Abstract
Molecular diagnostics currently has a crucial role in neuro-oncological patient care. (Epi)genetic assays testing for point mutations, copy number variations, gene fusions, translocations, and methylation status are of main diagnostic interest in neuro-oncology. Multiple assays have been developed for this purpose, ranging from single gene tests to high-throughput, integrated techniques enabling detection of multiple genetic aberrations in a single workflow. This review describes the nature of the simpler and more complex assays for molecular diagnostics of tumors of the central nervous system and briefly discusses their strengths and weaknesses.
Keywords: diagnosis, epigenetics, genetic alterations, molecular pathology, neuro-oncology
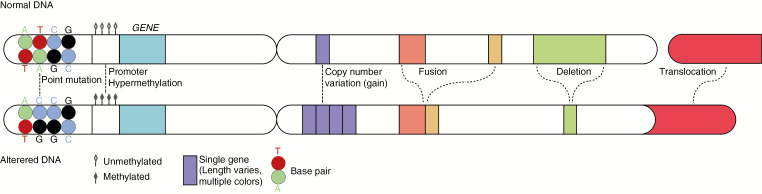
A wide spectrum of genetic aberrations is involved in the development of tumors of the central nervous system (CNS), including point mutations, copy number variations (CNVs, including gains/amplifications as well as losses/deletions), gene fusions, and translocations (Fig. 1). Integration of histopathologic data and information on such genetic aberrations allow for a precise and unequivocal diagnosis of many CNS tumors. Indeed, according to the revised fourth edition of the World Health Organization (WHO) Classification of Tumours of the Central Nervous System (published in 2016),1 the diagnosis of especially diffuse gliomas and embryonal CNS tumors is now ideally based on an integrated “histomolecular” approach. In some situations, information on epigenetic characteristics of the tumor is helpful for CNS tumor diagnosis and/or for therapeutic decision making.
Fig. 1.
Simplified representation of the nature of the most relevant (epi)genetic alterations in (neuro-)oncology. Normal DNA is displayed at the top, with part of another chromosome to the right. At the bottom, altered DNA is represented. The depicted alterations are examples. The mutation shows a change from a cytosine (C)-guanine (G) base pair to a thymine (T)-adenosine (A) base pair. A copy number change may involve a loss/deletion or a gain/amplification, of which the latter is shown here (from 1 to 4 copies of a single gene). The depicted deletion now involves loss of part of 1 single gene, but a deletion may also concern loss of a partial or whole chromosome arm. A gene fusion, depicted as a fusion between genes that were already on the same chromosome arm, may also concern fusion between genes originating on different chromosome arms. The translocation shows the addition of part of 1 arm of a chromosome to the arm of another chromosome.
Diffuse gliomas in adults are by far the most frequent tumors originating from the brain parenchyma. For this tumor group, integration of histopathologic and genetic data has led to recognition of 3 large, clinically relevant molecular subgroups: isocitrate dehydrogenase (IDH)-wildtype; IDH-mutant and 1p/19q-noncodeleted; IDH-mutant and 1p/19q-codeleted.2 IDH-mutant diffuse gliomas show a mutation in the IDH1 or IDH2 gene. The “hotspot” mutations for these genes result in amino acid substitution at codon R132 in or at codon R172, respectively. Presence of CNVs in the form of combined loss of the complete chromosome (chr) arms 1p and 19q in addition to an IDH mutation is now required for the diagnosis of “canonical” oligodendroglioma. Also, the therapy of choice for diffuse gliomas today relies in part on their genetic characteristics. For example, complete 1p/19q codeletion in diffuse gliomas predicts benefit from PCV (combined procarbazine, lomustine (CCNU), vincristine chemotherapy), and hypermethylation of the promoter of the methyl guanine methyl transferase gene (MGMT) has predictive value for response to the alkylating agent temozolomide in patients with glioblastoma.
Other genetic alterations that may be helpful for the molecular characterization of diffuse gliomas are the combination of point mutations in tumor protein 53 (TP53) and α-thalassemia/mental retardation syndrome X-linked (ATRX), which typically occur in IDH-mutant astrocytomas. Likewise, the combined gain of a complete chr 7 and loss of a complete chr 10, as well as amplification of epidermal growth factor receptor (EGFR), are highly indicative for IDH-wildtype glioblastoma (cIMPACT-NOW).3 Mutation in the promoter of telomerase reverse transcriptase (TERT) is very frequent in oligodendroglioma and in IDH-wildtype glioblastoma, but rare in IDH-mutant diffuse astrocytomas and glioblastomas. For the differential diagnosis of nondiffuse gliomas, molecular markers such as the v-RAF murine sarcoma viral oncogene homolog B1 (BRAF) V600E mutation, KIAA-BRAF fusion, and v-rel avian reticuloendotheliosis viral oncogene homolog A (RELA) fusion are of importance. BRAF V600E mutation is frequently present in gangliogliomas, pleomorphic xanthoastrocytomas, and in some pilocytic astrocytomas. KIAA-BRAF fusion is frequent in pilocytic astrocytomas, and a RELA fusion is a defining feature of a subgroup of supratentorial ependymomas. For concise overviews of the genetic aberrations found in glial and other primary and metastatic CNS tumors, see recent reviews.4–7
Given the poor prognosis of many malignant CNS tumors, there is an urgent need for new treatment options. An increasing number of clinical trials are based on molecularly selected or stratified patient cohorts, including trials with targeted agents in patients with EGFR-amplified glioblastoma, or stratification of glioblastoma patients by MGMT status. Thus, molecular analysis is becoming increasingly important for diagnostic accuracy and clinical management. Meanwhile, the molecular “toolbox” for the assessment of the clinically relevant molecular markers is expanding and becoming highly refined. There are often several possible molecular methods for the analysis of 1 potential (epi)genetic change. This review provides an overview of the mode of operation of the more common molecular tools for CNS tumor diagnosis today and briefly summarizes their strengths and limitations.
Assessment of Genetic Alterations
Elementary (“Simple”) Molecular Assays
Sanger sequencing
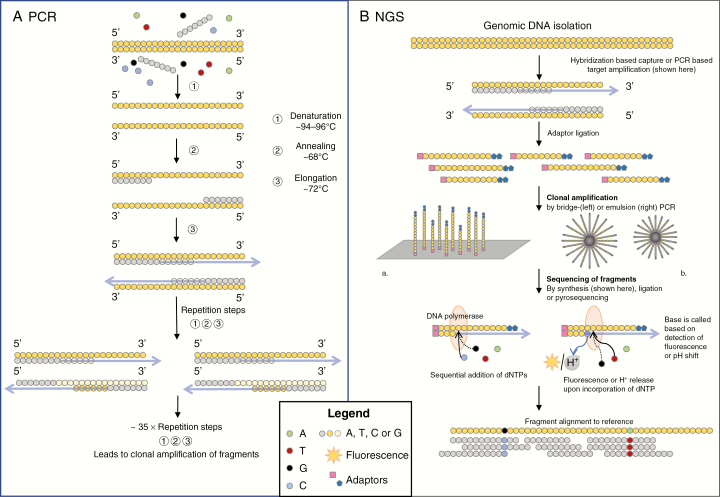
DNA sequencing is the process of determining the order of nucleotides (A—adenosine; C—cytosine; G—guanine; T—thymine) in a strand of DNA. The basis of most sequencing methods is the polymerase chain reaction (PCR), which is the most widely used method of nucleic acid amplification. PCR amplifies copies of a region of interest through the creation of a reaction mix with template DNA, primers that target the region of interest, DNA polymerase, dideoxynucleotide triphosphates (dNTPs), and reaction buffer (Fig. 2). The reaction depends on cyclic temperature changes that denature DNA, anneal primers, and subsequently duplicate the DNA. The products of each reaction act as a template in the next reaction, leading to an exponential increase in the number of copies of the DNA region of interest.
Fig. 2.
A, Simplified diagram showing the basic steps in polymerase chain reaction (PCR). A mixture of DNA, primers, enzymes, and dideoxynucleotide triphosphates is made. Then, a PCR cycle is started in which the temperature regulates the reactions. DNA is denatured (~95oC), primers anneal (~55 oC-60oC), and primers are elongated (~72oC) using the original DNA as a template. Next, PCR cycles are repeated ~35× with the new PCR products and DNA as a template. Repetition of PCR cycles leads to an exponential increase in DNA fragments with the same sequence. Next, the fragments can be analyzed. B, A simplified diagram showing the essentials of targeted next-generation sequencing technology. Genomic DNA is fragmented and adaptors are ligated, which help to anchor the DNA fragments to a solid surface (a plate in bridge PCR or a bead in emulsion PCR). DNA fragments are amplified through a PCR reaction. DNA polymerase causes elongation of the complementary sequence, and the release of fluorescence or hydrogen upon the incorporation of a nucleotide is analyzed to find the sequence of each fragment. To identify a mutation, the sequences of the fragments are digitally aligned to the sequence of reference DNA.
Sanger sequencing relies on this generation of many copies of the target DNA region and the incorporation of chain terminating dNTPs that are labeled with a fluorescent dye. After the incorporation of these dideoxynucleotides, no further elongation of the DNA strand is possible, resulting in termination of the chain. The DNA fragments are sorted based on their length through capillary electrophoresis and the dye at the end of the strand is detected by a laser, allowing the read out of the DNA sequence.
The first paper describing the Sanger sequencing technique was published in 1977.8 Since then, Sanger sequencing has been the gold standard and indeed represents a robust method that is still in use for especially “smaller”-scale projects. Sanger sequencing is primarily suitable for the analysis of relatively long stretches of DNA (up to 900 base pairs [bp]) and can be used for the detection of point mutations in a specific gene. Disadvantages of Sanger sequencing are the relatively high costs and inefficiency when multiple genes need to be sequenced. Furthermore, especially in samples with low tumor cell percentage, the relatively low sensitivity of 15% to 20% variant allele frequency (VAF) can be a limitation.9 In this case, the limit of detection is high and mutations in samples with low tumor cell percentage can be missed.
Fluorescent In Situ Hybridization
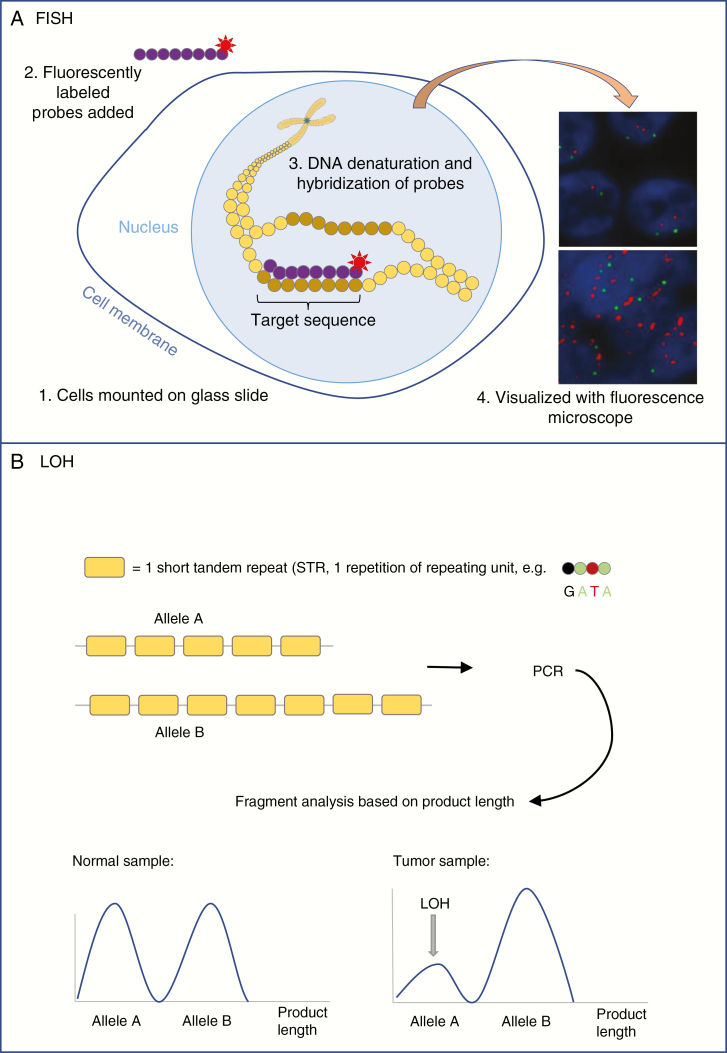
First published in 1969,10 in situ hybridization is a technique to visualize the presence of a DNA strand (representing a gene or a [larger part of a] chr). In situ means that the location of the DNA sequence is identified in its “natural position” within the chr (Fig. 3). After denaturation of the DNA through heat or chemicals, a labeled DNA or RNA sequence is used as a probe to identify and quantify the complementary target sequence. Generally, fluorescently labelled probes are hybridized (bound) to the tissue slide, allowing easy visualization, hence the name “fluorescent in situ hybridization” (FISH). In situ hybridization was one of the first methods used to measure gene amplification in malignant gliomas. See, for example, EGFR amplification as described in 1988 by Bigner et al.11
Fig. 3.
A, Simplified diagram showing the essential technique of fluorescent in situ hybridization (FISH). Tissue is mounted on a glass slide and cells are treated to make cell membranes and nuclei permeable to enzymes and probes. DNA is denatured and fluorescently labeled probes (eg, complementary to target sequence [red label] and centromere [green label]) are hybridized to the DNA. The fluorescent labels can be visualized with a fluorescence microscope. Normal signal for EGFR (upper panel) and EGFR amplification (bottom panel) are shown. B, Simplified diagram showing the basis of STR-based LOH analysis. For a certain gene, allele A carries 5 STRs while allele B carries 7 copies of that STR. Polymerase chain reaction is performed (see Figure 2), after which the fragments are analyzed based on their length. The ratio between fragments of different lengths gives information on the presence or absence of the alleles. LOH is declared when the presence of 1 of the alleles is lower than expected from the matched normal sample, and when the ratio between the signals for fragments of both alleles is disturbed.
EGFR indicates epidermal growth factor receptor; LOH, loss of heterozygosity; STR, short tandem repeat.
FISH is especially suitable for the detection of CNVs and gene fusions or translocations. For example, in case of a fusion, 2 genes that are normally located at a significant distance from each another are fused in neighboring loci on the chr (shown in Fig. 1). To detect such a fusion through FISH, probes for the 2 fusion partners are used that are labeled with different fluorescent colors, for example green and red. In case of a fusion, juxtaposition of the partners will result in 1 yellow/orange signal instead of a separate red and green signal. On the same basis, it is possible to demonstrate translocation by using “break-apart probes,” which will show separate signals when different parts of the investigated gene are translocated. In case of amplification of a gene, the probe for this gene will show multiple signals within the contours of a single nucleus.
An advantage of FISH is that single nuclei can be analyzed, allowing for the identification of subclones within a tumor. Furthermore, the exact number of copies in case of a gain or low-level amplification can be determined (though there are no set cutoff values for gene amplifications, and the clinical relevance of CNVs differs per gene, on average >2-5 gene copies can be considered gain and at >5-10 copies amplification can be called). For high copy amplifications (>20-30 copies), however, the exact number of gene copies becomes difficult to determine. FISH can be performed on formalin-fixed, paraffin-embedded (FFPE) of fresh-frozen material. A drawback of FISH is that its labeling intensity can vary within an investigated tissue area, and there is often some nonspecific “background hybridization,” making unequivocal assessment of the results challenging.12 Also, it is generally not possible to make a distinction between complete and partial loss of a chr arm, such as in case of a 1p/19q codeletion, because the probe set consists of a centromere probe and 1 probe located on a specific spot on the chr arm. When this latter signal is lost, it could still concern partial instead of complete deletion of the chr arm. Because of this limitation, FISH is not an ideal method for the determination of 1p/19q status. Lastly, FISH analysis can be more difficult on decalcified tissue13 and, in our experience, also on cytological specimens.
Loss of Heterozygosity Analysis
Except for the X and Y chr, for every gene there are 2 alleles (1 on the chr of maternal origin and 1 on the chr of paternal origin). Loss of heterozygosity (LOH) is a genetic event in which 1 allele of a gene is lost, leading to, for example, haploinsufficiency or the “uncovering” of a recessive mutated tumor suppressor gene in the remaining allele. It is important to note that LOH can lead to a decrease in the number of copies of a gene (from 2 to 1), but also copy number-neutral LOH can occur if the unaffected gene is duplicated (before or after the loss of the other allele).
The original method to evaluate LOH events consists of the analysis of microsatellites or short tandem repeats (STRs), which can be performed on fresh-frozen or FFPE samples (Figure 3). STRs are tens to hundreds of sequential repetitions of a series of 2-13 bases on a DNA strand. In STR analysis, the exact number of repeating units is measured and compared at a single locus for both alleles. LOH is declared when 1 allele is reduced in intensity in the tumor DNA compared to a matching DNA of nonneoplastic cells (eg, from leukocytes or oral mucosa) from the same patient. The requirement of normal DNA from the same patient is a disadvantage of this technique. Newer methods to detect LOH tackle this problem by comparing the allelic ratio to, among others, neighboring markers in the tumor sample rather than a normal DNA sample.14 A drawback of this alternative method is the fact that neighboring markers can also be lost, giving a false impression of a normal allelic ratio. Furthermore, in case of low tumor cell percentage in the sample, it can be difficult to recognize LOH, as the difference between both alleles is minimized by contamination with normal DNA. When the interrogated STRs are homozygous in the germline (ie, when the maternal and paternal STRs for the investigated DNA region are the same), the LOH probe for that region is uninformative.
Complex Molecular Assays
Targeted next-generation sequencing
Next-generation sequencing (NGS) is an umbrella term for sequencing techniques developed after Sanger sequencing. The term generally refers to short-read sequencing by synthesis methods, which means that the DNA sequence is read out (sequenced) based on the sequential incorporation of nucleotides during primer extension (synthesis). The nucleotides are detected while they are incorporated in the chain and a characteristic fluorescent or pH signal (different for each base) is emitted (Fig. 2). This can be performed using different platforms, such as Illumina or IonTorrent. Targeted NGS is also based on PCR, and the gene-specific primers that are used in the reaction determine the targets of the NGS panel. NGS methods are highly parallel, which means that many sequencing reactions take place at the same time. Compared to Sanger sequencing, shorter reads are generated, usually 100-200 nucleotides per sequence. Bioinformatics tools are generally used to identify changes in the order of nucleotides, which are then compared to known mutations, variants, and single-nucleotide polymorphisms (SNPs) as listed in online databases and/or the databases of the local laboratory, alleviating the need for a matched normal tissue sample from the same patient.
One of the main advantages of NGS is the amount of information that can be obtained in a single workflow, thereby increasing the chance of finding clinically relevant/actionable variants of mutant genes in a tumor. It has been reported that performing 1 NGS assay costs 15 times less compared to the total costs of older tests assessing all these genetic modifications separately.15 The option to multiplex several samples in 1 assay leads to increased throughput and further reduction of costs.16 Furthermore, NGS is a very sensitive technique that can find VAFs of 1% to 5%17,18 (for Sanger sequencing the VAF needs to be at least 10% to 20%9,19,20). This makes NGS better suited for samples with low tumor cell percentage.16 In addition, NGS was found to be reliable when performed on FFPE material, which is valuable when no fresh-frozen material is available.15,21 As routine CNS diagnostics often have to be performed on material from small biopsy specimens, it is advantageous that only a very small amount of DNA (20 ng)15,21,22 may be sufficient to prepare the NGS library. Furthermore, since NGS panels are customizable, laboratories have the opportunity to optimize panels (for example, in case of special genes of interest for specific trials) and to continue updating panels based on relevant new research findings.
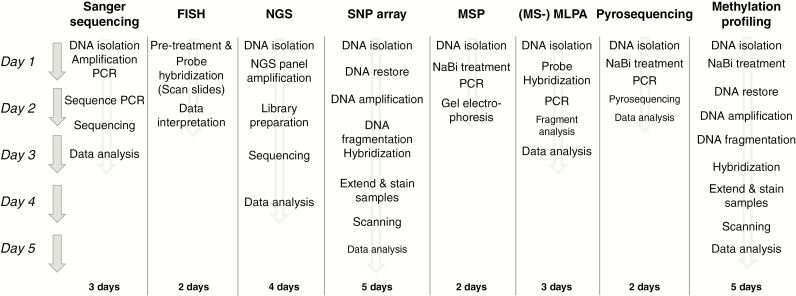
Disadvantages of NGS compared to conventional techniques are the turnaround time of 4 (our experience, Fig. 4) to 7 days,15,16,22 the need for access to bioinformatics resources for CNVs and fusion detection, interinstitutional variability related to the use of laboratory-specific gene sets, the costs and the influence of the length of the PCR product on amplicon coverage, especially for FFPE material (amplicons larger than 160 bp are not amplified efficiently).22 In addition, because NGS allows the analysis of many genes in 1 assay there is an increased chance of identifying unsolicited findings23 or variants of unknown clinical relevance. Based on coverage it may remain challenging to reliably detect deletions, for instance the 1p/19q codeletion and more subtle copy number differences such as CDKN2A deletion, gain of chr 7 and loss of chr 10. Most methods are based on the quantification of differences in depths of coverage per amplicon of a tumor sample compared to a matched normal sample24 or a normal pool dataset.22 In this way, partial amplification of a gene (such as EGFR in glioblastoma) can also be detected.22 NGS allows an alternative method to analyze these subtle differences through SNP analysis.25 SNPs are variations in nucleotides at a single point in the DNA that are conserved throughout evolution and within populations. By including SNPs in the amplicons that are sequenced, the frequency of the SNPs can be used to analyze potential imbalances between the presence of both alleles (B-allele frequency), allowing the detection of subtle CNVs.25
Fig. 4.
Flow diagram providing an indication of turnaround times (in working days) for several of the techniques described in this review article. These are the usual turnaround times of the authors’ laboratory or institution. Turnaround times may vary from laboratory to laboratory, for example, depending on the possibility of performing certain steps overnight.
FISH indicates fluorescent in situ hybridization; MS-MLPA, methylation-specific multiplex ligation-dependent probe amplification; MSP, methylation-specific polymerase chain reaction; NaBi, sodium bisulfite; NGS, next-generation sequencing; PCR, polymerase chain reaction; SNP, single-nucleotide polymorphism.
Single-nucleotide polymorphism array
SNP array analysis is a method that can be used to detect genomewide LOH and other chromosomal imbalances, including 1p/19q codeletion. In SNP array analysis, nucleic acid sequences are immobilized on a solid surface or bead and each SNP is interrogated by 1 or several labeled probes. After hybridization of the probe, the probe is elongated at the position of the SNP with fluorescently labeled nucleotides. Subsequent scanning of the slide reveals the genotype of the SNP. Heterozygous SNPs provide information on potential imbalances in which 1 allele is overrepresented compared to the other. An estimation of the likelihood of LOH is achieved by comparison of the results to standards derived from normal reference cases.26 This is an improvement compared to microarray-based LOH analysis, as it obviates the need for normal tissue from the same patient. High-resolution SNP array techniques are available and are especially suitable for detection of breakpoints and of LOH without loss of chr material.27 The main benefit compared with traditional LOH analysis is that high-resolution, genomewide information is obtained and that a single assay can therefore be used to detect different CNVs. Also, SNP array analysis provides information on both single genes as well as chr arms, which is of primary interest in glioma diagnostics.
Multiplex ligation-dependent probe amplification
Multiplex ligation-dependent probe amplification (MLPA) is a PCR-based method that uses 2 oligonucleotide probes that hybridize to the target sequence and are then ligated to each other. All MLPA probes in the mix have a specific sequence at the 5’ or 3’ end that are compatible with just 1 set of PCR primers. Only when the MLPA probes hybridize at the target sequences and are ligated will the probes be amplified with these primers. Because of different lengths of stuffer sequences incorporated in each probe, each MLPA probe pair will give rise to an amplification product of unique size, allowing multiplex analysis of different genes and gene fragments.27
This method is suitable to detect, among others, IDH1/2 mutation status, EGFR amplification, and 1p/19q codeletion. CNVs are detectable because it leads to a change in the relative amount of amplification product, which can be compared to the amount of amplification product of reference samples.28 Furthermore, control probes located in genomically stable regions for the specific tumor type are provided in the probe mix. Different probe mixes were evaluated for the detection of losses of 1p/19q, IDH1/2 mutation status, EGFR amplification, loss of CDKN2A, and loss of PTEN (P105) and were found to be robust and reliable.29,30
One of the main strengths of MLPA is that it allows for the simultaneous detection of different types of genetic aberrations in a single, relatively simple workflow that can provide results within 3 days (Fig. 4) and up to as fast as 24 hours28 (depending on the laboratory/workflow possibilities). Also, the method is suitable both for fresh-frozen and FFPE material, the costs are relatively low, and the analysis software is free.28 Limitations are that the method can detect only relative CNVs and will not detect CNVs when they affect the whole genome (ie, no distinction can be made among polyploidy, haploidy, and diploidy).28
Assessment of Epigenetic Alterations
Epigenetics concerns the study of functional changes to the genome with an effect on gene expression that do not involve changes in the DNA sequence. One example is DNA methylation, in which methyl groups are bound to nucleotides, most commonly C. A CpG site is a region in the DNA where a C nucleotide occurs next to a G nucleotide, connected via 1 phosphate molecule. CpG islands are stretches of DNA (~500-1500 bp) containing many CpG sites with a CG ratio of >0.6 vs the other nucleotides, which occur in regulatory areas of the genome such as promoter regions of genes. CpG sites can be methylated or unmethylated. Hypermethylation of the promoter of a gene generally results in inactivation of the gene due to inaccessibility of the DNA for transcription factors.
MGMT promoter methylation status is 1 of the main epigenetic markers of interest in clinical neuro-oncology. One of the bottle necks for reliable detection of this marker is that most techniques rely on sodium bisulfite conversion. During sodium bisulfite treatment, unmethylated C is changed to uracil (U), while methylated C is maintained. Especially when using FFPE material, this sodium bisulfite conversion process can be inefficient (leaving some unmethylated C unchanged) and cause unreliable results.29,31,32 In addition, most techniques interrogate only a limited number of CpG sites, which can be a problem as, in the case of hypermethylation, all CpG sites are not always methylated and there can be variation in CpG methylation patterns.33 Method-specific difficulties in determining MGMT status are described below.
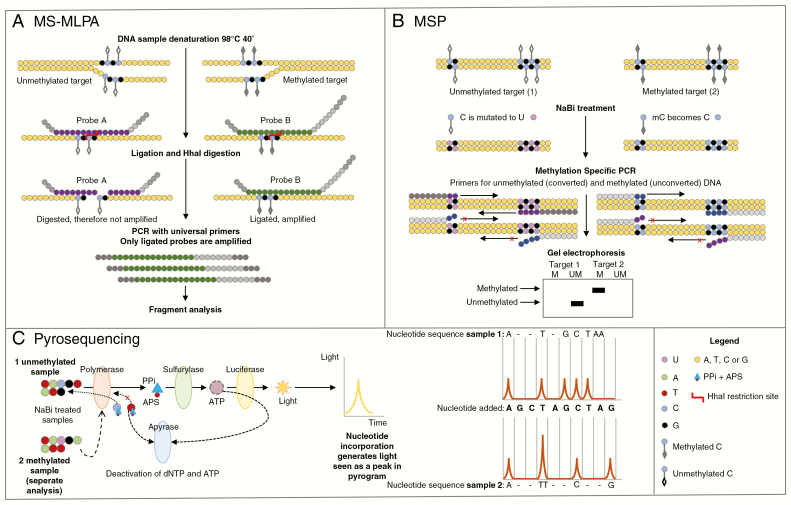
(Nested) Methylation-Specific-Polymerase Chain Reaction
Methylation-specific polymerase chain reaction (MSP, Fig. 5) is widely used to assess epigenetic silencing of genes in general, and it is the most commonly used assay to test for MGMT promoter methylation in gliomas.34 Hegi et al have investigated MGMT promoter methylation, as assessed by MSP, in patients with newly diagnosed glioblastoma. They found that MGMT promoter hypermethylation is an independent favorable prognostic factor (irrespective of treatment) and is associated with the benefit of adding temozolomide chemotherapy to radiotherapy.35
Fig. 5 .
Simplified diagrams of 3 techniques that can be used to assess methyl guanine methyl transferase gene (MGMT) promoter methylation status. A, In MS-MLPA DNA is denatured and gene-specific probes are hybridized to the genomic DNA. The probes contain stuffer sequences of different length, allowing multiplex analysis. Every reaction consists of 2 routes. One route is incubated with ligase and the other with ligase and the methylation-specific restriction enzyme HhaI. The first reaction allows for the identification of copy number variations, whereas the second allows for methylation analysis as the restriction enzyme digests only unmethylated DNA (because unmethylated DNA and methylated DNA are added). B, The methylation-specific PCR genomic DNA is treated with sodium bisulfite (NaBi). Next, a PCR reaction with specific primers for either completely methylated or unmethylated DNA is performed. After gel electrophoresis, presence of bands for the methylated primers indicates the presence of DNA methylation. C, For pyrosequencing DNA is treated with NaBi. Next, a PCR reaction is performed to amplify the region of interest. Finally, the PCR product is sequenced. Upon incorporation of a nucleotide by DNA polymerase, a pyrophosphate (PPi) molecule is released that can be converted into light by sulfurylase and luciferase. As single nucleotides are added to the reaction in a specific order, the presence of a light signal indicates the presence of that nucleotide in the sequence.
MS-MLPA indicates methylation-specific multiplex ligation-dependent probe amplification; PCR, polymerase chain reaction.
Nested MSP is a method in which the primers are added in 2 rounds (rather than 1 round in “normal” MSP). After an initial amplification of the MGMT promotor, a second round of PCR is performed that allows a more specific amplification of target sequences that are located within the products of the first round of PCR. Through this 2-step method, the likelihood of creating nonspecific PCR products is decreased.36 Two different PCRs are performed, 1 specifically for methylated and 1 for unmethylated MGMT promoter. The latter is a control of the assay as it should always provide a positive result. The assay creates a binary readout after gel electrophoresis: a signal (band) for methylated MGMT promoter is either present or not.
While MSP is considered a relatively accurate method,37 some studies show suboptimal reliability and reproducibility of this method.38 Also, the technique is challenging as it depends highly on the specificity of the selected primers, the PCR conditions, and the adequacy of the bisulfite conversion,39 illustrated by the observation of Hegi and colleagues that the success rate of the MSP on FFPE samples was highly variable and center dependent (median success rate 75%, range, 0-100%).35 In addition, MSP interrogates only a limited number of CpG sites within the promoter,40 which could affect sensitivity and specificity. The required amount of template DNA depends greatly on the nature and quality of the sample (eg, formalin fixation has a negative effect) and can vary from 75 ng to 150 ng of DNA.40 Also, it is not known exactly how methylation of only a subset of the CpG sites within the primer sequence might affect primer binding and thus the outcome of the MSP assay.
Quantitative Real-Time Methylation-Specific-Polymerase Chain Reaction
Quantitative real-time MSP (qMSP) is a PCR-based method that uses, for example, TaqMan (MethyLight) or Sybr Green40 technologies to allow for the quantification of methylation percentage. qMSP also relies on bisulfite modification of DNA, as described above. Primers (specific for bisulfite-converted DNA) for CpG sites in the MGMT promoter and a control (housekeeping) gene are used.41 The primers carry a fluorescent label and a quencher label. When the DNA is amplified, the quencher is removed from the primer sequence and the fluorescence signal can be detected. The methylation percentage is calculated by dividing the MGMT promoter:control ratio of a sample by the MGMT promoter:control ratio of CpG Methyltransferase (M.SssI)-treated human genomic DNA and multiplying by 100.42 The treatment of DNA with M.SssI should cause full methylation of cytosine in all CpG sites.41
qMSP is reported to be about 10 times more sensitive than conventional MSP,40 although it is not completely clear what this sensitivity assessment is based on. The primer set influences the sensitivity of the method. Post-PCR time of running a gel is eliminated by qMSP, resulting in a more time-efficient analysis. Because this method can quantify methylation percentages, it is possible to determine cutoff points for hypermethylation. A possible source of false-negative results is the fact that 1 of the housekeeping genes that is frequently used, actin beta (ACTB), is located on chr 7, which is frequently gained, especially in glioblastomas. This problem can be avoided by use of collagen type II alpha 1 (COL2A1) as the control gene, which is located on chr 12. Indeed, when COL2A1 instead of ACTB was used as the reference gene, the reproducibility of the qMSP results improved.41
Methylation-specific multiplex ligation-dependent probe amplification
Methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) is a modified version of MLPA that can be used to assess MGMT promoter methylation status (Fig. 5).43 This test is based on a mix containing 6 MGMT promoter probes with a methylation-sensitive restriction site (unmethylated DNA is sensitive to HhaI digestion) and 10 control probes without an HhaI restriction site. The tumor sample is split and 2 PCR amplifications are performed: 1 with an undigested sample and 1 with an HhaI digested sample (which leaves only the methylated fraction of the DNA intact). Subsequently, the ratio between the peak pattern of the digested and the undigested sample gives information about the percentage of DNA methylation.43
A drawback of MS-MLPA is that only CpG sites that are located within an HhaI restriction site can be analyzed. However, 1 of the CpG sites that was shown to correlate with response to temozolomide44 is included in the MS-MLPA probe kit. In addition, there are no clearly defined cutoff values for clinically significant methylation levels. A major advantage of MS-MLPA is that no bisulfite treatment is required. The MS-MLPA assay is suitable for FFPE tissue and only a small amount of DNA is required (20-100 ng). Also, MS-MLPA allows for simultaneous analysis of different CpG sites and the detection of copy numbers of the analyzed loci as well as IDH1/2 mutations29 (described above). The software required for MS-MLPA data analysis (Coffalyzer.Net) is openly available via the website www.mlpa.com. Accessed October 9, 2018.
Pyrosequencing
Pyrosequencing is a method that has been described and developed since 1993.45 For CNS tumors, pyrosequencing can be used for the detection of MGMT promoter hypermethylation but also of point mutations (Fig. 5). Pyrosequencing is also based on the “sequencing by synthesis” principle and relies on the detection of light that is emitted when pyrophosphate is released during PCR. The intensity of the light signal correlates to the number of nucleotides of a single type that are incorporated. To use pyrosequencing for MGMT promoter methylation detection, bisulfite-treated DNA is amplified and a kit is applied that detects the level of methylation of 5 CpG sites.46 Subsequent pyrosequencing analysis of the MGMT promoter methylation status provides statistically highly significant results that show good correlation with survival data.47 Disadvantages are the necessity for bisulfite conversion, and the requirement for specialized and relatively costly equipment.47
Methylation Profiling
In addition to gene-specific approaches to studying epigenetics, a (more) genomewide approach is now also available and is finding its way into the diagnostic workflow. Genomewide methylation profiling may serve several purposes: It can be used to assess MGMT promoter methylation status, but it can also help to classify CNS tumors through pattern analysis based on methylation profile44,48 and thus aid the diagnosis. In addition, methylation profiling gives indirect information about copy number alterations, including 1p/19q codeletion, combination of gain of chr 7 and loss of chr 10, and EGFR amplification, as well as loss of heterozygosity. Essentially this tool includes a SNP array-based data analysis. Methylome data can be obtained with the Infinium Methylation Beadchip (Illumina), of which the most recent (“850”) version interrogates >850 000 CpG sites.
For targeted use, Bady et al have previously shown that methylation profiling through the HumanMethylation450 Beadchip can be used to assess MGMT promotor status, as the chip interrogates 176 CpG sites within the MGMT gene, of which 14 are located in the promoter region.44 They describe a model (MGMT-STP27) that determines a probability of MGMT promoter methylation based on 2 probes (cg12434587 and cg12981137), the first of which is also included in the MS-MLPA assay described above. The MGMT-SP27 model was shown to have good performance in 2 external datasets when comparing the results to pyrosequencing data of 47 glioblastoma cases and MS-MLPA data of 62 WHO grade III glioma cases.44
One disadvantage of using the MGMT-SP27 model to predict MGMT promoter hypermethylation is the possible effect of gene loss on the ratio of methylated to unmethylated DNA. Of note, while most tumors usually carry both MGMT alleles, glioblastomas often show loss of chr 10, on which the MGMT gene is located (10q26.3), which may have consequences for the actual level of MGMT expression. Indeed, in 2016 Bady and colleagues described that in low-grade glioma a combination of MGMT promoter hypermethylation and loss of 10q showed lower MGMT expression compared to tumors with only MGMT promoter hypermethylation. The authors hypothesized that this could be because in this latter group only 1 of the 2 MGMT alleles that are present is hypermethylated. The remaining MGMT allele may cause residual MGMT expression, which would have a negative impact on the effectiveness of alkylating agents.49 As the STP-27 model is based on a set of mainly glioblastoma samples, many of which can be expected to have only one MGMT allele, using this model may lead to overestimation of MGMT gene inactivation in nonglioblastoma tumors.44 For these tumors, the optimal threshold for assessment of clinically relevant MGMT promoter hypermethylation would have to be determined using a reference set with tumors carrying both MGMT alleles.
Recently, an additional application of methylation profiling in the context of neuro-oncology was published by Capper et al.48 They describe the use of a classifier tool that matches the methylation profile of a sample to a CNS tumor entity. The classifier tool was developed by feeding the methylation profile, generated with the Infinium Human Methylation450K BeadChip array, of at least 8 cases per WHO-defined CNS tumor group as well as the profiles of nonneoplastic CNS tissue samples (including samples with inflammatory and other reactive changes) to a deep learning approach. Next, the methylation profiles were clustered and 91 methylation classes were identified. The identified classes can be used to investigate whether the methylation profile of a tumor sample matches a defined DNA methylation class and if the threshold value (calibrated score) of >0.9 is met. In the study by Capper et al the DNA methylation profile class matched the histopathologic diagnosis in 76% of the cases, while in 12% the DNA methylation profile class was reason to reconsider and revise the original histopathologic diagnosis. Obviously, in some situations such a change in diagnosis can have important clinical/therapeutic impact.
Advantages of the genomewide methylation profiling analysis are the large amount of information it provides and the open availability of the online classifier tool (www.molecularneuropathology.org Accessed October 9, 2018.). Current disadvantages of the tool are the relatively high amount of DNA required (100-200 ng), costs, and limited experience with interpretation of the results, especially when the outcome of the classifier tool suggests a diagnosis that does not match the histopathologic diagnosis or clinical presentation of a patient.50 Studies aiming to reveal how diagnoses based on DNA methylation profiling correlate with survival data are ongoing.
Conclusions and Future Perspectives
Current neuro-oncological practice is increasingly dependent on molecular diagnostics of tumor tissue. Molecular data are essential for an integrated histomolecular diagnosis and to inform treatment decisions, including the indication for targeted therapy. Sanger sequencing, FISH, and LOH analysis are established techniques that can provide very valuable molecular information (Table 1). Each of these methods, however, has its shortcomings, and a laboratory may need to combine several techniques to get the complete “package” of the required molecular information. Especially when there is a small amount of tissue available for analysis, it may be preferable to use 1 reliable high-throughput method that allows for the analysis of multiple markers in 1 assay.
Table 1.
Overview of Techniques and Their Uses as Described in this Review
| Sanger Sequencing | FISH | LOH Analysis | Next- Generation Sequencing (DNA) | SNP Array | MLPA | WGS/WES | Nanopore Sequencing | (Nested) MSP | RT-MSP | MS-MLPA | Pyrosequencing | Methylation Profiling | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Costs per assay | - | - | - | ~ | ~ | - | + | ~ | - | ~ | - | ~ | + |
| Sensitivity | ~ | ~ | ~ | + | + | ~ | + | - | + | ~ | + | + | + |
| Point mutations (eg, IDH1/2, ATRX, TP53) | • | ∆ | • | • | ø | • | • | ||||||
| Codeletion 1p/19q | ø | ø | • | ∆ | • | • | • | • | |||||
| Other CNVs (eg, EGFR amplification) | • | ø | • | ∆ | • | • | • | ||||||
| Translocations/fusions (eg, KIAA1549-BRAF) | ∆ | • | |||||||||||
| Methylation (eg, MGMT promoter/profile) | • | • | ∆ | • | ∆ |
Abbreviations: ATRX, α-thalassemia/mental retardation syndrome X-linked; BRAF, v-RAF murine sarcoma viral oncogene homolog B; CNV, copy number variation; EGFR, epidermal growth factor receptor; FISH, fluorescent in situ hybridization; IDH1/2, isocitrate dehydrogenase; LOH, loss of heterozygosity; MGMT, methyl guanine methyl transferase; MLPA, multiplex ligation-dependent probe amplification; MS, methylation specific; PCR, polymerase chain reaction; RT-MSP, reverse-transcriptase–methylation-specific polymerase chain reaction; TP53, tumor protein 53; WES, whole-exome sequencing; WGS, whole-genome sequencing.
LEGEND of symbols used: +: High; ~: Medium; -: Low; Δ: Preferred method; •: Possible alternative; ø: Least recommended.
Terms such as “preferred method,” “possible alternative,” and “least recommended” are based on the knowledge and experience of the authors; other laboratories may have a different point of view.
NGS is a very attractive and sensitive technique for combined assessment of many genetic markers such as somatic mutations, CNVs, and fusions in a single workflow, while inclusion of SNPs in the primer set enables detection of more subtle CNVs.25 For assessment of MGMT promoter methylation, a focused method such as MSP, MS-MLPA, or pyrosequencing may be preferred. MS-MLPA has the advantage of being a quantitative method that allows for the determination of a threshold for hypermethylation. As promoter methylation-mediated gene silencing is strongly dependent on the location of the methylated CpG sites,51 choosing the primers for the sites that correlate best with clinical outcome is important. Future research should focus on identifying probes/CpG sites that correlate best with clinical outcome, and give further insight in the most reliable way to assess MGMT promoter methylation status.
Methylation profiling is a very powerful platform for detection of epigenetic changes.48 At present, this tool is still relatively costly, but has a significant added value as it can contribute to diagnostic fine-tuning by suggesting a diagnosis based on the methylation profile, and at the same time provide information on important diagnostic, prognostic, and/or predictive markers (especially CNVs and MGMT promoter methylation status).48 How exactly methylation profiling performs with respect to prognostication and to what extent it aids in improving patient care need further investigation.
We expect that molecular diagnostics for CNS tumors will increasingly be performed with a combination of NGS and methylation profiling, providing an almost complete package of information on mutations, CNVs, fusions, MGMT promoter methylation status, and overall methylation pattern associated with a specific tumor entity or subgroup. NGS panels will likely be expanded as novel therapeutic targets and gene alterations of diagnostic importance are identified. Meanwhile, immunohistochemistry is a quick, easy, relatively inexpensive, and very reliable alternative for demonstration of the mutant protein that is the result of important molecular markers (eg, IDH1 R132H, BRAF V600E, H3K27M)52–54 or of surrogate markers that indicate a particular molecular makeup of the CNS neoplasm (eg, p53, ATRX, L1 cell adhesion molecule [L1CAM]).55,56
At the same time, innovative sequencing techniques such as RNA sequencing, whole-exome sequencing (WES; in which about 1% to 2% of the whole genome is sequenced), whole-genome sequencing (WGS), and nanopore sequencing may in the near future contribute to an even more in-depth understanding of the molecular underpinnings of CNS tumors. By analyzing RNA through NGS, it is possible to detect gene translocations/fusions (next to the somatic mutations and CNVs that can be detected by analyzing a DNA sample)57 and gene expression levels.58 There is currently limited application for WES and WGS for routine diagnostics because of high costs, long turnaround time, the need for a relatively large amount of high-quality DNA, the possibility of unsolicited findings (especially finding germline mutations), and the complicated data analysis required to sort out the (clinical) relevance of the findings.15 Nanopore sequencing has several advantages, including fast (±0.1× genome coverage in 6 hours) analysis of CNVs, methylation profile(s) as well as point mutations;59 deep amplicon sequencing of specific genes of importance by fragmenting 200 ng of tumor DNA from a fresh-frozen sample; and it does not require extensive laboratory equipment or lab technician expertise.59 The technique may not yet be ready for routine diagnostic use, however, because the error rate is still reported to be high (between ∼5% and 40%)60 unless the data can be aligned to, for example, NGS data.60–62
In conclusion, integration of histopathologic data and information on genetic aberrations allows for a much more precise and unequivocal diagnosis of many CNS tumors. As a consequence, molecular diagnostics is today a cornerstone in neuro-oncological patient care. Relatively simple tests such as Sanger sequencing, FISH, and LOH may already provide very helpful information on the clinically most relevant markers. More complex tools such as NGS, SNP array, and MLPA, however, allow a more comprehensive and robust assessment of relevant markers and may in the end be more cost effective. Methylation profiling has great potential as a molecular diagnostic tool for CNS tumors, but further study is needed to assess its optimal role in the clinical diagnostic process. While there is at present still limited use in this process for innovative sequencing techniques such as WES, WGS, and nanopore sequencing, this may change as costs decrease and the necessary bioinformatics pipelines become more widely available. For detection of MGMT promoter methylation, most of the presently used techniques involve a capricious sodium bisulfite conversion step, and MS-MLPA (in which this step is not needed) may be an attractive alternative, but the optimal threshold for clinically relevant MGMT promoter hypermethylation still needs to be determined. To provide the best patient care possible, and for the assessment of other molecular markers, it is important to carefully select the assays used as well as to monitor their validity and accuracy.
Funding
None declared.
Conflict of interest statement. None declared.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. World Health Organization Histological Classification of Tumours of the Central Nervous System. France: International Agency for Research on Cancer; 2016. [Google Scholar]
- 2. The Cancer Genome Atlas Research Network, Brat DJ, Verhaak RG, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. doi:10.1056/NEJMoa1402121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brat DJ, Aldape K, Colman H, et al. cIMPACT-NOW update 3: grading of IDH-wildtype diffuse astrocytic gliomas [published online ahead of print September 26, 2018]. Acta Neuropathol. doi:10.1007/s00401-018-1913-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wesseling P, Capper D. WHO 2016 classification of gliomas. Neuropathol Appl Neurobiol. 2018;44(2):139–150. doi:10.1111/nan.12432 [DOI] [PubMed] [Google Scholar]
- 5. Pickles JC, Hawkins C, Pietsch T, Jacques TS. CNS embryonal tumours: WHO 2016 and beyond. Neuropathol Appl Neurobiol. 2018;44(2):151–162. doi:10.1111/nan.12443 [DOI] [PubMed] [Google Scholar]
- 6. Sahm F, Reuss DE, Giannini C. WHO 2016 classification: changes and advancements in the diagnosis of miscellaneous primary CNS tumours. Neuropathol Appl Neurobiol. 2018;44(2):163–171. doi:10.1111/nan.12397 [DOI] [PubMed] [Google Scholar]
- 7. Berghoff AS, Bartsch R, Wöhrer A, et al. Predictive molecular markers in metastases to the central nervous system: recent advances and future avenues. Acta Neuropathol. 2014;128(6):879–891. doi:10.1007/s00401-014-1350-7 [DOI] [PubMed] [Google Scholar]
- 8. Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74(12):5463–5467. doi:10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsiatis AC, Norris-Kirby A, Rich RG, et al. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn. 2010;12(4):425–432. doi:10.2353/jmoldx.2010.090188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pardue ML, Gall JG. Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proc Natl Acad Sci U S A. 1969;64(2):600–604. doi:10.1073/pnas.64.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bigner SH, Burger PC, Wong AJ, et al. Gene amplification in malignant human gliomas: clinical and histopathologic aspects. J Neuropathol Exp Neurol. 1988;47(3):191–205. [DOI] [PubMed] [Google Scholar]
- 12. McKay JA, Murray GI, Keith WN, McLeod HL.. Amplification of fluorescent in situ hybridisation signals in formalin fixed paraffin wax embedded sections of colon tumour using biotinylated tyramide. Mol Pathol. 1997;50(6):322–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schrijver WA, van der Groep P, Hoefnagel LD, et al. Influence of decalcification procedures on immunohistochemistry and molecular pathology in breast cancer. Mod Pathol. 2016;29(12):1460–1470. doi:10.1038/modpathol.2016.116 [DOI] [PubMed] [Google Scholar]
- 14. Gorringe KL. Loss of heterozygosity. In: eLS. John Wiley & Sons, Ltd; september 2016. doi:10.1002/9780470015902.a0026643 [Google Scholar]
- 15. Nikiforova MN, Wald AI, Melan MA, et al. Targeted next-generation sequencing panel (GlioSeq) provides comprehensive genetic profiling of central nervous system tumors. Neuro Oncol. 2016;18(3):379–387. doi:10.1093/neuonc/nov289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sahm F, Schrimpf D, Jones DT, et al. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol. 2016;131(6):903–910. doi:10.1007/s00401-015-1519-8 [DOI] [PubMed] [Google Scholar]
- 17. Mafficini A, Amato E, Fassan M, et al. Reporting tumor molecular heterogeneity in histopathological diagnosis. PLoS One. 2014;9(8):e104979. doi:10.1371/journal.pone.0104979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ihle MA, Fassunke J, König K, et al. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p.V600E and non-p.V600E BRAF mutations. BMC Cancer. 2014;14:13. doi:10.1186/1471-2407-14-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin MT, Mosier SL, Thiess M, et al. Clinical validation of KRAS, BRAF, and EGFR mutation detection using next-generation sequencing. Am J Clin Pathol. 2014;141(6):856–866. doi:10.1309/AJCPMWGWGO34EGOD [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Portier BP, Kanagal-Shamanna R, Luthra R, et al. Quantitative assessment of mutant allele burden in solid tumors by semiconductor-based next-generation sequencing. Am J Clin Pathol. 2014;141(4):559–572. doi:10.1309/AJCP1JUGQMW7ZNTL [DOI] [PubMed] [Google Scholar]
- 21. de Leng WW, Gadellaa-van Hooijdonk CG, Barendregt-Smouter FA, et al. Targeted next generation sequencing as a reliable diagnostic assay for the detection of somatic mutations in tumours using minimal DNA amounts from formalin fixed paraffin embedded material. PLoS One. 2016;11(2):e0149405. doi:10.1371/journal.pone.0149405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoogstraat M, Hinrichs JW, Besselink NJ, et al. Simultaneous detection of clinically relevant mutations and amplifications for routine cancer pathology. J Mol Diagn. 2015;17(1):10–18. doi:10.1016/j.jmoldx.2014.09.004 [DOI] [PubMed] [Google Scholar]
- 23. Bijlsma RM, Bredenoord AL, Gadellaa-Hooijdonk CG, et al. Unsolicited findings of next-generation sequencing for tumor analysis within a Dutch consortium: clinical daily practice reconsidered. Eur J Hum Genet. 2016;24(10):1496–1500. doi:10.1038/ejhg.2016.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grasso C, Butler T, Rhodes K, et al. Assessing copy number alterations in targeted, amplicon-based next-generation sequencing data. J Mol Diagn. 2015;17(1):53–63. doi:10.1016/j.jmoldx.2014.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dubbink HJ, Atmodimedjo PN, van Marion R, et al. Diagnostic detection of allelic losses and imbalances by next-generation sequencing: 1p/19q co-deletion analysis of gliomas. J Mol Diagn. 2016;18(5):775–786. doi:10.1016/j.jmoldx.2016.06.002 [DOI] [PubMed] [Google Scholar]
- 26. Lo KC, Bailey D, Burkhardt T, Gardina P Turpaz Y Cowell JK. Comprehensive analysis of loss of heterozygosity events in glioblastoma using the 100K SNP mapping arrays and comparison with copy number abnormalities defined by BAC array comparative genomic hybridization. Genes Chromosomes Cancer. 2008;47(3):221–237. doi:10.1002/gcc.20524. [DOI] [PubMed] [Google Scholar]
- 27. Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D Diepvens F Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30(12):e57. doi:10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hömig-Hölzel C, Savola S. Multiplex ligation-dependent probe amplification (MLPA) in tumor diagnostics and prognostics. Diagn Mol Pathol. 2012;21(4):189–206. doi:10.1097/PDM.0b013e3182595516 [DOI] [PubMed] [Google Scholar]
- 29. Jeuken JW, Cornelissen SJ, Vriezen M, et al. MS-MLPA: An attractive alternative laboratory assay for robust, reliable, and semiquantitative detection of MGMT promoter hypermethylation in gliomas. Lab Invest. 2007;87(10):1055–1065. doi:10.1038/labinvest.3700664. [DOI] [PubMed] [Google Scholar]
- 30. Natté R, van Eijk R, Eilers P, et al. Multiplex ligation-dependent probe amplification for the detection of 1p and 19q chromosomal loss in oligodendroglial tumors. Brain Pathol. 2005;15:192–197. doi:10.1111/j.1750-3639.2005.tb00520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Warnecke PM, Stirzaker C, Song J, Grunau C Melki JR Clark SJ. Identification and resolution of artifacts in bisulfite sequencing. Methods. 2002;27(2):101–107. doi:10.1016/S1046-2023(02)00060-9. [DOI] [PubMed] [Google Scholar]
- 32. Dikow N, Nygren AO, Schouten JP, et al. Quantification of the methylation status of the PWS/AS imprinted region: comparison of two approaches based on bisulfite sequencing and methylation-sensitive MLPA. Mol Cell Probes. 2007;21(3):208–215. doi:10.1016/j.mcp.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 33. Dunn J, Baborie A, Alam F, et al. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer. 2009;101(1):124–131. doi:10.1038/sj.bjc.6605127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brell M, Ibáñez J, Tortosa A. O6-Methylguanine-DNA methyltransferase protein expression by immunohistochemistry in brain and non-brain systemic tumours: systematic review and meta-analysis of correlation with methylation-specific polymerase chain reaction. BMC Cancer. 2011;11:35. doi:10.1186/1471-2407-11-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi:10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 36. Haff LA. Improved quantitative PCR using nested primers. PCR Methods Appl. 1994; 3(6):332–337 http://genome.cshlp.org/content/3/6/332.abstract [DOI] [PubMed] [Google Scholar]
- 37. Vlassenbroeck I, Califice S, Diserens AC, et al. Validation of real-time methylation-specific PCR to determine O6-methylguanine-DNA methyltransferase gene promoter methylation in glioma. J Mol Diagn. 2008;10(4):332–337. doi:10.2353/jmoldx.2008.070169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Preusser M, Elezi L, Hainfellner JA. Reliability and reproducibility of PCR-based testing of O6-methylguanine-DNA methyltransferase gene (MGMT) promoter methylation status in formalin-fixed and paraffin-embedded neurosurgical biopsy specimens. Clin Neuropathol. 2008;27(6):388–390. doi:10.5414/NPP27388. [DOI] [PubMed] [Google Scholar]
- 39. Yip S, Iafrate AJ, Louis DN. Molecular diagnostic testing in malignant gliomas: a practical update on predictive markers. J Neuropathol Exp Neurol. 2008;67(1):1–15. doi:10.1097/nen.0b013e31815f65fb. [DOI] [PubMed] [Google Scholar]
- 40. Kagan J, Srivastava S, Barker PE, Belinsky SA, Cairns P. Towards clinical application of methylated DNA sequences as cancer biomarkers: a joint NCI’s EDRN and NIST workshop on standards, methods, assays, reagents and tools. Cancer Res. 2007;67(10):4545–4549. doi:10.1158/0008-5472.CAN-06-2888. [DOI] [PubMed] [Google Scholar]
- 41. Ogino S, Kawasaki T, Brahmandam M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8(2):209–217. doi:10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quillien V, Lavenu A, Karayan-Tapon L, et al. Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, MethyLight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients. Cancer. 2012;118(17):4201–4211. doi:10.1002/cncr.27392 [DOI] [PubMed] [Google Scholar]
- 43. Nygren AO, Ameziane N, Duarte HM, et al. Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res. 2005;33(14):e128. doi:10.1093/nar/gni127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bady P, Sciuscio D, Diserens AC, et al. MGMT methylation analysis of glioblastoma on the infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124(4):547–560. doi:10.1007/s00401-012-1016-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nyrén P, Pettersson B, Uhlén M. Solid phase DNA minisequencing by an enzymatic luminometric inorganic pyrophosphate detection assay. Anal Biochem. 1993;208(1):171–175. doi:10.1006/abio.1993.1024. [DOI] [PubMed] [Google Scholar]
- 46. Karayan-Tapon L, Quillien V, Guilhot J, et al. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J Neurooncol. 2010;97(3):311–322. doi:10.1007/s11060-009-0031-1 [DOI] [PubMed] [Google Scholar]
- 47. Wang L, Li Z, Liu C, et al. Comparative assessment of three methods to analyze MGMT methylation status in a series of 350 gliomas and gangliogliomas. Pathol Res Pract. 2017;213(12):1489–1493. doi:10.1016/j.prp.2017.10.007 [DOI] [PubMed] [Google Scholar]
- 48. Capper D, Engel NW, Stichel D, et al. DNA methylation-based reclassification of olfactory neuroblastoma. Acta Neuropathol. 2018;136(2):255–271. doi:10.1007/s00401-018-1854-7 [DOI] [PubMed] [Google Scholar]
- 49. Bady P, Delorenzi M, Hegi ME. Sensitivity analysis of the MGMT-STP27 model and impact of genetic and epigenetic context to predict the MGMT methylation status in gliomas and other tumors. J Mol Diagn. 2016;18(3):350–361. doi:10.1016/j.jmoldx.2015.11.009 [DOI] [PubMed] [Google Scholar]
- 50. Capper D, Stichel D, Sahm F, et al. Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: the Heidelberg experience. Acta Neuropathol. 2018;136(2):181–210. doi:10.1007/s00401-018-1879-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van Vlodrop IJ, Niessen HE, Derks S, et al. Analysis of promoter CpG island hypermethylation in cancer: location, location, location!Clin Cancer Res. 2011;17(13):4225–4231. doi:10.1158/1078-0432.CCR-10-3394 [DOI] [PubMed] [Google Scholar]
- 52. Capper D, Zentgraf H, Balss J, Hartmann C, von Deimling A. Monoclonal antibody specific for IDH1 R132H mutation. Acta Neuropathol. 2009;118(5):599–601. doi:10.1007/s00401-009-0595-z [DOI] [PubMed] [Google Scholar]
- 53. Capper D, Preusser M, Habel A, et al. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011;122(1):11–19. doi:10.1007/s00401-011-0841-z [DOI] [PubMed] [Google Scholar]
- 54. Venneti S, Santi M, Felicella MM, et al. A sensitive and specific histopathologic prognostic marker for H3F3A K27M mutant pediatric glioblastomas. Acta Neuropathol. 2014;128(5):743–753. doi:10.1007/s00401-014-1338-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reuss DE, Sahm F, Schrimpf D, et al. ATRX and IDH1-R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an “integrated” diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol. 2015;129(1):133–146. doi:10.1007/s00401-014-1370 [DOI] [PubMed] [Google Scholar]
- 56. Parker M, Mohankumar KM, Punchihewa C, et al. C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature. 2014;506(7489):451–455. doi:10.1038/nature13109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hovelson DH, McDaniel AS, Cani AK, et al. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia. 2015;17(4):385–399. doi:10.106/j.neo.2015.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rogawski DS, Vitanza NA, Gauthier AC, Ramaswamy V, Koschmann C. Integrating RNA sequencing into neuro-oncology practice. Transl Res. 2017;189:93–104. doi:10.1016/j.trsl.2017.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Euskirchen P, Bielle F, Labreche K, et al. Same-day genomic and epigenomic diagnosis of brain tumors using real-time nanopore sequencing. Acta Neuropathol. 2017;134(5):691–703. doi:10.1007/s00401-017-1743-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Goodwin S, Gurtowski J, Ethe-Sayers S, Deshpande P, Schatz MC, McCombie WR. Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res. 2015;25(11):1750–1756. doi:10.1101/gr.191395.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Madoui MA, Engelen S, Cruaud C, et al. Genome assembly using Nanopore-guided long and error-free DNA reads. BMC Genomics. 2015;16:327. doi:10.1186/s12864-015-1519-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Deschamps S, Mudge J, Cameron C, et al. Characterization, correction and de novo assembly of an Oxford Nanopore genomic dataset from Agrobacterium tumefaciens. Sci Rep. 2016;6:28625. doi:10.1038/srep28625 [DOI] [PMC free article] [PubMed] [Google Scholar]