Abstract
Over the past decade, advances in molecular biology and genomics techniques have revolutionized the diagnosis and treatment of cancer. The technological advances in tissue profiling have also been applied to the study of cell-free nucleic acids, an area of increasing interest for molecular pathology. Cell-free nucleic acids are released from tumour cells into the surrounding body fluids and can be assayed non-invasively. The repertoire of genomic alterations in circulating tumour DNA (ctDNA) is reflective of both primary tumours and distant metastatic sites, and ctDNA can be sampled multiple times, thereby overcoming the limitations of the analysis of single biopsies. Furthermore, ctDNA can be sampled regularly to monitor response to treatment, to define the evolution of the tumour genome, and to assess the acquisition of resistance and minimal residual disease. Recently, clinical ctDNA assays have been approved for guidance of therapy, which is an exciting first step in translating cell-free nucleic acid research tests into clinical use for oncology. In this review, we discuss the advantages of cell-free nucleic acids as analytes in different body fluids, including blood plasma, urine, and cerebrospinal fluid, and their clinical applications in solid tumours and haematological malignancies. We will also discuss practical considerations for clinical deployment, such as preanalytical factors and regulatory requirements.
Keywords: liquid biopsy, cell-free nucleic acids, circulating tumour DNA, genomics, PCR, DNA sequencing, molecular pathology, cancer
Introduction
Molecular pathology is critical for cancer management, owing to the expanding application of targeted treatments that are prescribed on the basis of tumour-specific mutations. Although many methods exist to examine these mutations, next-generation sequencing (NGS) of tumour samples has demonstrated tremendous utility in the clinic. Ranging from small [1] or large [2] gene panels to whole exome sequencing (WES) [3], NGS analysis leads not only to new insights into the genomic landscape of cancer [4–7], but also to the identification of clinically actionable genetic alterations and biomarkers that predict treatment outcomes for specific therapies; for example, the detection of microsatellite instability [8–10] or overall tumour mutation burden [11,12] that predict responses to immunotherapy, or epidermal growth factor receptor (EGFR) mutations that confer sensitivity to EGFR inhibitors [13].
Despite their many successes, tissue-based assays have limitations in the clinical setting [14]. Clinical samples such as formalin-fixed paraffin-embedded tissue may harbour chemically induced artefacts that require filtering with proper post-sequencing quality control metrics [14,15]. Additionally, biopsies can miss important drivers due to tumour heterogeneity or distant metastatic lesions. Biopsies are also sometimes not available, as a result of inaccessibility or widespread metastasis. In this regard, cell-free DNA (cfDNA) has created new possibilities for non-invasive diagnosis and therapy monitoring [16,17].
Advantages of cfDNA as an analyte for molecular testing
cfDNA refers to DNA fragments present outside of cells in body fluids such as plasma, urine, and cerebrospinal fluid (CSF). In plasma, the majority of cfDNA originates from leukocytes, and only a small fraction is tumour-derived, known as circulating tumour DNA (ctDNA) [18,19]. ctDNA can contain mutations missed in biopsy studies because of tumour heterogeneity or lesions in distant sites, and is generally found in larger quantities in the bloodstream than circulating tumour cells [20]. The concentration of ctDNA varies among patients, and differs according to the type, location and stage of cancer, with some producing extremely low concentrations [21]. The half-life of ctDNA is still unclear, although fetal cfDNA studies have highlighted its short duration (16 min to 2 h) [22,23]. This instability, which might be an issue at the preanalytical level, can be used to our advantage by providing a very dynamic tool for tracking treatment response within hours [22,24].
The relationship between tumour biology and ctDNA release into the circulation is still unclear. Experimental data have shown that cfDNA is highly fragmented [25–28], in a largely chromatosomal pattern – indicating potential associations with nucleosomes and transcription factors [29,30]. ctDNA fragments are slightly shorter, with the majority being <167 bp [19,26,27,31].
Clinical applications of cfDNA
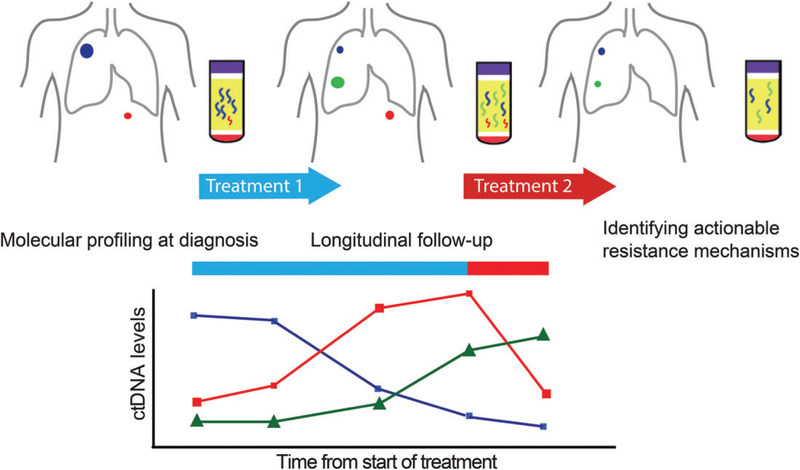
Extensive research studies and trials have demonstrated the clinical applications of cfDNA profiling at multiple stages of treatment: prognosis, molecular stratification at diagnosis, detecting resistance mechanisms at relapse, and detecting minimal residual disease (MRD). Several key applications are summarized below and in Figure 1.
Figure 1.
Clinical applications of cfDNA during the treatment process. Serial monitoring of ctDNA in plasma allows non-invasive identification of clinically relevant genomic alterations at different stages: molecular stratifications at diagnosis, tracking tumour responses during treatment, and identifying genetic mechanisms of resistance at progression. Colours represent tumour subclones with different mutations, which release ctDNA at different frequencies, as depicted in the graph below.
Prognostic value
cfDNA has been shown to predict prognosis and treatment response. The proportion of ctDNA correlates with both stage [21] and size [19,32–34] of the tumour. A study across multiple cancer types found that advanced tumours give 10–100 times the amount of ctDNA in plasma than tumours in early stages [21], and additional increases can be observed in patients with metastatic disease [35]. As expected with high tumour load, patients with high proportions of ctDNA have worse survival outcomes [36–41]. This correlation suggests that ctDNA levels can be a measure of disease burden and prognosis. For instance, in a post-treatment study of stage I–III colorectal cancer (CRC) patients, those with detectable ctDNA levels had a 48% 2-year recurrence-free survival rate, whereas those without had 100% survival [36], and, in a separate CRC study, ctDNA analysis had 100% sensitivity and specificity for detecting postsurgery relapse [42]. Pretreatment cfDNA levels are also prognostic, as shown in another CRC study, in which patients with low ctDNA levels (<25% quartile) had a disease control rate of 42%, whereas patients with high ctDNA levels (>75% quartile) had a 0% rate after 9 weeks[43]. High pretreatment ctDNA levels have also been associated with resistance in numerous cancer types [20,21,35,44–51]. Conversely, low ctDNA levels or an absence of ctDNA have been shown to correlate with treatment response [20,22,24,41,52,53], a change that can be observed earlier than with other clinical detection methods [20,24,54,55]. ctDNA analysis has great potential prognostic value in cancer management.
Molecular stratification
Detection of specific mutations can be used to stratify patients and guide therapy, including adjuvant therapy [44,56], endocrine therapy [57], and targeted therapies. For example, ctDNA testing with polymerase chain reaction (PCR) for EGFR mutations is now used clinically in non-small-cell lung cancer (NSCLC) [58–61]. Obtaining biopsy samples from NSCLC is difficult [62], and the high correlation between tumour and plasma mutations has accelerated the clinical implementation of ctDNA testing in this cancer type to detect EGFR hotspot mutations that confer sensitivity to EGFR inhibitors [63,64]. Two forms of the test have been approved by the European Medicines Agency and the US Food and Drug Administration (FDA): Therascreen EGFR [65] and Cobas EGFR [60]. PCR-based assays are ideal for situations in which mutations are known, either because they commonly occur or because they have been identified during tumour sequencing, and in which positive mutation identification will lead to enrolment for therapy, but are unable to identify additional informative mutations outside the regions and/or mutations covered in the assay.
More comprehensive assays interrogating multiple genes have been developed, using NGS to guide treatment [66–69] or enrolment in clinical trials [58,70–72]. The size of the panels varies from a small number of genes relevant for a specific cancer type, to large panels encompassing hundreds of genes [73], or WES [74]. Smaller panels can be sequenced deeply to detect mutations with a mutant allele fraction (MAF) of ~1% [73,75,76], whereas larger panels are generally sequenced to a lower depth, and therefore require a MAF of >5% for de novo mutation detection [77]. WES offers hypothesis-free comprehensive screening, but requires a MAF in the ~10% range [74], which is rarely seen outside progression or metastatic settings. Confidently calling mutations that can be present at <1% of reads, near the level of sequencing noise, remains difficult and a limitation for implementing cfDNA NGS assays clinically. Methodological advances have led to improved sensitivity; for example, the incorporation of molecular barcodes, also known as unique molecular indexes or unique identifiers, into sequencing libraries allows de novo mutation detection down to a MAF of 0.1% [78–81]. Additional improvements in detection rates have been obtained by applying size selection to enrich for ctDNA [27,82]. Further developments in technology, e.g. through the use of long-read sequencers [83,84] or via CRISPR-based diagnostics [85], may continue to improve detection. Together, these advances expand the applicability of cfDNA testing to a wider patient population across diseases status.
Detecting resistance mechanisms
cfDNA can be used to monitor acquisition of resistance, through screening for known resistance mutations [86–94], or searching for novel mechanisms of resistance [95–97]. Serial sampling can be performed to identify resistant clones prior to the onset of clinical progression [88,89]. For example, in a recent study of CRC patients receiving anti-EGFR therapy, KRAS resistance mutations emerged 5–6 months before radiographic disease became evident [90]. Timely identification of resistance mechanisms allows earlier clinical intervention.
Detecting MRD
The highly sensitive methods described above also have promise for the detection of MRD. In a recent study of localized breast cancer patients, postsurgery ctDNA levels after apparently curative treatment were predictive of metastatic relapse [98]. A similar study, performed on patients with early-stage CRC, found that the absence of ctDNA after resection was predictive of recurrence-free survival at 3 years, whereas those with ctDNA detection after serial sampling had 100% recurrence [99]. Highly sensitive approaches are required for MRD monitoring, and these assays are typically patient-specific and mutation-specific. This has been a challenge to implement clinically; however, with increasing sensitivity, broader panels could be used in the future.
cfDNA profiling in haematological malignancies
The emerging clinical benefits of ctDNA analysis in solid tumours have led to an expansion of groups using ctDNA in haematological malignancies. The value of ctDNA analysis was first demonstrated in haematological cancers in 1994, when tumour-specific NRAS mutations were identified in the plasma of patients with myelodysplasia and acute myeloid leukaemia [100]. Over the last few years, research in this area has rapidly expanded, with many studies now exploring the role of ctDNA for risk stratification and monitoring tumour burden and response to treatment in both lymphoid and myeloid malignancies.
In the management of lymphoma, patient-specific ctDNA analysis could be used to help monitor MRD and guide clinical decisions. In non-Hodgkin’s lymphoma, patient-specific IgH rearrangements are readily detectable through cfDNA analysis [101–104]. Tumour-specific V(D)J recombination of the immunoglobulin receptor genes can be used as an MRD monitoring strategy to identify individuals at increased risk of relapse, and to detect relapse before clinical evidence of disease [105,106]. In addition, comprehensive analysis of the ctDNA of lymphoma patients has emerged as an important clinical tool with which to identify distinct biological subtypes and provide insights into the patterns of genomic evolution throughout treatment [107–110]. In parallel, several studies have shown that ctDNA analysis allows more comprehensive genomic characterization of classic Hodgkin’s lymphoma, a disease that has traditionally been difficult to study, owing to the rarity of Hodgkin/Reed–Sternberg cells in tumour biopsies [111,112].
The use of ctDNA for disease monitoring has also recently been demonstrated in chronic lymphocytic leukaemia (CLL) [113]. In CLL, ctDNA analysis can reveal changes in the disease across tissue compartments, including the bone marrow and lymph nodes, providing additional information that cannot be obtained by monitoring the circulating disease alone [113]. Moreover, serial ctDNA analysis in CLL can allow monitoring of clonal dynamics and identify genomic changes associated with Richter’s syndrome [113]. ctDNA analysis is also a powerful tool in patients with myeloid malignancies, particularly myelodysplastic syndromes [114–116], and in those with plasma cell disorders, such as multiple myeloma [117–120], as ctDNA analysis can detect underlying genomic changes in the bone marrow and be used as a monitoring strategy to limit invasive bone marrow biopsies [114,117]. The potential of these approaches is now coming to the forefront of research, with clinical applications likely to expand across the breadth of haematological malignancies.
cfDNA profiling in other body fluids
Urine
Research findings suggest that cfDNA enters urine from blood after it passes through the renal filtration system [31,121,122]. Additionally, for patients with disease affecting the renal or genitourinary tract, urine may serve as a more concentrated sample source for disease-related cfDNA. The utility of urinary DNA analysis was initially evaluated through studies of urinary cells. In an early study, Sidransky et al showed that p53 mutations could be identified within the urine sediment of three patients with invasive bladder cancers [123]. Later studies in prenatal medicine have shown that Y chromosome DNA can be detected in the supernatants of urine samples from pregnant women carrying male fetuses [122]. Studies have shown that urinary DNA has two size profiles, i.e. <100 bp and 150–250 kb, which are hypothesized to represent the cfDNA and genomic DNA arising from urinary tract cells, respectively [124,125].
In oncology, urine ctDNA has been studied in multiple cancer types, particularly cancers of the genitourinary tract. For example, urine ctDNA showed 88% concordance with primary tumour tissue for EGFR mutations in NSCLC [126], and, in hepatocellular carcinoma, Hann et al showed that urinary biomarkers could constitute a promising tool for monitoring for relapse [127]. More recently, longitudinal sampling of urine ctDNA in patients with muscle-invasive bladder cancer before and after neoadjuvant chemotherapy was shown to help predict outcome [128]. Urine collection offers the advantages of being truly non-invasive and allowing large sample volumes to be obtained. It is particularly beneficial in the field of paediatric oncology, in which obtaining tumour samples or large volumes of plasma is more challenging.
As noted above, a proportion of the ctDNA found in urine originates from the circulatory system and is filtered through the kidneys [31,122,129]. Biological factors such as hydration status, kidney function and primary tumour site may all contribute to the amount of ctDNA that can be detected in urine. The effects of these parameters on the performance of urine-based cfDNA tests would require further evaluation before clinical implementation.
Cerebrospinal fluid
CSF circulates throughout the central nervous system (CNS), and is protected from the systemic circulation by the blood–brain barrier [130]; as a consequence, it possesses a low background level of normal cfDNA. Additionally, in cancers involving the CNS, the concentration of plasma ctDNA is much lower than in other solid tumours [21], so CSF has been proposed as an alternative potential reservoir for ctDNA.
Initial investigations identified mutations within tumours prior to evaluation of the CSF for mutations [131–134]. Subsequent studies have adopted a more clinically feasible paradigm, whereby mutations are de novo-called through targeted mutation panels or WES [135,136]. The improvement in sensitivity associated with these techniques has enabled CNS tumour mutations to be identified in the majority of samples from patients with CNS cancers, and at a greater concentration than in plasma samples [133].
Many of these studies have relied on mixed populations of primary brain tumour patients and those with metastatic disease within the CNS. The detection rate and the concentration of cfDNA vary between these pathological entities, with primary brain tumours having lower levels than secondary metastatic lesions [135,136]. The largest study on primary brain tumours demonstrated a higher detection rate and a higher MAF in high-grade tumours than in low-grade tumours [134]. Interestingly, in the same analysis, tumour size was not a significant factor in ctDNA detectability, although the position of the tumour influenced mutation detectability: those entirely encapsulated within the CNS parenchyma had no detectable ctDNA in the CSF [134].
Although obtaining CSF is less invasive than a surgical tumour biopsy, it remains a procedure that involves clinical risk and the potential for significant patient discomfort. The clinical utility of CSF analysis will ultimately be reliant upon the balance between the efficacy of the test and the importance of the clinical question. In this context, the molecular diagnosis of inoperable lesions or those with a non-surgical management paradigm may provide appropriate indications.
Other cell-free nucleic acids and strategies
cfDNA methylation studies
Analysis of DNA methylation can be applied to cfDNA to uncover methylation changes known to be important in cancer by the use of methylation-specific PCR [137], microarrays [138], or sequencing [139–143]. An example is uncovering gene methylation status, such as for BRCA1 in breast cancer [144]. Additionally, methylation deconvolution can be used to determine the source tissues of cfDNA fragments [145–147], providing information on tissue dynamics in various disease states [146]. This technique has great potential in oncology for determining the tissue of origin in cancer with an unknown primary [148] or identifying tumour subtypes. However, these techniques require a high tumour fraction, and a large proportion of the already limited starting material can be lost during bisulphite conversion.
Viral and bacterial cfDNA
Persistent infection with viruses such as human papillomavirus (HPV) and Epstein–Barr virus (EBV) is associated with certain cancers, such as cervical cancer and nasopharyngeal carcinoma (NPC), respectively. Viral DNA is often profiled from tumour samples or used in screening for precancerous states, but it can also be found in cfDNA samples, such as plasma [149], urine [150], and saliva [151]. For instance, HPV DNA has been detected in the plasma of patients with cervical cancer [152] and head and neck squamous cell carcinoma [151], and could potentially be used as a marker of the disease.
Similarly, short fragments of bacterial DNA have also been identified in plasma originating from commensal or infectious bacteria from throughout the body. Studies of bacterial cfDNA have led to the identification of hundreds of new species [153], and is of particular interest for identifying and monitoring infection post-transplantation or after therapy-related immunosuppression [154]. It may also offer a tool with which to monitor change in the microbiome during cancer therapy.
Cell-free RNA (cfRNA)
Previous work has shown that cfRNA can be isolated from maternal plasma to non-invasively screen for fetal-specific transcripts [155]. In the oncology setting, several reports have demonstrated the isolation of cell-free microRNA from CSF of cancer patients [156–159] and cell-free mRNA from plasma [160,161]. One area of great potential for cfRNA is fusion gene detection. Genomic and transcriptomic sequencing has led to the identification of many gene fusions in solid and haematological cancers [162,163]. Gene fusions can have diagnostic and prognostic, but also therapeutic, implications when they involve targetable genes [164]. Unfortunately, DNA sequencing has limited ability to detect gene fusions or identify fusion partners, owing to potentially large intronic sequences. For instance, in a study of advanced-stage NSCLC patients with ALK fusions, the sensitivity for patients with localized disease was only 28.6% [165]. Additionally, expressed driver events may not be easily distinguished from passenger events [166].
RNA-based detection methods provide a direct readout of expressed gene fusions in tumour cells [167]. Furthermore, gene fusions in low-purity tumours are more likely to be detected by RNA sequencing, owing to the higher abundance of fusion-derived RNA transcripts [168]. Therefore, RNA sequencing has been extensively used for the detection of gene fusions in tumour tissue [166,167]. The same strategy can be applied to cfRNA [169]; however, as RNA is more unstable, systematic optimization of preanalytical processing is required to ensure reproducibility and stability.
Early screening for cancer
Early screening for cancer has become increasingly effective through the collection of information from multiple different biospecimens. A recent study, combining protein and cfDNA detection across eight cancer types, achieved 73% sensitivity for stage II cancers and 49% sensitivity for stage I cancers with >99% specificity; although the study involved patients already diagnosed, it demonstrates the potential benefits of combining cfDNA with other markers for cancer screening [170].
Cell-based methods, such as the Papanicolaou test (Pap smear), have been tested for the diagnosis and screening of endometrial cancers [171,172], but have proved challenging. A recent study identified tumour-derived mutations in DNA extracted from Pap smears in 100% of endometrial cancer patients and 41% of ovarian cancer patients [173], offering the potential for early screening. In ovarian cancer, for which the sensitivity of Pap smears is low, perhaps combined Pap smear and mutation analyses may provide better performance for screening to compensate for the limitations of either approach alone [92,174,175].
Another strategy to improve the screening performance of cfDNA-based assays is to target biomarkers that are highly specific to the tumour and nearly absent in background cfDNA, e.g. targeting EBV in NPC. EBV DNA was first identified in plasma of NPC patients in 1999 [149], and is cleared from plasma after treatment [150,176], making it an ideal biomarker [177]. In a longitudinal study, >20 000 asymptomatic participants were screened for EBV DNA in their plasma, and 34 with NPC were identified [178]. These patients were diagnosed earlier and had better 3-year progression-free survival than historical cohorts, and only one participant with a negative EBV result developed NPC a year after testing [178]. As EBV DNA is highly specific for NPC, this test offers a sensitivity of 97.1% and specificity of 98.6%. This example demonstrates the potential of targeting highly specific tumour-derived information in cfDNA for early screening of cancer.
Practical considerations for clinical implementation
To date, the majority of clinical applications of cfDNA analysis have been demonstrated in prenatal testing and cancer diagnostics, but it has potential in many other physiological conditions, such as trauma, stroke, sepsis, epilepsy, autoimmune diseases, and post-transplantation monitoring [45–49,179]. In prenatal testing, multiple molecular tests have been developed for fetal aneuploidy [180–183] or single-gene disorders [184–186], with some having been clinically implemented worldwide [187–190]. In oncology, there is tremendous clinical potential for ctDNA to be incorporated in molecular diagnostics. However, before broad clinical implementation, practical considerations such as preanalytical factors and regulatory requirements must be addressed in order to achieve consistent and reproducible results.
Preanalytical considerations
Preanalytical factors have different effects on cfDNA yield, quality, and downstream molecular applications. One factor is the storage time between blood draw and processing to isolate plasma. Delayed processing results in blood clotting and lysis, which results in the release of large amounts of background genomic cfDNA, introducing additional challenges for identifying low-level somatic mutations [191]. Preservation tubes, such as EDTA tubes, that prevent blood clotting and minimize the release of genomic DNA from blood cells can be used to address this issue [191]; however, processing needs to be performed within 2–24 h [192–194], which is challenging to implement in a clinical setting. One potential solution is to use specialist blood collection tubes, which can extend storage for many days before processing [195–197]. Other preanalytical factors, such as centrifugation protocols, number of freeze–thaw cycles, and extraction methods, are also important [192,198]. Guidelines pertaining to preanalytical factors must be put in place to ensure accurate and efficient genomic profiling.
Considerations for clinical test development
The two most important initial decisions are the choice of platform and the scale of the analysis required. The choice of clinical sequencing platform and method depends on the sensitivity, the type and complexity of the test, the expected volume of testing, the turnaround time, the costs, the laboratory infrastructure, and the computational and human resources available to validate and perform the clinical testing.
The specific requirements for validation vary according to the intended use: single-locus, low multiplex panel testing, targeted NGS, WES, or genome-wide analysis. Single-locus or low multiplex assays, such as digital PCR, allow rapid detection and quantification of recurrent hotspot mutations and monitoring of well-established resistance mutations, with a rapid turnaround time for a relatively low cost. These assays are highly sensitive, although restricted in the number of loci assessed. Clinical validation encompasses the establishment of accuracy as compared with a gold standard, inter-assay and intra-assay reproducibility, and the establishment of sensitivity levels for each patient population.
For more comprehensive assays such as targeted NGS approaches, platform selection is pivotal to all further decisions regarding testing, validation, and capability for future expansion. Several important considerations include the size of the panel, selection of genes, depth of sequencing, coverage, sensitivity of the assay, and complexity of analytical and clinical interpretation. All of these will factor into the cost of running the assay, which, for large panels or WES, can be prohibitively high for clinical implementation. According to current guidelines for NGS testing, the laboratory should determine gene content on the basis of the available scientific evidence, clinical validity of the variants, and utility of the NGS assay [199], with the scientific evidence being documented in the validation protocol.
Considerations for test validation and clinical implementation
Although specific guidelines for cfDNA testing are not yet established, regulatory requirements under Clinical Laboratory Improvement Amendments call for all non-FDA-approved tests to address accuracy, precision, reportable and reference ranges, analytical sensitivity, analytical specificity, and any other parameters that may be relevant to the assay performance. All clinical validations should be included in the outlined validation protocol, including the entire range of clinical samples expected for the patient population, and, if appropriate, validation of the bioinformatics pipelines. Given the wide range of clinical platforms used for cfDNA testing, a comprehensive evaluation that would cover all of them is outside the scope of this review.
The wide variability in cfDNA content among samples introduces a high degree of complexity and more potential sources of error. Assessing all potential sources of error at every level of assay design, method validation and quality control is critical to avoid potential harm to the patient being caused by both false-positive and false-negative results. Special attention must be given to the qualification and quantification of input cfDNA. Dedicated standard operating procedures that outline the preanalytical steps for optimal collection, handling, extraction, isolation and storage of cfDNA samples must be validated with the same rigour as the analytical platform.
Conclusions
Cell-free nucleic acids have tremendous potential for molecular pathology in cancer. Advances in the sensitivity and specificity of cfDNA methodologies open up possibilities for the early detection of recurrence and cancer screening at asymptomatic stages. In the molecular diagnostic setting, we envision that tumour analysis will continue to play a critical role in diagnosis by revealing histological and genomic profiles. cfDNA will provide genomic information that may not be available if a single biopsy is performed or when tumour is not available, as well as enabling longitudinal monitoring of the evolving genome for timely treatment intervention. Further work needs will further define the specifications for broad clinical implementation, but, undoubtedly, cell-free nucleic acid profiling is creating a new paradigm of molecular pathology to improve cancer care through precision oncology.
Acknowledgements
We acknowledge the Department of Pathology, Department of Pediatrics and Marie-José and Henry R. Kravis Center for Molecular Oncology of the Memorial Sloan Kettering Cancer Center, and the Memorial Sloan Kettering Cancer Center Support Grant (NIH/NCI, Grant No. P30CA008748), as well as University of Cambridge, Cancer Research UK, the National Breast Cancer Foundation and the Victorian Cancer Agency, Australia, for their support of the respective authors.
Footnotes
Conflict of interest statement: DWYT is a former consultant of Inivata Ltd. DWYT is a contributor to a patent on cell-free DNA detection methodologies, and may receive royalties related to the licences of those patents to Inivata Ltd; the terms of these royalties are managed by Cancer Research Technology and Cambridge Enterprise. DWYT has received travel sponsorship and honoraria from AstraZeneca. MFB receives research support from Illumina and is a consultant for Sequenom. The remaining authors declare no competing financial interests. The sponsors had no involvement in the preparation of the manuscript or decision to submit.
References
- 1.Boland GM, Piha-Paul SA, Subbiah V, et al. Clinical next generation sequencing to identify actionable aberrations in a phase I program. Oncotarget 2015; 6: 20099–20110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017; 23: 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghazani AA, Oliver NM, St Pierre JP, et al. Assigning clinical meaning to somatic and germ-line whole-exome sequencing data in a prospective cancer precision medicine study. Genet Med 2017; 19: 787–795. [DOI] [PubMed] [Google Scholar]
- 4.AACR Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov 2017; 7: 818–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013; 499: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014; 505: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013; 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Middha S, Zhang L, Nafa K, et al. Reliable pan-cancer microsatellite instability assessment by using targeted next-generation sequencing data. JCO PO 2017; 1: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niu B, Ye K, Zhang Q, et al. MSIsensor: microsatellite instability detection using paired tumor–normal sequence data. Bioinformatics 2014; 30: 1015–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hause RJ, Pritchard CC, Shendure J, et al. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med 2016; 22: 1342–1350. [DOI] [PubMed] [Google Scholar]
- 11.Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther 2017; 11: 2598–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014; 371: 2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004; 304: 1497–1500. [DOI] [PubMed] [Google Scholar]
- 14.Damodaran S, Berger MF, Roychowdhury S. Clinical tumor sequencing: opportunities and challenges for precision cancer medicine. American Society of Clinical Oncology Educational Book 2015; e175–e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong SQ, Li J, Tan AY, et al. Sequence artefacts in a prospective series of formalin-fixed tumours tested for mutations in hotspot regions by massively parallel sequencing. BMC Med Genom 2014; 7: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017; 14: 531–548. [DOI] [PubMed] [Google Scholar]
- 17.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017; 17: 223–238. [DOI] [PubMed] [Google Scholar]
- 18.Stroun M, Anker P, Maurice P, et al. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology 1989; 46: 318–322. [DOI] [PubMed] [Google Scholar]
- 19.Thierry AR, Mouliere F, Gongora C, et al. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res 2010; 38: 6159–6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013; 368: 1199–1209. [DOI] [PubMed] [Google Scholar]
- 21.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014; 6: 224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008; 14: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo YM, Zhang J, Leung TN, et al. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet 1999; 64: 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riediger AL, Dietz S, Schirmer U, et al. Mutation analysis of circulating plasma DNA to determine response to EGFR tyrosine kinase inhibitor therapy of lung adenocarcinoma patients. Sci Rep 2016; 6: 33505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang P, Chan CWM, Chan KCA, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A 2015; 112: E1317–E1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mouliere F, Robert B, Arnau Peyrotte E, et al. High fragmentation characterizes tumour-derived circulating DNA. PLoS One 2011; 6: e23418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Underhill HR, Kitzman JO, Hellwig S, et al. Fragment length of circulating tumor DNA. PLoS Genet 2016; 12: e1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mouliere F, El Messaoudi S, Pang D, et al. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol Oncol 2014; 8: 927–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lo YM, Chan KC, Sun H, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med 2010; 2: 61ra91. [DOI] [PubMed] [Google Scholar]
- 30.Snyder MW, Kircher M, Hill AJ, et al. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 2016; 164: 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thierry AR, El Messaoudi S, Gahan PB, et al. Origins, structures, and functions of circulating DNA in oncology. Cancer Metast Rev 2016; 35: 347–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamat AA, Bischoff FZ, Dang D, et al. Circulating cell-free DNA: a novel biomarker for response to therapy in ovarian carcinoma. Cancer Biol Ther 2006; 5: 1369–1374. [DOI] [PubMed] [Google Scholar]
- 33.Nawroz H, Koch W, Anker P, et al. Microsatellite alterations in serum DNA of head and neck cancer patients. Nat Med 1996; 2: 1035–1037. [DOI] [PubMed] [Google Scholar]
- 34.Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017; 545: 446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parkinson CA, Gale D, Piskorz AM, et al. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. PLoS Med 2016; 13: e1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lecomte T, Berger A, Zinzindohoue F, et al. Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int J Cancer 2002; 100: 542–548. [DOI] [PubMed] [Google Scholar]
- 37.Zhuang R, Li S, Li Q, et al. The prognostic value of KRAS mutation by cell-free DNA in cancer patients: a systematic review and meta-analysis. PLoS One 2017; 12: e0182562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S, An T, Wang J, et al. Potential clinical significance of a plasma-based KRAS mutation analysis in patients with advanced non-small cell lung cancer. Clin Cancer Res 2010; 16: 1324–1330. [DOI] [PubMed] [Google Scholar]
- 39.Gautschi O, Huegli B, Ziegler A, et al. Origin and prognostic value of circulating KRAS mutations in lung cancer patients. Cancer Lett 2007; 254: 265–273. [DOI] [PubMed] [Google Scholar]
- 40.Santiago-Walker A, Gagnon R, Mazumdar J, et al. Correlation of BRAF mutation status in circulating-free DNA and tumor and association with clinical outcome across four BRAFi and MEKi clinical trials. Clin Cancer Res 2016; 22: 567–574. [DOI] [PubMed] [Google Scholar]
- 41.Gray ES, Rizos H, Reid AL, et al. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget 2015; 6: 42008–42018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reinert T, Scholer LV, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016; 65: 625–634. [DOI] [PubMed] [Google Scholar]
- 43.Spindler K-LG, Pallisgaard N, Vogelius I, et al. Quantitative cell-free DNA, KRAS and BRAF; mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res 2012; 18: 1177–1185. [DOI] [PubMed] [Google Scholar]
- 44.Romanel A, Tandefelt DG, Conteduca V, et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med 2015; 7: 312re310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altrichter J, Zedler S, Kraft R, et al. Neutrophil-derived circulating free DNA (cf-DNA/NETs), a potential prognostic marker for mortality in patients with severe burn injury. Eur J Trauma Emerg Surg 2010; 36: 551–557. [DOI] [PubMed] [Google Scholar]
- 46.Dwivedi DJ, Toltl LJ, Swystun LL, et al. Prognostic utility and characterization of cell-free DNA in patients with severe sepsis. Crit Care (Lond) 2012; 16: R151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alapirtti T, Jylhava J, Raitanen J, et al. The concentration of cell-free DNA in video-EEG patients is dependent on the epilepsy syndrome and duration of epilepsy. Neurol Res 2016; 38: 45–50. [DOI] [PubMed] [Google Scholar]
- 48.Bustamante A, Mancha F, Macher HC, et al. Circulating cell-free DNA is a predictor of short-term neurological outcome in stroke patients treated with intravenous thrombolysis. J Circ Biomark 2016; 5: 1849454416668791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karlas T, Weise L, Kuhn S, et al. Correlation of cell-free DNA plasma concentration with severity of non-alcoholic fatty liver disease. J Transl Med 2017; 15: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lipson EJ, Velculescu VE, Pritchard TS, et al. Circulating tumor DNA analysis as a real-time method for monitoring tumor burden in melanoma patients undergoing treatment with immune checkpoint blockade. J Immunother Cancer 2014; 2: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oshiro C, Kagara N, Naoi Y, et al. PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Breast Cancer Res Treat 2015; 150: 299–307. [DOI] [PubMed] [Google Scholar]
- 52.Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012; 4: 136ra168. [DOI] [PubMed] [Google Scholar]
- 53.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014; 20: 548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schreuer M, Meersseman G, Van Den Herrewegen S, et al. Quantitative assessment of BRAF V600 mutant circulating cell-free tumor DNA as a tool for therapeutic monitoring in metastatic melanoma patients treated with BRAF/MEK inhibitors. J Transl Med 2016; 14: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marchetti A, Palma JF, Felicioni L, et al. Early prediction of response to tyrosine kinase inhibitors by quantification of EGFR mutations in plasma of NSCLC patients. J Thorac Oncol 2015; 10: 1437–1443. [DOI] [PubMed] [Google Scholar]
- 56.Scherer F, Kurtz DM, Newman AM, et al. Distinct biological sub-types and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med 2016; 8: 364ra155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Laere B, van Dam P-J, Whitington T, et al. Comprehensive profiling of the androgen receptor in liquid biopsies from castration-resistant prostate cancer reveals novel intra-AR structural variation and splice variant expression patterns. Eur Urol 2017; 72: 192–200. [DOI] [PubMed] [Google Scholar]
- 58.US National Library of Medicine. Blood Sample Monitoring of Patients With EGFR Mutated Lung Cancer ClinicalTrialsgov. [Accessed 24 October 2017]. Available from: https://clinicaltrials.gov/ct2/show/NCT02284633
- 59.European Medicines Agency. Iressa: Public Assessment Report – Product Information EMA EMA; [Accessed 21 October 2017]. Available from: http://www.ema.europa.eu/docs/enGB/documentlibrary/EPAR-Product_Information/human/001016/WC500036358.pdf [Google Scholar]
- 60.US Food and Drug Administration. Premarket Approval P150044 – Cobas EGFR MUTATION TEST V2. [Accessed 21 October 2017]. Available from: https://www.accessdata.fda.gov/cdrh_docs/pdf15/P150047a.pdf
- 61.European Medicines Agency. Tagrisso: Public Assessment Report – Product Information EMA. EMA; [Accessed 21 October 2017]. Available from: http://www.ema.europa.eu/docs/enGB/documentlibrary/EPAR-Product_Information/human/004124/WC500202022.pdf [Google Scholar]
- 62.Vanderlaan PA, Yamaguchi N, Folch E, et al. Success and failure rates of tumor genotyping techniques in routine pathological samples with non-small-cell lung cancer. Lung Cancer 2014; 84: 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qian X, Liu J, Sun Y, et al. Circulating cell-free DNA has a high degree of specificity to detect exon 19 deletions and the single-point substitution mutation L858R in non-small cell lung cancer. Oncotarget 2016; 7: 29154–29165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Normanno N, Denis MG, Thress KS, et al. Guide to detecting epidermal growth factor receptor (EGFR) mutations in ctDNA of patients with advanced non-small-cell lung cancer. Oncotarget 2017; 8: 12501–12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qiagen. Therascreen EGFR Plasma RGQ PCR Kit Handbook – Version 2. [Accessed 21 October 2017]. Available from: https://www.qiagen.com/at/resources/resourcedetail?id=d217c9ca-b9f6-4891-8c46-32b71e83d88a&lang=en
- 66.Remon J, Soria JC, Planchard D, et al. Liquid biopsies for molecular profiling of mutations in non-small cell lung cancer (NSCLC) patients lacking tissue samples Inivata. [Accessed 23 October 2017]. Available from: http://inivata.com/wp-content/uploads/2016/09/MAP2016posterFINAL.pdf
- 67.Schwaederle M, Husain H, Fanta PT, et al. Use of liquid biopsies in clinical oncology: pilot experience in 168 patients. Clin Cancer Res 2016; 22: 5497–5505. [DOI] [PubMed] [Google Scholar]
- 68.Talasaz A, Sebisanovic D, Mei G, et al. Ultra-high quality sequencing assay for comprehensive genetic panel analysis of rare tumor-derived circulating cell-free DNA. Poster. Program Number: 3478T. American Society of Human Genetics Annual Conference, Boston, 2013. [Google Scholar]
- 69.Lanman RB, Mortimer SA, Zill OA, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One 2015; 10: e0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.US National Library of Medicine. Next Generation pErsonalized tX(Therapy) With Plasma DNA Trial-2 in Refractory Solid Tumors (The NEXT-2 Trial). [Accessed 21 October 2017]. Available from: https://clinicaltrials.gov/ct2/show/NCT02140463
- 71.US National Library of Medicine. Monitoring Plasma Tumor DNA in Early-Stage Breast Cancer. [Accessed 24 October 2017]. Available from: https://clinicaltrials.gov/ct2/show/NCT02743910
- 72.International Standard Randomized Controlled Trials Number Registry. plasmaMATCH: A clinical trial aiming to assess the safety and activity of targeted treatments in patients with advanced breast cancer where the targetable mutation is identified through circulating tumour DNA screening. [Accessed 24 October 2017]. Available from: http://www.isrctn.com/ISRCTN16945804
- 73.De Mattos-Arruda L, Weigelt B, Cortes J, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol 2014; 25: 1729–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adalsteinsson VA, Ha G, Freeman SS, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 2017; 8: 1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lebofsky R, Decraene C, Bernard V, et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol Oncol 2015; 9: 783–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frenel JS, Carreira S, Goodall J, et al. Serial next-generation sequencing of circulating cell-free DNA evaluating tumor clone response to molecularly targeted drug administration. Clin Cancer Res 2015; 21: 4586–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Murtaza M, Dawson S-J, Tsui DWY, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013; 497: 108–112. [DOI] [PubMed] [Google Scholar]
- 78.Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016; 34: 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kinde I, Wu J, Papadopoulos N, et al. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A 2011; 108: 9530–9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gale D, Plagnol V, Lawson A, et al. Analytical performance and validation of an enhanced TAm-Seq circulating tumor DNA sequencing assay. Inviata, American Association of Cancer Research, New Orleans, LA, 2016. [Google Scholar]
- 81.Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017; 9: pii: eaan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boles C, Popkie A, Hoda S, et al. Efficient generation of cfDNA libraries that are highly enriched in short fragments Advances in Genome Biology and Technology, Scottsdale, Arizona, 2016. [Google Scholar]
- 83.Norris AL, Workman RE, Fan Y, et al. Nanopore sequencing detects structural variants in cancer. Cancer Biol Ther 2016; 17: 246–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheng SH, Jiang P, Sun K, et al. Noninvasive prenatal testing by nanopore sequencing of maternal plasma DNA: feasibility assessment. Clin Chem 2015; 61: 1305–1306. [DOI] [PubMed] [Google Scholar]
- 85.Gootenberg JS, Abudayyeh OO, Lee JW, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017; 356: 438–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012; 486: 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chandarlapaty S, Chen D, He W, et al. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 Clinical Trial. JAMA Oncol 2016; 2: 1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sorensen BS, Wu L, Wei W, et al. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer 2014; 120: 3896–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 2014; 20: 1698–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morelli MP, Overman MJ, Dasari A, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol 2015; 26: 731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Christie EL, Fereday S, Doig K, et al. Reversion of BRCA1/2 germline mutations detected in circulating tumor DNA from patients with high-grade serous ovarian cancer. J Clin Oncol 2017; 35: 1274–1280. [DOI] [PubMed] [Google Scholar]
- 92.Weigelt B, Comino-Mendez I, de Bruijn I, et al. Diverse BRCA1 and BRCA2 reversion mutations in circulating cell-free DNA of therapy-resistant breast or ovarian cancer. Clin Cancer Res 2017; 23: 6708–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Quigley D, Alumkal JJ, Wyatt AW, et al. Analysis of circulating cell-free DNA identifies multiclonal heterogeneity of BRCA2 reversion mutations associated with resistance to PARP inhibitors. Cancer Discov 2017; 7: 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Goodall J, Mateo J, Yuan W, et al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov 2017; 7: 1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thierry AR, Pastor B, Jiang ZQ, et al. Circulating DNA demonstrates convergent evolution and common resistance mechanisms during treatment of colorectal cancer. Clin Cancer Res 2017; 23: 4578–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goyal L, Saha SK, Liu LY, et al. Polyclonal secondary FGFR2 mutations drive acquired resistance to FGFR inhibition in patients with FGFR2 fusion-positive cholangiocarcinoma. Cancer Discov 2017; 7: 252–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Russo M, Misale S, Wei G, et al. Acquired resistance to the TRK inhibitor entrectinib in colorectal cancer. Cancer Discov 2016; 6: 36–44. [DOI] [PubMed] [Google Scholar]
- 98.Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015; 7: 302ra133. [DOI] [PubMed] [Google Scholar]
- 99.Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016; 8: 346ra392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vasioukhin V, Anker P, Maurice P, et al. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodys-plastic syndrome or acute myelogenous leukaemia. Br J Haematol 1994; 86: 774–779. [DOI] [PubMed] [Google Scholar]
- 101.He J, Wu J, Jiao Y, et al. IgH gene rearrangements as plasma biomarkers in non-Hodgkin’s lymphoma patients. Oncotarget 2011; 2: 178–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kurtz DM, Green MR, Bratman SV, et al. Noninvasive monitoring of diffuse large B-cell lymphoma by immunoglobulin high-throughput sequencing. Blood 2015; 125: 3679–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Armand P, Oki Y, Neuberg DS, et al. Detection of circulating tumour DNA in patients with aggressive B-cell non-Hodgkin lymphoma. Br J Haematol 2013; 163: 123–126. [DOI] [PubMed] [Google Scholar]
- 104.Frickhofen N, Muller E, Sandherr M, et al. Rearranged Ig heavy chain DNA is detectable in cell-free blood samples of patients with B-cell neoplasia. Blood 1997; 90: 4953–4960. [PubMed] [Google Scholar]
- 105.Roschewski M, Dunleavy K, Pittaluga S, et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol 2015; 16: 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Herrera AF, Kim HT, Kong KA, et al. Next-generation sequencing-based detection of circulating tumour DNA after allogeneic stem cell transplantation for lymphoma. Br J Haematol 2016; 175: 841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bohers E, Viailly PJ, Dubois S, et al. Somatic mutations of cell-free circulating DNA detected by next-generation sequencing reflect the genetic changes in both germinal center B-cell-like and activated B-cell-like diffuse large B-cell lymphomas at the time of diagnosis. Haematologica 2015; 100: e280–e284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rossi D, Diop F, Spaccarotella E, et al. Diffuse large B-cell lymphoma genotyping on the liquid biopsy. Blood 2017; 129: 1947–1957. [DOI] [PubMed] [Google Scholar]
- 109.Camus V, Sarafan-Vasseur N, Bohers E, et al. Digital PCR for quantification of recurrent and potentially actionable somatic mutations in circulating free DNA from patients with diffuse large B-cell lymphoma. Leuk Lymphoma 2016; 57: 2171–2179. [DOI] [PubMed] [Google Scholar]
- 110.Scherer F, Kurtz DM, Newman AM, et al. Distinct biological sub-types and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med 2016; 8: 364ra155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vandenberghe P, Wlodarska I, Tousseyn T, et al. Non-invasive detection of genomic imbalances in Hodgkin/Reed–Sternberg cells in early and advanced stage Hodgkin’s lymphoma by sequencing of circulating cell-free DNA: a technical proof-of-principle study. Lancet Haematol 2015; 2: e55–e65. [DOI] [PubMed] [Google Scholar]
- 112.Camus V, Stamatoullas A, Mareschal S, et al. Detection and prognostic value of recurrent exportin 1 mutations in tumor and cell-free circulating DNA of patients with classical Hodgkin lymphoma. Haematologica 2016; 101: 1094–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yeh P, Hunter T, Sinha D, et al. Circulating tumour DNA reflects treatment response and clonal evolution in chronic lymphocytic leukaemia. Nat Commun 2017; 8: 14756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yeh P, Dickinson M, Ftouni S, et al. Molecular disease monitoring using circulating tumor DNA in myelodysplastic syndromes. Blood 2017; 129: 1685–1690. [DOI] [PubMed] [Google Scholar]
- 115.Albitar F, Ma W, Diep K, et al. Deep sequencing of cell-free peripheral blood DNA as a reliable method for confirming the diagnosis of myelodysplastic syndrome. Genet Test Mol Biomark 2016; 20: 341–345. [DOI] [PubMed] [Google Scholar]
- 116.Suzuki Y, Tomita A, Nakamura F, et al. Peripheral blood cell-free DNA is an alternative tumor DNA source reflecting disease status in myelodysplastic syndromes. Cancer Sci 2016; 107: 1329–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kis O, Kaedbey R, Chow S, et al. Circulating tumour DNA sequence analysis as an alternative to multiple myeloma bone marrow aspirates. Nat Commun 2017; 8: 15086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mithraprabhu S, Khong T, Ramachandran M, et al. Circulating tumour DNA analysis demonstrates spatial mutational heterogeneity that coincides with disease relapse in myeloma. Leukemia 2017; 31: 1695–1705. [DOI] [PubMed] [Google Scholar]
- 119.Oberle A, Brandt A, Voigtlaender M, et al. Monitoring multiple myeloma by next-generation sequencing of V(D)J rearrangements from circulating myeloma cells and cell-free myeloma DNA. Haematologica 2017; 102: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rustad EH, Coward E, Skytoen ER, et al. Monitoring multiple myeloma by quantification of recurrent mutations in serum. Haematologica 2017; 102: 1266–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bryzgunova OE, Laktionov PP. Extracellular nucleic acids in urine: sources, structure, diagnostic potential. Acta Naturae 2015; 7: 48–54. [PMC free article] [PubMed] [Google Scholar]
- 122.Botezatu I, Serdyuk O, Potapova G, et al. Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin Chem 2000; 46: 1078–1084. [PubMed] [Google Scholar]
- 123.Sidransky D, Von Eschenbach A, Tsai YC, et al. Identification of p53 gene mutations in bladder cancers and urine samples. Science 1991; 252: 706–709. [DOI] [PubMed] [Google Scholar]
- 124.Tsui NBY, Jiang P, Chow KCK, et al. High resolution size analysis of fetal DNA in the urine of pregnant women by paired-end massively parallel sequencing. PLoS One 2012; 7: e48319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cheng THT, Jiang P, Tam JCW, et al. Genomewide bisulfite sequencing reveals the origin and time-dependent fragmentation of urinary cfDNA. Clin Biochem 2017; 50: 496–501. [DOI] [PubMed] [Google Scholar]
- 126.Chen S, Zhao J, Cui L, et al. Urinary circulating DNA detection for dynamic tracking of EGFR mutations for NSCLC patients treated with EGFR-TKIs. Clin Trans Oncol 2017; 19: 332–340. [DOI] [PubMed] [Google Scholar]
- 127.Hann HW, Jain S, Park G, et al. Detection of urine DNA markers for monitoring recurrent hepatocellular carcinoma. Hepatoma Res 2017; 3: 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Patel KM, van der Vos KE, Smith CG, et al. Association of plasma and urinary mutant DNA with clinical outcomes in muscle invasive bladder cancer. Sci Rep 2017; 7: 5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Su YH, Wang M, Brenner DE, et al. Human urine contains small, 150 to 250 nucleotide-sized, soluble DNA derived from the circulation and may be useful in the detection of colorectal cancer. J Mol Diagn 2004; 6: 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mathiisen TM, Lehre KP, Danbolt NC, et al. The perivascular astroglial sheath provides a complete covering of the brain microvessels: an electron microscopic 3D reconstruction. Glia 2010; 58: 1094–1103. [DOI] [PubMed] [Google Scholar]
- 131.Harker Rhodes C, Honsinger C, Sorenson GD. PCR-detection of tumor-derived p53 DNA in cerebrospinal fluid. Am J Clin Pathol 1995; 103: 404–408. [DOI] [PubMed] [Google Scholar]
- 132.Swinkels DW, de Kok JB, Hanselaar A, et al. Early detection of leptomeningeal metastasis by PCR examination of tumor-derived K-ras DNA in cerebrospinal fluid. Clin Chem 2000; 46: 132–133. [PubMed] [Google Scholar]
- 133.Pan W, Gu W, Nagpal S, et al. Brain tumor mutations detected in cerebral spinal fluid. Clin Chem 2015; 61: 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang J, Bettegowda C. Applications of DNA-based liquid biopsy for central nervous system neoplasms. J Mol Diagn 2017; 19: 24–34. [DOI] [PubMed] [Google Scholar]
- 135.De Mattos-Arruda L, Mayor R, Ng CK, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun 2015; 6: 8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pentsova EI, Shah RH, Tang J, et al. Evaluating cancer of the central nervous system through next-generation sequencing of cerebrospinal fluid. J Clin Oncol 2016; 34: 2404–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001; 61: 1659–1665. [PubMed] [Google Scholar]
- 138.Melnikov AA, Scholtens D, Talamonti MS, et al. Methylation profile of circulating plasma DNA in patients with pancreatic cancer. J Surg Oncol 2009; 99: 119–122. [DOI] [PubMed] [Google Scholar]
- 139.Kadam SK, Farmen M, Brandt JT. Quantitative measurement of cell-free plasma DNA and applications for detecting tumor genetic variation and promoter methylation in a clinical setting. J Mol Diagn 2012; 14: 346–356. [DOI] [PubMed] [Google Scholar]
- 140.Chan KC, Jiang P, Chan CW, et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci U S A 2013; 110: 18761–18768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhao Y, Xue F, Sun J, et al. Genome-wide methylation profiling of the different stages of hepatitis B virus-related hepatocellular carcinoma development in plasma cell-free DNA reveals potential biomarkers for early detection and high-risk monitoring of hepatocellular carcinoma. Clin Epigenet 2014; 6: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li Z, Guo X, Tang L, et al. Methylation analysis of plasma cell-free DNA for breast cancer early detection using bisulfite next-generation sequencing. Tumour Biol 2016; 37: 13111–13119. [DOI] [PubMed] [Google Scholar]
- 143.Xu R-h, Wei W, Krawczyk M, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mat 2017; 16: 1155–1161. [DOI] [PubMed] [Google Scholar]
- 144.Sturgeon SR, Balasubramanian R, Schairer C, et al. Detection of promoter methylation of tumor suppressor genes in serum DNA of breast cancer cases and benign breast disease controls. Epigenetics 2012; 7: 1258–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sun K, Jiang P, Chan KCA, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A 2015; 112: E5503–E5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lehmann-Werman R, Neiman D, Zemmour H, et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci U S A 2016; 113: E1826–E1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Guo S, Diep D, Plongthongkum N, et al. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat Genet 2017; 49: 635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kang S, Li Q, Chen Q, et al. CancerLocator: non-invasive cancer diagnosis and tissue-of-origin prediction using methylation profiles of cell-free DNA. Genome Biol 2017; 18: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lo KW, Lo YM, Leung SF, et al. Analysis of cell-free Epstein–Barr virus associated RNA in the plasma of patients with nasopharyngeal carcinoma. Clin Chem 1999; 45: 1292–1294. [PubMed] [Google Scholar]
- 150.Chan KCA, Leung SF, Yeung SW, et al. Quantitative analysis of the transrenal excretion of circulating EBV DNA in nasopharyngeal carcinoma patients. Clin Cancer Res 2008; 14: 4809–4813. [DOI] [PubMed] [Google Scholar]
- 151.Wang Y, Springer S, Mulvey CL, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med 2015; 7: 293ra104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pornthanakasem W, Shotelersuk K, Termrungruanglert W, et al. Human papillomavirus DNA in plasma of patients with cervical cancer. BMC Cancer 2001; 1: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kowarsky M, Camunas-Soler J, Kertesz M, et al. Numerous uncharacterized and highly divergent microbes which colonize humans are revealed by circulating cell-free DNA. Proc Natl Acad Sci U S A 2017; 114: 9623–9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Burnham P, Kim MS, Agbor-Enoh S, et al. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci Rep 2016; 6: 27859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tsui NB, Jiang P, Wong YF, et al. Maternal plasma RNA sequencing for genome-wide transcriptomic profiling and identification of pregnancy-associated transcripts. Clin Chem 2014; 60: 954–962. [DOI] [PubMed] [Google Scholar]
- 156.Pacifici M, Delbue S, Kadri F, et al. Cerebrospinal fluid microRNA profiling using quantitative real time PCR. J Vis Exp 2014; 83: e51172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008; 105: 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Burgos K, Malenica I, Metpally R, et al. Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS One 2014; 9: e94839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Burgos KL, Javaherian A, Bomprezzi R, et al. Identification of extracellular miRNA in human cerebrospinal fluid by next-generation sequencing. RNA 2013; 19: 712–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Terrin L, Rampazzo E, Pucciarelli S, et al. Relationship between tumor and plasma levels of hTERT mRNA in patients with colorectal cancer: implications for monitoring of neoplastic disease. Clin Cancer Res 2008; 14: 7444–7451. [DOI] [PubMed] [Google Scholar]
- 161.Pucciarelli S, Rampazzo E, Briarava M, et al. Telomere-specific reverse transcriptase (hTERT) and cell-free RNA in plasma as predictors of pathologic tumor response in rectal cancer patients receiving neoadjuvant chemoradiotherapy. Ann Surg Oncol 2012; 19: 3089–3096. [DOI] [PubMed] [Google Scholar]
- 162.Aman P, Panagopoulos I, Lassen C, et al. Expression patterns of the human sarcoma-associated genes FUS and EWS and the genomic structure of FUS. Genomics 1996; 37: 1–8. [DOI] [PubMed] [Google Scholar]
- 163.Mertens F, Johansson B, Fioretos T, et al. The emerging complexity of gene fusions in cancer. Nat Rev Cancer 2015; 15: 371–381. [DOI] [PubMed] [Google Scholar]
- 164.Shaw AT, Yeap BY, Solomon BJ, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol 2011; 12: 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cui S, Zhang W, Xiong L, et al. Use of capture-based next-generation sequencing to detect ALK fusion in plasma cell-free DNA of patients with non-small-cell lung cancer. Oncotarget 2017; 8: 2771–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Reeser JW, Martin D, Miya J, et al. Validation of a targeted RNA sequencing assay for kinase fusion detection in solid tumors. J Mol Diagn 2017; 19: 682–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kumar-Sinha C, Kalyana-Sundaram S, Chinnaiyan AM. Landscape of gene fusions in epithelial cancers: seq and ye shall find. Genome Med 2015; 7: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zarnegar S, Durham BH, Khattar P, et al. Novel activating BRAF fusion identifies a recurrent alternative mechanism for ERK activation in pediatric Langerhans cell histiocytosis. Pediatr Blood Cancer 2018; 65: e26699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Alexander KE, Jackson LP, Mellert HS, et al. Optimization of blood-based liquid biopsy assays for the rapid identification of 19 actionable RNA fusion variants in NSCLC. Cancer Genet 2017; 214: 34. [Google Scholar]
- 170.Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018; 359: 926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gu M, Shi W, Barakat RR, et al. Pap smears in women with endometrial carcinoma. Acta Cytol 2001; 45: 555–560. [DOI] [PubMed] [Google Scholar]
- 172.Lai CR, Hsu CY, Hang JF, et al. The diagnostic value of routine Papanicolaou smears for detecting endometrial cancers: an update. Acta Cytol 2015; 59: 315–318. [DOI] [PubMed] [Google Scholar]
- 173.Kinde I, Bettegowda C, Wang Y, et al. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med 2013; 5: 167ra164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Harris FR, Kovtun IV, Smadbeck J, et al. Quantification of somatic chromosomal rearrangements in circulating cell-free DNA from ovarian cancers. Sci Rep 2016; 6: 29831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhou Q, Li W, Leng B, et al. Circulating cell free DNA as the diagnostic marker for ovarian cancer: a systematic review and meta-analysis. PLoS One 2016; 11: e0155495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.To EW, Chan KC, Leung SF, et al. Rapid clearance of plasma Epstein–Barr virus DNA after surgical treatment of nasopharyngeal carcinoma. Clin Cancer Res 2003; 9: 3254–3259. [PubMed] [Google Scholar]
- 177.Le QT, Zhang Q, Cao H, et al. An international collaboration to harmonize the quantitative plasma Epstein–Barr virus DNA assay for future biomarker-guided trials in nasopharyngeal carcinoma. Clin Cancer Res 2013; 19: 2208–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chan KCA, Woo JKS, King A, et al. Analysis of plasma Epstein–Barr virus DNA to screen for nasopharyngeal cancer. N Engl J Med 2017; 377: 513–522. [DOI] [PubMed] [Google Scholar]
- 179.Yamanouchi S, Kudo D, Yamada M, et al. Plasma mitochondrial DNA levels in patients with trauma and severe sepsis: time course and the association with clinical status. J Crit Care 2013; 28: 1027–1031. [DOI] [PubMed] [Google Scholar]
- 180.Lo YM, Lun FM, Chan KC, et al. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc Natl Acad Sci U S A 2007; 104: 13116–13121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kinde I, Papadopoulos N, Kinzler KW, et al. FAST-SeqS: a simple and efficient method for the detection of aneuploidy by massively parallel sequencing. PLoS One 2012; 7: e41162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sehnert AJ, Rhees B, Comstock D, et al. Optimal detection of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell-free fetal DNA from maternal blood. Clin Chem 2011; 57: 1042–1049. [DOI] [PubMed] [Google Scholar]
- 183.Yu SCY, Chan KCA, Zheng YWL, et al. Size-based molecular diagnostics using plasma DNA for noninvasive prenatal testing. Proc Natl Acad Sci U S A 2014; 111: 8583–8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hudecova I, Jiang P, Davies J, et al. Noninvasive detection of F8 int22h-related inversions and sequence variants in maternal plasma of hemophilia carriers. Blood 2017; 130: 340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hyett JA, Gardener G, Stojilkovic-Mikic T, et al. Reduction in diagnostic and therapeutic interventions by non-invasive determination of fetal sex in early pregnancy. Prenat Diagn 2005; 25: 1111–1116. [DOI] [PubMed] [Google Scholar]
- 186.Saito H, Sekizawa A, Morimoto T, et al. Prenatal DNA diagnosis of a single-gene disorder from maternal plasma. Lancet 2000; 356: 1170. [DOI] [PubMed] [Google Scholar]
- 187.Chiu RW, Akolekar R, Zheng YW, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. BMJ Clin Res 2011; 342: c7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Palomaki GE, Kloza EM, Lambert-Messerlian GM, et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet Med 2011; 13: 913–920. [DOI] [PubMed] [Google Scholar]
- 189.Hill M, Wright D, Daley R, et al. Evaluation of non-invasive prenatal testing (NIPT) for aneuploidy in an NHS setting: a reliable accurate prenatal non-invasive diagnosis (RAPID) protocol. BMC Pregnancy Childbirth 2014; 14: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ehrich M, Deciu C, Zwiefelhofer T, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am J Obstet Gynecol 2011; 204: 205.e201–205.e211. [DOI] [PubMed] [Google Scholar]
- 191.Barra GB, Santa Rita TH, de Almeida Vasques J, et al. EDTA-mediated inhibition of DNases protects circulating cell-free DNA from ex vivo degradation in blood samples. Clin Biochem 2015; 48: 976–981. [DOI] [PubMed] [Google Scholar]
- 192.Sherwood JL, Corcoran C, Brown H, et al. Optimised pre-analytical methods improve KRAS mutation detection in circulating tumour DNA (ctDNA) from patients with non-small cell lung cancer (NSCLC). PLoS One 2016; 11: e0150197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Barrett AN, Zimmermann BG, Wang D, et al. Implementing prenatal diagnosis based on cell-free fetal DNA: accurate identification of factors affecting fetal DNA yield. PLoS One 2011; 6: e25202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Parpart-Li S, Bartlett B, Popoli M, et al. The effect of preservative and temperature on the analysis of circulating tumor DNA. Clin Cancer Res 2017; 23: 2471–2477. [DOI] [PubMed] [Google Scholar]
- 195.Warton K, Yuwono NL, Cowley MJ, et al. Evaluation of Streck BCT and PAXgene stabilised blood collection tubes for cell-free circulating DNA studies in plasma. Mol Diagn Ther 2017; 21: 563–570. [DOI] [PubMed] [Google Scholar]
- 196.Medina Diaz I, Nocon A, Mehnert DH, et al. Performance of Streck cfDNA blood collection tubes for liquid biopsy testing. PLoS One 2016; 11: e0166354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bowen RA, Adcock DM. Blood collection tubes as medical devices: the potential to affect assays and proposed verification and validation processes for the clinical laboratory. Clin Biochem 2016; 49: 1321–1330. [DOI] [PubMed] [Google Scholar]
- 198.Page K, Guttery DS, Zahra N, et al. Influence of plasma processing on recovery and analysis of circulating nucleic acids. PLoS One 2013; 8: e77963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jennings LJ, Arcila ME, Corless C, et al. Guidelines for Validation of Next-Generation Sequencing-Based Oncology Panels: a Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn 2017; 19: 341–365. [DOI] [PMC free article] [PubMed] [Google Scholar]