The potential of circulating tumor DNA (ctDNA) detection in early stage lung cancer is explored. This study investigated whether bronchial washing, a minimally invasive procedure that yields fluids that may contain ctDNA, can reflect genetic profiles of primary tumors using next‐generation sequencing.
Abstract
A blood‐based approach such as circulating tumor DNA remains challenging in diagnosis for early‐stage disease. Bronchial washing (BW) is a minimally invasive procedure that yields fluids that may contain tumor DNA. Therefore, we prospectively enrolled 12 patients with early‐stage non‐small cell lung cancer without endoscopically visible tumors. Somatic mutations were analyzed using ultra‐deep next‐generation sequencing in 48 paired specimens (primary tumor tissue, normal tissue, BW supernatant, and BW precipitate). In primary tumors, 130 missense mutations/indels (5–16 per patient) and 20 driver mutations (0–3 per patient) were found. Concordance of driver mutations between BW fluids and primary tumors was 95.0%. The allele frequencies for missense mutations/indels in BW supernatants significantly correlated with those in primary tumors and were higher than those in BW precipitates. These findings suggest that BW supernatants are reflective of tumor‐associated mutations and could be used for early‐stage lung cancer diagnosis.
摘要
早期疾病的诊断中,难以采用循环肿瘤 DNA 等以血液为基础的诊断方法。支气管冲洗 (BW) 是一种微创手术,可产生可能含有肿瘤 DNA 的冲洗液。因此,本研究前瞻性分析了 12 例内镜下无可见肿瘤的早期非小细胞肺癌患者临床资料。采用超深新一代测序技术对 48 对标本(原发肿瘤组织、正常组织、BW 上清液和 BW 沉淀物)进行了体细胞突变分析。在原发性肿瘤中,发现 130 处错义突变/插入缺失(每位患者 5‐16 处)和 20 处驱动突变(每位患者 0‐3处)。BW 液与原发肿瘤的驱动突变一致性为 95.0%。BW 上清液中错义突变/插入缺失的等位基因频率与原发肿瘤的等位基因频率显著相关,且高于 BW 沉淀物。结果表明,BW 上清液可反映出肿瘤相关突变,可用于早期肺癌的诊断。
Lung cancer screening with low‐dose computed tomography (CT) markedly increases the incidental identification of lung lesions, most of which are benign but require pathological diagnosis via invasive procedures. Because lung cancers identified in these screenings reflect largely early‐stage disease without endobronchial lesions, the potential utility of circulating tumor DNA (ctDNA) detection in early‐stage lung cancer should be explored. However, ctDNA is not currently detectable in most early‐stage lung cancers [1], [2], [3], [4]; thus, alternative tools are needed for the ancillary diagnosis.
Bronchial washing (BW) has a low diagnostic yield of 10%–20% in cytological examinations but is minimally invasive, repeatable, and rarely causes complications [5]. BW fluid may contain non‐ctDNA shed from tumor cells [6]. Recently, epidermal growth factor receptor (EGFR) mutations were effectively detected in bronchoalveolar lavage fluid from patients with advanced‐stage non‐small cell lung cancer (NSCLC) [7]. However, no studies showing the landscape of somatic mutations in BW fluids and their concordance with primary tumor mutations in early‐stage NSCLC have been published. Here, we investigated whether BW fluid can reflect genetic profiles of primary tumors using next‐generation sequencing.
One hundred forty‐one consecutive patients who underwent bronchoscopic examinations for lung nodules or masses by chest CT at Inha University Hospital from January 2016 to March 2017 were initially considered for this study. Patients who had endoscopically visible tumors (n = 85), had acute inflammation (n = 6), did not undergo surgical resection (n = 30), had a benign diagnosis upon pathological examination (n = 4), or were lost to follow‐up (n = 4) were excluded. Finally, 48 paired specimens (primary tumor tissue, normal tissue, BW supernatant, and BW precipitate) from patients with early‐stage NSCLC were enrolled after written informed consent and local ethics approval were obtained. During initial bronchoscopic examination, BW was performed on each patient by placing 30 mL sterile saline in the subsegmental bronchus where the lung nodule or mass was thought to be located. BW fluid was collected in 15 mL centrifuge tubes, and half of each fluid specimen was centrifuged at 1,000g for 20 minutes at 4°C. The BW supernatants and precipitates were separated and stored at −80°C. The other half of each BW fluid specimen was used for cytological examination. Upon tumor resection, the pathologist prepared the tumor tissue and adjacent normal tissue, and aliquots were stored at −80°C. The amount and quality of DNA extracted from the specimens were evaluated with the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). The custom panel was designed with 113 genes with more than 5% sample frequency in each sample group with reference to previous reports in Korean populations and The Cancer Genome Atlas database [8], [9]. The panel covered 1.705 Mbp and contained 27,480 probes. The targeted DNA library was sequenced on an Illumina HiSeq2500 (Illumina, Inc., San Diego, CA, USA) with 100 base‐pair paired‐end reads. Tissues and BW fluids were sequenced at 1,000× and 10,000× depths, respectively. Missense mutations, insertions/deletions (indels), or driver mutations in BW supernatants and precipitates were identified by comparing primary tumor and normal tissue sequences. Finally, the prevalence and allele frequencies (AFs) of mutations in primary tumors and BW supernatants and precipitates were determined.
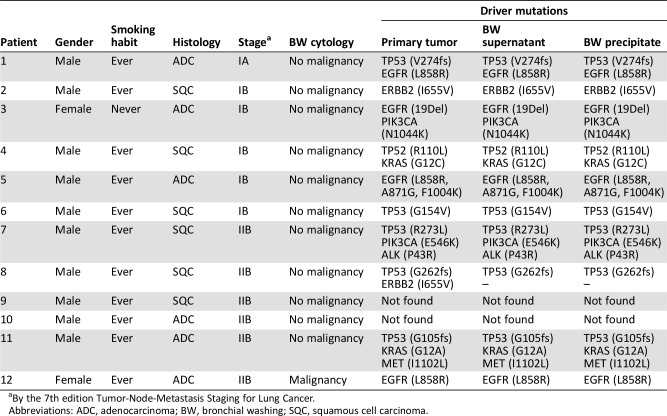
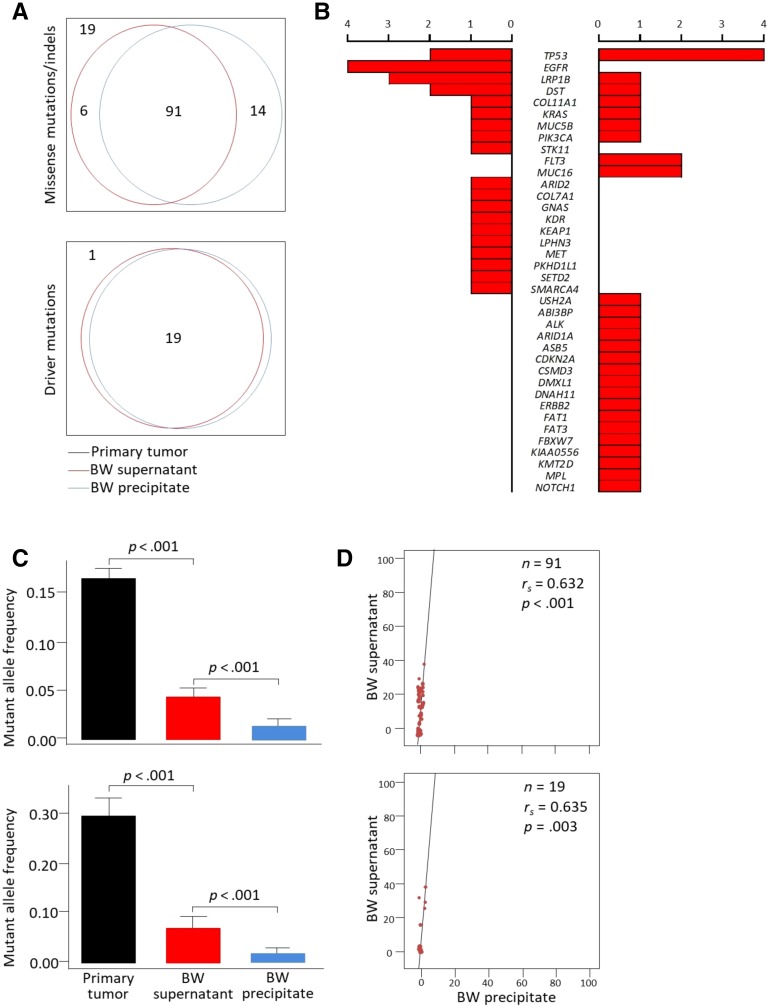
Of the 12 patients in this study, squamous cell carcinomas were found in 6. Ten patients had a history of cigarette smoking, and four patients carried EGFR activating mutations (Table 1). Malignant cells were not identified upon BW cytological examination in 11 (92.7%) patients. A total of 130 missense mutations/indels (5–16 per patient) and 20 driver mutations (0–3 per patient) were found in primary tumors. In BW supernatants, 105 (80.8%) of the missense mutations/indels and 19 (95.0%) of the driver mutations were detected; 97 (74.6%) and 19 (95.0%) were found in BW precipitates, respectively (Fig. 1A). More missense mutations/indels were detected in BW supernatants than in precipitates, but statistical significance was marginal (p = .07). All driver mutations identified in primary tumors were also detected in BW supernatants and BW precipitates except for the ERBB2I655V mutation. Concordance of driver mutations between BW fluids and primary tumors was 95.0% (κ = 0.96; 95% confidence interval = 0.89–1.00; p < .001). More interestingly, six (50%) patients had two or more concurrent driver mutations that could be associated with different tumor behaviors [10]. EGFR and STK11 were exclusively mutated in adenocarcinoma, and FLT3 and MUC16 in squamous cell carcinoma (Fig. 1B).
Table 1. Driver mutations in primary tumors, BW supernatants, and BW precipitates from individual patients.
By the 7th edition Tumor‐Node‐Metastasis Staging for Lung Cancer.
Abbreviations: ADC, adenocarcinoma; BW, bronchial washing; SQC, squamous cell carcinoma.
Figure 1.
Mutational profiles identified in primary tumors, BW supernatants, and BW precipitates using ultra‐deep next‐generation sequencing. (A): Venn diagrams showing the overlap of mutations in primary tumors, BW supernatants, and BW precipitates. (B): Representative missense mutations identified in BW supernatants (left, adenocarcinoma; right, squamous cell carcinoma). (C): Comparison of mutant allele frequencies in primary tumors, BW supernatants, and BW precipitates. p values by Mann‐Whitney U test. (D): Correlations of mutant allele frequencies in BW supernatants and BW precipitates.
Abbreviation: BW, bronchial washing.
The AFs for missense mutations/indels and driver mutations in BW supernatants were significantly higher than those in BW precipitates (all p < .001; Fig. 1C). The AFs of mutations in BW supernatants significantly correlated with those in primary tumors and BW precipitates (Fig. 1D). The AFs of missense mutations/indels in BW supernatants were higher in squamous cell carcinoma (vs. adenocarcinoma), wild‐type EGFR (vs. mutant type), and tumors greater than 3 cm in diameter (vs. ≤3 cm; all p < .001; data not shown).
This study is the first attempt to characterize the landscape of tumor‐derived somatic mutations in BW fluids using next‐generation sequencing. We found that most missense mutations/indels and driver mutations in primary tumors were also detected in BW fluids. Interestingly, substantial concordance of driver mutations between BW fluids and primary tumors was found. Thus, our study suggests that detection of somatic mutations in BW fluids can be used for the diagnosis, especially in patients who are concerned with procedure‐related complications. In addition, BW supernatants can effectively be used in the practical application because it had the high AFs of the mutations. However, some limitations are present in this pilot study. First, we could not evaluate the quantitative relationships between the mutations and biological characteristics of primary tumors including spread through air spaces because of the small sample size. However, the AFs of missense mutations/indels in BW fluids significantly correlated with those in primary tumors and were affected by histology, EGFR mutation, and tumor size. Second, the diagnostic performance of the mutations was not presented because of a lack of control group. Therefore, future research is required to test the performance of BW supernatants for early‐stage NSCLC through artificial intelligence and clinical and radiological data‐driven machine‐learning mechanisms.
In conclusion, this study demonstrates that BW fluids, especially supernatants, can be reflective of tumoral mutations and can potentially be used as an ancillary diagnosis for early‐stage lung cancer.
Acknowledgments
This research was supported by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), which was funded by the Ministry of Health & Welfare (HI16C0286) and Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT and Future Planning (NRF‐2017R1E1A1A01074863).
Contributed equally.
Disclosures
The authors indicated no financial relationships.
References
- 1.Cohen JD, Li L, Wang Y et al. Detection and localization of surgically resectable cancers with a multi‐analyte blood test. Science 2018;359:926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bettegowda C, Sausen M, Leary RJ et al. Detection of circulating tumor DNA in early‐ and late‐stage human malignancies. Sci Transl Med 2014;6:224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rolfo C, Mack PC, Scagliotti GV et al. IASLC statement paper: Liquid biopsy for advanced non‐small cell lung cancer (NSCLC). J Thorac Oncol 2018;13:1248–1268. [DOI] [PubMed] [Google Scholar]
- 4.Abbosh C, Birkbak NJ, Wilson GA et al. Phylogenetic ctDNA analysis depicts early‐stage lung cancer evolution. Nature 2017;545:446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solomon DA, Solliday NH, Gracey DR. Cytology in fiberoptic bronchoscopy. Comparison of bronchial brushing, washing and post‐bronchoscopy sputum. Chest 1974;65:616–619. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt B, Carstensen T, Engel E et al. Detection of cell‐free nucleic acids in bronchial lavage fluid supernatants from patients with lung cancer. Eur J Cancer 2004;40:452–460. [DOI] [PubMed] [Google Scholar]
- 7.Park S, Hur JY, Lee KY et al. Assessment of EGFR mutation status using cell‐free DNA from bronchoalveolar lavage fluid. Clin Chem Lab Med 2017;55:1489–1495. [DOI] [PubMed] [Google Scholar]
- 8.Seo JS, Ju YS, Lee WC et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res 2012;22:2109–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell JD, Alexandrov A, Kim J et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shepherd FA, Lacas B, Le Teuff G et al. Pooled analysis of the prognostic and predictive effects of TP53 comutation status combined with KRAS or EGFR mutation in early‐stage resected non‐small‐cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol 2017;35:2018–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]