Abstract
Spinal Muscular Atrophy (SMA) is a disorder characterized by the degeneration of motor neurons in the spinal cord, leading to muscular atrophy. In the majority of cases, SMA is caused by the homozygous absence of the SMN1 gene. The disease severity of SMA is strongly influenced by the copy number of the closely related SMN2 gene. In addition, an SMN variant lacking exons 7 and 8 has been reported in 8% and 23% of healthy Swedish and Spanish individuals respectively. We tested 1255 samples from the 1000 Genomes Project using a new version of the multiplex ligation-dependent probe amplification (MLPA) P021 probemix that covers each SMN exon. The SMN variant lacking exons 7 and 8 was present in up to 20% of individuals in several Caucasian populations, while being almost completely absent in various Asian and African populations. This SMN1/2Δ7–8 variant appears to be derived from an ancient deletion event as the deletion size is identical in 99% of samples tested. The average total copy number of SMN1, SMN2 and the SMN1/2Δ7–8 variant combined was remarkably comparable in all populations tested, ranging from 3.64 in Asian to 3.75 in African samples.
Introduction
Spinal Muscular Atrophy (SMA) is one of the most prevalent genetic disorders in Caucasians with an incidence of approximately 1:10.000 newborns. SMA is a recessive disorder with a population dependent carrier frequency ranging from 1:35 in Caucasians to 1:91 in African Americans [1, 2]. Based on age of onset and maximum motor milestones achieved, SMA is divided into different disease categories, distinguished as type 0 to type IV [3–5]. Type 0 and I are the most severe types with SMA onset beginning at birth, patients achieving minimal motor milestones, and a life expectancy of around two years. Intermediate types, type II and III, are milder forms of the disorder where patients are able to sit or walk and most individuals survive beyond 10 years of age. Type IV is the mildest form of SMA with an adult-onset of the disorder and patients having a normal life expectancy [4, 5]. With the recent availability of treatment options, newborn screening and pre-symptomatic treatment of SMA is now being considered in several countries [6].
In more than 95% of SMA cases, the disease is caused by the complete absence of the SMN1 gene or by SMN1 to SMN2 gene conversions [7–11]. In the majority of the remaining cases, intragenic mutations or partial deletions in the SMN1 gene cause the disease [12, 13]. The copy number of both SMN1 and SMN2 is very variable due to instability of the 5q13.2 region, leading to a high frequency of deletion/duplication and gene conversion events [11, 14–17]. In the majority of populations, most individuals have two copies of both SMN1 and the almost identical SMN2 gene [18]. There is a single clinically relevant nucleotide difference between SMN1 and SMN2, c.840C>T located in exon 7 [19]. This single nucleotide difference affects splicing of the last coding exon, resulting in 90% of SMN2 transcripts forming a dysfunctional SMN protein [20, 21]. The complete absence of SMN2 has no effect on healthy individuals. However, for patients who have no functional SMN1 gene copies, approximately 10% of SMN functionality is retained per copy of SMN2. This results in an inverse correlation between the number of SMN2 copies and disease severity [8, 13, 22–24].
Both SMN1 and SMN2 consist of nine exons, historically named exons 1, 2a, 2b, 3, 4, 5, 6, 7 and 8. Using multiplex ligation-dependent probe amplification (MLPA), the presence of an SMN1 or SMN2 gene variant lacking exons 7 and 8 has been reported to have a frequency of 8% in healthy Swedish individuals [18], and 23% in healthy Spanish individuals [25]. During initial validation tests of a new version, B1, of the SALSA MLPA Probemix P021 SMA at MRC Holland the copy number status of both SMN1 and SMN2 were determined in a large number of blood derived DNA samples from Dutch individuals. Results showed that approximately 20% of the samples had a lower exon 7 and 8 copy number for the SMN1 and SMN2 genes combined as compared to the exon 1–6 copy number. This validation study led us to investigate the prevalence of an SMN variant lacking exons 7–8 in multiple populations. Our study showed a strong inverse correlation between the copy number of SMN gene variants lacking exons 7 and 8 and the SMN2 copy number, corroborating a report by Alias et al. [26]. This suggests the SMN1/2Δ7–8 variant in the general population to be predominantly derived from SMN2 deletion events, in concordance with the difference in deletion frequency between SMN1 and SMN2 [24].
In this study 1255 samples from the 1000 Genomes Project comprising of multiple ethnic groups were analyzed using the SALSA MLPA Probemix P021-B1 SMA, which can readily detect the SMN1/2Δ7–8 variant (Fig 1). The copy numbers for SMN1, SMN2 and SMN1/2Δ7–8 were determined for each sample and compared for each ethnic group. Our results show that the frequency of the SMN1/2Δ7–8 variant is strongly population dependent.
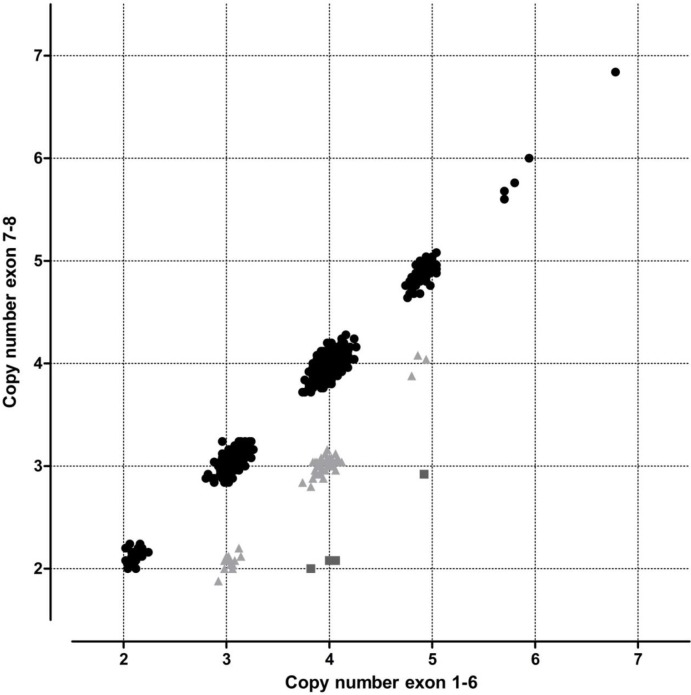
Fig 1. SMN copy number distribution.
The distribution of the total copy number of exons 1–6 and exon 7–8 of the SMN1 and SMN2 genes combined. SMN1/2Δ7–8 copies can be identified by a higher copy number of exons 1–6 as compared to exons 7 and 8. Black dots represent samples not carrying SMN1/2Δ7–8 copies, grey triangles samples containing one copy of SMN1/2Δ7–8 and dark grey squares samples containing two copies of SMN1/2Δ7–8.
Results
We tested 1255 DNA samples from the 1000 Genomes Project collection with a new B1 version of the SALSA MLPA Probemix P021 SMA. This new P021 probemix has increased gene coverage for the SMN genes as compared to previous versions and covers each exon of the SMN genes with one or more probes. Sequence differences in exon 7 (c.840C>T) and 8 (c.*239G>A) of both SMN1 and SMN2 are covered by four probes, in addition 17 probes detect sequences present in both SMN1 and SMN2. Of the latter, ten probes detect sequences in the exon 1–6 region and seven probes detect sequences in the exon 7–8 region.
Upon data analysis, a high frequency of SMN1/2Δ7–8 variant alleles were observed in specific populations (Table 1), which is in line with previous reports [18, 25]. No other partial gene deletions or duplications were observed. Using synthetic MLPA probes, we located the breakpoint of the SMN1/2Δ7–8 variant to be in intron 6, between nucleotides 4679 and 2363 before exon 7 (Fig 2). 13 samples were tested and all showed the same probes deviating, indicating that the breakpoint is in the same 2.3 kb region.
Table 1. Number of samples containing the SMN1/2Δ7–8 variant per tested population.
| 1000 Genomes Project samples | Sample count | One copy SMN1/2Δ7–8 | Two copies SMN1/2Δ7–8 | At least one copy SMN1/2Δ7–8 |
|---|---|---|---|---|
| Country of origin | Count (%) | Count (%) | Count (%) | |
| Italy | 100 | 18 (18.0) | 1 (1.0) | 19 (19.0) |
| Spain | 96 | 13 (13.5) | 2 (2.1) | 15 (15.6) |
| England & Scotland | 100 | 19 (19.0) | 1 (1.0) | 20 (20.0) |
| Finland | 100 | 8 (8.0) | 0 | 8 (8.0) |
| Sum Europe | 396 | 58 (14.6) | 4 (1.0) | 62 (15.7) |
| Japan | 100 | 0 | 0 | 0 |
| China | 100 | 0 | 0 | 0 |
| Bangladesh | 104 | 1 (1.0) | 0 | 1 (1.0) |
| Sum Asia | 304 | 1 (0.3) | 0 | 1 (0.3) |
| Kenya | 99 | 1 (1.0) | 0 | 1 (1.0) |
| Sierra Leone & Gambia | 150 | 0 | 0 | 0 |
| Nigeria | 97 | 0 | 0 | 0 |
| Sum Africa | 346 | 1 (0.3) | 0 | 1 (0.3) |
| Gujarati Indians | 101 | 7 (6.9) | 0 | 7 (6.9) |
| Peru & Colombia | 108 | 11 (10.2) | 0 | 11 (10.2) |
| Sum Americas | 209 | 18 (8.6) | 0 | 18 (8.6) |
| Total samples | 1255 | 78 (6.2) | 4 (0.3) | 82 |
Fig 2. Visualization of both SMN1/2Δ7–8 and exon 1–6 deletion of SMN1 breakpoint locations within intron 6.
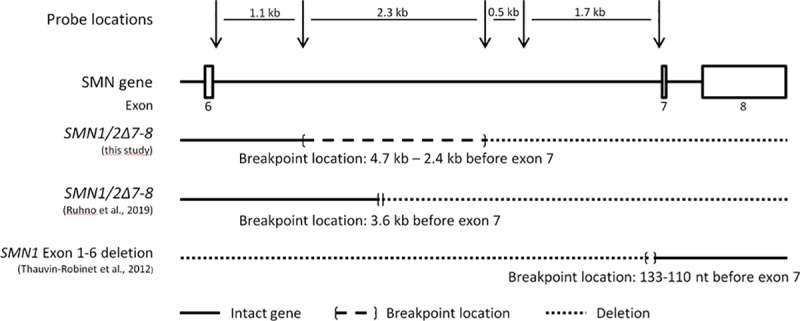
Arrows in the top line indicate the location of the five additional probes designed to map the breakpoint of the SMN variant lacking exons 7 and 8. The third and fourth lines show the breakpoints of the truncated SMN genes.
Gap-PCR was used to determine if the deletion of exons 7 and 8 of the 82 SMN1/2Δ7–8 positive samples was of equal size to the exon 7 and 8 deletion described by Ruhno et al [27]. A deletion of equal size was found in 99% of positive samples (81/82). None of 24 negative samples tested positive by this Gap-PCR.
The total number of copies for SMN1, SMN2 and SMN1/2Δ7–8 in a sample could reliably be quantified by using the median value of the ten MLPA probes for the exon 1–6 region (Fig 1). The total copy number for SMN1, SMN2 and SMN1/2Δ7–8 ranged from 2 to 7 in our sample cohort. Samples that contained the SMN1/2Δ7–8 variant could clearly be identified by subtracting the median copy number obtained from the seven MLPA probes detecting exons 7 and 8 of both SMN1 and SMN2 from the median copy number obtained from the ten MLPA probes detecting exons 1–6 of both SMN1 and SMN2 (Fig 1). In total, we observed 78 samples with one copy and four samples with two copies of the SMN1/2Δ7–8 gene variant among the 1255 samples tested.
Marked differences in the frequency of the SMN1/2Δ7–8 gene variant were observed between different populations, as shown in Tables 1 and 2. The frequency of SMN1/2Δ7–8 copies in the European populations tested was between 8–20%, in line with previously reported frequencies, whereas the number of African and Asian samples with an SMN1/2Δ7–8 copy was virtually zero (1 in 304 Asian samples, 1 in 346 African samples, 0.3%, Tables 1 and 2). In addition, the average number of either SMN1 or SMN2 copies in each individual was found to be strongly population dependent with African populations showing much higher SMN1 and lower SMN2 copy numbers in comparison to the other populations examined (Table 2). The number of SMN1 copies ranged from an average of 2.70 in African samples to 2.03 in European samples. SMN2 copy numbers were highest in Asian samples with an average of 1.56 and lowest in African samples with an average of 1.05. The total number of SMN copies (SMN1, SMN2 and SMN1/2Δ7–8 combined) was very similar among the different ethnic populations, ranging from 3.64 in Asian to 3.75 in African samples. However, when excluding the SMN1/2Δ7–8 copies from the average combined SMN1 and SMN2 copy number the European population shows a marked difference, with the average copy number dropping from 3.70 to 3.54 while the other populations remain essentially unchanged (Table 2).
Table 2. Average number of SMN copies per population.
Asterisks (*) indicate a significant difference in prevalence.
| SMN1 copy number | Europe n = 396 Count (%) |
Asia n = 304 Count (%) |
Africa n = 346 Count (%) |
Americas n = 209 Count (%) |
||||
|---|---|---|---|---|---|---|---|---|
| 1 | 11 (2.8) | 2 (0.7) | 1 (0.3) | 4 (1.9) | ||||
| 2 | 364 (91.9) | 279 (91.8) | 152 (43.9) | 177 (84.7) | ||||
| 3 | 21 (5.3) | 22 (7.2) | 143 (41.3) | 27 (12.9) | ||||
| 4 | 0 | 1 (0.3) | 50 (14.5) | 1 (0.5) | ||||
| Average | 2.03 | 2.07 | 2.70 | 2.12 | ||||
| SMN2 copy number | ||||||||
| 0 | 27 (6.8) | 12 (3.9) | 80 (23.1) | 15 (7.2) | ||||
| 1 | 158 (39.9) | 117 (38.4) | 174 (50.3) | 82 (39.2) | ||||
| 2 | 195 (49.2) | 168 (55.3) | 86 (24.9) | 107 (51.2) | ||||
| 3 | 15 (3.8) | 7 (2.3) | 6 (1.7) | 5 (2.4) | ||||
| 4 | 1 (0.3) | 0 | 0 | 0 | ||||
| Average | 1.51 | 1.56 | 1.05 | 1.49 | ||||
| SMN1/2Δ7–8 copy number | ||||||||
| 1 | 58 (14.6) | 1 (0.3) | 1 (0.3) | 18 (8.6) | ||||
| 2 | 4 (1.0) | 0 | 0 | 0 | ||||
| Average | 0.17* | 0.003* | 0.003* | 0.09 | ||||
| Average copy number of SMN1,SMN2 and SMN1/2Δ7–8 | 3.70 | 3.64 | 3.75 | 3.69 | ||||
| Average copy number of SMN1 and SMN2 | 3.54 | 3.63 | 3.75 | 3.61 | ||||
* p < 0.001
The presence of a SMN1/2Δ7–8 gene variant is strongly linked to the SMN2 copy number as shown in Table 3. A significant difference in SMN1/2Δ7–8 presence was found between individuals carrying two or more copies of either SMN1 or SMN2 (p = 1.48e-12, OR: 39.6 [95%CI: 6.9–1573.9]). Presence of SMN1/2Δ7–8 was nearly absent (0.17%) within the group of two or more SMN2 copies while more frequently found in carriers of two or more SMN1 copies (6.73%). When comparing the presence of SMN1/2Δ7–8 between the groups of one or less copies of either SMN1 or SMN2 no significant difference was found (p = 0.39). Samples without SMN1/2Δ7–8 copies had on average 2.25 SMN1 copies and 1.44 SMN2 copies. While samples that contained one or two SMN1/2Δ7–8 copies had on average 2.02 SMN1 copies and 0.76 SMN2 copies. 50.2% of samples (589 out of 1173) without the SMN1/2Δ7–8 variant had two or more SMN2 copies, while in the 82 samples that contained one or two SMN1/2Δ7–8 copies, only one sample had two SMN2 copies.
Table 3. SMN1 and SMN2 copy numbers in samples with and without SMN1/2Δ7–8.
| SMN1 copy number | All samples Count (%) |
Samples without SMN1/2Δ7–8 copies Count (%) |
Samples with one SMN1/2Δ7–8 copy Count (%) |
Samples with two SMN1/2Δ7–8 copies Count (%) |
|||
|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | |||
| 1 | 18 (1.4) | 14 (1.2) | 2 (2.6) | 2 (50) | |||
| 2 | 972 (77.5) | 900 (76.7) | 70 (89.7) | 2 (50) | |||
| 3 | 215 (17.1) | 209 (17.8) | 6 (7.7) | 0 | |||
| 4 | 50 (4.0) | 50 (4.3) | 0 | 0 | |||
| Total samples | 1255 | 1173 | 78 | 4 | |||
| Average SMN1 copy # | 2.24 | 2.25 | 2.05 | 1.50 | |||
| SMN2 copy number | |||||||
| 0 | 134 (10.7) | 113 (9.6) | 18 (23.1) | 3 (75) | |||
| 1 | 531 (42.3) | 471 (40.2) | 59 (75.6) | 1 (25) | |||
| 2 | 556 (44.3) | 555 (47.3) | 1 (1.3) | 0 | |||
| 3 | 33 (2.6) | 33 (2.8) | 0 | 0 | |||
| 4 | 1 (0.1) | 1 (0.1) | 0 | 0 | |||
| Total samples | 1255 | 1173 | 78 | 4 | |||
| Average SMN2 copy # | 1.39 | 1.44 | 0.78 | 0.25 | |||
Discussion
Our results show the frequency of having at least one SMN1/2Δ7–8 copy to be 15–20% in three European populations (Italian, Spanish, and English/Scottish), which is in line with previous reports [25]. Interestingly, only two out of 650 Asian and African samples tested contained an SMN1/2Δ7–8 copy, which is approximately 50 times lower than in the tested European populations (Table 1). Furthermore, 99% of 82 SMN1/2Δ7–8 positive samples had a deletion of equal size as shown by gap-PCR. This strongly suggests that the great majority of SMN1/2Δ7–8 gene variant copies are the result of an ancient founder event. We noticed an intermediate frequency of SMN1/2Δ7–8 copies in samples from native North American (Gujarati Indians from Houston; 7%), Peruvian and Colombian (combined 10%), and Finnish individuals (8%).
Our results also confirm previous reports of several African populations having a higher average SMN1 copy number [1, 2], with an average SMN1 copy number of 2.70 in African populations and 2.03–2.19 in non-African populations. The increased SMN1 copy number in Africans is accompanied by a lower average SMN2 copy number and essentially complete absence of SMN1/2Δ7–8 copies. Interestingly, our results show that the combined copy number of SMN1, SMN2 and SMN1/2Δ7–8 is remarkably constant across all populations tested (Table 2), ranging from 3.64 in Asian samples to 3.75 in African samples.
The frequent occurrence of an SMN1 or SMN2 variant lacking exons 7–8 was reported first by Arkblad et al in 2006 [18]. Thus far, little attention has been given to this SMN gene variant, likely due to technical difficulties in variant detection [28]. The development of an improved version of the SALSA MLPA Probemix P021 SMA has made the identification and copy number determination of SMN genes, including the SMN1/2Δ7–8 variant, simple and robust. Our results on 1255 samples from the 1000 Genomes Project show that the presence of this SMN1/2Δ7–8 variant is almost completely restricted to DNA samples that have an SMN2 copy number of zero or one (Table 3). Contrary to our results Ruhno et al [27] presented evidence showing that SMN1/2Δ7–8 can also originate from SMN1. This discrepancy could be due to the absence of patients and the small number of SMA carriers within our cohort resulting from the lower frequency of SMN1 deletion events compared to that of SMN2 [24]. Only one out of the 590 samples with two SMN2 copies contained an SMN1/2Δ7–8 copy, while 81 out of the 665 samples (12%) with zero or only one SMN2 copy had at least one SMN1/2Δ7–8 copy. This finding is in agreement with earlier reports [26] where a strong correlation between SMN2 copy number and the presence of one or two copies of the SMN1/2Δ7–8 variant was found, further suggesting that most SMN1/2Δ7–8 variant copies are originally derived from an SMN2 deletion event.
Using additional MLPA probes, we localized the 3’ breakpoint of SMN1/2Δ7–8 to intron 6 between nucleotides 4679 and 2363 before exon 7, in 13 samples examined (Fig 2). This is in line with a recent report from Ruhno et al [27] who describe a 6310 bp exon 7–8 deletion that has breakpoints at 3643 nucleotides before exon 7 and 2164 nucleotides after the start of exon 8. The SMN1/2Δ7–8 breakpoint, located in intron 6, is more than 2 kb from the 3’ breakpoint of a known SMN1 exon 1–6 deletion that is located between 133 and 110 nucleotides before exon 7 [15]. This latter breakpoint does not contain the recurring repeat sequence identified by Ruhno et al that contains the breakpoint of SMN1/2Δ7–8. Thus, the SMN1/2Δ7–8 copies are not the reciprocal of the recurrent SMN1 exon 1–6 deletion [15, 18]. It should be noted that the presence of an SMN1/2Δ7–8 copy could mask an SMN1 exon 1–6 deletion, although the occurrence would be very rare this could have clinical relevance.
SMN2 copy number is the most important modifier of SMA disease severity. Next to this, the presence of the c.859G>C variant and/or the intron 6 A-44G, A-549G and C-1897T variants in SMN2 also seem to be correlated with a milder phenotype [27, 29]. One example of a disease modifier located outside the SMN region is the PLS3 gene, located on the X chromosome. In siblings, with identical SMN genotypes but discordant phenotypes, the expression level of PLS3, is found to influence the severity of the SMA phenotype. High expression levels were found in lymphoblastoid cell lines of asymptomatic but not in symptomatic siblings [30]. The effect of PLS3 expression is gender specific and not completely penetrant [30, 31] therefore there must be additional disease modifiers inside or outside the SMN region. Several articles have reported healthy adults that lack all SMN1 exon 7 copies [18, 30, 32–41]. In some cases, the absence of symptoms was attributed to a high SMN2 copy number (4–5 copies). However, the identification of individuals with homozygous SMN1 loss and a physique described as “extremely well-muscled” [36] or “athletically built” [32]. As these unaffected individuals had the same SMN2 copy number as their affected siblings, additional disease modifiers outside the SMN genes are expected. The start of SMA newborn screening and pre-symptomatic treatment makes further research on SMA disease modifiers urgent. The SMN1/2Δ7–8 copy number is unlikely to be a strong disease modifier as its presence was not related to disease severity in the patient cohort studied by Ruhno et al [27]. Besides, Calucho et al. [25] identified 13 SMA patient samples with an SMN1/2Δ7–8 copy (2%, 13/625), and of these, 11 patients were diagnosed as type I.
The SALSA MLPA Probemix P021-B1 version provides accurate copy number determination of the SMN1 and SMN2 genes and the SMN1/2Δ7–8 variant and will likely result in a more clear picture of the functionality of the SMN1/2Δ7–8 variant in the future when more SMA patient samples are analyzed.
Methods
Subjects
Cell line derived DNA samples from the 1000 Genomes Project [42] were obtained from Coriell laboratories (https://www.coriell.org/). Subsets from the following panels were tested: Bengali in Bangladesh (panel MGP00022); British from England and Scotland (MGP00003); Colombian in Medellin (MGP00005); Esan in Nigeria (MGP00023); Finnish in Finland (MGP00001); Gambian in Western Division-Mandinka (MGP00019); Gujarati Indians in Houston, Texas (MGP00018); Han Chinese South (MGP00002); Iberian populations in Spain (MGP00010); Japanese in Tokyo, Japan (MGP00009); Luhya in Webuye, Kenya (MGP00008); Mende in Sierra Leone (MGP00021); Peruvian in Lima, Peru (MGP00011); Toscani in Italia (MGP00007) and Yoruba in Ibadan, Nigeria (MGP00013). A total of 1255 samples were tested, for which the cell line catalog IDs are listed in S1 Table. The number of samples per subset are shown in Table 1.
Copy number analysis
A new version, B1, of the SALSA MLPA Probemix P021 SMA was developed at MRC Holland (MRC Holland, Amsterdam, The Netherlands). In contrast to previous P021 versions all exons of the SMN genes are covered by at least one probe in this B1 version. In addition to four SMN1 and SMN2 specific probes, each detecting a single nucleotide variation in exon 7 and 8, the probemix contains seven probes that detect a sequence present in, or close to, exon 7 or exon 8 of both SMN1 and SMN2, as well as ten probes that detect sequences present in exons 1–6 of both SMN1 and SMN2.
All MLPA reactions were performed according to the General MLPA protocol (www.mlpa.com). Reactions were analyzed by capillary electrophoresis using the Applied Biosystems 3130 (Life Technologies/Applied Biosystems, Carlsbad, CA). Data analysis was performed using Coffalyser.Net software (www.mlpa.com).
SMN1 and SMN2 copy numbers were determined with the use of the SMN1 and SMN2 specific probes for exon 7. The SMN1/2Δ7–8 copy number was determined by subtracting the median copy number obtained for the seven MLPA probes that detect a sequence present in exons 7 and 8 of both SMN1 and SMN2 from the median copy number obtained for the ten MLPA probes that detect a sequence present in exons 1–6 of both SMN1 and SMN2.
Five additional probes were designed to investigate the 3’ breakpoint of the SMN1/2Δ7–8 variant. These probes are specific for sequences located in intron 6 of the SMN genes (Fig 2). Out of the 82 samples containing a SMN1/2Δ7–8 variant a random subset of 13 samples were tested using these probes.
Hot start gap-PCR was performed in order to determine whether the size of the deletion in SMN1/2Δ7–8 positive samples is equal to the deletion of 6310 nt described by Ruhno et al [27]. Primers (Forward: /56FAM/CAGTTATCTGACTGTAACACTGTAGGC, Reverse: GTTGTTGCTTATGCTGGTCTTG) were designed based on breakpoints described by Ruhno et al [27] and positive samples generated an amplicon of 583 nucleotides.
Statistical analysis
Statistics analysis was performed using R version 3.5.1 [43] and Rstudio version 1.1.463 [44]. SMN1 copy numbers, SMN2 copy numbers and SMN1/2Δ7–8 copy numbers were compared using the Fischer’s exact test with the basic R function fisher.test. SMN1/2Δ7–8 results were dichotomized in a group with either 0 copies or in a group with one or more copies. Presence of the SMN1/2Δ7–8 was compared within two groups. In the first group SMN1/2Δ7–8 presence was compared between carriers of one or less copies of either SMN1 or SMN2. In the second group SMN1/2Δ7–8 presence was compared between individuals carrying two or more copies of either SMN1 or SMN2. Individuals carrying one copy of both SMN1 and SMN2 were excluded from the analysis (n = 2). Furthermore, continent data was dichotomized per continent and subsequently compared using the Fischer’s exact test. Bonferroni correction was applied to correct for multiple testing.
Supporting information
The median copy number value based on both the SMN exon 1–6 probes and SMN exon 7–8 probes is provided for each DNA sample tested, as shown in Fig 1. For each DNA sample, the Coriell Institute calatog ID is provided.
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work. RV, RS, MC, AGM, MR, MZ, NM, RB, RY, RK, SS, SL are employees at MRC Holland. JS is owner of MRC Holland. MRC Holland provided support in the form of salaries for authors and employees of MRC Holland RV, RS, MC, AGM, MR, MZ, NM, RB, RY, RK, SS, SL, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section. JS (the owner of MRC Holland) played a role in study design, decision to publish and preparation of the manuscript.
References
- 1.Hendrickson BC, Donohoe C, Akmaev VR, Sugarman EA, Labrousse P, Boguslavskiy L, et al. Differences in SMN1 allele frequencies among ethnic groups within North America. Journal of medical genetics. 2009;46(9):641–4. 10.1136/jmg.2009.066969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugarman EA, Nagan N, Zhu H, Akmaev VR, Zhou Z, Rohlfs EM, et al. Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: clinical laboratory analysis of >72,400 specimens. European journal of human genetics: EJHG. 2012;20(1):27–32. 10.1038/ejhg.2011.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolb SJ, Kissel JT. Spinal Muscular Atrophy. Neurologic clinics. 2015;33(4):831–46. 10.1016/j.ncl.2015.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finkel RS, Mercuri E, Meyer OH, Simonds AK, Schroth MK, Graham RJ, et al. Diagnosis and management of spinal muscular atrophy: Part 2: Pulmonary and acute care; medications, supplements and immunizations; other organ systems; and ethics. Neuromuscular disorders: NMD. 2018;28(3):197–207. 10.1016/j.nmd.2017.11.004 . [DOI] [PubMed] [Google Scholar]
- 5.Mercuri E, Finkel RS, Muntoni F, Wirth B, Montes J, Main M, et al. Diagnosis and management of spinal muscular atrophy: Part 1: Recommendations for diagnosis, rehabilitation, orthopedic and nutritional care. Neuromuscular disorders: NMD. 2018;28(2):103–15. 10.1016/j.nmd.2017.11.005 . [DOI] [PubMed] [Google Scholar]
- 6.Wirth B, Barkats M, Martinat C, Sendtner M, Gillingwater TH. Moving towards treatments for spinal muscular atrophy: hopes and limits. Expert opinion on emerging drugs. 2015;20(3):353–6. 10.1517/14728214.2015.1041375 . [DOI] [PubMed] [Google Scholar]
- 7.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–65. 10.1016/0092-8674(95)90460-3 . [DOI] [PubMed] [Google Scholar]
- 8.McAndrew PE, Parsons DW, Simard LR, Rochette C, Ray PN, Mendell JR, et al. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. American journal of human genetics. 1997;60(6):1411–22. 10.1086/515465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell L, Potter A, Ignatius J, Dubowitz V, Davies K. Genomic variation and gene conversion in spinal muscular atrophy: implications for disease process and clinical phenotype. American journal of human genetics. 1997;61(1):40–50. 10.1086/513886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiDonato CJ, Ingraham SE, Mendell JR, Prior TW, Lenard S, Moxley RT 3rd, et al. Deletion and conversion in spinal muscular atrophy patients: is there a relationship to severity? Annals of neurology. 1997;41(2):230–7. 10.1002/ana.410410214 . [DOI] [PubMed] [Google Scholar]
- 11.Hahnen E, Schonling J, Rudnik-Schoneborn S, Zerres K, Wirth B. Hybrid survival motor neuron genes in patients with autosomal recessive spinal muscular atrophy: new insights into molecular mechanisms responsible for the disease. American journal of human genetics. 1996;59(5):1057–65. [PMC free article] [PubMed] [Google Scholar]
- 12.Alias L, Bernal S, Fuentes-Prior P, Barcelo MJ, Also E, Martinez-Hernandez R, et al. Mutation update of spinal muscular atrophy in Spain: molecular characterization of 745 unrelated patients and identification of four novel mutations in the SMN1 gene. Human genetics. 2009;125(1):29–39. 10.1007/s00439-008-0598-1 . [DOI] [PubMed] [Google Scholar]
- 13.Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. American journal of human genetics. 2002;70(2):358–68. 10.1086/338627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markowitz JA, Singh P, Darras BT. Spinal muscular atrophy: a clinical and research update. Pediatr Neurol. 2012;46(1):1–12. 10.1016/j.pediatrneurol.2011.09.001 . [DOI] [PubMed] [Google Scholar]
- 15.Thauvin-Robinet C, Drunat S, Saugier Veber P, Chantereau D, Cossee M, Cassini C, et al. Homozygous SMN1 exons 1–6 deletion: pitfalls in genetic counseling and general recommendations for spinal muscular atrophy molecular diagnosis. American journal of medical genetics Part A. 2012;158A(7):1735–41. 10.1002/ajmg.a.35402 . [DOI] [PubMed] [Google Scholar]
- 16.Wirth B, Schmidt T, Hahnen E, Rudnik-Schoneborn S, Krawczak M, Muller-Myhsok B, et al. De novo rearrangements found in 2% of index patients with spinal muscular atrophy: mutational mechanisms, parental origin, mutation rate, and implications for genetic counseling. American journal of human genetics. 1997;61(5):1102–11. 10.1086/301608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Steege G, Grootscholten PM, Cobben JM, Zappata S, Scheffer H, den Dunnen JT, et al. Apparent gene conversions involving the SMN gene in the region of the spinal muscular atrophy locus on chromosome 5. American journal of human genetics. 1996;59(4):834–8. [PMC free article] [PubMed] [Google Scholar]
- 18.Arkblad EL, Darin N, Berg K, Kimber E, Brandberg G, Lindberg C, et al. Multiplex ligation-dependent probe amplification improves diagnostics in spinal muscular atrophy. Neuromuscular disorders: NMD. 2006;16(12):830–8. 10.1016/j.nmd.2006.08.011 . [DOI] [PubMed] [Google Scholar]
- 19.Burglen L, Lefebvre S, Clermont O, Burlet P, Viollet L, Cruaud C, et al. Structure and organization of the human survival motor neurone (SMN) gene. Genomics. 1996;32(3):479–82. 10.1006/geno.1996.0147 . [DOI] [PubMed] [Google Scholar]
- 20.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(11):6307–11. 10.1073/pnas.96.11.6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wirth B, Garbes L, Riessland M. How genetic modifiers influence the phenotype of spinal muscular atrophy and suggest future therapeutic approaches. Curr Opin Genet Dev. 2013;23(3):330–8. 10.1016/j.gde.2013.03.003 . [DOI] [PubMed] [Google Scholar]
- 22.Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, et al. The survival motor neuron protein in spinal muscular atrophy. Human molecular genetics. 1997;6(8):1205–14. 10.1093/hmg/6.8.1205 . [DOI] [PubMed] [Google Scholar]
- 23.Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nature genetics. 1997;16(3):265–9. 10.1038/ng0797-265 . [DOI] [PubMed] [Google Scholar]
- 24.Mailman MD, Heinz JW, Papp AC, Snyder PJ, Sedra MS, Wirth B, et al. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genetics in medicine: official journal of the American College of Medical Genetics. 2002;4(1):20–6. 10.1097/00125817-200201000-00004 . [DOI] [PubMed] [Google Scholar]
- 25.Calucho M, Bernal S, Alias L, March F, Vencesla A, Rodriguez-Alvarez FJ, et al. Correlation between SMA type and SMN2 copy number revisited: An analysis of 625 unrelated Spanish patients and a compilation of 2834 reported cases. Neuromuscular disorders: NMD. 2018;28(3):208–15. 10.1016/j.nmd.2018.01.003 . [DOI] [PubMed] [Google Scholar]
- 26.Alias L, Bernal S, Barcelo MJ, Also-Rallo E, Martinez-Hernandez R, Rodriguez-Alvarez FJ, et al. Accuracy of marker analysis, quantitative real-time polymerase chain reaction, and multiple ligation-dependent probe amplification to determine SMN2 copy number in patients with spinal muscular atrophy. Genet Test Mol Biomarkers. 2011;15(9):587–94. 10.1089/gtmb.2010.0253 . [DOI] [PubMed] [Google Scholar]
- 27.Ruhno C, McGovern VL, Avenarius MR, Snyder PJ, Prior TW, Nery FC, et al. Complete sequencing of the SMN2 gene in SMA patients detects SMN gene deletion junctions and variants in SMN2 that modify the SMA phenotype. Human genetics. 2019;138(3):241–56. 10.1007/s00439-019-01983-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheng-Yuan Z, Xiong F, Chen YJ, Yan TZ, Zeng J, Li L, et al. Molecular characterization of SMN copy number derived from carrier screening and from core families with SMA in a Chinese population. European journal of human genetics: EJHG. 2010;18(9):978–84. 10.1038/ejhg.2010.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prior TW, Krainer AR, Hua Y, Swoboda KJ, Snyder PC, Bridgeman SJ, et al. A positive modifier of spinal muscular atrophy in the SMN2 gene. American journal of human genetics. 2009;85(3):408–13. 10.1016/j.ajhg.2009.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oprea GE, Krober S, McWhorter ML, Rossoll W, Muller S, Krawczak M, et al. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320(5875):524–7. 10.1126/science.1155085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernal S, Also-Rallo E, Martinez-Hernandez R, Alias L, Rodriguez-Alvarez FJ, Millan JM, et al. Plastin 3 expression in discordant spinal muscular atrophy (SMA) siblings. Neuromuscular disorders: NMD. 2011;21(6):413–9. 10.1016/j.nmd.2011.03.009 . [DOI] [PubMed] [Google Scholar]
- 32.Cobben JM, van der Steege G, Grootscholten P, de Visser M, Scheffer H, Buys CH. Deletions of the survival motor neuron gene in unaffected siblings of patients with spinal muscular atrophy. American journal of human genetics. 1995;57(4):805–8. [PMC free article] [PubMed] [Google Scholar]
- 33.Brahe C, Servidei S, Zappata S, Ricci E, Tonali P, Neri G. Genetic homogeneity between childhood-onset and adult-onset autosomal recessive spinal muscular atrophy. Lancet. 1995;346(8977):741–2. 10.1016/s0140-6736(95)91507-9 . [DOI] [PubMed] [Google Scholar]
- 34.Jedrzejowska M, Borkowska J, Zimowski J, Kostera-Pruszczyk A, Milewski M, Jurek M, et al. Unaffected patients with a homozygous absence of the SMN1 gene. European journal of human genetics: EJHG. 2008;16(8):930–4. 10.1038/ejhg.2008.41 . [DOI] [PubMed] [Google Scholar]
- 35.Wang CH, Xu J, Carter TA, Ross BM, Dominski MK, Bellcross CA, et al. Characterization of survival motor neuron (SMNT) gene deletions in asymptomatic carriers of spinal muscular atrophy. Human molecular genetics. 1996;5(3):359–65. 10.1093/hmg/5.3.359 . [DOI] [PubMed] [Google Scholar]
- 36.Prior TW, Swoboda KJ, Scott HD, Hejmanowski AQ. Homozygous SMN1 deletions in unaffected family members and modification of the phenotype by SMN2. American journal of medical genetics Part A. 2004;130A(3):307–10. 10.1002/ajmg.a.30251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jedrzejowska M, Szczaluba K, Sielska D. Homozygous deletion in the SMN1 gene in asymptomatic individual—genetic counselling issues in SMA-risk families. Medycyna wieku rozwojowego. 2011;15(2):126–31. . [PubMed] [Google Scholar]
- 38.Chong JX, Ouwenga R, Anderson RL, Waggoner DJ, Ober C. A population-based study of autosomal-recessive disease-causing mutations in a founder population. American journal of human genetics. 2012;91(4):608–20. 10.1016/j.ajhg.2012.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arkblad E, Tulinius M, Kroksmark AK, Henricsson M, Darin N. A population-based study of genotypic and phenotypic variability in children with spinal muscular atrophy. Acta paediatrica. 2009;98(5):865–72. 10.1111/j.1651-2227.2008.01201.x . [DOI] [PubMed] [Google Scholar]
- 40.Helmken C, Hofmann Y, Schoenen F, Oprea G, Raschke H, Rudnik-Schoneborn S, et al. Evidence for a modifying pathway in SMA discordant families: reduced SMN level decreases the amount of its interacting partners and Htra2-beta1. Human genetics. 2003;114(1):11–21. 10.1007/s00439-003-1025-2 . [DOI] [PubMed] [Google Scholar]
- 41.Hahnen E, Forkert R, Marke C, Rudnik-Schoneborn S, Schonling J, Zerres K, et al. Molecular analysis of candidate genes on chromosome 5q13 in autosomal recessive spinal muscular atrophy: evidence of homozygous deletions of the SMN gene in unaffected individuals. Human molecular genetics. 1995;4(10):1927–33. 10.1093/hmg/4.10.1927 . [DOI] [PubMed] [Google Scholar]
- 42.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Team RC. R: A language and environment for statistical computing. 2013. [Google Scholar]
- 44.Allaire J. RStudio: integrated development environment for R. Boston, MA. 2012;537:538. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The median copy number value based on both the SMN exon 1–6 probes and SMN exon 7–8 probes is provided for each DNA sample tested, as shown in Fig 1. For each DNA sample, the Coriell Institute calatog ID is provided.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.