Abstract
Sage-grouse are two closely related iconic species of the North American West, with historically broad distributions across sagebrush-steppe habitat. Both species are dietary specialists on sagebrush during winter, with presumed adaptations to tolerate the high concentrations of toxic secondary metabolites that function as plant chemical defenses. Marked range contraction and declining population sizes since European settlement have motivated efforts to identify distinct population genetic variation, particularly that which might be associated with local genetic adaptation and dietary specialization of sage-grouse. We assembled a reference genome and performed whole-genome sequencing across sage-grouse from six populations, encompassing both species and including several populations on the periphery of the species ranges. Population genomic analyses reaffirmed genome-wide differentiation between greater and Gunnison sage-grouse, revealed pronounced intraspecific population structure, and highlighted important differentiation of a small isolated population of greater sage-grouse in the northwest of the range. Patterns of genome-wide differentiation were largely consistent with a hypothesized role of genetic drift due to limited gene flow among populations. Inferred ancient population demography suggested persistent declines in effective population sizes that have likely contributed to differentiation within and among species. Several genomic regions with single-nucleotide polymorphisms exhibiting extreme population differentiation were associated with candidate genes linked to metabolism of xenobiotic compounds. In vitro activity of enzymes isolated from sage-grouse livers supported a role for these genes in detoxification of sagebrush, suggesting that the observed interpopulation variation may underlie important local dietary adaptations, warranting close consideration for conservation strategies that link sage-grouse to the chemistry of local sagebrush.
Keywords: local adaptation, evolutionarily significant units, cytochrome P450, Artemisia, PSM
Introduction
Identifying and preserving potentially unique genetic adaptations is an important component for developing sound conservation and management strategies (Allendorf et al. 2010; Oyler-McCance et al. 2016), especially in species with highly constrained ecological niches such as host or dietary specialization. Both theory (Garcia-Ramos and Kirkpatrick 1997) and empirical studies (Power 1979; Petren et al. 2005) suggest that populations isolated on the periphery of a species’ range are particularly prone to rapid adaptive change in response to local selection in the absence of gene flow, highlighting the importance of such populations as potential reservoirs of adaptive diversity (Macdonald et al. 2017). Yet disentangling the relative contributions of adaptive versus neutral evolutionary processes to interpopulation genetic variation can be challenging for species with complex population structure or unknown demographic histories (Städler et al. 2009). Here, studies at the genome-wide level can prove useful by simultaneously providing inferences regarding the underlying population dynamics and also the ability to parse potential targets of selection from neutral variation in the genome (Oleksyk et al. 2010; Foote et al. 2016).
Sage-grouse (Centrocercus spp.) are endemic to the distinct sagebrush-steppe habitat of western North America and have recently been the focus of intensive research and conservation concern due to dramatic and ongoing population declines since European settlement (fig. 1) (Connelly and Braun 1997; Schroeder et al. 2004) which threaten long-term population persistence (Aldridge et al. 2008; Garton et al. 2011). Several greater sage-grouse (Centrocercusurophasianus) populations existing on the periphery of the species ranges, along with entire range of the Gunnison sage-grouse (Centrocercusminimus) have received particular interest due to small population sizes and lack of gene flow associated with geographic isolation (Schroeder et al. 2004; Crist et al. 2017). Both species are sagebrush (Artemisia spp.) obligates, relying entirely on these shrubs as winter forage despite the high concentrations of plant secondary metabolites (PSMs) such as monoterpenes, sesquiterpene lactones, and phenolics produced in high concentrations on the sagebrush epidermal surface (Kelsey et al. 1982). These compounds are known to inhibit palatability and digestibility in vertebrate digestive tracts (Personius et al. 1987; Striby et al. 1987) and are thus thought to have evolved as chemical defenses to herbivory. Yet sagebrush specialists like the sage-grouse possess both behavioral (Ulappa et al. 2014) and biochemical counter-adaptations (Kohl et al. 2015) that mitigate such effects. Among these, metabolic detoxification by hepatic enzymes in the cytochrome P450 superfamily (hereafter, CYP) in general, and the CYP1 and CYP2 family in particular, are considered among the most critical adaptations (Liukkonen-Anttila et al. 2003; Goldstone et al. 2007), and marked intra- and inter-specific diversity in the associated protein-coding genes are thought to reflect the results of a coevolutionary arms race between plants and herbivores (Gonzalez and Nebert 1990). Furthermore, considerable geographic variation in both composition and concentration of PSM among and within sagebrush varieties is well documented (Welch and McArthur 1981), suggesting that sage-grouse populations across the species’ ranges may be subject to distinct natural selection imposed by the chemistry of local sagebrush varieties.
Fig. 1.

—Current and estimated pre-European settlement geographic species range for sage-grouse (data adapted from Schroeder et al. [2004]). Sampling locations for populations included in this study indicated by red circles.
In this study, we sequenced and assembled the first reference genome for sage-grouse, along with 90 individual genomes representing both species and several small and relatively isolated populations of interest for conservation (fig. 1). From these data, we used a high-density set of single-nucleotide polymorphisms (SNPs) to probe the genomic landscape of differentiation among and within species and generate inference regarding the evolutionary mode of divergence. Whole-genome diploid sequences were used to reconstruct ancestral demographic trends in both species. Finally, we evaluated evidence for elevated interpopulation divergence in regions associated with putative CYP loci as a potential local adaptation to PSMs, coupled with an in vitro assay of microsomes of sage-grouse livers to determine relative CYP enzyme activity. This study provides valuable new insights regarding population differences at the whole-genome level for these species which have been the focus of intense study and ongoing public debate.
Materials and Methods
Sequencing, Genome Assembly, and Mapping
Genomic DNA was extracted from a whole blood sample of a single Gunnison sage-grouse male and genomic libraries were prepared and submitted for both short-read (2× 125-bp paired-end Illumina HiSeq SBS) and long-read (Pacific Biosciences SMRT) sequencing, yielding a combined total of ∼204 billion bp of sequence data. De novo genome assembly was performed via a hybrid approach (Ye et al. 2016) that leverages the high accuracy of the dense short-read data set along with the repeat-spanning benefits afforded by the long-read sequences (supplementary section S1.1, Supplementary Material online). A draft genome annotation was generated using homology-based methods (Cantarel et al. 2008), utilizing existing genetic and protein databases from other galliform species (supplementary section S1.4, Supplementary Material online).
Genomic DNA from an additional 90 samples was extracted as above, with 15 samples from each of five greater sage-grouse populations (AL, southeastern Alberta, Canada; BI, Bi-state population from the southern border of Nevada and California, USA; JH, Jackson Hole, WY, USA; PI, Piceance basin in northwestern Colorado, USA; WA, from southern Washington, USA) along with 15 C. minimus samples from the Gunnison basin and the nearby Crawford population were submitted for 2× 150-bp whole-genome shotgun sequencing (supplementary section S2.1, Supplementary Material online).
Quality Assessment and Site Filtering
Initial quality assessment of mapped resequencing data was carried out in ANGSD v0.917 (Korneliussen et al. 2014). Following adjustment for indels (-bap option) and excessive mismatches (-C 50), the distributions of quality scores and per-site depths were visualized using the R scripts in ngsTools v1.0.1 (Fumagalli et al. 2014). Final filters selected in ANGSD included removal of low-quality reads and bases (-remove_bads, -minMapQ 20, -minQ 20), triallelic sites (-skipTriallelic), and reads with unmapped pairs (-only_proper_pairs). Only sites with global read depths (i.e., total across all samples) ranging from 75 to 8,000× and those present in at least 75 samples were included. The final data set was restricted to sites with strong evidence for a SNP presence (-SNP_pval 1e-6).
Genomic Distance and Neighbor-Joining Tree
A matrix of pairwise genetic distances among all samples was generated using ngsDist v1.0.2 (Vieira et al. 2016), which utilizes a probabilistic approach to estimate genetic distances while accounting for genotyping uncertainty. As input, we used the posterior probabilities estimated from ANGSD, using the same filters as above. A neighbor-joining phylogenetic tree was estimated from this distance matrix in FastME v2.1.4 (Lefort et al. 2015) utilizing the BIONJ agglomerative clustering algorithm (Gascuel 1997), and the resulting tree visualized using FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/ last accessed 1 December 2018).
Principal Component Analysis
Principal component analysis (PCA) was carried out without calling genotypes following the procedure outlined in Fumagalli et al. (2013). Genotype likelihoods were estimated in ANGSD v0.917 (Korneliussen et al. 2014) using the SAMtools (Li 2011) model (“GL -1” option) with the default filter settings, along with options as indicated above. These filtered genotype likelihoods were then used to infer major and minor alleles and calculate per-site allele frequencies. Allele frequency estimates for these sites were in turn provided as priors (assuming Hardy Weinberg equilibrium) for estimating genotype posterior probabilities for all loci mapping to putative autosomes. A covariance matrix between all samples was calculated based on the genotype probabilities using the ngsCovar utility in ngsTools (Fumagalli et al. 2014) and subjected to eigenvector decomposition to estimate the first two principal components.
Population Structure
Pairwise estimates of per-site FST were calculated among populations using ANGSD v0.917 (Korneliussen et al. 2014), using the corresponding pairwise site frequency spectra (2D-SFS) as priors for the joint allele frequency probabilities at each site. Sites mapping to putative Z chromosome were analyzed separately from those mapping to putative autosomes. The realSFS utility in ANGSD was used to calculate the weighted global FST values for autosomal and Z-linked sites, as well as perform a 50-kb sliding window analysis (with 25-kb intervals). To further investigate patterns of differentiation, sliding window analyses were also performed between species, and also contrasting C. urophasianus populations that clustered strongly together in the PCA (i.e., “northeast” populations: AL, JH, and PI) versus the more distinct BI and WA populations.
Genetic Admixture
Individual admixture proportions were estimated using NGSadmix (Skotte et al. 2013), which utilizes a core model (e.g., Pritchard et al. 2000) and maximum-likelihood framework (Alexander et al. 2009) similar to other approaches, but operates on genotype likelihoods as input, which makes it well suited for our low-coverage resequencing data set. Genotype likelihoods for autosomal loci were first calculated in ANGSD, and admixture proportions were estimated for a range of ancestral populations K = 2–6, and a minimum minor allele frequency of 0.01. Admixture plots were generated using the R package pophelper (Francis 2017).
To test for potential bias on inferred population structure due to isolation-by-distance (Meirmans 2012), a partial Mantel test was carried out using the vegan R package (Oksanen et al. 2017). Pearson’s correlation coefficient was calculated between a matrix of pairwise FST values and a matrix of cluster membership with WA and BI populations in distinct clusters apart for a third cluster composed of the other three C. urophasianus populations (AL, JH, and PI), while controlling for straight-line geographic distance (Euclidean) between populations. Significance was evaluated by 100 permutations of the FST matrix.
Outlier Detection
We performed genomic scans for potential targets of adaptive divergence using BayPass v2.1 (Gautier 2015) which uses Bayesian approach to identify loci with extreme allelic divergence among populations. This is indicated by elevated values of XTX (Günther and Coop 2013), a statistic that can be viewed as a measure of population allelic differentiation (e.g., FST) that explicitly controls for observed background population genetic covariance. Briefly, this approach first estimates the scaled covariance matrix of population allele frequencies (Ω) directly from the data. A VCF file including all samples was first generated from ANGSD (-doVcf 1) using the same filters as above. Sites mapping to putative autosomes and Z chromosome were analyzed separately. Genotypes were set based on the genotype likelihood scores using the vcfkeepgeno and vcfglxgt scripts in vcflib library (https://github.com/vcflib/vcflib last accessed 1 December 2018), and the data set was converted to the BayPass input format using a Python script (https://github.com/chrishah/genomisc/blob/master/popogeno/vcf_2_div.py last accessed 1 December 2018). XTX outlier thresholds were estimated by first generating a pseudo-observed data set of the equivalent sample size and containing 100,000 SNPs, by providing the posterior estimate of Ω and hyperparameters of the prior distribution for SNP allele frequencies (β) to the simulate.baypass() R function. This data set was then analyzed in BayPass, and the 99% probability value from the distribution of resulting XTX values was selected as the outlier threshold. Classification according to genomic region (intron, exon, intergenic, and 25-kb flanking) for SNPs within each quintile of the XTX distribution was carried out using SnpEff (Cingolani et al. 2012); under/overrepresentation of SNPs within each region was evaluated using a chi-square goodness of fit test in comparison to genome-wide expected proportions, which was significant for all quintiles (all P < 2e−16). Subsequent post hoc tests were performed using exact binomial tests in R for each region with Bonferroni corrections applied to account for multiple tests (P < 0.0125). To evaluate patterns of population divergence at outlier loci, we performed PCA using ANGSD v0.97 as above, but only including the 1% outlier loci.
Ancestral Demography
Estimates of ancestral effective population sizes (Ne) for both species were inferred using a pairwise sequentially Markovian coalescent (PSMC) model as described in Li and Durbin (2011) and implemented in the accompanying software package (https://github.com/lh3/psmc last accessed 1 December 2018). Diploid consensus sequences were generated using functions in SAMtools v1.3 (Li et al. 2009) and BCFtools (Li 2011) for the single male C. minimus sample (from the Utah population) utilized in the reference genome assembly as well as the single C. urophasianus male (collected from the Piceance Basin population) that was sequenced at 27× coverage. To facilitate comparison between the species, the original sequence data for the C. minimus were downsampled to the same mean coverage value (27×) prior to consensus calling and only regions mapping to the chicken autosomes were included in the final data sets. Consensus sequences were converted to psmcfa format using the fq2psmcfa utility and pilot runs of the PSMC model were carried out to determine appropriate parameters according to the authors’ recommendations. The final model for each species was run for 20 iterations and inferred Ne across 84 atomic time intervals and 34 free interval parameters (-p option specified as “4 + 30*2 + 4 + 6 + 10”), such that the first parameter spanned the first four atomic time intervals, each of the next 30 parameters spanned two time intervals, and the final three free intervals spanned four, six, and ten time intervals. The upper limit for the time to most recent common ancestor (-t option) and the initial θ/ρ value (-r option) were set at 12 and 1, respectively, which were determined to result in ∼10 inferred recombination events occurring within each parameter interval (https://github.com/lh3/psmc/blob/master/README last accessed 1 December 2018). Variance in the inferred population history was estimated by a bootstrapping procedure wherein the sequences were first split into smaller (500,000-bp maximum) segments, followed by 100 rounds of random sampling (with replacement) to create new data sets which were analyzed using the same model parameters as above (20 iterations each round). Final results were scaled based on a mean generation of 2.1 years (Stiver et al. 2008) and a galliform lineage-specific mutation rate of 1.91 × 10−9 site−1 year−1 (based on divergence at 4-fold degenerate sites) (Nam et al. 2010).
Intraspecific Variation of Candidate Metabolic Genes
We assembled a panel of candidate genes for dietary adaptations (supplementary table S2, Supplementary Material online) which included 45 cytochrome P450 genes along with 22 genes highlighted as “very important pharmacogenes” by the Pharamacogenomics Research Network (www.pharmgkb.org) that were annotated in the Gunnison sage-grouse genome. Intersections between outlier SNPs and candidate gene regions (BLAT best hit location ± 10-kb flanking region) were obtained using BEDOPS v2.4.30 (Neph et al. 2012). Analysis of adaptive differentiation in genomic regions containing these loci was performed in Gowinda v1.12 (Kofler and Schlötterer 2012), which uses randomization tests to evaluate associations between gene sets and SNPs while accounting for gene length, multiple gene overlap, as well as multiple SNPs per gene. Specifically, we identified gene overlap between the XTX outlier SNPs (identified in the genome scans above) and tested for overrepresentation of gene sets composed of 1) all 67 candidate genes, 2) the 45 cytochrome P450 genes, or 3) only the CYP1 and CYP2 candidate genes, using 100,000 simulation iterations each.
In Vitro Cytochrome P450 Activity Assays
Cytochrome P450 activity in liver microsomes from five wild C. urophasianus samples (supplementary section S4.1 and S4.2, Supplementary Material online) and commercially available composite microsomes from chicken (Sekisui XenoTech, LLC, Kansas City, KS) were measured using P450-Glo luminescent assays (Promega Corp., Madison, WI). Two different pro-luciferin substrates were utilized: Luciferin-MultiCYP, which is known to cross-react with at least 21 human CYP enzymes (Sobol et al. 2008); and Luciferin-ME, which cross-reacts more specifically with human CYP1A2 (orthologous to the avian CYP1A5 gene, Goldstone and Stegeman 2006), CYP2C8, CYP2C9, CYP2J2, CYP4A11, CYP4F3B, and CYP19. All assays included a negative control (no microsomes) and were run in quadruplicate according to the manufacturer’s protocols in 96-well plates (with modifications as described in supplementary section S4.3, Supplementary Material online). Estimates of enzyme activity were calculated as pmol luciferin/mg microsomal protein. Negative estimates were set to zero and activity levels were scaled and centered prior to statistical analysis in a mixed linear model using the lme4 package (Bates et al. 2014) in R v3.4.3 (R Development Core Team 2016), wherein individual sample ID was included as a random effect and species/control as a fixed effect. Significance of the fixed effect was assessed using a likelihood ratio test comparing full and reduced models.
Results
A high-quality reference genome (Cmin_1.0) was assembled from a high-depth (146× Illumina SBS short-read and 58X Pacific Biosciences SMRT long-read) sequencing data set for a single male Gunnison sage-grouse from southeastern Utah, USA (supplementary section S1, Supplementary Material online). The de novo scaffolded assembly was composed of 985 scaffolds with a N50 value of 3.85 Mb and a total assembly length of 1.00 Gb. Whole-genome syntenic alignment to the chicken genome (galGal4) (Grabherr et al. 2010) allowed us to join scaffolds into near-chromosome-length sequences (N50 = 131 Mb), reducing the total number of scaffolds to 294, while retaining low levels (0.02%) of ambiguous bases (supplementary table S1, Supplementary Material online). We also assembled a complete sequence for the mitochondrial genome (supplementary section S1.2, Supplementary Material online). Subsequent homology-based gene prediction and annotation resulted in a final set of 18,565 protein-coding nuclear genes.
Whole-genome resequencing of 90 individual genomes and subsequent mapping to the Gunnison sage-grouse reference (supplementary section S2.2, Supplementary Material online) yielded a mean sequence read depth of 4.46× (S.D. = 3.51). We also performed additional sequencing for one greater sage-grouse sample to a mean depth of 27.7×, which was utilized to create a draft greater sage-grouse reference. Population genomic analyses were carried out in a probabilistic statistical framework using ANGSD v0.917 (Korneliussen et al. 2014), which is based on genotype likelihoods to carry uncertainty regarding sequence and mapping quality through downstream analyses, thereby reducing inherent error associated with calling genotypes from low-coverage data sets. Initial filtering for read and mapping quality resulted in a data set including 1,500,781 nuclear sites.
Population Genomic Structure and Divergence
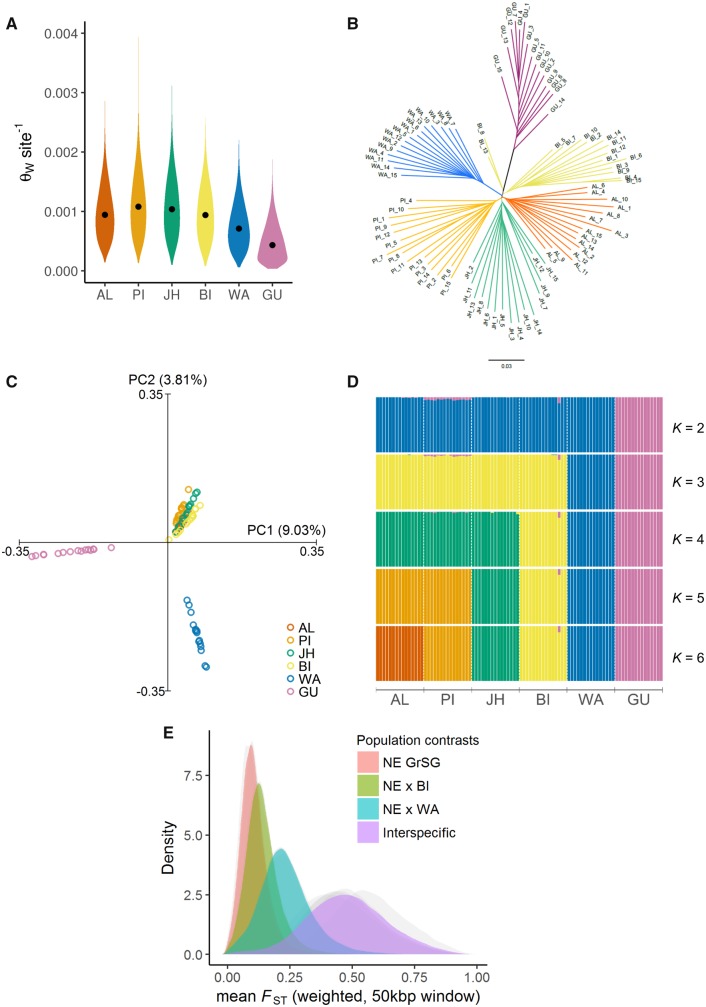
Evaluation of genome-wide diversity (fig. 2A) indicated significant differences among populations in nucleotide diversity (ANOVA: F(5,616019) = 26,123, P < 0.001; post hoc Tukey test: all P < 0.001). Notably, there were markedly low levels of nucleotide diversity in Gunnison sage-grouse and greater sage-grouse from Washington compared with other populations. A neighbor-joining tree based on pairwise genomic distances (fig. 2B) supported clear interspecific divergence between greater and Gunnison sage-grouse and also provided evidence of pronounced population structure within greater sage-grouse, with all samples unambiguously clustering within each population. Greater sage-grouse from Washington exhibited elevated divergence with respect to other conspecific populations, as evidence by the comparatively longer branch length for these samples. PCA of whole-genome SNP data sets (fig. 2C) suggested a similar pattern, with the leading eigenvector (PC1) capturing interspecific divergence (9.03% of variance in the data set), whereas PC2 (3.81%) differentiated all Washington samples from other greater sage-grouse. PCA including only greater sage-grouse populations (supplementary fig. S1, Supplementary Material online) also showed distinct clustering by population, with PC1 differentiating Washington sage-grouse from all other greater sage-grouse, and PC2 distinguishing Bi-State samples. Additional tests using a maximum-likelihood clustering approach (fig. 2D) (Skotte et al. 2013) showed very little evidence of genetic admixture at K = 6 populations, and inspection of clustering at lower numbers of ancestral populations (K = 2–5) were indicative of a hierarchical population structure, with well-delineated clustering nesting at successive levels. Estimation of pairwise genomic FST values (fig. 2E, supplementary fig. S2, Supplementary Material online) provided quantitative corroboration of this pattern, with a spectrum of whole-genome divergence ranging from relatively low divergence among greater sage-grouse populations in the northeast of the range (mean pairwise autosomal FST = 0.119) up to relatively high levels of interspecific divergence (mean autosomal FST = 0.460). Among greater sage-grouse populations, comparisons involving the Washington population exhibited the highest levels of pairwise divergence, with mean FST values approximately double the magnitude observed among other conspecific population comparisons (fig. 2E, supplementary fig. S2, Supplementary Material online).
Fig. 2.
—Inter- and intra-specific population genomic analyses of sage-grouse based on whole-genome resequencing. (A) Violin plots of mean per-site autosomal nucleotide diversity (Watterson’s estimator, θW) across populations. Values were estimated from 50-kb sliding windows. Median values are plotted within each kernel density curve. (B) Neighbor-joining tree based on pairwise genetic distances at autosomal SNPs, with branches colored by population. (C) Principal components analysis of complete data set, based on 1,500,781 nuclear SNPs. Axes represent first (PC1) and second (PC2) principal components, with percentage of total genetic variance explained by each component shown in parentheses. (D) Hierarchical population genetic structure illustrated by admixture proportions (y axis) for 90 individuals (vertical columns) across populations, based on genotype likelihoods assuming different numbers of ancestral populations (K = 2–6). Solutions with greatest likelihood for each K value are shown. (E) Distribution of genome-wide mean FST values (calculated as weighted average within 50-kb sliding windows with 25-kb steps) from all population comparisons. Distributions from individual pairwise tests (gray) tended to fall into four distinct intervals, representing a spectrum of genetic differentiation ranging from relatively genetically similar populations in the northeastern portion of the Centrocercus urophasianus range (“NE GrSG”: Alberta, Jackson Hole, and Piceance Basin), to the interspecific divergence observed between C. urophasianus populations and Centrocercus minimus. Superimposed colored distributions represent the average FST values taken across pairwise tests within each of these intervals. FST distributions for each pairwise test are provided in the supplementary figure S1, Supplementary Material online. Population abbreviations: C. urophasianus: AL, Alberta; BI, Bi-State; JH, Jackson Hole; PI, Piceance Basin; C. minimus: GU, Gunnison Basin and Crawford.
Results of a partial Mantel test controlling for interpopulation geographic distance confirmed significant population structure (Pearson’s ρ = 0.625, P = 0.05) wherein both Washington and Bi-State populations were distinct from a cluster composed of the three other C. urophasianus populations (Alberta, Jackson Hole, and Piceance Basin).
Ancestral Population Demography
Analysis of diploid full genome sequences from both sage-grouse species using PSMC models (Li and Durbin 2011) yielded insight into ancestral population demographic trends (fig. 3). Although the inferred effective population sizes (Ne) of both species converge at ∼200,000 at ∼4.5 Ma, demographic decoupling is evident during a subsequent period that coincides with the estimated time of species divergence (median = 1.24 Ma, range = 0.58–1.64 Ma) (Kumar et al. 2017), suggesting reproductive isolation between ancestral populations and the initial stages of speciation. Additionally, both species appear to have undergone persistent declines in effective population size, starting at ∼1 Ma for Gunnison sage-grouse and 2 Ma for greater sage-grouse. The estimated period for these declines overlaps with the last glacial period in North America, interrupted only by a relatively brief spike in population size for greater sage-grouse from 15 to 60 ka, before dropping again to Ne = 50,000 within the most recent time interval (10 ka). Inferred effective population size for Gunnison sage-grouse during this same period was considerably lower (Ne = 5,000), which is consistent with extant population sizes in this species that are orders of magnitude lower than its congener (Schroeder et al. 2004), though we note the increased uncertainty in model performance in the most contemporary time intervals.
Fig. 3.

—Historical changes in effective population size, from PSMC analysis of whole-genome sequences for a single Centrocercus urophasianus (blue) male and a single Centrocercus minimus (red) male. Lighter lines represent bootstrap replicates. x axis indicates (log-scale) years before present, model results were scaled based on sage-grouse generation time (g) and lineage-specific mutation rate (µ).
Genomic Scans of Allelic Divergence
Patterns of divergence were relatively uniform across the genome (fig. 4A). Moreover, evaluation of the genomic regions in which SNPs were observed across the distribution of XTX values (fig. 4B) showed no disproportionate occurrence of highly divergent loci (i.e., highest quintile of the XTX distribution) in exons or flanking (i.e., putative gene regulatory) regions, which would suggest a dominant role of natural or sexual selection in driving patterns of population genetic differentiation. Instead, SNPs with low levels of population differentiation were more likely to occur in exons or flanking regions, most likely due to the effects of stabilizing selection for conserved gene function across populations. As expected, a PCA of the 1% outlier SNPs (fig. 4C) produced distinct population clustering, with a topology similar to that observed with the whole-genome data set (fig. 2C), except that the leading eigenvector (PC1, which accounted for 10.8% of the total genetic variation at these sites) differentiated Washington greater sage-grouse from all other conspecific and heterospecific populations.
Fig. 4.
—Identification and characterization of potential genomic targets of selection. (A) Manhattan plot of SNP outlier tests from whole-genome resequencing of six sage-grouse populations. Elevated XTX values are indicative of loci with extreme population differentiation, while controlling for underlying neutral population structure. Dashed red line represents the 1% probability threshold as estimated from analysis of pseudo-observed data sets. Contrasting gray points correspond to different chromosomes based on syntenic alignment to the chicken genome. Highlighted are SNPs that occur in regions (gene ± 10-kb flanking region) containing candidate genes for metabolic adaptation (green = cytochrome P450 genes, yellow = other important pharmacogenes). (B) Proportion of SNP effects in different genomic regions across each quintile of the observed XTX distribution. Asterisks indicate significant overrepresentation of SNP effects in a given genomic region. (C) PCA of 1% XTX outlier SNPs (8,592 sites). Axes represent first (PC1) and second (PC2) principal components, with percentage of total genetic variance explained by each component shown in parentheses.
Despite these overall patterns suggesting predominantly nonadaptive differentiation among populations, our assessment of candidate genes underlying xenobiotic metabolism (supplementary table S2, Supplementary Material online) revealed evidence of potential adaptive variation. Specifically, multiple XTX outlier SNPs overlapped with genomic regions containing candidate genes underlying adaptation to sagebrush diets (fig. 4A), including 16 putative cytochrome P450 genes, and five regions with genes identified from pharmacological research to harbor important polymorphisms influencing xenobiotic metabolism and drug responses (Peters and McLeod 2008). Analysis using randomization tests found that outlier XTX SNPs were significantly more likely to occur in association with CYP1 and CYP2 genes than expected by chance (false discovery rate-corrected P = 0.024), but not for gene sets containing all CYP genes (P = 0.130) or the entire 67 candidate genes (P = 1.00).
Metabolic Enzyme Activity
Assessment of enzyme activity from in vitro assays (fig. 5) utilizing the Luciferin-ME substrate indicated elevated cytochrome P450 activity in greater sage-grouse compared with a nonsagebrush specialist galliform species (chicken) (χ2(2) = 6.77, P = 0.034). The difference between species was not significant for assays utilizing a relatively nonselective cytochrome P450 substrate assay (Luciferin-MultiCYP) (χ2(2) = 2.87, P = 0.239), though the effect was in the same direction.
Fig. 5.

—In vitro assays of sage-grouse liver enzymes. Boxplot of cytochrome P450 enzyme activity of liver microsomes from greater sage-grouse (green, n = 5 individuals), chicken (blue, n = 1 composite), and negative control (orange, n = 1, no microsomes), from in vitro assays (four replicates per category) utilizing a nonselective cytochrome P450 substrate (Luciferin-MultiCYP) (left) and a more specific substrate (Luciferin-ME, right) that targets putative detoxification enzyme activity (CYP1 and CYP2, see Materials and Methods). Different letters above plots indicate significant differences among categories within each assay type. To aid in visual comparison, enzyme activity values have been scaled and centered within each comparison. Diagram for assay reaction (inset) wherein cytochrome P450 enzymes convert luciferin substrate (top) to luciferin methyl ester, which then produces luminescence in the presence of luciferin detecting reagent (LDR, Promega Corp.).
Discussion
Dramatic declines in sage-grouse populations over the past century (Braun 1998) have prompted significant interest in understanding population connectivity and the distributions of genetic variation across the species ranges. Previous population genetic studies utilizing relatively small numbers of anonymous markers (e.g., Oyler-Mccance et al. 2005) yielded important insights regarding taxonomic status and population structure. More recently, genomic approaches have been employed to investigate fine-scale population differentiation (Oyler-McCance et al. 2015), and a low-coverage (∼3.53×) reference-guide genome for Gunnison sage-grouse was assembled by Card et al. (2014) to estimate repeat content. However, inference regarding genome-wide patterns of divergence, ancient demographic events, and the relative influence of adaptive versus neutral processes has been limited by the lack of species-specific genomic resources.
In this study, de novo assembly of a Gunnison sage-grouse reference genome, along with whole-genome resequencing of an additional 90 sage-grouse genomes, yielded a data set composed of over 1.5 million SNPs. Evaluation of genetic variation at these loci revealed pronounced population genetic structure with little evidence of contemporary admixture. Levels of interpopulation divergence were relatively consistent across the genome, with no indication of genomic “islands” of elevated differentiation. Inferred ancestral demography of both sage-grouse species suggests that divergence was likely accompanied by increasing geographic isolation and cessation of gene flow as the species underwent substantial declines in effective population sizes during the last glacial period, which also coincided with the hypothesized origin of the two extant sage-grouse species. Taken together, these results support a scenario of population differentiation mostly by genetic drift (Foote et al. 2016), wherein limited dispersal is likely to have interacted simultaneously with substantial range contraction during the Late Pleistocene, as has been inferred from genomic analyses in other North America galliform species (Halley et al. 2014). Such a demographic scenario is consistent with the pattern of hierarchical population structure observed in this study, and in previous genetic analyses (Oyler-McCance et al. 2014).
Although overall patterns indicate a predominant role of genetic drift in genetic differentiation among sage-grouse populations, this does not exclude a role for adaptive processes in shaping patterns of variation across the genome. Indeed, the lack of connectivity to peripheral populations included in this study (Crist et al. 2017) may favor local adaptation by reducing the homogenizing effects of gene flow (Garcia-Ramos and Kirkpatrick 1997). Our analysis of SNPs with extreme levels of divergence relative to background population structure highlighted several genes crucial to dietary adaptation and detoxification of plant PSMs, suggesting a potential genetic basis to local adaptation to distinct sagebrush defensive compounds. At least eight of these genes (CYP2AC2, CYP2U1, CYP1C1, CYP2H2, CYP2J23, CYP2J19, CYP2AC7, and CYP2W1) belong to CYP gene families known to have central roles in xenobiotic metabolism in birds (Watanabe et al. 2013). More crucially for the present study, a recent phylogenetic analysis of CYP2 gene sequences across genomes representing all extant avian clades found evidence of recurring positive selection at substrate binding sites for both CYP2D and CYP2H subfamilies (Almeida et al. 2016), as well as associations between polymorphisms in CYP2 genes and feeding habits and habitat resources used by birds. The observed rapid evolutionary diversification in CYP2D6 among vertebrate herbivores in particular has been hypothesized to reflect metabolic adaptation to toxic plant alkaloids (Yasukochi and Satta 2011), though recent work suggests that the relationship between dietary diversification and evolution of this gene family at higher taxonomic levels is likely complex (Feng and Liu 2018).
The identification of these CYP genes as potential targets of selection in our whole-genome scans was corroborated by in vitro assays that found increased metabolic activity in these gene products in sage-grouse compared with a nonspecialist relative. The reason for the disparity between the two substrate assays is unclear, though we note the relatively large error in the control samples used in the MultiCYP assay, which likely reflects the nonspecificity of this reaction. Moreover, these assays are designed to cross-react with human CYP isoforms, and thus the translation into substrate specificity for avian enzymes has not been resolved and warrants a closer study of enzymatic kinetics. As the sage-grouse included in these assays were all sampled from the same population in Idaho, the degree to which such enzymatic or substrate specificity may differ among populations is currently unknown and represents a critical future research objective. However, by demonstrating elevated activity in hepatic CYP enzymes of sage-grouse (compared with a generalist galliform relative) our results suggest an important role for these phenotypes in the unique dietary adaptation to sagebrush, and point to a potential basis for local adaptation to different sagebrush varieties.
Utilizing a combination of genomic and experimental evidence, we have identified candidate genes with known adaptive significance for dietary specialization in general, and detoxification of plant PSM in particular. We acknowledge, however, that due to the remarkable evolutionary lability in gene sequence and copy number commonly observed for the CYP gene superfamily across vertebrate genomes (Thomas 2007), and that our annotation was based on homology with congeneric orthologs, the identification of specific CYP isoforms and substrates should be considered preliminary at this stage. Robust support for such metabolic adaptations will require further characterization of CYP sequence variation, classification of enzyme substrate specificity, as well as phenotypic associations. Nevertheless, our results raise the possibility of distinct sage-grouse ecotypes specialized to different sagebrush varieties. Interestingly, a recent study of sage-grouse cecal metagenome suggested that gut microbiota may also play an important role in detoxification of sagebrush PSM (Kohl et al. 2016). An important step for future research is to understand the synergistic interactions between these gut microbial communities and sage-grouse metabolic adaptation to detoxification of local sagebrush populations with distinct PSM profiles.
Two other CYP genes of note associated with highly divergent SNPs include CYP2J19, which is known to underlie polymorphisms affecting carotenoid color deposition in feathers (Lopes et al. 2016), and CYP19A1, which codes for the enzyme aromatase, a key sex-determining gene in birds (Smith et al. 2007). The potential adaptive significance among sage-grouse populations is currently unknown, though we note that divergence due to sexual selection on male display traits (plumage and behavioral displays) is thought to have been key in reproductive isolation between Gunnison and greater sage-grouse (Young et al. 1994).
Our results also shed light on the genetic characteristics of isolated populations on the margins of a species range. The marked reductions in nucleotide diversity we observed in greater sage-grouse in Washington and Gunnison sage-grouse are similar to results from previous genetic studies (Oyler-Mccance et al. 2005); and likely reflect extreme geographic isolation and small population sizes. Washington greater sage-grouse were the most genetically distinct among conspecific populations, suggesting a greater magnitude of divergence in this population than recognized by earlier studies (Benedict et al. 2003). This pronounced genetic differentiation of the Washington population is consistent with patterns of genetic divergence observed among diverse taxa in the Pacific northwest United States (e.g., Miller et al. 2006), where Pleistocene glacial refugia and vicariance are hypothesized to have shaped genetic structure for codistributed species (Brunsfeld et al. 2001). Greater sage-grouse in Washington occur in the Columbia Basin, where arid steppe habitat is largely surrounded by mesic coniferous forests that represent significant barriers to gene flow (Row et al. 2018), thus likely contributing to the elevated rates of genetic differentiation observed here.
Results from the analysis of population structure based only on the outlier SNPs, which largely recapitulated patterns with the whole-genome data set, suggest that adaptive processes have also contributed to the divergence of the Washington population. This finding implies that isolated peripheral populations of sage-grouse might possess adaptive genetic variation in ecologically relevant metabolic phenotypes. This raises the possibility that such populations might serve as important genetic reservoirs of adaptive diversity that might be utilized, via “targeted gene flow” (i.e., translocation) (Kelly and Phillips 2016) by managers to augment vulnerable populations facing rapid ecological or climate change (Macdonald et al. 2017). Thus, highly differentiated populations like the Washington greater sage-grouse may warrant recognition and protection as a genetically distinct conservation unit. At the same time, efforts to restore or rehabilitate sage-grouse habitat through revegetation of sagebrush should carefully consider the local source of plant materials to avoid potentially deleterious mismatches in PSMs of reseeded or restored sagebrush with the local sage-grouse populations.
Overall, this work provides important genomic resources for future research efforts in these species of current conservation concern. More generally, this study highlights the value of whole-genome approaches for discerning potentially adaptive significance of population genetic variation that can inform management strategies and practices.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors thank A. Apa, M. Casazza, J. Connelly, C. Hagen, M. Holloran, M. Schroeder, and numerous field personnel for contributing samples to the archives at the U.S. Geological Survey (USGS) Fort Collins Science Center (FORT) Molecular Ecology Laboratory. We also thank S. Vasilchenko, N. Wiggins, falconers T. Maechtle, D. Skinner, and H. Quade as well as Gus, the German short-haired pointer for assistance with collecting tissues in the field used in the enzyme activity analysis. J. Fike assisted with sample archiving and preparation along with molecular laboratory procedures. S. Cornman provided helpful suggestions for bioinformatics analyses. S. Pedigo and D. Wild provided computing support at FORT and J. Falgout provided technical support and access to the Yeti high-performance computing cluster at the USGS Core Science Systems Advanced Research Computing Center in Denver, CO. Funding: K.P.O. was funded through a USGS Mendenhall Fellowship. C.L.A. was supported by Colorado State University and USGS FORT. J.S.F. was supported by the Idaho Department of Fish and Game, Idaho Governor’s Office for Species Conservation, the National Science Foundation (DEB-1146194, IOS-1258217, OIA-1757324, and OIA-1826801), and Idaho INBRE Program-National Institutes of Health grant P20 GM103408. C.Y.D. was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant P20GM103408. S.J.O.-M. was supported by USGS FORT. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Author Contributions
K.P.O., C.L.A., and S.J.O.-M. conceived the study. K.P.O. performed all genomic analyses and wrote the manuscript with input from the coauthors. J.S.F. and C.Y.D. collected liver specimens and carried out in vitro assays.
Data deposition: This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession SPOS00000000. The version described in this article is version SPOS01000000. The resequencing data have been deposited at the NCBI Sequence Read Archive under BioProject accession PRJNA531321. Sample metadata are available in the U.S. Geological Survey ScienceBase repository at https://doi.org/10.5066/P9G9CCQE.
Literature Cited
- Aldridge CL, et al. 2008. Range-wide patterns of greater sage-grouse persistence. Divers Distrib. 14(6):983–994. [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorf FW, Hohenlohe PA, Luikart G.. 2010. Genomics and the future of conservation genetics. Nat Rev Genet. 11(10):697–709. [DOI] [PubMed] [Google Scholar]
- Almeida D, et al. 2016. Whole-genome identification, phylogeny, and evolution of the cytochrome P450 family 2 (CYP2) subfamilies in birds. Genome Biol Evol. 8(4):1115–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S.. 2014. Fitting linear mixed-effects models using lme4. J Stat Softw. 67:1–48. [Google Scholar]
- Benedict NG, Oyler MS, Taylor SE, Braun CE, Quinn TW.. 2003. Evaluation of the eastern (Centrocercus urophasianus urophasianus) and western (Centrocercus urophasianus phaios) subspecies of Sage-grouse using mitochondrial control-region sequence data. Conserv Genet. 4(3):301–310. [Google Scholar]
- Braun CE. 1998. Sage grouse declines in western North America: what are the problems? Proc West Assoc Fish Wildl Agencies 78:139–156. [Google Scholar]
- Brunsfeld SJ, Sullivan JM, Soltis DE, Soltis PS.. 2001. Comparative phylogeography of northwestern North America: a synthesis In: Silvertown J, Antonovics J, editors. Integrating ecology and evolution in a spatial context. Oxford: Blackwell Science; p. 319–340. [Google Scholar]
- Cantarel BL, et al. 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18(1):188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card DC, et al. 2014. Two low coverage bird genomes and a comparison of reference-guided versus de novo genome assemblies. PLoS One 9(9):e106649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, et al. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JW, Braun CE.. 1997. Long-term changes in sage grouse Centrocercus urophasianus populations in western North America. Wildlife Biol. 3(1):229–234. [Google Scholar]
- Crist MR, Knick ST, Hanser SE.. 2017. Range-wide connectivity of priority areas for greater sage-grouse: implications for long-term conservation from graph theory. Condor 119(1):44–57. [Google Scholar]
- Feng P, Liu Z.. 2018. Complex gene expansion of the CYP2D gene subfamily. Ecol Evol. 8(22):11022–11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote AD, et al. 2016. Genome-culture coevolution promotes rapid divergence of killer whale ecotypes. Nat Commun. 7:11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis RM. 2017. pophelper: an R package and web app to analyse and visualize population structure. Mol Ecol Resour. 17(1):27–32. [DOI] [PubMed] [Google Scholar]
- Fumagalli M, Vieira FG, Linderoth T, Nielsen R.. 2014. ngsTools: methods for population genetics analyses from next-generation sequencing data. Bioinformatics 30(10):1486–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, et al. 2013. Quantifying population genetic differentiation from next-generation sequencing data. Genetics 195(3):979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ramos G, Kirkpatrick M.. 1997. Genetic models of adaptation and gene flow in peripheral populations. Evolution 51(1):21–28. [DOI] [PubMed] [Google Scholar]
- Garton EO, et al. 2011. Greater sage-grouse population dynamics and probability of persistence In: Knick ST, Connelly JW, editors. Greater sage-grouse ecology and conservation of a landscape species and its habitats. Berkeley and Los Angeles, California: University of California Press; p. 292–379. [Google Scholar]
- Gascuel O. 1997. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 14(7):685–695. [DOI] [PubMed] [Google Scholar]
- Gautier M. 2015. Genome-wide scan for adaptive divergence and association with population-specific covariates. Genetics 201(4):1555–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone HMH, Stegeman JJ.. 2006. A revised evolutionary history of the CYP1A subfamily: gene duplication, gene conversion, and positive selection. J Mol Evol. 62(6):708–717. [DOI] [PubMed] [Google Scholar]
- Goldstone JV, et al. 2007. Cytochrome P450 1 genes in early deuterostomes (tunicates and sea urchins) and vertebrates (chicken and frog): origin and diversification of the CYP1 gene family. Mol Biol Evol. 24(12):2619–2631. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Nebert DW.. 1990. Evolution of the P450 gene superfamily: animal-plant ‘warfare’, molecular drive and human genetic differences in drug oxidation. Trends Genet. 6(6):182–186. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2010. Genome-wide synteny through highly sensitive sequence alignment: satsuma. Bioinformatics 26(9):1145–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther T, Coop G.. 2013. Robust identification of local adaptation from allele frequencies. Genetics 195(1):205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halley YA, et al. 2014. A draft de novo genome assembly for the northern bobwhite (Colinus virginianus) reveals evidence for a rapid decline in effective population size beginning in the Late Pleistocene. PLoS One 9(3):e90240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly E, Phillips BL.. 2016. Targeted gene flow for conservation. Conserv Biol. 30(2):259–267. [DOI] [PubMed] [Google Scholar]
- Kelsey RG, Stephens JR, Shafizadeh F.. 1982. The chemical constituents of sagebrush foliage and their isolation. J Range Manage. 35(5):617–622. [Google Scholar]
- Kofler R, Schlötterer C.. 2012. Gowinda: unbiased analysis of gene set enrichment for genome-wide association studies. Bioinformatics 28(15):2084–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl KD, Connelly JW, Dearing MD, Forbey JS.. 2016. Microbial detoxification in the gut of a specialist avian herbivore, the greater sage-grouse. FEMS Microbiol Lett. 363(14):fnw144. [DOI] [PubMed] [Google Scholar]
- Kohl KD, et al. 2015. Monoterpenes as inhibitors of digestive enzymes and counter-adaptations in a specialist avian herbivore. J Comp Physiol B 185(4):425–434. [DOI] [PubMed] [Google Scholar]
- Korneliussen T, Albrechtsen A, Nielsen R.. 2014. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics 15:356.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB.. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34(7):1812–1819. [DOI] [PubMed] [Google Scholar]
- Lefort V, Desper R, Gascuel O.. 2015. FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol Biol Evol. 32(10):2798–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27(21):2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2011. Inference of human population history from individual whole-genome sequences. Nature 475(7357):493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liukkonen-Anttila T, Honkanen H, Peltokangas P, Pelkonen O, Hohtola E.. 2003. Cytochrome P450 enzyme activity in five herbivorous, non-passerine bird species. Comp Biochem Physiol C Toxicol Pharmacol. 134(1):69–77. [DOI] [PubMed] [Google Scholar]
- Lopes RJJ, et al. 2016. Genetic basis for red coloration in birds. Curr Biol. 26(11):1427–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald SL, Llewelyn J, Moritz C, Phillips BL.. 2017. Peripheral isolates as sources of adaptive diversity under climate change. Front Ecol Evol. 5:88. [Google Scholar]
- Meirmans PG. 2012. The trouble with isolation by distance. Mol Ecol. 21(12):2839–2846. [DOI] [PubMed] [Google Scholar]
- Miller MP, Bellinger MR, Forsman ED, Haig SM.. 2006. Effects of historical climate change, habitat connectivity, and vicariance on genetic structure and diversity across the range of the red tree vole (Phenacomys longicaudus) in the Pacific Northwestern United States. Mol Ecol. 15(1):145–159. [DOI] [PubMed] [Google Scholar]
- Nam K, et al. 2010. Molecular evolution of genes in avian genomes. Genome Biol. 11(6):R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, et al. 2012. BEDOPS: high-performance genomic feature operations. Bioinformatics 28(14):1919–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, et al. 2017. vegan: community ecology package. R package version 2.4-4.
- Oleksyk TK, Smith MW, O’Brien SJ.. 2010. Genome-wide scans for footprints of natural selection. Philos Trans R Soc Lond B Biol Sci. 365(1537):185–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyler-McCance SJ, Casazza ML, Fike JA, Coates PS.. 2014. Hierarchical spatial genetic structure in a distinct population segment of greater sage-grouse. Conserv Genet. 15(6):1299–1311. [Google Scholar]
- Oyler-McCance SJ, Cornman RS, Jones KL, Fike JA.. 2015. Genomic single-nucleotide polymorphisms confirm that Gunnison and Greater sage-grouse are genetically well differentiated and that the Bi-State population is distinct. Condor 117(2):217–227. [Google Scholar]
- Oyler-McCance SJ, Oh KP, Langin KM, Aldridge CL.. 2016. A field ornithologist’s guide to genomics: practical considerations for ecology and conservation. Auk 133(4):626–648. [Google Scholar]
- Oyler-McCance SJ, St. John J, Taylor SE, Apa AD, Quinn TW.. 2005. Population genetics of Gunnison sage-grouse: implications for management. J Wildl Manage. 69(2):630–637. [Google Scholar]
- Oyler-Mccance SJ, Taylor SE, Quinn TW.. 2005. A multilocus population genetic survey of the greater sage-grouse across their range. Mol Ecol. 14(5):1293–1310. [DOI] [PubMed] [Google Scholar]
- Personius TL, Wambolt CL, Stephens JR, Kelsey RG.. 1987. Crude terpenoid influence on mule deer preference for sagebrush. J Range Manage. 40(1):84–88. [Google Scholar]
- Peters EJ, McLeod HL.. 2008. Ability of whole-genome SNP arrays to capture ‘must have’ pharmacogenomic variants. Pharmacogenomics 9(11):1573–1577. [DOI] [PubMed] [Google Scholar]
- Petren K, Grant PR, Grant BR, Keller LF.. 2005. Comparative landscape genetics and the adaptive radiation of Darwin’s finches: the role of peripheral isolation. Mol Ecol. 14(10):2943–2957. [DOI] [PubMed] [Google Scholar]
- Power DM. 1979. Evolution in peripheral isolated populations: Carpodacus finches on the California Islands. Evolution 33(3):834–847. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P.. 2000. Inference of population structure using multilocus genotype data. Genetics 155(2):945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2016. R: a language and environment for statistical computing team. R Found Stat Comput. 1:409. [Google Scholar]
- Row JR, et al. 2018. Quantifying functional connectivity: the role of breeding habitat, abundance, and landscape features on range-wide gene flow in sage-grouse. Evol Appl. 11(8):1305–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MA, et al. 2004. Distribution of sage-grouse in North America. Condor 106(2):363–376. [Google Scholar]
- Skotte L, Korneliussen TS, Albrechtsen A.. 2013. Estimating individual admixture proportions from next generation sequencing data. Genetics 195(3):693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Roeszler KN, Hudson QJ, Sinclair AH.. 2007. Avian sex determination: what, when and where? Cytogenet Genome Res. 117(1-4):165–173. [DOI] [PubMed] [Google Scholar]
- Sobol M, Ma D, Unch J, Cali JJ. Nonselective cytochrome P450 enzyme assay using a novel bioluminescent probe substrate that cross-reacts with multiple P450 enzymes. Promega Corporation Web site. http://www.promega.com/resources/pubhub/enotes/nonselective-cyp450-assay-using-a-biolum-probe-substrate-that-cross-reacts-with-multiple-p450s/ Updated 2008. Last accessed 1 December 2018.
- Städler T, Haubold B, Merino C, Stephan W, Pfaffelhuber P.. 2009. The impact of sampling schemes on the site frequency spectrum in nonequilibrium subdivided populations. Genetics 182(1):205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiver JR, Apa AD, Remington TE, Gibson RM.. 2008. Polygyny and female breeding failure reduce effective population size in the lekking Gunnison sage-grouse. Biol Conserv. 141(2):472–481. [Google Scholar]
- Striby KD, Wambolt CL, Kelsey RG, Havstad KM.. 1987. Crude terpenoid influence on in vitro digestibility of sagebrush. J Range Manage. 40(3):244–248. [Google Scholar]
- Thomas JH. 2007. Rapid birth-death evolution specific to xenobiotic cytochrome P450 genes in vertebrates. PLoS Genet. 3:720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulappa AC, et al. 2014. Plant protein and secondary metabolites influence diet selection in a mammalian specialist herbivore. J Mammal. 95(4):834–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira FG, Lassalle F, Korneliussen TS, Fumagalli M.. 2016. Improving the estimation of genetic distances from Next-Generation Sequencing data. Biol J Linn Soc. 117(1):139–149. [Google Scholar]
- Watanabe KP, et al. 2013. Avian cytochrome P450 (CYP) 1-3 family genes: isoforms, evolutionary relationships, and mRNA expression in chicken liver. PLoS One 8(9):e75689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch BL, McArthur ED.. 1981. Variation of monoterpenoid content among subspecies and accessions of Artemisia tridentata grown in a uniform garden. J Range Manage. 34(5):380–384. [Google Scholar]
- Yasukochi Y, Satta Y.. 2011. Evolution of the CYP2D gene cluster in humans and four non-human primates. Genes Genet Syst. 86(2):109–116. [DOI] [PubMed] [Google Scholar]
- Ye C, et al. 2016. DBG2OLC: efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci Rep 6:31900. [DOI] [PMC free article] [PubMed]
- Young JR, Hupp JW, Bradbury JW, Braun CE.. 1994. Phenotypic divergence of secondary sexual traits among sage grouse, Centrocercus urophasianus, populations. Anim Behav. 47(6):1353–1362. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.