrated the importance of this process in the homeostasis of adipose tissue macrophages. Autophagic proteins such as Beclin 1 and LC3B in macrophages act as critical regulators of caspase-1-mediated immune responses and IL-1β and IL-18 secretion, in vitro and in vivo, in NALP3 inflammasome-dependent manner (90). Importantly, macrophage autophagy is downregulated by inflammatory stimuli, which is attested by decreasing LC3-II conversion and increasing p62 accumulation in mice on HFD (45). Under HFD conditions, Atg7 regulate M1 and M2 macrophage polarization, reactive oxygen species production, proinflammatory cytokines (IL-1β and TNF-α) secretion by M1 macrophage population, and lipid-induced inflammasome activation in adipose tissue (45, 61). Concomitantly, increased proinflammatory cytokines such as IL1β, IL18, and TNFα produced by adipose tissue macrophages inhibit Akt, which impairs insulin signaling, resulting in insulin resistance (45) (Fig. 6). Interestingly, macrophage-specific deletion of Atg5 and Atg16L increases hepatic inflammation, but does not affect adipose tissue inflammation or body mass in mice-fed HFD (67, 70). However, the role of autophagy in obesity, insulin resistance, and adipose tissue inflammation is still controversial and appears to be context dependent. Different studies show distinct results depending on the diet, age, and sex of mice; the duration of HFD; the mediators of obesity in the different models; the autophagy genes deletion; and the tissue where autophagy proteins were depleted. In almost all the studies, lipopolysaccharides (LPSs) were used and when it comes to inflammation studies, addition of LPS may complicate the interpretation of the results related to adipose tissue inflammation (8).
FIG. 6.
The role of autophagy in adipocytes–macrophages cross-talk. In physiological conditions, autophagy regulates inflammation, ROS levels, and M1 to M2 shift in macrophages population. During inflammatory conditions, such as diabetes or HFD, autophagy is impaired in macrophages, which impair ROS regulation, cytokine production, and M1/M2 ratio. Concomitantly, increased inflammatory cytokines such as IL1β, IL18, and TNFα produced by M1 macrophages inhibit Akt signaling pathway in adipocytes, resulting in the development of insulin resistance. Leptin produced by adipocytes inhibits autophagy, which impact ER stress response and inflammation in adipocytes. ER, endoplasmic reticulum; HFD, high-fat diet; ROS, reactive oxygen species. Color images are available online.
In humans, a defective regulation of adipose tissue autophagy is observed in adipocytes from T2D and obese individuals, which exhibit autophagosome accumulation and increased autophagy assessed by higher LC3-II/LC3-I ratio, upregulation of autophagy gene (LC3 and Atg5), along with decreased p62 and mTOR protein levels when compared with lean individuals without diabetes (52, 54). This upregulated adipose tissue autophagy during obesity modulates proinflammatory cytokine expression and secretion, and involves autophagy-related protein Atg7 (42). Another study involving obese-diabetic subjects undergoing calorie restriction suggests that autophagy and inflammation are regulated independently (94).
Altogether, data from animal and human studies suggest that autophagy in both adipose tissue macrophage and adipocytes is important for inflammation, which participates in the regulation of systemic glucose homeostasis and metabolic disease. Modulation of autophagy might lead to therapeutic interventions for diseases associated with chronic inflammation such as metabolic diseases.
Summary and Conclusions
Autophagy has originally been viewed as an adaptive response to cellular stress, but in recent years this process was shown to regulate important cellular processes. In adipose tissue, autophagy is a key regulator of WAT and BAT adipogenesis, and dysregulated autophagy impairs fat accumulation both in vitro and in vivo. Furthermore, autophagic regulation of mitochondrial clearance is important for the conversion of beige fat into white fat, and it is also possible that suppression of autophagy may be necessary to prevent BAT whitening during obesity. What is still not so clear is the role of autophagy in mature adipocytes beyond development. Furthermore, is autophagy beneficial or detrimental to adipocytes in a state of inflammation? More comprehensive studies are needed to define cell type-specific contribution to autophagy impairment in adipose tissue of obese humans and animals. A careful characterization of autophagy function in adipose tissue function postdevelopment and its role in inflammation and metabolic homeostasis can certainly lead to a better understanding of obesity and its metabolic complications, and can help provide a rationale for targeting this process for the treatment of obesity and metabolic diseases.
Acknowledgments
This work is supported by the National Institute of Diabetes and Digestive and Kidney Disease (NIDDK) grant R01DK098646 to S.B and grant R01DK107397 to K.F. We thank Mrs. Diana Lim for her assistance in the preparation of the figures.
Abbreviations Used
- AMPK
adenosine monophosphate-activated protein kinase
- ATGL
adipose triglyceride lipase
- ATGs
autophagy related genes
- ATP
adenosine triphosphate
- BAT
brown adipose tissue
- BMI
body mass index
- Bnip3l
Bcl2/adenovirus E1B-interacting protein 3-like
- CL
cardiolipin
- CNS
central nervous system
- CREB
cAMP response element binding
- DAG
diacylglycerol
- DIO
diet-induced obesity
- ER
endoplasmic reticulum
- HFD
high-fat diet
- iWAT
inguinal white adipose tissue
- LDs
lipid droplets
- LPS
lipopolysaccharides
- MEF
mouse embryonic fibroblast
- mRNA
messenger RNA
- mTOR
mammalian target of rapamycin
- mTORC1
mTOR complex 1
- NE
norepinephrine
- PA
phosphatidic acid
- PE
phosphoethanolamine
- PI
phosphatidylinositol
- PI3K3C
phosphatidylinositol-4,5-bisphosphate 3-kinase
- Pink1
PTEN-induced putative kinase 1
- PKA
protein kinase A
- POMC
pro-opiomelanocortin
- T2D
type 2 diabetes
- Tfeb
transcription factor EB
- TG
triglyceride
- TLR
toll-like receptor
- UCP
uncoupling protein
- ULK1
Unc-51-like kinase 1
- WAT
white adipose tissue
References
- 1. Akabane S, Uno M, Tani N, Shimazaki S, Ebara N, Kato H, Kosako H, and Oka T. PKA regulates PINK1 stability and parkin recruitment to damaged mitochondria through phosphorylation of MIC60. Mol Cell 62: 371–384, 2016 [DOI] [PubMed] [Google Scholar]
- 2. Altshuler-Keylin S, Shinoda K, Hasegawa Y, Ikeda K, Hong H, Kang Q, Yang Y, Perera RM, Debnath J, and Kajimura S. Beige adipocyte maintenance is regulated by autophagy-induced mitochondrial clearance. Cell Metab 24: 402–419, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anding AL. and Baehrecke EH. Cleaning house: selective autophagy of organelles. Dev Cell 41: 10–22, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arner P. and Rydén M. Fatty acids, obesity and insulin resistance. Obes Facts 8: 147–155, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baerga R, Zhang Y, Chen PH, Goldman S, and Jin S. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy 5: 1118–1130, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bell A, Grunder L, and Sorisky A. Rapamycin inhibits human adipocyte differentiation in primary culture. Obes Res 8: 249–254, 2000 [DOI] [PubMed] [Google Scholar]
- 7. Bersuker K, Peterson CWH, To M, Sahl SJ, Savikhin V, Grossman EA, Nomura DK, and Olzmann JA. A proximity labeling strategy provides insights into the composition and dynamics of lipid droplet proteomes. Dev Cell 44: 97.e7–112.e7, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boutagy NE, McMillan RP, Frisard MI, and Hulver MW. Metabolic endotoxemia with obesity: is it real and is it relevant? Biochimie 124: 11–20, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boutant M, Kulkarni SS, Joffraud M, Ratajczak J, Valera-Alberni M, Combe R, Zorzano A, and Cantó C. Mfn2 is critical for brown adipose tissue thermogenic function. EMBO J 36: 1543–1558, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cairó M, Villarroya J, Cereijo R, Campderrós L, Giralt M, and Villarroya F. Thermogenic activation represses autophagy in brown adipose tissue. Int J Obes (Lond) 40: 1591–1599, 2016 [DOI] [PubMed] [Google Scholar]
- 11. Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, Tyurin VA, Yanamala N, Shrivastava IH, Mohammadyani D, Wang KZQ, Zhu J, Klein-Seetharaman J, Balasubramanian K, Amoscato AA, Borisenko G, Huang Z, Gusdon AM, Cheikhi A, Steer EK, Wang R, Baty C, Watkins S, Bahar I, Bayir H, and Kagan VE. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol 15: 1197–1205, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cui C, Chen S, Qiao J, Qing L, Wang L, He T, Wang C, Liu F, Gong L, Chen L, and Hou X. PINK1-Parkin alleviates metabolic stress induced by obesity in adipose tissue and in 3T3-L1 preadipocytes. Biochem Biophys Res Commun 498: 445–452, 2018 [DOI] [PubMed] [Google Scholar]
- 13. Cummins TD, Holden CR, Sansbury BE, Gibb AA, Shah J, Zafar N, Tang Y, Hellmann J, Rai SN, Spite M, Bhatnagar A, and Hill BG. Metabolic remodeling of white adipose tissue in obesity. Am J Physiol Endocrinol Metab 307: E262–E277, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deter RL, Baudhuin P, and De Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol 35: C11–C16, 1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diao J, Liu R, Rong Y, Zhao M, Zhang J, Lai Y, Zhou Q, Wilz LM, Li J, Vivona S, Pfuetzner RA, Brunger AT, and Zhong Q. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 520: 563–566, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci 15: 305–309, 1990 [DOI] [PubMed] [Google Scholar]
- 17. Dikic I. and Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol 19: 349–364, 2018 [DOI] [PubMed] [Google Scholar]
- 18. Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, and Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331: 456–461, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, and Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 294: 1942–1945, 2001 [DOI] [PubMed] [Google Scholar]
- 20. Fujioka Y, Noda NN, Nakatogawa H, Ohsumi Y, and Inagaki F. Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. J Biol Chem 285: 1508–1515, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, Choi AM, Chu CT, Codogno P, Colombo MI, Cuervo AM, Debnath J, Deretic V, Dikic I, Eskelinen EL, Fimia GM, Fulda S, Gewirtz DA, Green DR, Hansen M, Harper JW, Jäättelä M, Johansen T, Juhasz G, Kimmelman AC, Kraft C, Ktistakis NT, Kumar S, Levine B, Lopez-Otin C, Madeo F, Martens S, Martinez J, Melendez A, Mizushima N, Münz C, Murphy LO, Penninger JM, Piacentini M, Reggiori F, Rubinsztein DC, Ryan KM, Santambrogio L, Scorrano L, Simon AK, Simon HU, Simonsen A, Tavernarakis N, Tooze SA, Yoshimori T, Yuan J, Yue Z, Zhong Q, and Kroemer G. Molecular definitions of autophagy and related processes. EMBO J 36: 1811–1836, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gan L, Liu Z, Luo D, Ren Q, Wu H, Li C, and Sun C. Reduced endoplasmic reticulum stress-mediated autophagy is required for leptin alleviating inflammation in adipose tissue. Front Immunol 8: 1507, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gatica D, Lahiri V, and Klionsky DJ. Cargo recognition and degradation by selective autophagy. Nat Cell Biol 20: 233–242, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ghosh AK, Mau T, O'Brien M, Garg S, and Yung R. Impaired autophagy activity is linked to elevated ER-stress and inflammation in aging adipose tissue. Aging (Albany NY) 8: 2525–2537, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ghosh AK, O'Brien M, Mau T, and Yung R. Toll-like receptor 4 (TLR4) deficient mice are protected from adipose tissue inflammation in aging. Aging (Albany NY) 9: 1971–1982, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goldman SJ, Zhang Y, and Jin S. Autophagic degradation of mitochondria in white adipose tissue differentiation. Antioxid Redox Signal 14: 1971–1978, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gospodarska E, Nowialis P, and Kozak LP. Mitochondrial turnover: a phenotype distinguishing brown adipocytes from interscapular brown adipose tissue and white adipose tissue. J Biol Chem 290: 8243–8255, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, Yan QW, Miyoshi H, and Mashek DG. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest 121: 2102–2110, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo L, Huang JX, Liu Y, Li X, Zhou SR, Qian SW, Liu Y, Zhu H, Huang HY, Dang YJ, and Tang QQ. Transactivation of Atg4b by C/EBPbeta promotes autophagy to facilitate adipogenesis. Mol Cell Biol 33: 3180–3190, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo Y, Cordes KR, Farese RV, Jr., and Walther TC. Lipid droplets at a glance. J Cell Sci 122: 749–752, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haim Y, Blüher M, Slutsky N, Goldstein N, Klöting N, Harman-Boehm I, Kirshtein B, Ginsberg D, Gericke M, Guiu Jurado E, Kovsan J, Tarnovscki T, Kachko L, Bashan N, Gepner Y, Shai I, and Rudich A. Elevated autophagy gene expression in adipose tissue of obese humans: a potential non-cell-cycle-dependent function of E2F1. Autophagy 11: 2074–2088, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Haim Y, Tarnovscki T, Bashari D, and Rudich A. A chromatin immunoprecipitation (ChIP) protocol for use in whole human adipose tissue. Am J Physiol Endocrinol Metab 305: E1172–E1177, 2013 [DOI] [PubMed] [Google Scholar]
- 33. Hamacher-Brady A, Brady NR, Logue SE, Sayen MR, Jinno M, Kirshenbaum LA, Gottlieb RA, and Gustafsson AB. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ 14: 146–157, 2007 [DOI] [PubMed] [Google Scholar]
- 34. Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, and Yoshimori T. Autophagosomes form at ER-mitochondria contact sites. Nature 495: 389–393, 2013 [DOI] [PubMed] [Google Scholar]
- 35. Harno E, Cottrell EC, and White A. Metabolic pitfalls of CNS Cre-based technology. Cell Metab 18: 21–28, 2013 [DOI] [PubMed] [Google Scholar]
- 36. He C. and Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43: 67–93, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Heden TD, Neufer PD, and Funai K. Looking beyond structure: membrane phospholipids of skeletal muscle mitochondria. Trends Endocrinol Metab 27: 553–562, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, and Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20: 1981–1991, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Inoki K, Kim J, and Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu Rev Pharmacol Toxicol 52: 381–400, 2012 [DOI] [PubMed] [Google Scholar]
- 40. Itakura E, Kishi-Itakura C, and Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151: 1256–1269, 2012 [DOI] [PubMed] [Google Scholar]
- 41. Jaishy B, Zhang Q, Chung HS, Riehle C, Soto J, Jenkins S, Abel P, Cowart LA, Van Eyk JE, and Abel ED. Lipid-induced NOX2 activation inhibits autophagic flux by impairing lysosomal enzyme activity. J Lipid Res 56: 546–561, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jansen HJ, van Essen P, Koenen T, Joosten LA, Netea MG, Tack CJ, and Stienstra R. Autophagy activity is up-regulated in adipose tissue of obese individuals and modulates proinflammatory cytokine expression. Endocrinology 153: 5866–5874, 2012 [DOI] [PubMed] [Google Scholar]
- 43. Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, and Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20: 1992–2003, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, and Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol 205: 143–153, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kang YH, Cho MH, Kim JY, Kwon MS, Peak JJ, Kang SW, Yoon SY, and Song Y. Impaired macrophage autophagy induces systemic insulin resistance in obesity. Oncotarget 7: 35577–35591, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Karanasios E, Stapleton E, Manifava M, Kaizuka T, Mizushima N, Walker SA, and Ktistakis NT. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J Cell Sci 126: 5224–5238, 2013 [DOI] [PubMed] [Google Scholar]
- 47. Kaushik S. and Cuervo AM. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol 19: 365–381, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim KH, Jeong YT, Oh H, Kim SH, Cho JM, Kim YN, Kim SS, Kim DH, Hur KY, Kim HK, Ko T, Han J, Kim HL, Kim J, Back SH, Komatsu M, Chen H, Chan DC, Konishi M, Itoh N, Choi CS, and Lee MS. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med 19: 83–92, 2013 [DOI] [PubMed] [Google Scholar]
- 49. Koga H, Kaushik S, and Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J 24: 3052–3065, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, and Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441: 880–884, 2006 [DOI] [PubMed] [Google Scholar]
- 51. Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M, Knebel A, Alessi DR, and Muqit MM. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol 2: 120080, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kosacka J, Kern M, Klöting N, Paeschke S, Rudich A, Haim Y, Gericke M, Serke H, Stumvoll M, Bechmann I, Nowicki M, and Blüher M. Autophagy in adipose tissue of patients with obesity and type 2 diabetes. Mol Cell Endocrinol 409: 21–32, 2015 [DOI] [PubMed] [Google Scholar]
- 53. Kosacka J, Nowicki M, Paeschke S, Baum P, Blüher M, and Klöting N. Up-regulated autophagy: as a protective factor in adipose tissue of WOKW rats with metabolic syndrome. Diabetol Metab Syndr 10: 13, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kovsan J, Blüher M, Tarnovscki T, Klöting N, Kirshtein B, Madar L, Shai I, Golan R, Harman-Boehm I, Schön MR, Greenberg AS, Elazar Z, Bashan N, and Rudich A. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab 96: E268–E277, 2011 [DOI] [PubMed] [Google Scholar]
- 55. Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, and Mizushima N. The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036, 2004 [DOI] [PubMed] [Google Scholar]
- 56. Kuma A, Mizushima N, Ishihara N, and Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem 277: 18619–18625, 2002 [DOI] [PubMed] [Google Scholar]
- 57. Labbé SM, Mouchiroud M, Caron A, Secco B, Freinkman E, Lamoureux G, Gélinas Y, Lecomte R, Bossé Y, Chimin P, Festuccia WT, Richard D, and Laplante M. mTORC1 is required for brown adipose tissue recruitment and metabolic adaptation to cold. Sci Rep 6: 37223, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lam T, Harmancey R, Vasquez H, Gilbert B, Patel N, Hariharan V, Lee A, Covey M, and Taegtmeyer H. Reversal of intramyocellular lipid accumulation by lipophagy and a p62-mediated pathway. Cell Death Discov 2: 16061, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Las G, Serada SB, Wikstrom JD, Twig G, and Shirihai OS. Fatty acids suppress autophagic turnover in beta-cells. J Biol Chem 286: 42534–42544, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, and Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524: 309–314, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee HY, Kim J, Quan W, Lee JC, Kim MS, Kim SH, Bae JW, Hur KY, and Lee MS. Autophagy deficiency in myeloid cells increases susceptibility to obesity-induced diabetes and experimental colitis. Autophagy 12: 1390–1403, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lee PL, Tang Y, Li H, and Guertin DA. Raptor/mTORC1 loss in adipocytes causes progressive lipodystrophy and fatty liver disease. Mol Metab 5: 422–432, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lee YH, Petkova AP, Konkar AA, and Granneman JG. Cellular origins of cold-induced brown adipocytes in adult mice. FASEB J 29: 286–299, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res 8: 3–5, 2005 [DOI] [PubMed] [Google Scholar]
- 65. Li Y, Zong WX, and Ding WX. Recycling the danger via lipid droplet biogenesis after autophagy. Autophagy 13: 1995–1997, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lim YM, Lim H, Hur KY, Quan W, Lee HY, Cheon H, Ryu D, Koo SH, Kim HL, Kim J, Komatsu M, and Lee MS. Systemic autophagy insufficiency compromises adaptation to metabolic stress and facilitates progression from obesity to diabetes. Nat Commun 5: 4934, 2014 [DOI] [PubMed] [Google Scholar]
- 67. Litwinoff EMS, Gold MY, Singh K, Hu J, Li H, Cadwell K, and Schmidt AM. Myeloid ATG16L1 does not affect adipose tissue inflammation or body mass in mice fed high fat diet. Obes Res Clin Pract 12: 174–186, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liu D, Bordicchia M, Zhang C, Fang H, Wei W, Li JL, Guilherme A, Guntur K, Czech MP, and Collins S. Activation of mTORC1 is essential for beta-adrenergic stimulation of adipose browning. J Clin Invest 126: 1704–1716, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu HY, Han J, Cao SY, Hong T, Zhuo D, Shi J, Liu Z, and Cao W. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem 284: 31484–31492, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liu K, Zhao E, Ilyas G, Lalazar G, Lin Y, Haseeb M, Tanaka KE, and Czaja MJ. Impaired macrophage autophagy increases the immune response in obese mice by promoting proinflammatory macrophage polarization. Autophagy 11: 271–284, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu Y, Takahashi Y, Desai N, Zhang J, Serfass JM, Shi YG, Lynch CJ, and Wang HG. Bif-1 deficiency impairs lipid homeostasis and causes obesity accompanied by insulin resistance. Sci Rep 6: 20453, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lizaso A, Tan KT, and Lee YH. beta-adrenergic receptor-stimulated lipolysis requires the RAB7-mediated autolysosomal lipid degradation. Autophagy 9: 1228–1243, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lu X, Altshuler-Keylin S, Wang Q, Chen Y, Henrique Sponton C, Ikeda K, Maretich P, Yoneshiro T, and Kajimura S. Mitophagy controls beige adipocyte maintenance through a Parkin-dependent and UCP1-independent mechanism. Sci Signal 11:pii: , 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lu Y, Fujioka H, Joshi D, Li Q, Sangwung P, Hsieh P, Zhu J, Torio J, Sweet D, Wang L, Chiu SY, Croniger C, Liao X, and Jain MK. Mitophagy is required for brown adipose tissue mitochondrial homeostasis during cold challenge. Sci Rep 8: 8251, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lynes MD, Shamsi F, Sustarsic EG, Leiria LO, Wang CH, Su SC, Huang TL, Gao F, Narain NR, Chen EY, Cypess AM, Schulz TJ, Gerhart-Hines Z, Kiebish MA, and Tseng YH. Cold-activated lipid dynamics in adipose tissue highlights a role for cardiolipin in thermogenic metabolism. Cell Rep 24: 781–790, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mahdaviani K, Benador IY, Su S, Gharakhanian RA, Stiles L, Trudeau KM, Cardamone M, Enríquez-Zarralanga V, Ritou E, Aprahamian T, Oliveira MF, Corkey BE, Perissi V, Liesa M, and Shirihai OS. Mfn2 deletion in brown adipose tissue protects from insulin resistance and impairs thermogenesis. EMBO Rep 18: 1123–1138, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Maixner N, Bechor S, Vershinin Z, Pecht T, Goldstein N, Haim Y, and Rudich A. Transcriptional dysregulation of adipose tissue autophagy in obesity. Physiology (Bethesda) 31: 270–282, 2016 [DOI] [PubMed] [Google Scholar]
- 78. Marcher AB, Loft A, Nielsen R, Vihervaara T, Madsen JG, Sysi-Aho M, Ekroos K, and Mandrup S. RNA-seq and mass-spectrometry-based lipidomics reveal extensive changes of glycerolipid pathways in brown adipose tissue in response to cold. Cell Rep 13: 2000–2013, 2015 [DOI] [PubMed] [Google Scholar]
- 79. Martinez-Lopez N, Athonvarangkul D, Sahu S, Coletto L, Zong H, Bastie CC, Pessin JE, Schwartz GJ, and Singh R. Autophagy in Myf5+ progenitors regulates energy and glucose homeostasis through control of brown fat and skeletal muscle development. EMBO Rep 14: 795–803, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Martinez-Lopez N, Garcia-Macia M, Sahu S, Athonvarangkul D, Liebling E, Merlo P, Cecconi F, Schwartz GJ, and Singh R. Autophagy in the CNS and periphery coordinate lipophagy and lipolysis in the brown adipose tissue and liver. Cell Metab 23: 113–127, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Masaki T, Yoshimatsu H, Chiba S, and Sakata T. Impaired response of UCP family to cold exposure in diabetic (db/db) mice. Am J Physiol Regul Integr Comp Physiol 279: R1305–R1309, 2000 [DOI] [PubMed] [Google Scholar]
- 82. May FJ, Baer LA, Lehnig AC, So K, Chen EY, Gao F, Narain NR, Gushchina L, Rose A, Doseff AI, Kiebish MA, Goodyear LJ, and Stanford KI. Lipidomic adaptations in white and brown adipose tissue in response to exercise demonstrate molecular species-specific remodeling. Cell Rep 18: 1558–1572, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. McWilliams TG, Prescott AR, Allen GF, Tamjar J, Munson MJ, Thomson C, Muqit MM, and Ganley IG. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J Cell Biol 214: 333–345, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Meng Q. and Cai D. Defective hypothalamic autophagy directs the central pathogenesis of obesity via the IkappaB kinase beta (IKKbeta)/NF-kappaB pathway. J Biol Chem 286: 32324–32332, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Menon D, Salloum D, Bernfeld E, Gorodetsky E, Akselrod A, Frias MA, Sudderth J, Chen PH, DeBerardinis R, and Foster DA. Lipid sensing by mTOR complexes via de novo synthesis of phosphatidic acid. J Biol Chem 292: 6303–6311, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mikami K, Okita N, Tokunaga Y, Ichikawa T, Okazaki T, Takemoto K, Nagai W, Matsushima S, and Higami Y. Autophagosomes accumulate in differentiated and hypertrophic adipocytes in a p53-independent manner. Biochem Biophys Res Commun 427: 758–763, 2012 [DOI] [PubMed] [Google Scholar]
- 87. Mizunoe Y, Sudo Y, Okita N, Hiraoka H, Mikami K, Narahara T, Negishi A, Yoshida M, Higashibata R, Watanabe S, Kaneko H, Natori D, Furuichi T, Yasukawa H, Kobayashi M, and Higami Y. Involvement of lysosomal dysfunction in autophagosome accumulation and early pathologies in adipose tissue of obese mice. Autophagy 13: 642–653, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Müller TD, Lee SJ, Jastroch M, Kabra D, Stemmer K, Aichler M, Abplanalp B, Ananthakrishnan G, Bhardwaj N, Collins S, Divanovic S, Endele M, Finan B, Gao Y, Habegger KM, Hembree J, Heppner KM, Hofmann S, Holland J, Küchler D, Kutschke M, Krishna R, Lehti M, Oelkrug R, Ottaway N, Perez-Tilve D, Raver C, Walch AK, Schriever SC, Speakman J, Tseng YH, Diaz-Meco M, Pfluger PT, Moscat J, and Tschöp MH. p62 links beta-adrenergic input to mitochondrial function and thermogenesis. J Clin Invest 123: 469–478, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Mullican SE, Tomaru T, Gaddis CA, Peed LC, Sundaram A, and Lazar MA. A novel adipose-specific gene deletion model demonstrates potential pitfalls of existing methods. Mol Endocrinol 27: 127–134, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, and Choi AM. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol 12: 222–230, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Namkoong S, Lee KI, Lee JI, Park R, Lee EJ, Jang IS, and Park J. The integral membrane protein ITM2A, a transcriptional target of PKA-CREB, regulates autophagic flux via interaction with the vacuolar ATPase. Autophagy 11: 756–768, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Nishimura T, Tamura N, Kono N, Shimanaka Y, Arai H, Yamamoto H, and Mizushima N. Autophagosome formation is initiated at phosphatidylinositol synthase-enriched ER subdomains. EMBO J 36: 1719–1735, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Novikoff AB, Novikoff PM, Rosen OM, and Rubin CS. Organelle relationships in cultured 3T3-L1 preadipocytes. J Cell Biol 87: 180–196, 1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nuñez CE, Rodrigues VS, Gomes FS, Moura RF, Victorio SC, Bombassaro B, Chaim EA, Pareja JC, Geloneze B, Velloso LA, and Araujo EP. Defective regulation of adipose tissue autophagy in obesity. Int J Obes (Lond) 37: 1473–1480, 2013 [DOI] [PubMed] [Google Scholar]
- 95. Ohsumi Y. Historical landmarks of autophagy research. Cell Res 24: 9–23, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ost A, Svensson K, Ruishalme I, Brännmark C, Franck N, Krook H, Sandström P, Kjolhede P, and Strålfors P. Attenuated mTOR signaling and enhanced autophagy in adipocytes from obese patients with type 2 diabetes. Mol Med 16: 235–246, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Oude Elferink RP, Harms E, Strijland A, and Tager JM. The intralysosomal pH in cultured human skin fibroblasts in relation to cystine accumulation in patients with cystinosis. Biochem Biophys Res Commun 116: 154–161, 1983 [DOI] [PubMed] [Google Scholar]
- 98. Pickles S, Vigié P, and Youle RJ. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr Biol 28: R170–R185, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pires KM, Buffolo M, Schaaf C, David Symons J, Cox J, Abel ED, Selzman CH, and Boudina S. Activation of IGF-1 receptors and Akt signaling by systemic hyperinsulinemia contributes to cardiac hypertrophy but does not regulate cardiac autophagy in obese diabetic mice. J Mol Cell Cardiol 113: 39–50, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Polak P, Cybulski N, Feige JN, Auwerx J, Rüegg MA, and Hall MN. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab 8: 399–410, 2008 [DOI] [PubMed] [Google Scholar]
- 101. Qian M, Fang X, and Wang X. Autophagy and inflammation. Clin Transl Med 6: 24, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Reue K. A thematic review series: lipid droplet storage and metabolism: from yeast to man. J Lipid Res 52: 1865–1868, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ro SH, Jung CH, Hahn WS, Xu X, Kim YM, Yun YS, Park JM, Kim KH, Seo M, Ha TY, Arriaga EA, Bernlohr DA, and Kim DH. Distinct functions of Ulk1 and Ulk2 in the regulation of lipid metabolism in adipocytes. Autophagy 9: 2103–2114, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Rockenfeller P, Koska M, Pietrocola F, Minois N, Knittelfelder O, Sica V, Franz J, Carmona-Gutierrez D, Kroemer G, and Madeo F. Phosphatidylethanolamine positively regulates autophagy and longevity. Cell Death Differ 22: 499–508, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, and Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature 454: 232–235, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sarraf SA. and Youle RJ. Parkin mediates mitophagy during beige-to-white fat conversion. Sci Signal 11, 2018 [DOI] [PubMed] [Google Scholar]
- 107. Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, Schrier D, Falb D, Kirkland JL, Wagers AJ, and Tseng YH. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci U S A 108: 143–148, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, Kundu M, Opferman JT, Cleveland JL, Miller JL, and Ney PA. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A 104: 19500–19505, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scimè A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, and Spiegelman BM. PRDM16 controls a brown fat/skeletal muscle switch. Nature 454: 961–967, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Settembre C, Fraldi A, Medina DL, and Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 14: 283–296, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Shahnazari S, Namolovan A, Mogridge J, Kim PK, and Brumell JH. Bacterial toxins can inhibit host cell autophagy through cAMP generation. Autophagy 7: 957–965, 2011 [DOI] [PubMed] [Google Scholar]
- 112. Shatz O, Holland P, Elazar Z, and Simonsen A. Complex relations between phospholipids, autophagy, and neutral lipids. Trends Biochem Sci 41: 907–923, 2016 [DOI] [PubMed] [Google Scholar]
- 113. Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, and Flier JS. TLR4 links innate immunity and fatty acid-induced insulin resistance. J Clin Invest 116: 3015–3025, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Shiba-Fukushima K, Imai Y, Yoshida S, Ishihama Y, Kanao T, Sato S, and Hattori N. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep 2: 1002, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Shibata M, Yoshimura K, Furuya N, Koike M, Ueno T, Komatsu M, Arai H, Tanaka K, Kominami E, and Uchiyama Y. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem Biophys Res Commun 382: 419–423, 2009 [DOI] [PubMed] [Google Scholar]
- 116. Shimizu I, Aprahamian T, Kikuchi R, Shimizu A, Papanicolaou KN, MacLauchlan S, Maruyama S, and Walsh K. Vascular rarefaction mediates whitening of brown fat in obesity. J Clin Invest 124: 2099–2112, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Sica V, Galluzzi L, Bravo-San Pedro JM, Izzo V, Maiuri MC, and Kroemer G. Organelle-specific initiation of autophagy. Mol Cell 59: 522–539, 2015 [DOI] [PubMed] [Google Scholar]
- 118. Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, and Czaja MJ. Autophagy regulates lipid metabolism. Nature 458: 1131–1135, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, and Czaja MJ. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest 119: 3329–3339, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Slobodkin MR. and Elazar Z. The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy. Essays Biochem 55: 51–64, 2013 [DOI] [PubMed] [Google Scholar]
- 121. Slutsky N, Vatarescu M, Haim Y, Goldstein N, Kirshtein B, Harman-Boehm I, Gepner Y, Shai I, Bashan N, Blüher M, and Rudich A. Decreased adiponectin links elevated adipose tissue autophagy with adipocyte endocrine dysfunction in obesity. Int J Obes (Lond) 40: 912–920, 2016 [DOI] [PubMed] [Google Scholar]
- 122. Soussi H, Clément K, and Dugail I. Adipose tissue autophagy status in obesity: expression and flux—two faces of the picture. Autophagy 12: 588–589, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Soussi H, Reggio S, Alili R, Prado C, Mutel S, Pini M, Rouault C, Clément K, and Dugail I. DAPK2 downregulation associates with attenuated adipocyte autophagic clearance in human obesity. Diabetes 64: 3452–3463, 2015 [DOI] [PubMed] [Google Scholar]
- 124. Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, and Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci U S A 106: 17049–17054, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Stolz A, Ernst A, and Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 16: 495–501, 2014 [DOI] [PubMed] [Google Scholar]
- 126. Stotland A. and Gottlieb RA. alpha-MHC MitoTimer mouse: in vivo mitochondrial turnover model reveals remarkable mitochondrial heterogeneity in the heart. J Mol Cell Cardiol 90: 53–58, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Sun N, Malide D, Liu J, Rovira II, Combs CA, and Finkel T. A fluorescence-based imaging method to measure in vitro and in vivo mitophagy using mt-Keima. Nat Protoc 12: 1576–1587, 2017 [DOI] [PubMed] [Google Scholar]
- 128. Sustarsic EG, Ma T, Lynes MD, Larsen M, Karavaeva I, Havelund JF, Nielsen CH, Jedrychowski MP, Moreno-Torres M, Lundh M, Plucinska K, Jespersen NZ, Grevengoed TJ, Kramar B, Peics J, Hansen JB, Shamsi F, Forss I, Neess D, Keipert S, Wang J, Stohlmann K, Brandslund I, Christensen C, Jørgensen ME, Linneberg A, Pedersen O, Kiebish MA, Qvortrup K, Han X, Pedersen BK, Jastroch M, Mandrup S, Kjær A, Gygi SP, Hansen T, Gillum MP, Grarup N, Emanuelli B, Nielsen S, Scheele C, Tseng YH, Faergeman NJ, and Gerhart-Hines Z. Cardiolipin synthesis in brown and beige fat mitochondria is essential for systemic energy homeostasis. Cell Metab 28: 159.e11–174.e11, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Taylor D. and Gottlieb RA. Parkin-mediated mitophagy is downregulated in browning of white adipose tissue. Obesity (Silver Spring) 25: 704–712, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Tran CM, Mukherjee S, Ye L, Frederick DW, Kissig M, Davis JG, Lamming DW, Seale P, and Baur JA. Rapamycin blocks induction of the thermogenic program in white adipose tissue. Diabetes 65: 927–941, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Tsukada M. and Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333: 169–174, 1993 [DOI] [PubMed] [Google Scholar]
- 132. Vance JE. and Vance DE. Phospholipid biosynthesis in mammalian cells. Biochem Cell Biol 82: 113–128, 2004 [DOI] [PubMed] [Google Scholar]
- 133. Virgin HW. and Levine B. Autophagy genes in immunity. Nat Immunol 10: 461–470, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Wang L, Zhou J, Yan S, Lei G, Lee CH, and Yin XM. Ethanol-triggered lipophagy requires SQSTM1 in AML12 hepatic cells. Sci Rep 7: 12307, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Wikstrom JD, Mahdaviani K, Liesa M, Sereda SB, Si Y, Las G, Twig G, Petrovic N, Zingaretti C, Graham A, Cinti S, Corkey BE, Cannon B, Nedergaard J, and Shirihai OS. Hormone-induced mitochondrial fission is utilized by brown adipocytes as an amplification pathway for energy expenditure. EMBO J 33: 418–436, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Xiao Y, Deng Y, Yuan F, Xia T, Liu H, Li Z, Liu Z, Ying H, Liu Y, Zhai Q, Chen S, and Guo F. ATF4/ATG5 signaling in hypothalamic proopiomelanocortin neurons regulates fat mass via affecting energy expenditure. Diabetes 66: 1146–1158, 2017 [DOI] [PubMed] [Google Scholar]
- 137. Yang L, Li P, Fu S, Calay ES, and Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab 11: 467–478, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Yeh WC, Bierer BE, and McKnight SL. Rapamycin inhibits clonal expansion and adipogenic differentiation of 3T3-L1 cells. Proc Natl Acad Sci U S A 92: 11086–11090, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Yoon MS, Zhang C, Sun Y, Schoenherr CJ, and Chen J. Mechanistic target of rapamycin controls homeostasis of adipogenesis. J Lipid Res 54: 2166–2173, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Yoshizaki T, Kusunoki C, Kondo M, Yasuda M, Kume S, Morino K, Sekine O, Ugi S, Uzu T, Nishio Y, Kashiwagi A, and Maegawa H. Autophagy regulates inflammation in adipocytes. Biochem Biophys Res Commun 417: 352–357, 2012 [DOI] [PubMed] [Google Scholar]
- 141. Zaffagnini G. and Martens S. Mechanisms of selective autophagy. J Mol Biol 428: 1714–1724, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, and Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A 106: 19860–19865, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Zhou L, Zhang J, Fang Q, Liu M, Liu X, Jia W, Dong LQ, and Liu F. Autophagy-mediated insulin receptor down-regulation contributes to endoplasmic reticulum stress-induced insulin resistance. Mol Pharmacol 76: 596–603, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]