SUMMARY
A defining feature of early embryogenesis is the transition from maternal to zygotic control. This transition requires embryo-wide zygotic genome activation (ZGA), but the extent of spatiotemporal coordination of ZGA between individual cells is unknown. Multiple interrelated parameters, including elapsed time, completed cycles of cell division, and cell size may impact ZGA onset; however, the principle determinant of ZGA during vertebrate embryogenesis is debated. Here, we perform single-cell imaging of large-scale ZGA in whole-mount Xenopus embryos. We find a striking new spatiotemporal pattern of ZGA whose onset tightly correlates with cell size but not with elapsed time or number of cell divisions. Further, reducing cell size induces premature ZGA, dose-dependently. We conclude that large-scale ZGA is not spatially uniform and that its onset is determined at the single-cell level, primarily by cell size. Our study suggests that spatial patterns of ZGA onset may be a common feature of embryonic systems.
Keywords: Zygotic genome activation, single-cell, spatiotemporal pattern, cell size, nascent transcription, early embryogenesis, sizer, timer, counter, miniature embryo
eTOC
Zygotic genome activation (ZGA) is a hallmark of early embryonic development. Chen et al. imaged nascent transcripts at the single-cell level in Xenopus whole-mount embryos, discovering a stereotypic spatiotemporal pattern of large-scale ZGA. This patterned onset is dependent on cells reaching a threshold size, not time or cell cycle count.
INTRODUCTION
Following fertilization, metazoan embryogenesis proceeds autonomously, undergoing multiple rounds of cell division in the absence of zygotic transcription. Early cell divisions are governed by maternal factors, including mRNAs and proteins, loaded into the egg. After a defined interval, cleavage-stage embryos undergo zygotic genome activation (ZGA), initiating the transcription of hundreds to thousands of genes in a period called the maternal-to-zygotic transition (MZT) (Jukam et al., 2017; Lee et al., 2014; Schier, 2007; Tadros and Lipshitz, 2009; Zhang et al., 2017). Activation of zygotic gene expression is essential for gastrulation, germ-layer specification and cell differentiation, and dysregulation of ZGA impairs development (Lee et al., 2014). Although ZGA is a process universal to early embryo development, the timing of ZGA varies dramatically between species. For example, in human embryos widespread ZGA occurs at the third cleavage (about 2 days post-fertilization, pf), whereas in model vertebrate embryos such as zebrafish and Xenopus, it occurs after > 10 rounds of cell division (at about 3 hours and 7 hours pf, respectively) (Jukam et al., 2017). Despite an essential role in development, the mechanisms underlying the systems level onset of ZGA within vertebrate embryos are still poorly understood.
Studies in model organisms have suggested three broad paradigms for determining ZGA onset: a clock or a timer, a cell cycle counter, and a cell sizer (Lee et al., 2014; Lu et al., 2009; Newport and Kirschner, 1982a; Satoh and Ikegami, 1981). A timer hypothesis proposes that embryos measure elapsed time by steady accumulation of transcriptional activators required for ZGA. In zebrafish, the translation of mRNAs encoding pluripotency factors, including Pou5f1 (Oct4), Sox and Nanog family members, is triggered following fertilization and knockdown of these factors delays ZGA (Lee et al., 2013; Leichsenring et al., 2013);. Translation of p300 may also constitute a timer (Chan et al., 2018). Independent of absolute time, it has been suggested that embryos can enumerate a defined number of divisions prior to initiating early developmental events (Satoh and Ikegami, 1981), constituting a counter.
Conversely, ZGA can occur independently of developmental timing or number of cell divisions. A cell sizer model postulates that ZGA is triggered by cells achieving a threshold size or DNA:cytoplasm ratio. In cleavage-stage vertebrate embryos, cells divide without growing and therefore cell volume reduces with each division during early development. Because DNA content is constant within each blastomere, the decrease in cell volume causes an increase in DNA concentration (or DNA:cytoplasm ratio). In the syncytial fly blastoderm, a single cell containing many nuclei within a shared cytoplasm, DNA:cytoplasm ratio – historically termed nucleocytoplasmic ratio (N:C) – increases due to rounds of nuclear division. Achievement of a DNA:cytoplasm ratio threshold correlates to the onset of events associated with the mid-blastula transition (MBT) in flies (Di Talia et al., 2013; Edgar et al., 1986; Lu et al., 2009), frogs (Amodeo et al., 2015; Newport and Kirschner, 1982a, b; Prioleau et al., 1994) and zebrafish (Dekens et al., 2003; Kane and Kimmel, 1993).
This DNA:cytoplasm ratio hypothesis is supported by studies within vertebrates in which DNA concentration was manipulated, via alteration of ploidy or by DNA injection, leading to premature or delayed induction of MBT activities (Jevtic and Levy, 2017; Kane and Kimmel, 1993; Newport and Kirschner, 1982a, b). Similarly, manipulation of DNA content within fly embryos shifts the onset of zygotic transcription and other MBT activities (Di Talia et al., 2013; Edgar et al., 1986; Lu et al., 2009). However, it may be challenging to interpret the data from haploids and polyploids or embryos containing extrachromosomal DNA, because their developmental timing can be dramatically altered (Edgar et al., 1986). Although there is widespread acceptance that a high DNA:cytoplasm ratio contributes to ZGA onset, to date there is no direct evidence that reaching a minimum cell size regulates the onset of zygotic gene expression. Furthermore, use of whole-embryo DNA:cytoplasm ratio as a predictor of the onset of genome activation overlooks the large variation in cell sizes (single-cell DNA:cytoplasm ratio) present in a single blastula embryo, such as those of Xenopus. Whole-embryo DNA:cytoplasm ratio would predict that all cells become activated relatively synchronously (Newport and Kirschner, 1982b), whereas a model based on a threshold single-cell DNA:cytoplasm ratio would predict distinct spatiotemporal pattern of ZGA (Figure 1A).
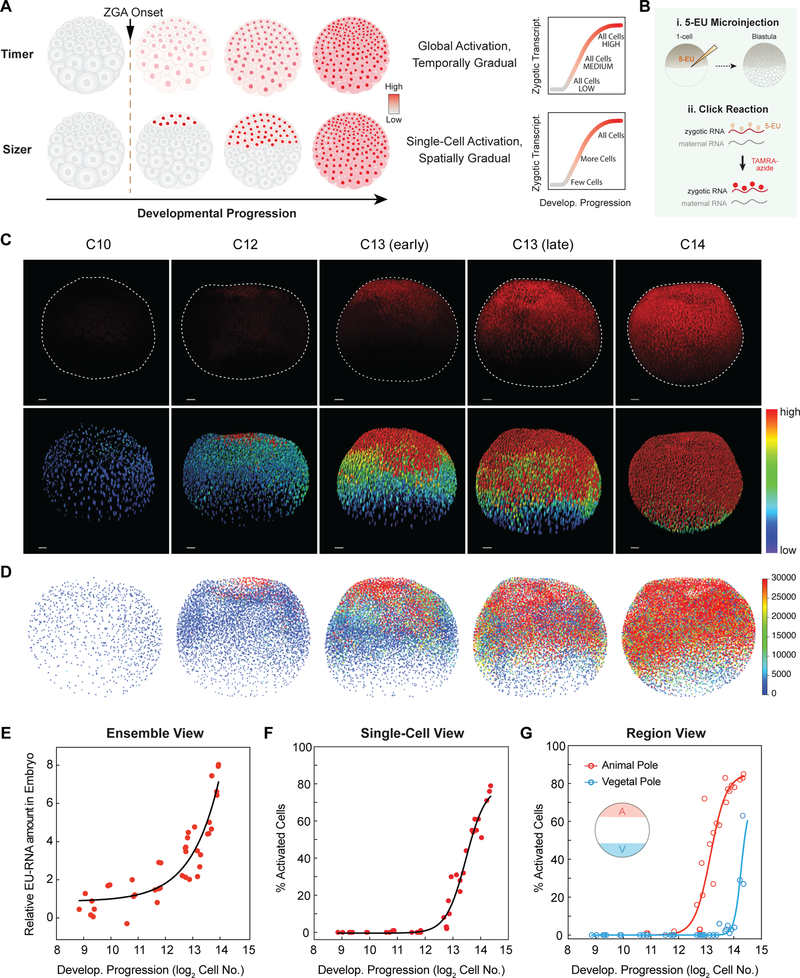
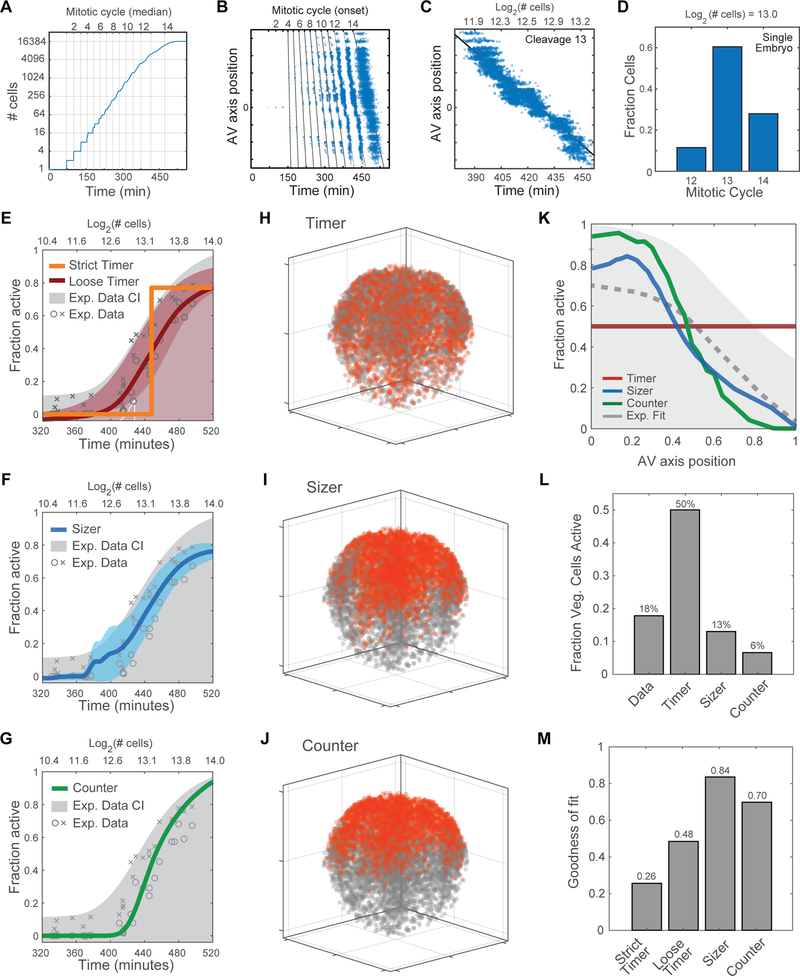
Figure 1. Spatiotemporal Patterning of Zygotic Genome Activation at A Single Cell Level in Xenopus Early Embryogenesis.
(A) Hypotheses for patterning of genome activation in blastula embryos based on a timer and sizer model, respectively. Color scale indicates low (gray) to high (red) transcription.
(B) Schematic of metabolic labeling of nascent zygotic transcripts in early Xenopus embryos.
(C) Confocal images of nascent EU-RNA (upper panel) and heatmap of its intensity (lower panel) in individual nucleus for blastula stage embryos from embryonic cleavage 10 (C10) to 14 (C14). Color scale indicates original EU-RNA intensity from low (blue) to high (red), without background subtraction. AP, animal pole; VP, vegetal pole. Dashed line demarcates individual embryos. Scale bar, 100 μm.
(D) 3D reconstruction and heatmap of nascent EU-RNA amount with background subtraction in individual nucleus of blastula embryos. Color scale indicates low (blue) to high (red) transcription. No significant EU-RNA signal until C12.
(E-G) Ensemble view (E), single-cell view (F) and regional view (G) of ZGA. Each point indicates one embryo. Exponential (E) or sigmoidal (F and G) fit to data as visual aid.
(E) Ensemble view of ZGA: total nascent EU-RNA amount with background subtraction within entire blastula embryos.
(F) Single-cell view of ZGA: percentage of cells above the threshold EU-RNA amount in nucleus of each blastula embryo.
(G) Regional view of ZGA: percentage of cells above the threshold EU-RNA amount in nucleus of the animal (A, red) and vegetal (V, blue) pole in each blastula embryo. Animal pole and vegetal pole at 200 μm depth from the top and the bottom, respectively.
See also Figure S1.
Within vertebrate embryos, DNA:cytoplasm ratio dependent regulation of ZGA is proposed to center on the presence of a transcriptional inhibitor whose level or activity is titrated away by DNA as cells reduce in volume. Potential inhibitors include core histones, which are responsible for packaging DNA into repressive chromatin that blocks transcription (Almouzni and Wolffe, 1995; Amodeo et al., 2015; Joseph et al., 2017), and DNA replication factors that restrict transcription activation by promoting DNA duplication in cell cycles of short duration (Collart et al., 2013). Also, by reaching a threshold size or DNA:cytoplasm ratio, the cell cycle appears to elongate, which may also contribute to ZGA onset (Collart et al., 2013; Kane and Kimmel, 1993; Wang et al., 2000), although a cause-effect relationship varies between species (Blythe and Wieschaus, 2015; Zhang et al., 2017).
At the embryo level, prior work using metabolic labeling or sequencing have demonstrated gradual accumulation of zygotic mRNAs at the onset of genome activation (Collart et al., 2014; Heyn et al., 2014; Paranjpe et al., 2013; Peshkin et al., 2015; Yanai et al., 2011). However, the degree of temporal and spatial coordination of ZGA between individual cells has been unknown. Gradual ZGA onset could be explained by incremental increase of transcription, synchronously in all cells, creating a uniform pattern of onset (Figure 1A). Alternatively, gradual onset could be spatially, in which first a subset of cells highly induces transcription, followed by ZGA in additional nuclei, producing a stereotypic spatial pattern.
Several techniques have been used to detect zygotic gene expression during early embryo development. These include single molecule fluorescent in situ hybridization (smFISH) in fixed samples (Stapel et al., 2017) and MS2 tagging in live embryos (Campbell et al., 2015; Garcia et al., 2013). A limitation of these methods is that they only detect single genes, not large-scale genome activation, and are not compatible with imaging through entire whole-mount embryos, particularly those from large vertebrates. Additionally, gene profiling studies of early embryogenesis that isolate RNA from whole embryos and perform sequencing are largely blind to cell-to-cell variability and spatial organization of ZGA (Collart et al., 2014; Heyn et al., 2014; Paranjpe et al., 2013; Peshkin et al., 2015; Yanai et al., 2011). Recent studies using single-cell RNA sequencing (scRNA-seq) have attempted to dissociate cells and later computationally reconstruct three-dimensional patterns of gene expression (Briggs et al., 2018; Farrell et al., 2018; Karaiskos et al., 2017; Satija et al., 2015; Wagner et al., 2018). However, the spatial position of individual cells cannot be localized with high confidence in a computationally reconstructed embryo containing thousands of cells. Moreover, prior approaches have not revealed how cell size, elapsed time, and cycles of cell divisions intersect to control the timing of ZGA. New approaches are required to gain fresh insights into mechanisms that regulate large-scale ZGA at the single-cell level.
In this study, we leveraged transcriptional imaging of whole-mount embryos to define the spatiotemporal patterns of large-scale ZGA in a model vertebrate. We injected 5-ethynyl uridine (EU) into fertilized eggs from Xenopus laevis to label nascent zygotic transcripts for imaging large-scale ZGA in space and time. We chose to image transcription within cleavage-stage embryos from Xenopus laevis for two reasons. First, at the onset of large-scale ZGA, a single embryo contains thousands of cells whose volumes vary by greater than 100-fold between the smallest cells at the animal pole and the largest cells at the vegetal pole. This heterogeneity in cell size allowed us to distinguish between timer and sizer models for ZGA without perturbing the embryo. Second, use of Xenopus embryos allowed us to manipulate cell size at the 1-cell stage without significantly affecting early embryo development, which enabled us to distinguish sizer and counter models for ZGA.
We discovered that the onset of large-scale ZGA is not uniform across a vertebrate embryo. The embryo undergoes a stereotypic pattern of single-cell activation in which large-scale transcription turns on first in the smallest cells at the animal pole and is significantly delayed in the largest cells located in the vegetal pole. A gradual onset of ZGA as a function of developmental progression is explained by a gradual spatial induction of ZGA in single cells, but not by globally ramping-up of embryo-wide transcriptional output. This spatial pattern of activation and our computational modeling falsify both a simple timer model for ZGA regulation and a regulatory model based on embryo-wide DNA:cytoplasm ratio. Our data instead strongly supports a model in which transcriptional repression is relieved by reaching a threshold cell size and single-cell DNA:cytoplasm ratio. Experiments in miniaturized embryos demonstrate that the pattern of genome activation cannot be explained by a simple cell cycle counter model and that cell size is sufficient to regulate the onset of large-scale ZGA in a dose-dependent manner.
RESULTS
Visualizing Large-scale Nascent Zygotic Transcription at the Single-cell Level
To detect nascent zygotic genes in individual cells in blastula embryos, we microinjected 1-cell Xenopus laevis embryos with 5-ethynyl uridine (5-EU). This analog of uridine is incorporated into newly transcribed RNAs, and contains an alkyne group that enables its conjugation with fluorescent molecules or biotin through a copper(I)-catalyzed alkyne-azide cycloaddition or ‘click reaction’ (Jao and Salic, 2008) (Figure 1B). The EU-injected embryos developed normally up to mid-blastula transition (MBT) (Figure S1A). We collected and fixed them beginning at the 8th embryonic division through the 15th embryonic division (C8–C15). To visualize nascent RNAs, we conjugated EU-RNA with tetramethylrhodamine (TAMRA)-azide (Figure 1B), enabling imaging of large-scale transcription in individual blastomeres. To stage embryos precisely, we used 3D image segmentation and automated object counting to define the number of nuclei (Joseph et al., 2017). In this strategy, developmental progression is defined by log2 of the number of nuclei within an embryo.
To validate that our assay specifically visualizes nascent zygotic transcription, we performed an array of experiments (Figure S1). As expected for zygotic transcripts, we could not detect significant new gene expression through the first ten embryonic cleavages (Figure S1B). EU-RNA was first detected in nuclei of embryos that had developed to ~ 4000-cell stage (log2 cell no. = 12), and thereafter continued to increase (Figure 1C and 1D). Inhibition of RNA polymerase II (RNAPII) using a low dose of α-amanitin (Herberg et al., 2015) almost entirely eliminated nascent transcription detectable by imaging at ~ 8000-cell stage (log2 cell no. = 13); it reduced EU-RNA levels by more than 80% (Figures 2A and 2B). Additionally, α-amanitin treatment eliminated nascent transcripts that are normally detected at ZGA onset (Figure 2C). Raising the dose, which also inhibits RNAPIII, did not significantly reduce transcription further, suggesting that EU-RNA imaging predominantly measures RNA polymerase II mediated transcription.
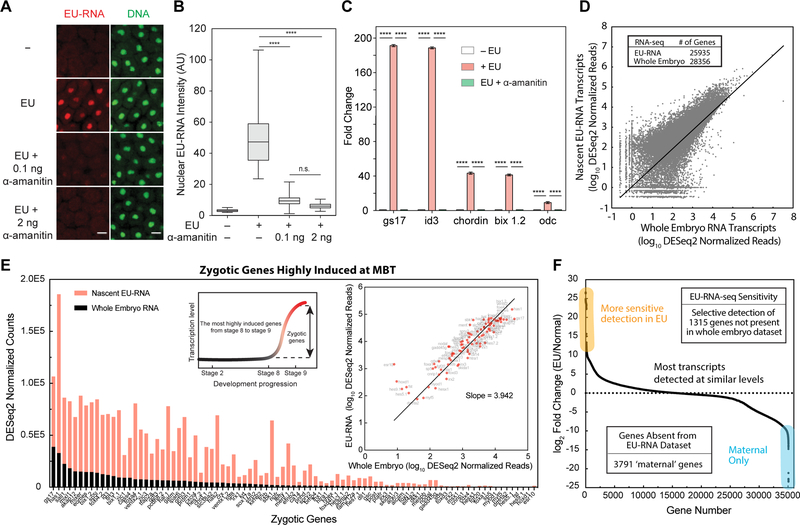
Figure 2. Nascent Zygotic Transcription During Early Embryogenesis.
(A) Confocal images of EU-RNA (red) and DNA (green) in the animal pole of embryos at ~ C13 with or without EU or indicated amount of α-amanitin. Scale bars, 20 μm.
(B) Box and whiskers plot of EU-RNA intensity in individual nunclei at the animal pole of embryos (N > 60 cells for each group). Statistical difference determined by one-way ANOVA. ****, p < 0.0001; n.s., not significant.
(C) qRT-PCR quantification for indicated genes with or without EU or α-amanitin. Statistical difference determined by one-way ANOVA. ****, p < 0.0001.
(D) RNA-seq for nascent EU-RNA and total RNA isolated from EU-injected and normal embryos, respectively, at ~ 9 hpf (mid-ZGA). Table shows number of genes with DESeq2 normalized reads > 10.
(E) Levels of highly induced zygotic genes from RNA-seq for nascent EU-RNA and total RNA. The inset schematic shows selection of highly induced zygotic genes. The inset plot shows correlation of highly induced genes between nascent EU-RNA and total RNA. Slope indicates ~ 4-fold higher read depth in nascent EU-RNA-seq. Linear fitting.
(F) Levels of all genes ranked by log2 fold change. The genes more sensitive to detection in EU are highlighted in yellow and the exclusively maternal genes are shown in light blue. Table shows number of genes with DESeq2 normalized reads > 10.
To validate that EU-RNA imaging provides a direct readout of zygotic transcription, we sought to identify the nascent transcriptome using RNA-seq. We biotinylated EU-RNA and purified it to generate cDNA libraries for sequencing. We found over 25,000 nascent genes in this EU-RNA dataset at mid-ZGA (~ 9 hours post-fertilization) (Figure 2D). The nascent transcriptome dataset captures nearly 90% of the transcripts present in the whole-embryo dataset and with similar or better read-depth, indicating that our EU-RNA imaging is truly representative of wide-spread zygotic transcription. Furthermore, of the known zygotic genes that are most highly induced at MBT (Collart et al., 2014; Yanai et al., 2011), we detected all of them and at much higher levels than in the whole-embryo dataset (Figure 2E). These results show that our labeling captures the nascent zygotic transcriptome. We found ~ 4-fold higher sensitivity for nascent transcripts in the EU-RNA dataset compared to the whole embryo dataset (Figure 2E), and as excepted we did not detect thousands of maternal-only transcripts in the nascent dataset (Figures 2F). We also used quantitative reverse transcription - polymerase chain reaction (qRT-PCR) to validate nascent transcripts that are highly expressed at the onset of ZGA; in the absence of EU these genes were not detectable (Figure 2C), suggesting that the click reaction is specific. Together, these results suggest that EU-labeling provides a visual readout of bona fide nascent zygotic transcripts.
Our large-scale transcriptional imaging method has several advantages over approaches that rely on purifying RNA from homogenized embryos. First, it provides a single-cell and single-nucleus view of large-scale transcriptional onset. Due to the rapid (~ 25 min) period of interphase, RNAs largely accumulate in the nucleus (Kimelman et al., 1987). This feature allowed us to distinguish between a cell’s current transcriptional activity (nuclear EU-RNA signal) and its transcriptional history (total EU-RNA present in cell) (Figure S1C). Traditional gene profiling approaches measure only the accumulation of transcripts but not the transcriptional activity at each cell division or developmental stage. Second, methods for detecting isolated bulk RNAs often do not take into account that the amount of DNA template increases exponentially in an embryo, nearly doubling after every embryonic division. Importantly, in our method we measure transcription inside single cells and single nuclei that have at most doubled their DNA content during interphase.
By quantifying total EU-RNA accumulated in an embryo we first detected large-scale zygotic gene expression at 4000–8000 cell stages (log2 cell no. = 12–13) (Figures 1E, S1D and S1E), consistent with previous studies (Newport and Kirschner, 1982a, b). By setting a threshold for minimal level of transcript accumulation, we calculated the percentage of cells activated within an embryo during early blastula development. Consistent with the ensemble view, we found little to no large-scale zygotic transcription until embryos had reached 4000-cell stage, whereas nearly 80% of cells became activated a few hours later at 16000-cell stage (log2 cell no. = 14). Importantly, our approach yields bulk data consistent with conventional metabolic labeling while also generating new insights on spatiotemporal patterning of ZGA at the single-cell level.
Onset of Large-Scale ZGA is Not Spatially Homogenous: The Earliest Transcribing Cells Are Localized to the Animal Pole
To determine the spatiotemporal pattern of large-scale ZGA, we imaged whole-mount EU-injected embryos containing ~ 500 cells (29) – ~ 23000 (214.5) cells (320 – 540 minutes post-fertilization, mpf). A timer model would predict homogenous activation of all cells at one timepoint, such as 6 hours 20 minutes at 23°C (embryonic cleavage 12), whereas a sizer model would predict single-cell activation and that distinct regions of the embryo would turn on at different times (Figure 1A). We first detected EU-RNAs in a small fraction of cells of the animal pole in embryos that had developed to ~ 4000 cells (212), which occurs soon after embryonic cleavage 12 (Figures 1C and S1F–S1I). To account for the effect of nuclear volume on EU-RNA intensity, we calculated the total integrated amount of nascent transcript (EU-RNA) in every nucleus of each embryo (Figures 1D and S1G). Importantly, we found a nearly identical trend when plotting nuclear EU-RNA intensity or amount – large scale ZGA occurs first in cells in the animal pole, then in more medial regions and finally in cells in the deepest vegetal regions (Figures 1C, 1D, 1F, 1G and S1G–S1I). These blastomeres are not yet motile because the embryo has not yet reached gastrulation; therefore, this pattern does not arise from cell movements. This stereotyped spatial pattern of activation falsifies a simple global timer hypothesis for explaining the onset of large-scale ZGA in Xenopus embryos.
Taken together, we found that the decision to activate large-scale zygotic gene expression occurs at the single-cell level and is not a synchronous embryo-wide event. Earlier studies showed that non-synchronous gene activation occurs at the single-gene level in other model organisms (Boettiger and Levine, 2009; Stapel et al., 2017). Our findings support a new view that transcriptional activation of many thousands of genes is determined at the level of an individual cell, independent of time elapsed since fertilization. Furthermore, we uncover a previously unknown spatial pattern of transcriptional activation at the mid-blastula transition: large-scale ZGA occurs first in cells localized to the animal pole and is delayed by up to two hours in vegetal cells (Figure S1G).
Cells That Achieve A Threshold Size Initiate Large-Scale Zygotic Transcription
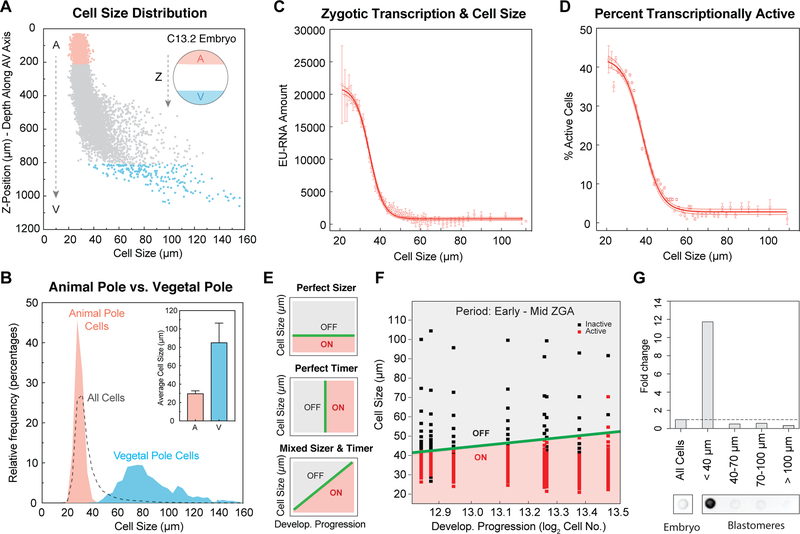
During early development, a single Xenopus embryo contains a gradient of cell sizes, and cell dimensions reduce concomitant with developmental progression (Figure S2A). At the onset of ZGA, a single embryo contains a greater than 100-fold range of cell volumes between the smallest cells at the animal pole and the largest cells at the vegetal pole (Newport and Kirschner, 1982a). This broad gradient of cell size led us to ask whether a cell sizer model could explain the pattern of single-cell genome activation. To systematically estimate the sizes of individual cells in embryos, we measured the average vector distance between the centroid of nucleus and its closest three nuclei, which provides an approximation of cell diameter. Cells are smallest and most homogenous in size in the animal pole and display the largest sizes and variation in the vegetal pole (Figures 3A and 3B), Intriguingly, we found that the onset of large-scale ZGA correlated with cells reaching a threshold size, as measured by nuclear EU-RNA accumulation (Figures 3C and S2B). Cells larger than 50 μm appear inactive: their average nuclear EU-RNA amount is ~ 1000 arbitrary units (A.U.), which is 20-fold less than the amount of EU-RNA in the nucleus of the smallest cells (25 mm cell, > 20,000 A.U.). Cells smaller than 45 μm show increasing accumulation of EU-RNA. By setting a minimum threshold for transcriptional activation we found that cells appear largely inactive until they achieve ~ 45 μm cell diameter (Figures 3D and S2C). Comparing cells that are 45 μm vs. 25 μm, there is a 1.8-fold difference in cell diameter, or about 6-fold difference in cell volume. Across this size difference, a cell displays ~ 20-fold increase in nuclear EU-RNA amount (Figure 3C). The cell cycle state of blastomeres includes a mixture of early, mid and late interphase as well as mitosis, which is why the percent active less than one hundred percent (Fig 3D). By analyzing cells in late interphase, excluding mitotic cells by averaging only those that have high nuclear sphericity, we observed much higher levels of nascent transcripts (Figure S2B) and nearly 100% of smallest cells are activated (Figure S2C). Importantly, the cell size threshold for activation is unchanged (Figure 3D and S2C). Thus, these data strongly support a sizer-based model for transcriptional regulation.
Figure 3. Activation of Zygotic Transcription Depends on Achieving a Critical Cell Size.
(A) Cell size distribution along the AV axis in a single embryo at ~ C13.2. The dashed arrows indicate the z-position from the animal pole (A, red) to vegetal pole (V, blue).
(B) Histogram of cell size distribution in the animal pole (red), vegetal pole (blue) and whole embryos (dashed line) at ~ C13.2. Data from 5 embryos binned by cell size in 4 μm increments. Inset plot shows average cell size in the animal pole (A) and vegetal pole (V); error bar, standard deviation.
(C and D) Nuclear EU-RNA amount (C) and percentage of transcriptionally active cells (D) as a function of cell diameter in embryos at ~ C13.2. Data from 5 embryos binned in 1 μm increments of cell diameter and represented as mean ± 95% confidence interval (CI). Sigmoidal fit to data with 95% CI band.
(E) Predicted decision boundaries for ZGA with a perfect sizer (top), perfect timer (middle) and mixed sizer and timer (bottom). Gray, inactive; red, active; green line, decision boundary for ZGA.
(F) Logistic regression boundary decision for ZGA as a function of cell size and developmental progression. Each point is a bin of 200 cells for each of 8 embryos at ~ C12.8-C13.5. Binary activation based on threshold amount EU-RNA. Black square, inactive cells; red square, active cells; green line, decision boundary for ZGA.
(G) Dot blot for accumulated nascent EU-RNA in blastomeres at various size ranges isolated and sorted from embryos at ~ C13. The fold change was normalized to whole embryos.
See also Figures S2 and S3.
The significant delay in the onset of large-scale ZGA between cells near the vegetal pole compared to cells near the animal pole can be explained by cell size. In an 8000-cell embryo with partial ZGA onset, the average cell size in the animal pole is ~ 29.5 μm, and the average cell size in the vegetal pole is ~ 85 μm (Figure 3B). This 2.88-fold difference in average cell diameter equates to a ~ 24-fold difference in spherical cell volume. In cleavage stage embryos, each time that a cell divides symmetrically, its volume is reduced by half, and therefore four consecutive divisions would a generate 16-fold (24) reduction in cell volume. Each blastomere from embryonic cleavages two to twelve divides on average approximately every thirty minutes (Anderson et al., 2017; Newport and Kirschner, 1982a). Therefore, it would take about 4.5 cell cycles (approximately 135 minutes) for the vegetal cells that are 24-fold larger than those in the animal pole to reach the critical cell size threshold. Based on this prediction, there should be a 135-min delay between ZGA onset at the two poles. Consistent with a cell sizer model, we observed at least a ~ 130-min delay of activation of cells in deepest vegetal regions compared to cells in the animal pole (Figure 1D and S1G).
A cell sizer is based on the idea that at a threshold DNA:cytoplasm ratio there is sufficient titration of repressive factors by DNA to allow transcriptional onset. However, it is necessary to distinguish between DNA:cytoplasm ratio of the whole embryo and that of single cells. Unlike a fly embryo which is a 1-cell syncytium at mitosis 13, the Xenopus blastula is cellularized and thus the cytoplasm is not continuous. Additionally, a Xenopus embryo contains broad variation in cell size and thus in DNA:cytoplasm ratio (Figures S2G and S2H). Based on our observations of spatially patterned single-cell activation, i.e., new transcription accumulates first in the small cells in the animal pole and later in large cells of the vegetal pole (Figures 1C, 1D and S1G), it appears that individual cells initiate ZGA based on their individual DNA:cytoplasm ratio but not embryo-wide DNA:cytoplasm ratio. We observed a stereotypic pattern of single-cell activation correlated to cell size and thus DNA:cytoplasm ratio of individual blastomeres (Figures S2H–S2J). Whole-embryo DNA:cytoplasm ratio fails to predict proper ZGA progression; the DNA:cytoplasm ratio required for half-max activation is off by ~ 4-fold (Figure S2I). If the summed DNA:cytoplasm of an entire Xenopus embryo regulated ZGA onset, we would have seen gradual synchronous activation of all nuclei (Figure S2J), which was not what we observed in this study. Taken together, our data indicate that the DNA:cytoplasm ratio of single-cells but not the whole embryo, explains the onset of ZGA, consistent with a cell sizer model.
Next, we asked whether large-scale ZGA can be regulated by both a sizer and timer. A perfect sizer predicts a binary switch from inactive to active transcription when cells reach a size threshold (Figure 3E). In a mixed sizer and timer, the sizer threshold would shift due to accumulated time or cleavage. Using logistic regression to determine a decision boundary fo ZGA, we found that the size threshold was largely unchanged from early-mid ZGA in a 90-minute period (Figure 3F), consistent with a nearly perfect sizer with a threshold at ~ 45 μm in cell diameter (Figure S2D). A hypothetical global timer model fails to explain the observed pattern of activation; it has a much higher error rate (Figures S2E and S2F). To further confirm that a sizer model regulates genome activation, and to rule out artifacts related to imaging, we biochemically quantified the level of nascent transcription. We dissociated blastomeres from embryos in early ZGA (~ 8000-cell stage), physically separated them on the basis of cell size and detected the accumulation of nascent RNAs. We found that cells whose sizes were smaller than 40 μm contained > 12-fold higher levels of nascent transcripts compared to larger cells (Figure 3G). In summary, cell size is a major predictor for large-scale ZGA onset, whereas absolute developmental time appears to have only a limited predictive value.
Consistent with previous studies, we observed a correlation between nucleocytoplasmic volume (N:C) ratio and the onset of ZGA (Figures S3A–S3C) (Jevtic and Levy, 2015, 2017). In this study, we emphasize DNA:cytoplasm ratio over nucleocytoplasmic ratio for a variety of reasons. First, a majority of titration models that explain derepression of ZGA assume titration of repressors relative to DNA. Second, cleavage stage blastomeres undergo divisions without growth and thus there is a near one-to-one correspondence between cell volume and DNA:cytoplasm ratio in a blastomere. Because nucleus volume is much smaller than cell volume - it represents less than ten percent of cytoplasmic volume in all blastomeres that are very small - changes in nucleus size alter cytoplasmic volume by only a small percentage. Third, N:C volume ratio is a composite parameter (Figure S3A) - it is regulated by cell size (Jorgensen et al., 2007; Neumann and Nurse, 2007), nucleus size (the numerator of N:C ratio), which is also regulated by cytoplasmic volume (Hara and Merten, 2015), and duration within interphase, tied to DNA replication (Figures S3D–S3I). Finally, an N:C volume ratio threshold, such as the reported 3.5% for Xenopus (Jevtic and Levy, 2015), inappropriately predicts high transcriptional activity in blastomeres of small size in early interphase (Figures S3J and S3K).
In summary, a size threshold for individual cells predicts the onset of large-scale zygotic transcription in the period of early-mid ZGA, consistent with regulation based on the DNA:cytoplasm ratio of a single cell but not that of a whole embryo.
Positional Regulation versus Size-Dependent Activation: The Spatial Coordinates of a Cell Alone Cannot Explain the Onset of ZGA
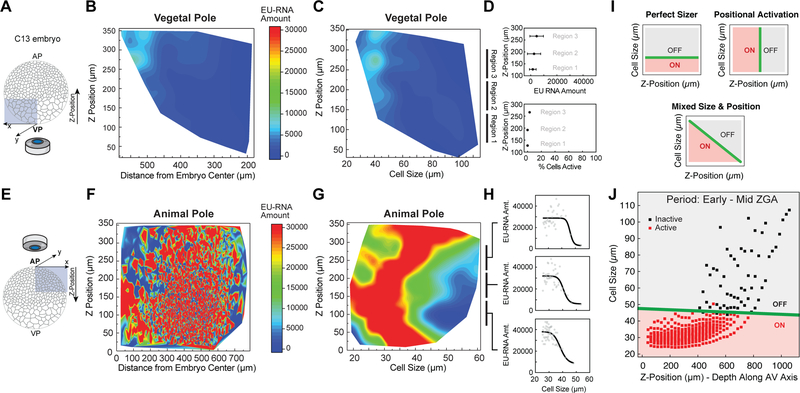
We analyzed the spatial patterns of ZGA in blastula embryos to determine whether additional mechanisms, independent of a cell sizer, regulate the onset of large-scale ZGA. We wondered whether a cell’s zip code (XYZ position) within an embryo would independently predict when zygotic gene expression was triggered in that cell. In particular, we wanted to answer two questions: i) whether lateral (XY) position or cell size is a strong predictor of nascent transcription, and ii) whether the delay in ZGA in the vegetal pole (Figure 1C, 1D, 1G and S1G–S1I) can be explained by polarized distribution of a ZGA inhibitor to the vegetal pole or by the endogenous cell size gradient (Figure 3A). To accomplish this, we imaged regions within the animal and vegetal poles at higher magnification (Figures 4A–4H). Strikingly, within both the animal and vegetal poles, transcription activation was not correlated to the spatial position of the cell. As expected, at ~ C13 cells in the vegetal pole were transcriptionally silent (Figures 4A–4D), while cells of the animal pole contained high levels of nascent transcripts (Figures 4E–4H). Within the animal pole, nascent transcription is uncorrelated with the lateral position (Figure 4F), but is strongly correlated with cell size (Figure 4G), similar to what we found in whole-mount embryos (Figure 3C). Additionally, this size-dependent behavior was largely independent of Z-position within the animal pole (Figure 4H).
Figure 4. A Cell Size Threshold, Not Spatial Position, Dictates ZGA Onset.
(A and E) Schematic of confocal imaging of the vegetal pole (VP) (A) and animal pole (AP) (E) in embryos at ~ C13. Z-position along the AV axis in the indicated directions.
(B, C, F and G) Heatmap contour plot of nuclear EU-RNA amount as a function of z-position and distance from embryo center within the vegetal pole (B) and animal pole (F), and of z-position and cell size within the vegetal pole (C) and animal pole (G). Color scales indicate low (blue) to high (red) transcription.
(D and H) EU-RNA amount and percent of transcriptionally active cells as a function of z-position within the indicated sub-regions of the vegetal pole (D) and animal pole (H).
(I) Predicted decision boundaries for ZGA with a perfect sizer (top), positional bias (middle) and mixed sizer and positional effect (bottom). Gray, inactive; red, active; green line, decision boundary for ZGA.
(J) Logistic regression boundary decision for ZGA as a function of cell size and z-position along the AV axis. Each point is a bin of 25 μm by z-position followed by a bin of 250 cells for each of 8 embryos at ~ C12.8-C13.5. Binary activation based on threshold amount EU-RNA. Black square, inactive cells; red square, active cells; green line, decision boundary.
See also Figure S4.
Oocytes and blastula embryos contain polarized distributions of components. Many macromolecules are asymmetrically distributed in oocytes and fertilized eggs, including proteins and mRNAs that are uniquely localized to either the animal (Kotani et al., 2013) or vegetal cortex (Heasman et al., 2001; Zhang and King, 1996). An alternative hypothesis to the sizer model is that large-scale ZGA delay in the vegetal pole is due to the presence of factors that block zygotic transcription. To determine whether a component asymmetrically distributed along the animal-vegetal (AV) axis regulates ZGA onset, we used logistic regression to map the decision boundary for transcriptional activation, comparing cell size and spatial location (Z-position) along the AV axis for embryos from early to mid-ZGA (Figures 4I and 4J). A positional effect model predicts a positional bias for ZGA (Figures 4I and S4B), while a sizer model predicts a size threshold for ZGA (Figure S4A). Our data show no positional effect, and that a cell sizer almost entirely explains the onset of zygotic transcription. Regardless of position along the AV axis, large-scale gene expression is triggered when a cell reaches a threshold diameter of ~ 45 mm (Figures 4J and S4C). Thus, the delay in zygotic genome activation in the vegetal pole can be explained by the additional time it takes for vegetal cells to undergo extra cell divisions and reach a critical size.
Our data thus far are consistent with a cell sizer model for ZGA regulation based on the DNA:cytoplasm ratio of an individual cell regulating large-scale transcriptional onset. We wondered whether the cell sizer threshold was universal, as indicated for early-mid ZGA (Figures 3F and 4J), or whether it might vary at later timepoints in the vegetal regions of the embryo. To answer this question, we analyzed ZGA in later development, in 14,000+ cell embryos, whose vegetal cells display transcriptional activation (Figure S4E). We found that these vegetal cells also had an inverse correlation between cell size and transcriptional activation. Intriguingly, the size threshold for ZGA in the vegetal pole is ~ 62 μm, higher than that for cells in the animal pole, i.e., ~ 45 μm (Figures S4D–S4F). Because DNA:cytoplasm ratio dependent regulation of ZGA is based on titration of repressors (Figure S4K), we wondered whether the concentration of core histones, repressors of ZGA (Almouzni and Wolffe, 1995; Amodeo et al., 2015; Joseph et al., 2017), were present at uniform concentrations throughout the embryo (Figure S4G). If histone levels in a cell were decreased, a DNA:cytoplasm ratio dependent model would predict that genome activation initiates at lower DNA concentration (larger cell sizes). Indeed, we found that the levels of core histones were ~ 1.6-fold lower in the vegetal pole than the animal pole (Figures S4H–S4J), which is consistent with a threshold for activation in the vegetal pole at larger cell sizes and lower DNA:cytoplasm ratios (Figure S4F). Whether alterations in histone levels are sufficient to alter the cell size threshold for large-scale ZGA in Xenopus remains to be determined.
A Computational Model Recapitulates the Timing and Patterning of ZGA
To better understand the mechanisms of ZGA regulation in blastula embryos, we computationally simulated Xenopus developmental progression and tested whether a timer, a sizer or a cell cycle counter could explain the spatiotemporal pattern of large-scale zygotic gene expression. For cell division timing, this simulation uses parameters for propagation speed of the mitotic wave along the animal-vegetal axis and within the dorsal-ventral/left-right plane (Anderson et al., 2017). The intervals between initiation of successive waves were set to match previous measurements (Anderson et al., 2017; Newport and Kirschner, 1982a). In addition to the measured mitotic wave speed, the values of model parameters were constrained so as to generate the observed distribution of cell sizes across a range of embryo ages (Figures S5A–S5J). The simulation thereby provides the sizes, cleavage number, times of mitosis, and relative positions along the AV axis for all cells, at all times following fertilization (Figure 5A).
Figure 5. Computational Model for Characterization of Zygotic Genome Activation.
(A) Developmental progression in silico: showing relationship between cell number, rounds of division (mitotic cycle) and time elapsed since fertilization.
(B) Successive mitotic waves generate asynchrony, resulting in an increase in the phase delay between the division of the first cell (on animal side) and the division of the last cell (on vegetal side).
(C) Delay of up to 80 minutes (2+ rounds of division), between onset of 13th division in cells at the animal pole and cells at the vegetal pole.
(D) A single embryo at ~ 8,000-cell stage contains a mixture of cells that have undergone 12, 13 or 14 divisions. Cell that have undergone more divisions are located in the animal pole.
(E-G) Predicted timing of ZGA onset for computationally simulated models compared to experimental data and confidence interval (CI).
(E) ‘Strict Timer’ (orange line) poorly fits experimental data (gray) (R2 = 0.758); ‘Loose Timer’ model (red line + confidence interval) better fits the experimental data (R2 = 0.96).
(F) Cell Sizer model (blue line + confidence interval) and experimental data (gray) (R2 = 0.959).
(G) Cell Cycle Counter model (green line + confidence interval) and experimental data (gray) (R2 = 0.939).
(H-J) Predicted spatial patterns of ZGA for an embryo in early ZGA (~ 9,000 cells, log2 = 13.1), based on timer, sizer, and counter models. Activated nuclei (red) and inactive nuclei (gray).
(H) Timer model 3D prediction: inaccurately predicts ZGA in the vegetal cells.
(I-J) Sizer model predicts a 3D spatial pattern of ZGA similar to the experimental data.
(J) Counter model predicts a 3D spatial pattern of ZGA similar to the experimental data.
(K) Predicted fraction of activated cells along the AV axis. Comparing experimental data (gray dotted line and confidence interval) vs. computationally simulated models: timer (red), sizer (blue), counter (green).
(L) Fraction of cells active in the vegetal pole in a 9000-cell embryo, comparing computational models to experimental data. Timer model incorrectly predicts high activity in vegetal region.
(M) Error of fit for model predictions to experimental data in subplot Figure 5K.
See also Figure S5.
Cell divisions occur faster in the animal pole than the vegetal pole, creating a phase delay in the number of divisions cells have occurred (Figure 5B) (Anderson et al., 2017). Within a single embryo, large cells in the vegetal region have divided fewer times than those in the animal pole (Figure 5B), and it takes up to ~ 80 additional minutes for vegetal cells to undergo the thirteenth mitotic division (Figure 5C). A single embryo that has progressed developmentally to contain ~ 8,000 cells is comprised of a mixture of those that have completed 12, 13 or 14 divisions (Figure 5D). Therefore, ZGA patterning – the observed delay in activation of cells along the AV axis - could potentially be regulated by a gradient in cell division timing along this same axis, rather than a gradient of cell size. Stated in another way, because cell size is largely dictated by the number of cell divisions that have occurred, a cell cycle counter mechanism could explain the patterning of ZGA that we previously ascribed to cell size.
To test each model for ZGA regulation, we applied a rule that determines when a cell undergoes large-scale ZGA, and compared the simulated data to our experimental data, analyzing ZGA as a function of time and spatial position. From EU-labeled embryos of different ages, we extracted a confidence interval for the fraction of transcriptionally active nuclei. For each of the models we asked what time, size, or cleavage number would provide the closest fit to the observed fraction of active cells and the distribution of active cells along the AV axis. A strict timer model, based on activation at 445 minutes post-fertilization, poorly fit the experimental data (Figure 5E). We also tested a two-parameter “loose timer” model in which transcription is activated non-synchronously in individual cells at an average time, allowing some cells to activate more slowly than others. Although this model could predict the fraction of cells activated as a function of developmental progression (Figure 5E), it could not accurately predict the spatial pattern of activation (Figures 5H, 5K–5M, S5K and S5L). This eliminates a model in which the mitotic wave generates the AV axis gradient, even downstream of an imprecise timer. In contrast, a cell sizer model accurately predicted ZGA as a function of both time (Figure 5F) and spatial position (Figures 5I, 5K–5M, S5K and S5N). Intriguingly, a cell cycle counter based on activation at cycle 13 also predicted ZGA onset in a pattern that matched our experimental data (Figures 5G, 5J–5M, S5K and S5M). Taken together, this computational simulation of Xenopus development suggests that both a cell sizer and cell cycle counter could predict the observed spatiotemporal pattern of genome activation in early embryogenesis. Given this result, we required an additional experiment to discriminate between sizer and counter models for regulation of ZGA.
Interrogating Cell Cycle Counter and Cell Sizer Mechanisms for ZGA Regulation Using Miniature Embryos
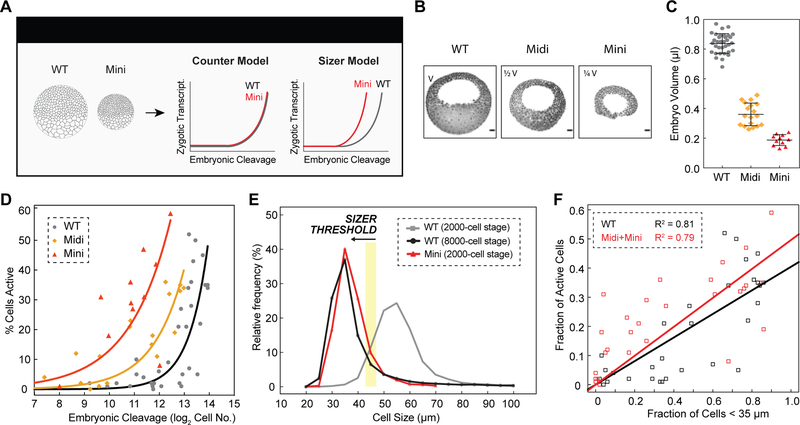
To distinguish experimentally between cell cycle counter and cell sizer models for ZGA regulation, we generated 1-cell stage embryos that have reduced cell volumes. Premature ZGA in these mini-embryos would support a cell sizer model, whereas activation at the same developmental stage or cleavage as wild-type embryos would support a counter model (Figure 6A). Xenopus embryos tolerate constriction via ligature at 1-cell stage, resulting in embryos of decreased size that nonetheless continue to develop (Figures S6A and S6B) (Clute and Masui, 1995). We used this method to create miniature embryos whose volumes were ~ ½ or ¼ of a wild-type embryo (Figures 6B and 6C). The cells in these miniature embryos divided with nearly identical timing to those in wild-type embryos, such that at early ZGA, wild-type or miniature embryos contained similar numbers of cells (Figure S6E). Because the nucleus is biased towards the animal pole in a 1-cell Xenopus embryo, the resulting miniature embryos are mainly derived from material in the animal hemisphere. Intriguingly, we found that mini embryos consistently activated large-scale ZGA earlier than wild-type (Figures S6C, S6F and S6G), ruling out a model for ZGA regulation based solely on counting 12 or 13 rounds of cell division.
Figure 6. Premature ZGA in Miniature Embryos Rules Out a Cell Cycle Counter Model and Supports a Cell Sizer Model for ZGA.
(A) Schematic predictions of ZGA onset in wild-type (WT) and miniature (Mini) embryos for a counter and sizer model.
(B) Representative sagittal slices of WT, Midi (half volume, 1/2 V) and Mini (quarter volume, 1/4 V) embryos stained for DNA (black pseudocolored). Scale bars, 100 μm.
(C) Volume measurements for WT (gray), Midi (orange) and Mini (red) embryos at blastula stages. Each point indicates one embryo. Data represented as mean ± standard deviation.
(D) Percentage of transcriptionally active cells as a function of embryonic cleavage for WT (gray), Midi (orange) and Mini (red) embryos. Exponential fits to data as visual aid.
(E) Histogram of cell size distributions for WT embryos at ~ 2000-cell stage (~ C11, gray) and ~ 8000-cell stage (~ C13, black), and Mini embryos at ~ 2000-cell stage (~ C11, red). Yellow stripe indicates a putative threshold of ~ 45 μm.
(F) Fraction of transcriptionally active cells as a function of cells smaller than 35 μm for WT (black) and Midi and Mini (red) embryos. Linear fits to data.
See also Figure S6.
If ZGA truly depends on achieving a threshold cell volume, as predicted by a cell ‘sizer’ model, then the onset of ZGA in half-sized embryos should occur earlier in development time (half the number of cells as wild-type, indicative of one fewer division) and quarter-sized embryos even earlier (two fewer divisions). We found exactly this predictive relationship (Figure 6D), consistent with a mechanism for ZGA regulation dependent on cell size (Figure 6A) and not the absolute number of cell divisions. This finding shows that cell size is causal for genome activation – reaching a critical size is sufficient to trigger large-scale zygotic transcription. With this regulation in mind, we can now explain the inactivity of wild-type embryos at ~ 2000-cell stage (211 cells) and their large-scale activation at ~ 8000-cell stage (213 cells), and the precocious activation of quarter-size mini embryos at ~ 2000-cell stage (211 cells). Wild-type embryos containing 211 cells do not contain sufficient numbers of cells whose sizes are smaller than the animal pole size threshold (Figure 6E). In contrast, quarter-sized embryos, which have undergone the same developmental progression (contain 211 cells) show 20–40% of their cells activated; a similar percent of their cells are smaller than the size threshold. Strikingly, the distribution of cell sizes is nearly identical in a miniature embryo containing 211 cells and a wild-type embryo containing 213 cells (Figure 6E). Reproducibly, the fraction of cells that have achieved a critical cell size is the parameter that best predicts the fraction of cells activated in embryo (Figures 6F and S6D). These findings clearly rule out models for ZGA regulation based exclusively on cell cycle counters or global timers. To our knowledge, this is the first demonstration that cell size is sufficient to regulate the onset of ZGA in a vertebrate embryo.
DISCUSSION
Our single-cell analysis of nascent zygotic transcription within an intact vertebrate embryo demonstrates that large-scale ZGA occurs asynchronously, at the single-cell level, in a stereotypic spatial pattern dictated by cell size and independent of absolute elapsed time or the number of cell divisions post fertilization. Our data are consistent with a cell sizer model for ZGA regulation based on achieving a threshold DNA:cytoplasm ratio within individual cells, but not within the whole embryo. Furthermore, reduction of cell size is sufficient to trigger premature ZGA in a dose dependent fashion. Because prior measurements of the onset of global transcription activation in vertebrate embryos have relied on analysis of whole embryos (Paranjpe et al., 2013; Peshkin et al., 2015), our findings offer a new view of ZGA. Complementary to the concept of stochastic gene activation during ZGA in zebrafish (Stapel et al., 2017), our work demonstrates asynchronous, single-cell initiation of large-scale ZGA, which may be a general phenomenon in vertebrate early development.
This study relied on technological development to advance our understanding of the control mechanisms governing ZGA in a model vertebrate embryo. We developed an approach to image nascent transcription in whole-mount vertebrate embryos using EU labeling of RNA (Figures 1 and S1). This method is highly specific – it is not affected by the large pool of maternal RNA in an embryo – and sensitive to provide signal for the very earliest period of large-scale zygotic transcription. Such sensitivity is difficult to achieve in whole-mount embryos using RNA in situ hybridization to individual zygotic genes. Additionally, this method allowed us to sequence the nascent transcriptome (Figures 2D–2F), which provides higher sensitivity in RNAseq analyses of early ZGA. In summary, EU-RNA labeling and imaging enabled the novel insight that these vertebrate embryos undergo a stereotypic spatiotemporal pattern of genome activation.
To distinguish between regulatory models for ZGA our experiments leveraged two special features of Xenopus. First, the blastula stage embryo contains a broad gradient of cell sizes (> 100-fold in volume between the smallest and the largest cells). This facet was instrumental to rule out a timer-only model for ZGA regulation. Second, Xenopus is amenable to generation of miniature embryos (Clute and Masui, 1995), enabling manipulation of DNA:cytoplasm ratio directly via alterations in cell volume rather than in DNA ploidy. These miniature embryos allowed us to test and rule out an alternate hypothesis in which a cell cycle counter controls the onset of ZGA.
Prior to our study it had not been tested whether cell size directly impacts DNA:cytoplasm ratio dependent ZGA within a vertebrate embryo in vivo. In previous studies, the impact of DNA:cytoplasmic ratio was tested by manipulating ploidy in different model organisms (Edgar et al., 1986; Jevtic and Levy, 2017; Kane and Kimmel, 1993; Lu et al., 2009; Newport and Kirschner, 1982b). However, there were limitations to many of these experiments. First, large-scale ZGA was not directly measured; instead a surrogate activity such as cell cycle elongation or transcription of a reporter gene or a subset of genes was measured. Second, alterations in ploidy may have pleiotropic or unintended effects on embryo development. By directly altering cell size (Figure 6), our study complements previous work while eliminating concerns associated with manipulations of DNA content. To our knowledge, this is the first demonstration that cell size is sufficient to dictate the onset of large-scale ZGA, and we identify the size thresholds at which this occurs in the animal pole and vegetal pole. This finding raises the possibility of a cell size checkpoint in early development, similar to that observed at G1/S in cultured cells (Schmoller et al., 2015; Zatulovskiy et al., 2018).
The differential onset of ZGA between the animal and vegetal pole cannot be explained simply by gradients of activators or repressors along the AV axis. Instead, this pattern correlates with cells achieving a threshold size. In Xenopus, a cell sizer is thought to depend on DNA:cytoplasm ratio and the concentration of histones in a cell (Almouzni and Wolffe, 1995; Amodeo et al., 2015). The prevailing theory is that histones function in part to repress ZGA and that repression is relieved once cell size has sufficiently decreased (DNA:cytoplasmic ratio has sufficiently increased). If histone concentrations were uniform throughout the embryo, the cell sizer threshold should be similar for all regions of embryo. Conversely, if the concentrations of histones differed in distinct regions of the embryo, the size threshold would vary. We found that the vegetal cells contain lower concentrations of core histones (Figures S4H and S4I). At the onset of ZGA, the vegetal pole contains ~ 1.6-fold lower concentrations of H3 and H4 (Figure S4J). Based on this asymmetry, a model dependent on cell size, DNA concentration, and histone concentration (Figure S4K) would predict a more relaxed cell size threshold in the vegetal pole cells compared to the animal pole cells. Blastomeres containing higher concentrations of histones would need to reach higher DNA:cytoplasm ratios (smaller cell sizes) to overcome the repressive effects of histones. Consistent with this hypothesis, we found that in late ZGA, when a sufficient fraction of vegetal blastomeres are activated, cells in the vegetal pole need to only reach ~ 62 μm in diameter to undergo large-scale ZGA, while cells in the animal pole achieve ~ 45 μm in diameter (Figures S4D–S4F). Further studies are warranted to confirm these observations the dose-dependent relationship between histone levels and the cell size threshold regulating large-scale ZGA.
The dependency of genome activation on cell size is likely present in embryos from other model organisms, such as zebrafish, which lack a cell size gradient. Because their cells are much more uniform in size, the parameters of elapsed time, cell size and number of rounds of cell division cannot be separated from one another in wild-type embryos. Future studies will be required to manipulate these parameters and perform single-cell imaging of large-scale nascent zygotic transcription in other vertebrate model organisms.
Cell volume predicts the timing of large-scale ZGA, but achieving a threshold cell volume alone may not be sufficient for ZGA onset when considering very early developmental times. In zebrafish, translation of pluripotency factors Pou5F1, Sox2 and Nanog is required for ZGA (Lee et al., 2013). Similarly, translation of p300 may constitute an early timer required for ZGA (Chan et al., 2018). However, the levels of these factors have not been measured in Xenopus for the period from 2-cell to 8000-cell stage embryos. If Xenopus genome activation also depends on translation of these factors, it may not be possible to force large-scale ZGA based solely on cell size, i.e., initiation of ZGA by generating a forty-micron diameter cell at 32-cell stage of development. Nevertheless, quarter-sized embryos display genome activation in ~ 2000-cell embryos compared to ~ 8000-cell wild-type embryos (two cycles earlier than wild-type embryos), which is consistent with a cell sizer. This result suggests that within this window of developmental progression, if pluripotency factors are required, they would have already accumulated to sufficient levels.
At the systems level, our study rules out simple timer and counter models for controlling the onset of large-ZGA and strongly supports a sizer model that is consistent with a mechanism in which DNA:cytoplasm ratio titrates repressive factors such as histones (Figure S4K). Our results provide new insights at the systems level and do not rule out specific molecular mechanisms related to a cell sizer and DNA:cytoplasm ratio dependent initiation of MBT event, such as titration of DNA replication factors (Collart et al., 2013), or the potential contribution of cell cycle lengthening to ZGA. Our methodology for nascent EU-RNA imaging requires fixed embryos and we therefore cannot measure cell cycle elongation. Previous work suggested that the cell cycle period increases at a threshold cell size (Wang et al., 2000). This mechanism would fall under the umbrella of a cell sizer model and is potentially consistent with the observed pattern of large-scale ZGA. Future studies will be required to falsify or support specific molecular pathways that can constitute a cell sizer.
N:C volume ratio correlates with ZGA onset and perturbation of N:C volume ratio can trigger early expression of some zygotic genes in Xenopus embryos (Jevtic and Levy, 2015, 2017). However, N:C volume ratio is a complex, hybrid parameter – it is governed by cell size, nucleus size, and duration within interphase (DNA content) (Figure S3). Changes in cell size alter nucleus size, and cytoplasmic volume can regulate nuclear growth (Hara and Merten, 2015); furthermore, cytoplasmic volume is the denominator of N:C volume ratio. Additionally, nuclei grow during interphase as DNA replication proceeds, and thus N:C volume ratio in a blastomere correlates with that cell’s duration within interphase. A threshold of N:C volume ratio (3.5%) (Jevtic and Levy, 2015) also mispredicts high activation in small cells, regardless of their duration in interphase. Given that cell size is the physical parameter directly altered by cleavage stage embryos – not zygotic DNA amount within the cell – and that cell size is upstream of N:C volume ratio, we believe cell size or single-cell DNA:cytoplasm ratio provides a clear and parsimonious prediction of the spatiotemporal pattern of large-scale ZGA within cellularized vertebrates.
The biological significance of the cell size gradient in Xenopus is not understood. A strong cell size gradient is also present in embryos from other organisms, including lamprey, axolotl and species of frogs. Our work demonstrates that the largest cells present in the vegetal pole significantly delay genome activation. These cells are part of the presumptive endoderm, and therefore the cell size gradient may have evolved to delay expression of endoderm factors in the vegetal pole, relative to expression of ectoderm factors in the animal pole. Future studies will be required to control the gradient of sizes in Xenopus and determine their impact on downstream developmental progression.
In summary, our work identifies an essential framework for controlling the spatial pattern of ZGA in a model vertebrate, and this regulatory paradigm may be a conserved feature of other embryonic systems.
STAR METHODS
EXPERIMENTAL MODEL AND SUBJECT DETAILS
African claw frogs Xenopus laevis were maintained at 16 °C and used for collecting embryos at 22 °C ± 0.5 °C.
METHOD DETAILS
Frogs
All animal experiments in this study were performed according to the Animal Use Protocol approved by the University of Pennsylvania Animal Care and Use Committee. Mature Xenopus laevis females and males were purchased from Nasco. No harm was caused to the animals during experimentation. To ovulate eggs, female frogs were first primed by injecting 100 U of pregnant mare serum gonadotropin (PMSG) into the dorsal lymph sac at 3–7 days prior to experiment. At 14–15 h before egg collection, the females were injected with 500 U of human chorionic gonadotropin (HCG) and kept at 16 °C in 1× Marc’s Modified Ringer’s (MMR: 100 mM HEPES pH 7.8, 2 mM EDTA, 2 M NaCl, 40 mM KCl, 20 mM MgCl2, and 40 mM CaCl2). Females were ovulated with at least 3-month rest intervals. To isolate testes, males were euthanized with 0.2% benzocaine for at least 20 min before dissection. The testes were kept in L-15 medium on ice for further use.
In vitro fertilization (IVF)
Eggs were collected in glass dishes by gently squeezing the females. Sperm slurry was prepared by crushing 1/2 of a testis in 1 ml of Milli-Q water inside a microcentrifuge tube using a plastic pestle and added directly onto eggs. Five minutes post-IVF, embryos were flooded with 0.1× MMR. At 30 min post-IVF, embryo jelly coats were removed by incubating with 2% L-cysteine in 0.1× MMR for 2–5min, followed by washing with 0.1× MMR for multiple times. The embryos were kept in 0.1× MMR for further use.
Microinjection
Prior to microinjection, embryos were transferred into chambers containing 3% Ficoll® in 0.5 × MMR. To track the spatiotemporal patterning of ZGA, embryos at 1-cell stage were microinjected with 10 nl of 50 mM 5-ethynyl uridine (EU) using a PLI-100 picoliter microinjector (Medical Systems Corp., Greenvale, NY). Embryos developed to 256-cell to 32000-cell stage (log2 cell no. = 8–15) were fixed in 4% paraformaldehyde / 1× MEM (100 mM MOPS pH 7.4, 2 mM EGTA, and 1 mM MgSO4) solution in 2 ml scintillation vials by rotating for at least 2 hours at room temperature. Embryos were completely dehydrated with methanol before being stored at - 20 °C.
To test whether EU-RNA is RNA polymerase II dependent, 0.1 ng or 2 ng α-Amanitin was co-microinjected with 5-EU into embryos at 2-cell stage and embryos were fixed at around 8000-cell stage (log2 cell no. = 13) as described above.
Mini-embryo preparation
Embryos were microinjected with 10 nl of 50 mM 5-EU from the vegetal side at ~ 40 minutes post fertilization (mpf). At 1 hour post fertilization (hpf), the vegetal pole of embryos was poked with a 30G needle so that some cytoplasm could be released by gently pressing the embryo. Next, the animal side of embryo was tied and constricted with a hair loop, which one side was attached into a P10 tip, by pulling the two ends of the loop in opposite directions. Different sized mini-embryos were generated by placing different positions of the hair loop on the embryo. The resulting size-reduced mini-embryos were maintained in 3% Ficoll® / 0.5 × MMR until around 4 hpf or up to the stages to be collected. The mini-embryos, as well as normal controls, were fixed in 4% paraformaldehyde in 1× MEM (100 mM MOPS pH 7.4, 2 mM EGTA, and 1 mM MgSO4) in 2 ml scintillation vials by rotating for at least 2 hours at room temperature. Embryos were completely dehydrated with methanol before being stored at −20 °C.
Blastomere dissociation and sorting by size
EU-microinjected embryos at ~ 4000-cell stage (log2 cell no. = 12) were incubated with 20 ml of calcium and magnesium-free medium (CMFM: 50.3 mM NaCl, 0.7 mM KCl, 9.2 mM Na2HPO4, 0.9 mM KH2PO4, 2.4 mM NaHCO3 and 1 mM EDTA, pH7.4) (Venkataraman et al., 2004) in glass dishes for about 1 h at room temperature with regular gentle shaking every 5–10 min. Blastomeres were sequentially filtered with Falcon® cell strainers (Corning) with pore sizes at 40 μm, 70 μm and 100 μm, repectively. Blastomeres are at different size ranges, i.e., < 40 μm, 40–70 μm, 70–100 μm and > 100 μm, as well as control intact embryos, were collected in 2-ml microcentrifuge tubes by gently using plastic transfer pipette. As much as medium was removed before quickly freezing blastomeres or control embryos in liquid nitrogen. The blastomeres and control embryos were stored at −80 °C before they were used for RNA isolation.
RNA isolation, EU-RNA biotinylation and dot blot assay
To isolate total RNA, embryos or blastomeres from −80 °C were added with 700 μl RLT buffer using the RNeasy Mini Kit (Qiagen), followed by gentle pipetting until they were homogenized. The lysates were transferred into the columns and RNA was eluted following the instructions provided by the kit. To biotinylated RNA, 6 μg of RNA was incubated with 25 μM biotin-azide, 100 mM Tris-HCl pH 8.5, 1 mM CuSO4, and 100 mM ascorbic acid for 30 min at room temperature. The biotinylated RNA was purified and cleaned following the instructions provided by the RNeasy MinElute Cleanup Kit (Qiagen). To perform dot blot assay, the biotinylated RNA was diluted in RNA dilution buffer (5 parts of RNase-free water, 3 parts of 20× SSC and 2 parts of formaldehyde), dotted on a nitrocellulose membrane and crosslinked with 2000 J of energy in a Stratalinker® UV Crosslinker Model 2400 (Stratagene). The membrane was blocked with blocking buffer (1× TBS with Western Blocking Reagent, Roche, 11921673001) for 1 h at room temperature, and incubated with Streptavidin-HRP (1:7500 dilution) for 1 h at room temperature. The membrane was washed five times with 15 mL 1× TBST for 1 hour. The membrane was exposed with chemiluminescent substrate (SuperSignal West PICO Plus, Invitrogen) and imaged with an Amersham™ imager 600 (GE Healthcare). Each dot appeared as round with a ring of maximum signal at the edges. The signal intensity inside the rings and background signal intensity at four different regions surrounding the rings were measured in Image J. The amount of signal in the rings were calculated by subtracting average background intensity of four regions surrounding the ring from the pixel intensity of the ring and multiplying by the area of the ring.
On-bead RT-qPCR
Total RNA from embryos microinjected with buffer (1× TBS), EU, and EU + α-amanitin was isolated using the RNeasy Mini Kit (Qiagen) and biotinylated and cleaned as described above. The biotinylated EU-RNA was pulled down with magnetic streptavidin beads following the instructions provided by the Click-iT™ Nascent RNA Capture Kit (Invitrogen, C10365). The EU-RNA conjugated to the beads was reverse transcribed with the SuperScript III First-Strand Synthesis kit (Invitrogen, 18080051). Briefly, the EU-RNA conjugated beads were added with 1 μl of 10 mM dNTPs, 1 μl random hexamers, and 3 μl of RNase-free H2O. The mixture was incubated at 65 °C for 5 min and put on ice. The following reaction was added for a total reaction of 20 μl: 2 μl of 0.1 M DTT, 2 μl of 10× RT Buffer, 4 μl of 25 mM MgCl2, 1 μl of RNaseOUT, and 1 μl of SuperScript III RT (or water for no RT reaction). The reaction was run on a thermocycler at 25 °C for 10 min, 50 °C for 50 min, and 85 °C for 5 min. The resulting cDNA was amplified and quantified with qPCR using Power SYBR Green qPCR Master Mix (Applied Biosystems, 4367659) on the StepOne qPCR thermal cycler (Applied Biosystems). Fold change in expression was quantified by ΔCt between buffer, EU, and EU+ α-amanitin.
Nascent EU-RNA-seq and analysis
Total RNA was isolated from EU-injected embryos (N = 10) at 9 hpf and EU-RNA was biotinylated and purified as described above. As control, total RNA was isolated from normal embryos at 9 hpf. Two duplicates were used for both EU-injected embryos and normal embryos. cDNA libraries were prepared from purified EU-RNA (from EU-injected embryos) or total RNA (from normal embryos) using the Ovation® Human FFPE RNA-Seq Library Systems (NuGEN) with minor modifications (Palozola et al., 2017). The quality of cDNA libraries was assayed using the Agilent High Sensitivity DNA Kit (Agilent, Cat. No. 5067–4626) in the Agilent 2100 Bioanalyzer System (Agilent Technologies, CA). The cDNA libraries were subjected to size selection using SPRIselect beads (Beckman Coulter, Cat. No. B23317) before quantification using the NEBNext® Library Quant Kit (NEB, Cat. No. E7630). The cDNA libraries were sequenced using the NSQ 500/550 Hi Output KT v2.5 (75 CYS) (Illumina, Cat. No. 20024906) in the NextSeq 500 sequencer (Illumina, CA). Raw sequence data (fastq files) were aligned to Xenopus laevis genome build 9.2 using salmon v0.12.0 (Patro et al., 2017). Data were normalized for sequencing depth using DESeq2 (bioconductor v3.8) (Love et al., 2014). The number of genes was counted with DESeq2 normalized reads > 10. Zygotic genes highly induced at MBT were selected based on the criteria of significant induction of gene expression from stage 8 to stage 9 following the dataset by Yanai et al. (Yanai et al., 2011).
Whole-mount click chemistry and immunofluorescence
Embryos were rehydrated by washing sequentially with 75%, 50% and 25% methanol in 0.5× SSC (75 mM NaCl and 7.5 mM sodium citrate) for 10 min each, followed by washing with 0.5× SSC for three times. Embryos were bleached in 0.5× SSC containing 5% formamide and 2% H2O2 for 5–6 h under light. Embryos were rinsed quickly with 0.5× SSC for three times, followed by washing with 1× TBST for 30 min. Repeat washing for at least six times. Embryos were washed three times with 1× TBS for 10 min each. Embryos were incubated with 25 μM TAMRA-azide, 100 mM Tris-HCl pH 8.5, 1 mM CuSO4, and 100 mM ascorbic acid for 8–12 h at room temperature. Embryos were extensively washed with 1× TBST all day at room temperature by changing buffer every 2 h, overnight at 4 °C, and all day at room temperature by changing buffer every 2 h. Embryos were incubated with TO-PRO-3 (1:500 dilution) overnight at 4°C. Embryos were washed with 1× TBST all day at room temperature by changing buffer every 2 h. Embryos were completely dehydrated with anhydrous methanol by replacing it for multiple times. Embryos were cleared in BABB (mixture of 1 part of benzyl alcohol and 2 parts of benzyl benzoate) for 12–24 h before confocal imaging.
Confocal microscopy
Confocal microscopy was performed with the ZEN software on Zeiss LSM710 confocal microscope. EU-RNA and TO-PRO-3 signals in Xenopus embryos were imaged with a frame size of 1,024 pixels × 1,024 pixels using lasers 561 nm (0.15% power), 633 nm (10% power) and 488 nm (10% power), respectively, without saturating signals. For tracking spatiotemporal patterning of ZGA, whole embryo Z-stacks were collected using the Plan-Apochromat 10× / 0.45 objective with 5 μm interval. For generating spatial maps of ZGA, Z-stacks were collected from animal pole and vegetal pole, respectively, using the Plan-Apochromat 25× / 0.8 immersion oil objective with 2 μm interval.
Image analysis
To count nuclei number in whole embryos and measure nuclear volume, cell diameter and embryo volume, and quantify the EU-RNA signal intensity in nucleus, the confocal z-stack images were processed using Imaris 9.2 (Bitplane, Switzland) (see Supplemental Methods). Before segmentation, the images the effect of BABB (refractive index ≈ 1.5) was corrected using a voxel size of 7.5 and signal attenuation of TO-PRO-3 was corrected using the plugin Attenuation Correction after measuring the intensity on the front and back images. For segmenting nuclei in whole embryos, the surfaces of nuclei were created based on the corrected TO-PRO-3 signal using the following settings: smooth detail, 1.5 μm; thresholding, background subtraction (local contrast) and diameter of largest sphere which fits into the object, 16 μm. Automatic thresholding was used for generating surfaces; these were then also inspected and adjusted manually. Objects with less than 10 voxels were filtered out. The goodness of fit of the segmentation was confirmed manually in comparison to an image stack containing nuclei that were masked manually. The total number of nuclei in embryo was counted directly from the objects of surfaces for TO-PRO-3 and the nuclei in interphase was identified based on the sphericity of nucleus over 0.82 and late interphase over 0.9. The log2 of total nuclei number was used for staging the developmental progression of embryos. The nuclear volume was obtained directly from the statistics generated by the software. The 3D positional information, i.e., X, Y and Z positions, were exported for calculating the vector distances between one nucleus and its three closest nuclei and the average vector distance was used as a proxy of cell diameter of that cell. The nucleocytoplasmic volume (N:C) ratio was calculated by dividing nucleus volume with cytoplasmic volume, following the method by Jevtic and Levy (Jevtic and Levy, 2015). Nuclear DNA intensity was exported from the statistics and subtracted with the cytoplasmic background. Nuclear DNA amount was calculated by multiplying corrected nuclear intensity with nuclear volume. The DNA:cytoplasmic (D:C) ratio was estimated using 3N for DNA content per nucleus (average from 2N to 4N before and after DNA duplication in each cleavage, respectively) and 3.4 pg for 1N (the Xenopus laevis haploid genome size is 18 chromosome and 3.1 billion bp, which is 3.4 pg per haploid genome) (Hellsten et al., 2010). Whole-embryo D:C ratio was calculated for individual embryos from C8–C15 (40 embryos) by multiplying DNA content per nucleus with nuclei number in the embryo and divided by total cytoplasmic volume of the embryo. Single-cell D:C ratio was calculated by dividing DNA content in each nucleus by cytoplasmic volume of the cell, and the data were binned by 40 increments.
The original nuclear EU-RNA intensity was measured by overlaying the segmented nuclei on the EU-RNA-TAMRA channel. The cytoplasmic background was measured by generating a 3–5 μm shell surrounding individual nucleus surfaces and overlaying the segmented nuclei on the EU-RNA-TAMRA channel. The net nuclear EU-RNA intensity was calculated by subtracting the cytoplasmic background from the original nuclear EU-RNA intensity, and the nuclear EU-RNA amount was calculated by multiplying the net nuclear EU-RNA intensity with the nuclear volume. Threshold of the mean EU-RNA intensity and amount at 8000-cell stage (log2 cell no = 13) was used to calculate the percentage of transcriptional active cells in individual embryos at various stages.
To measure the volume of whole embryos, an autofluorescence channel was segmented to generate the surfaces of the embryos and the volume statistics were exported. The total EU-RNA intensity inside whole embryos were measured by overlaying the segmented embryo channel with the EU-RNA-TAMRA channel. The net EU-RNA intensity in whole embryo was calculated by subtracting the background intensity measured from the embryos that were not injected with EU. The EU-RNA amount in whole embryos were calculated by multiplying the net nuclear EU-RNA intensity with the embryo volume.
Logistic regression boundary decision
To determine the logistic regression boundary, the transcriptional activity of each cell in embryos at early and mid-ZGA stages was first binned as 0 (inactive) or 1 (active) based on the threshold of EU-RNA amount. To distinguish sizer and timer model (Figure 3F), the data were sorted by cell size and then binned every 200 cells. To distinguish sizer and position effect (Figure 4J), the data were binned every 25 μm by Z-position followed by binning of every 250 cells. In both cases, for each bin only data with n > 20 were used. The logit function of glm (generalized linear model) in R was used to determine the slope and intercept for distinguishing the transcriptionally inactive and active populations. Alternatively, the logistic regression model in scikit-learn in Python was used, in which three hyper-parameters, i.e., penalty, “C” (inverse of regularization strength) and “dual” (dual or primal formulation), was tuned via exhaustive grid search to optimize hyper-parameter combination based on average F1 score in 5-fold cross validation. In the latter, the model was set to adjust class weights inversely proportional to class frequencies in each dataset. With the fitted model, the mlxtend package was used to plot decision regions of the model in 2 dimensions. To compare different models of ZGA, artificial decision boundaries were made by manually choosing a point near the decision boundary passing the point horizontally, vertically and in a 45-degree angle on the 2-dimension space. F1 scores were used to determine the robustness fitting of models of ZGA.
Computational model
A model of Xenopus embryo cleavage divisions was instituted to simulate cell volumes and the timing of mitosis for all cells during the first 14 cleavage divisions. Cell volumes are calculated based on the conservation of volume of a sphere of unit radius. The position of the division plane determined the volume of mother cell cytoplasm inherited by each daughter. The division plane of each mitosis was set by drawing positions from a Gaussian distribution of some width to introduce variability / imprecision in the sizes daughter cells. As noted below, we also imposed three asymmetric divisions. These features, the imprecision and the asymmetry, were both required to generate the smooth, long-tailed distribution of cell sizes observed in the data.
For simplicity, mitotic planes were imposed exactly perpendicular to one of the three major axes. The orientation of the planes of the first five divisions was determined by observation: the first and fourth planes were perpendicular to the dorsal-ventral (DV) axis, the second perpendicular to the left-right (LR) axis, and the third and fifth perpendicular to the animal-vegetal (AV) axis. For the remaining divisions, the simulation cycled through each orientation in the order [LR, AV, DV].
The timing of mitosis for individual cells depends on the speed of the mitotic wave, which was previously measured (Anderson et al., 2017). For simplicity, the wave was set to initiate at the animal pole. The initiation of each mitotic wave was calculated using the timing of mitosis reported in Figure 2 of Anderson et al., 2017 for the first 12 divisions and from Figure 2 of Newport and Kirschner 1982 for cleavages 13 and 14. For each individual cell, the simulation calculates the interval between the initiation of the mitotic signal and the arrival of that signal at the centroid of that cell. The signal propagation speed is 0.192 units per minute in the LR/DV plane, and 0.067 units per minute along the AV axis, corresponding to the measured speeds of 2 microns per second and 0.7 microns per second (Anderson et al., 2017), assuming an egg of diameter of 1.25 microns. The difference between this interval and the median interval for a given cleavage division was used to generate a list of division times relative to the median. This number was then added to the time of mitosis for each cell’s mother cell and the timing of the wave initiation. This generated the vectors of “birth” and “death” times (relative to the start of the simulation) for every cell, where a cell comes into existence at the moment of its mother’s mitosis and exits the simulation upon its own mitosis. By summing the division intervals in this manner, cells in the vegetal hemisphere inherit the mitotic delay of their mothers. As a consequence, vegetal pole cells become increasingly delayed with each cleavage relative to cells in the animal hemisphere. This generates the increasingly broad distribution of cell interphase numbers along the AV axis, and is consistent with the delays reported by Anderson et al. (Anderson et al., 2017). Cleavage 14 was not simulated as a wave but rather as a stochastic event occurring at times drawn from a Gaussian distribution (mean = 75 minutes, standard deviation = 15 minutes) after cleavage 13. Our experiments were performed at 23 °C, whereas the mitotic waves on which the simulation is based were measured at 18 °C. To confer the simulation with similar timing as in our experiments, we rescaled the times of the simulation by a factor of 0.75.
To generate the distribution of individual cell sizes over simulated time, we noted the long-tailed distribution of cells sizes in our data. This distribution results from both asynchronous divisions and asymmetric divisions. At least two cleavages are clearly asymmetric in actual embryos. During the third cleavage, for example, Anderson et al. (Anderson et al., 2017) estimated the position of the division plane at one-third of egg radius from the animal pole. From observation, the fifth division is also asymmetric, although the degree of asymmetry has not been measured. The degree of asymmetry at during these two divisions affects the distribution of cell sizes at all times after those divisions, since daughter cell sizes are dependent on mother cell size.
We sought to explore a range of values for two free parameters describing the degree of asymmetry in the third and fifth divisions as the fraction of mother cell volume inherited by the smaller of the two daughters. We also noted that cells undergoing symmetric divisions likely will not place their division planes at exactly the centroid of the cell, i.e. there is likely some degree of variability (or noise) in the placement of the division plane. Nor will the asymmetric division planes be perfectly positioned. Variability in mitotic plane position has not been measured and was left as a third free parameter in the model, represented as the fractional standard deviation. The best-fitting parameter values were determined by searching a range of values between 0.05 and 0.5 for the degree of asymmetry in the third and fifth divisions. Variability in mitotic plane placement was calculated so as to generate a coefficient of variation (ratio of standard deviation to mean) in daughter volume variability between 0.01 and 0.7. For every parameter set, we calculated the sum of the square of differences (chi-square minimization) between the cumulative distributions function (CDF, the probability that a random variable will a value less than or equal to x; Figure S5B) of simulated and actual cell sizes for 40 measured embryos spanning a range of ages, between 484 and 20521 cells.
We found that even the best-fitting values in the three-parameter model provided a poor match to the observed distribution of cell sizes across ages. The closest match to data was obtained using an inherited volume of the smaller cell following both the third and fifth divisions of 0.26, consistent with division plane placement at 33% of mother cell diameter ((Anderson et al., 2017) and a variability about the mean cleavage plane position of 0.5. However, this large variability generated small cells at inappropriately early times, as well as cells of unrealistically small sizes (e.g., diameters of 5 microns or less) at late times. Overall, we could not find values in the three-parameter model capable of reproducing the observed cell size distribution at all times, because it was not possible to generate distributions with a narrow peak of relatively small cells simultaneously with the broad, long distribution of large sizes.
To properly account for the size distribution required the introduction of a fourth parameter to impose asymmetry on another division. We chose the eighth mitosis as a third asymmetric division largely because its modeled division plane orientation (perpendicular to the animal-vegetal axis) is parallel to the third and fifth planes which also show asymmetric division. It is noted that this is an arbitrary choice rooted in finding a single additional parameter. Using the same minimization of CDF differences described above, we found that the following values for the fraction of mother cell cytoplasm inherited by the smaller of the two daughters resulted in the best fit to the data: 0.29 (cycle 3), 0.22 (cycle 5), 0.18 (cycle 8), with variability in mitotic plane position set to generate a C.V. (standard deviation / mean) in daughter volume variability of 0.12 for all mitoses (both symmetric and asymmetric). Following mitosis, the positions of centroids of the daughter cells are calculated by finding the position halfway between the division plane and the plane tangential to the edge of the mother cell. After finding the best-fitting parameters, we ran 100 independent simulations. The results shown in Figures 5B, 5C, 5H–5J, and S5C are representative single embryo simulations, whereas Figures 5A, 5D, 5L, 5M, S5A, S5B, S5D and S5E comprise 100 simulated embryos.
Note that this model does not account for the formation of the blastocoel. To account for this, we performed a renormalization of the simulated volumes. For each measured embryo, the total volume was calculated by taking the sum of the measured volumes for all cells. Total volume of a simulated embryo was calculated using a diameter of 1.25 mm, then the volumes of simulated cells rescaled by multiplying by the ratio of actual volume to simulated volume. Although the centroids of daughter cells are constrained by the boundaries of the mother cells, the model does not otherwise explicitly account for packing. This is likely a source of the mismatch of model and observation, since the observed cell diameters will be smaller than the modeled diameters. This is one explanation for the trend of the larger deviation of the model from observation at early times when cells are larger: packing of larger cells is expected to produce larger measurable differences in cell size compared to small changes for small cells at later times.
Western blot analysis
Embryos at various developmental stages, 512-cell to 16000-cell (log2 cell no. = 9–14) (N = 5 each) were trisected into three parts, namely animal poles, intermediate parts and vegetal poles, using a hair loop. The intermediate parts were discarded. The animal poles and vegetal poles were collected in 1.5 ml microcentrifuge tubes and snap frozen in liquid nitrogen and stored at −80 °C. The samples were added with 100 μl of sample buffer (1 part of lysis buffer, 1 part of 2× loading buffer, 5% 2-merpoletoethanol and protease inhibitor cocktail; lysis buffer: 20 mM Tris-HCl pH8, 50 mM NaCl, 2 mM EDTA, 0.1% NP-40; 2× loading buffer: 100 mM Tris-HCl pH 6.8, 20% glycerol, 4 % SDS and 0.01% bromophenol blue) and pipetted 20 times with P200 tips. The samples were heated for 2 min at 95 °C and centrifuged for 5 min at 13,000 rpm, 4 °C. Ten microliters of samples were loaded onto Novex™ 16% Tris-Glycine Mini Gels (Thermo Fisher Scientific) and run at 100 V for 2 h at room temperature. Proteins were transferred onto nitrocellulose membrane (0.2 μm pore size; GE Healthcare) in transfer buffer (10 mM CAPS and 20% methanol) at 100 V for 2 h at 4 °C. Membranes were blocked with 5% non-fat milk in 1× TBST (20 mM Tris pH 7.6, 150 mM NaCl and 0.1% Tween-20) for 1 h at room temperature and incubated with rabbit polyclonal anti-histone H2B (1:1000 dilution), H3 (1:1000 dilution), H4 (1:1000 dilution), respectively, and mouse monoclonal anti-tubulin (1:2000 dilution) overnight at 4 °C. Membranes were washed with 1× TBST four times for 10 min each and incubated with secondary rabbit polyclonal IRDye 680 (1:10,000 dilution)and mouse monoclonal IRD800 (1:10,000 dilution) for 1 h at room temperature. Membranes were washed with 1× TBST four times for 10 min each, rinsed three times with deionized water and scanned under LI-COR Odyssey Infrared Imager (LI-COR Biosciences). The intensity of bands for histones and β-tubulin was measured by drawing the same sized box covering individual bands in ImageJ. The background was measured by drawing the same sized box in the empty regions next to individual bands. The net intensity for histones and β-tubulin was calculated by subtracting the background intensity from the measured intensity for individual bands. The relative levels of histones were further calculated by normalizing to β-tubulin and the fold change in histone levels at animal pole and vegetal pole was calculated by animal pole/vegetal pole.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical parameters, including samples numbers, mean and standard deviation, were included in Figures and Figure legends. The significance was determined by ANOVA analysis for multiple groups and student t-test for comparing two groups. ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05; and n.s., not significant.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-histone H3 | Abcam | Cat# ab1791; RRID: AB_302613 |
| Rabbit polyclonal anti-histone H4 | Abcam | Cat# ab10158; RRID: AB_296888 |
| Rabbit polyclonal anti-histone H2B | Abcam | Cat# ab1790; RRID: AB_302612 |
| Mouse monoclonal anti-tubulin | DSHB | Cat# E7-c |
| Rabbit polyclonal Alexa Fluor® 488 | Thermo Fisher Scientific | Cat# A11034; RRID: AB_2576217 |
| Rabbit polyclonal IRDye® 680RD | LI-COR | Cat# 926–68071; RRID: AB_10956166 |
| Mouse monoclonal IRDye® 800RD | LI-COR | Cat# 926–32210; RRID: AB_621842 |
| Streptavidin-HRP | Pierce | Cat# 21130 |
| SuperSignal West POCO Plus | Invitrogen | Cat# 34577 |
| RNeasy Minelute Cleanup Kit | Qiagen | Cat# 74204 |
| Western Blocking Reagent | Roche | Cat# 11921673001 |
| Click-iT™ Nascent RNA Capture Kit | Thermo Fisher Scientific | Cat# C10365 |
| Ovation® Human FFPE RNA-Seq Multiplex System 1–8 | NuGEN | Cat# 0340 |
| Agilent High Sensitivity DNA Kit | Agilent | Cat# 5067–4626 |
| SPRIselect Reagent | Beckman Coulter | Cat# B23317 |
| NEBNext® Library Quant Kit for Illumina® | NEB | Cat# E7630 |
| NSQ 500/550 Hi Output KT v2.5 (75 CYS) | Illumina | Cat# 20024906 |
| Biological Samples | ||
| Xenopus laevis embryos | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 5-Ethynyl Uridine (5-EU) | Thermo Fisher Scientific | Cat# E 10345 |
| Tetramethylrhodamine (TAMRA)-azide | Abcam | Cat# ab146486 |
| Biotin azide (PEG4 carboxamide-6-azidohexanyl biotin) | Thermo Fisher Scientific | Cat# C 10365 (component in Click-iT™ Nascent RNA Capture Kit) |
| Ascorbic acid | Sigma | Cat# A7506 |
| CuSO4 | Sigma | Cat# 61230 |
| TO-PRO-3 | Thermo Fisher Scientific | Cat# T3605 |
| Paraformaldehyde | EMS | Cat#15710-S |
| Ficoll® 400 | Sigma | Cat# F4375 |
| Hydrogen peroxide | Sigma | Cat# H1009 |
| Formamide | Thermo Fisher Scientific | Cat# AC181090010 |
| Benzyl alcohol | Sigma | Cat# 305197 |
| Benzyl benzoate | ACROS Organics | Cat# 105860010 |
| Experimental Models: Organisms/Strains | ||
| Xenopus laevis embryos | This paper | N/A |
| Oligonucleotides | ||
| Ode Fwd 5’ GAT CAT GCA CAT GTC AAG CC 3’ | This paper | N/A |
| Ode Rev 5’ TCT ACG ATC GAT CCA GCC 3’ | This paper | N/A |
| Gs17 Fwd 5’ CAGCCATGGAAAGACTGGT 3’ | This paper | N/A |
| Gs17 Rev 5’ TGGGTTCTGGAGTACGTTTATG 3’ | This paper | N/A |
| Id3 Fwd GAGCAGAGTCTGAGCATTG | This paper | N/A |
| Id3 Rev GAGTAGCAGCCGTTCATATC | This paper | N/A |
| Bix 1.2 Fwd 5’ ACAGCAACAAGTCCAACCCA 3’ | This paper | N/A |
| Bix 1.2 Rev 5’ CCCATGAGGATTCAGGGCAA 3’ | This paper | N/A |
| Chordin Fwd 5’ TGGGAGCAGTATAGGGTTAG 3’ | This paper | N/A |
| Chordin Rev 5’ GGGAACCATTCGGAGTTATG 3’ | This paper | N/A |
| Software and Algorithms | ||
| ImageJ Version 2.0.0 | NIH | https://imagej.nih.gov/ij/ |
| GraphPad Prism 7 | GraphPad | N/A |
| Imaris 9.2 | Bitplane | N/A |
| Adobe Illustrator CC 2017 | Adobe | N/A |
| OriginPro2017 | OriginLab | N/A |
| Excel 2017 | Microsoft | N/A |
| RStudio | RStudio Inc | https://www.rstudio.com |
| Python 3 | Jupyter notebook | http://jupyter.org |
| MATLAB | MathWorks | N/A |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Matthew Good (mattgood@pennmedicine.upenn.edu).
Highlights.
Quantitative, single-cell imaging of nascent transcription during early embryogenesis
Large-scale ZGA onset is not globally uniform; it follows a stereotypic spatial pattern
Patterned ZGA onset is explained by a cell sizer, not a simple timer or cycle counter
Reducing cell size is sufficient to trigger precocious ZGA in Xenopus embryos
ACKNOWLEDGEMENTS
We thank Peter Klein and Mary Mullins for critical reading of the manuscript, members of the Good, Klein, Mullins and Zaret labs at the University of Pennsylvania for discussion of the work and technical advice, the Cell and Developmental Biology Microscopy Core for imaging support, the Penn Bioinformatics Core for analyses, and National Xenopus Resource (NXR) for training and guidance on frogs. We also thank Wenchao Qian for assistance in preparing EU-RNA-seq library and John Tobias (Penn Genomic Analysis Core) for analysis of RNA-seq data. This work was supported in part by Burroughs Wellcome Fund, Charles E. Kaufman Foundation, the March of Dimes and the National Institute of General Medical Sciences (R35GM12874) (M.C.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes six figures.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Almouzni G, and Wolffe AP (1995). Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopus embryogenesis. The EMBO journal 14, 1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodeo AA, Jukam D, Straight AF, and Skotheim JM (2015). Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proceedings of the National Academy of Sciences of the United States of America 112, E1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GA, Gelens L, Baker JC, and Ferrell JE Jr. (2017). Desynchronizing Embryonic Cell Division Waves Reveals the Robustness of Xenopus laevis Development. Cell reports 21, 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe SA, and Wieschaus EF (2015). Coordinating Cell Cycle Remodeling with Transcriptional Activation at the Drosophila MBT. Curr Top Dev Biol 113, 113–148. [DOI] [PubMed] [Google Scholar]
- Boettiger AN, and Levine M (2009). Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325, 471–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JA, Weinreb C, Wagner DE, Megason S, Peshkin L, Kirschner MW, and Klein AM (2018). The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PD, Chao JA, Singer RH, and Marlow FL (2015). Dynamic visualization of transcription and RNA subcellular localization in zebrafish. Development 142, 1368–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SH, Tang Y, Miao L, Darwich-Codore H, Vejnar CE, Beaudoin J-D, Musaev D, Fernandez JP, Moreno-Mateos MA, and Giraldez AJ (2018). Brd4 and P300 regulate zygotic genome activation through histone acetylation. bioRxiv. [Google Scholar]
- Clute P, and Masui Y (1995). Regulation of the appearance of division asynchrony and microtubule-dependent chromosome cycles in Xenopus laevis embryos. Developmental biology 171, 273–285. [DOI] [PubMed] [Google Scholar]
- Collart C, Allen GE, Bradshaw CR, Smith JC, and Zegerman P (2013). Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science 341, 893–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart C, Owens ND, Bhaw-Rosun L, Cooper B, De Domenico E, Patrushev I, Sesay AK, Smith JN, Smith JC, and Gilchrist MJ (2014). High-resolution analysis of gene activity during the Xenopus mid-blastula transition. Development 141, 1927–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekens MP, Pelegri FJ, Maischein HM, and Nusslein-Volhard C (2003). The maternal-effect gene futile cycle is essential for pronuclear congression and mitotic spindle assembly in the zebrafish zygote. Development 130, 3907–3916. [DOI] [PubMed] [Google Scholar]
- Di Talia S, She R, Blythe SA, Lu X, Zhang QF, and Wieschaus EF (2013). Posttranslational control of Cdc25 degradation terminates Drosophila’s early cell-cycle program. Current biology : CB 23, 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Kiehle CP, and Schubiger G (1986). Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell 44, 365–372. [DOI] [PubMed] [Google Scholar]
- Farrell JA, Wang Y, Riesenfeld SJ, Shekhar K, Regev A, and Schier AF (2018). Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia HG, Tikhonov M, Lin A, and Gregor T (2013). Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Current biology : CB 23, 2140–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y, and Merten CA (2015). Dynein-Based Accumulation of Membranes Regulates Nuclear Expansion in Xenopus laevis Egg Extracts. Developmental cell 33, 562–575. [DOI] [PubMed] [Google Scholar]
- Heasman J, Wessely O, Langland R, Craig EJ, and Kessler DS (2001). Vegetal localization of maternal mRNAs is disrupted by VegT depletion. Developmental biology 240, 377–386. [DOI] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, et al. (2010). The genome of the Western clawed frog Xenopus tropicalis. Science 328, 633–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herberg S, Simeone A, Oikawa M, Jullien J, Bradshaw CR, Teperek M, Gurdon J, and Miyamoto K (2015). Histone H3 lysine 9 trimethylation is required for suppressing the expression of an embryonically activated retrotransposon in Xenopus laevis. Scientific reports 5, 14236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn P, Kircher M, Dahl A, Kelso J, Tomancak P, Kalinka AT, and Neugebauer KM (2014). The earliest transcribed zygotic genes are short, newly evolved, and different across species. Cell reports 6, 285–292. [DOI] [PubMed] [Google Scholar]
- Jao CY, and Salic A (2008). Exploring RNA transcription and turnover in vivo by using click chemistry. Proceedings of the National Academy of Sciences of the United States of America 105, 15779–15784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevtic P, and Levy DL (2015). Nuclear size scaling during Xenopus early development contributes to midblastula transition timing. Current biology : CB 25, 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevtic P, and Levy DL (2017). Both Nuclear Size and DNA Amount Contribute to Midblastula Transition Timing in Xenopus laevis. Scientific reports 7, 7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, and Futcher B (2007). The size of the nucleus increases as yeast cells grow. Molecular biology of the cell 18, 3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SR, Palfy M, Hilbert L, Kumar M, Karschau J, Zaburdaev V, Shevchenko A, and Vastenhouw NL (2017). Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukam D, Shariati SAM, and Skotheim JM (2017). Zygotic Genome Activation in Vertebrates. Developmental cell 42, 316–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DA, and Kimmel CB (1993). The zebrafish midblastula transition. Development 119, 447–456. [DOI] [PubMed] [Google Scholar]
- Karaiskos N, Wahle P, Alles J, Boltengagen A, Ayoub S, Kipar C, Kocks C, Rajewsky N, and Zinzen RP (2017). The Drosophila embryo at single-cell transcriptome resolution. Science. [DOI] [PubMed] [Google Scholar]
- Kimelman D, Kirschner M, and Scherson T (1987). The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell 48, 399–407. [DOI] [PubMed] [Google Scholar]
- Kotani T, Yasuda K, Ota R, and Yamashita M (2013). Cyclin B1 mRNA translation is temporally controlled through formation and disassembly of RNA granules. The Journal of cell biology 202, 1041–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, and Giraldez AJ (2014). Zygotic genome activation during the maternal-to-zygotic transition. Annual review of cell and developmental biology 30, 581–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, and Giraldez AJ (2013). Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichsenring M, Maes J, Mossner R, Driever W, and Onichtchouk D (2013). Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science 341, 1005–1009. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Li JM, Elemento O, Tavazoie S, and Wieschaus EF (2009). Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development 136, 2101–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann FR, and Nurse P (2007). Nuclear size control in fission yeast. The Journal of cell biology 179, 593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, and Kirschner M (1982a). A Major Developmental Transition in Early Xenopus Embryos I: Characterization and Timing of Cellular Changes At the Midblastula Stage. Cell, 675–686. [DOI] [PubMed] [Google Scholar]
- Newport J, and Kirschner M (1982b). A Major Developmental Transition in Early Xenopus Embryos II: Control of the Onset of Transcription. Cell, 687–696. [DOI] [PubMed] [Google Scholar]
- Palozola KC, Donahue G, Liu H, Grant GR, Becker JS, Cote A, Yu H, Raj A, and Zaret KS (2017). Mitotic transcription and waves of gene reactivation during mitotic exit. Science 358, 119–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjpe SS, Jacobi UG, van Heeringen SJ, and Veenstra GJ (2013). A genome-wide survey of maternal and embryonic transcripts during Xenopus tropicalis development. BMC Genomics 14, 762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nature methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshkin L, Wuhr M, Pearl E, Haas W, Freeman RM Jr., Gerhart JC, Klein AM, Horb M, Gygi SP, and Kirschner MW (2015). On the Relationship of Protein and mRNA Dynamics in Vertebrate Embryonic Development. Developmental cell 35, 383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau MN, Huet J, Sentenac A, and Mechali M (1994). Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell 77, 439–449. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nature biotechnology 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh N, and Ikegami S (1981). On the ‘clock’ mechanism determining the time of tissue-specific enzyme development during ascidian embryogenesis. II. Evidence for association of the clock with the cycle of DNA replication. J Embryol Exp Morphol 64, 61–71. [PubMed] [Google Scholar]
- Schier AF (2007). The maternal-zygotic transition: death and birth of RNAs. Science 316, 406–407. [DOI] [PubMed] [Google Scholar]
- Schmoller KM, Turner JJ, Koivomagi M, and Skotheim JM (2015). Dilution of the cell cycle inhibitor Whi5 controls budding-yeast cell size. Nature 526, 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapel LC, Zechner C, and Vastenhouw NL (2017). Uniform gene expression in embryos is achieved by temporal averaging of transcription noise. Genes & development 31, 1635–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, and Lipshitz HD (2009). The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042. [DOI] [PubMed] [Google Scholar]
- Venkataraman T, Dancausse E, and King ML (2004). PCR-based cloning and differential screening of RNAs from Xenopus primordial germ cells: cloning uniquely expressed RNAs from rare cells. Methods Mol Biol 254, 67–78. [DOI] [PubMed] [Google Scholar]
- Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, and Klein AM (2018). Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Hayden S, and Masui Y (2000). Transition of the blastomere cell cycle from cell size-independent to size-dependent control at the midblastula stage in Xenopus laevis. J Exp Zool 287, 128–144. [DOI] [PubMed] [Google Scholar]
- Yanai I, Peshkin L, Jorgensen P, and Kirschner MW (2011). Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Developmental cell 20, 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatulovskiy E, Berenson DF, Topacio BR, and Skotheim JM (2018). Cell growth dilutes the cell cycle inhibitor Rb to trigger cell division. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, and King ML (1996). Xenopus VegT RNA is localized to the vegetal cortex during oogenesis and encodes a novel T-box transcription factor involved in mesodermal patterning. Development 122, 4119–4129. [DOI] [PubMed] [Google Scholar]
- Zhang M, Skirkanich J, Lampson MA, and Klein PS (2017). Cell Cycle Remodeling and Zygotic Gene Activation at the Midblastula Transition. Adv Exp Med Biol 953, 441–487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.