Can an application powered by artificial intelligence construct patient profiles in context and suggest evidence‐based treatment options? The Oncology Expert Advisor was designed for such a purpose. Preliminary results of a study using this technological innovation are reported.
Keywords: Artificial intelligence application in medicine, Virtual expert advisor, Clinical decision support, Closing the cancer care gap, Democratization of evidence‐based care
Abstract
Background.
Rapid advances in science challenge the timely adoption of evidence‐based care in community settings. To bridge the gap between what is possible and what is practiced, we researched approaches to developing an artificial intelligence (AI) application that can provide real‐time patient‐specific decision support.
Materials and Methods.
The Oncology Expert Advisor (OEA) was designed to simulate peer‐to‐peer consultation with three core functions: patient history summarization, treatment options recommendation, and management advisory. Machine‐learning algorithms were trained to construct a dynamic summary of patients cancer history and to suggest approved therapy or investigative trial options. All patient data used were retrospectively accrued. Ground truth was established for approximately 1,000 unique patients. The full Medline database of more than 23 million published abstracts was used as the literature corpus.
Results.
OEA's accuracies of searching disparate sources within electronic medical records to extract complex clinical concepts from unstructured text documents varied, with F1 scores of 90%–96% for non‐time‐dependent concepts (e.g., diagnosis) and F1 scores of 63%–65% for time‐dependent concepts (e.g., therapy history timeline). Based on constructed patient profiles, OEA suggests approved therapy options linked to supporting evidence (99.9% recall; 88% precision), and screens for eligible clinical trials on ClinicalTrials.gov (97.9% recall; 96.9% precision).
Conclusion.
Our results demonstrated technical feasibility of an AI‐powered application to construct longitudinal patient profiles in context and to suggest evidence‐based treatment and trial options. Our experience highlighted the necessity of collaboration across clinical and AI domains, and the requirement of clinical expertise throughout the process, from design to training to testing.
Implications for Practice.
Artificial intelligence (AI)‐powered digital advisors such as the Oncology Expert Advisor have the potential to augment the capacity and update the knowledge base of practicing oncologists. By constructing dynamic patient profiles from disparate data sources and organizing and vetting vast literature for relevance to a specific patient, such AI applications could empower oncologists to consider all therapy options based on the latest scientific evidence for their patients, and help them spend less time on information “hunting and gathering” and more time with the patients. However, realization of this will require not only AI technology maturation but also active participation and leadership by clincial experts.
Introduction
The exponential increase in cancer knowledge, coupled with the speed of advances, is creating a knowledge gap for practicing oncologists [1]. There is ever more to know about each patient and more to incorporate from the literature in providing evidence‐based cancer care. It has become humanly impossible to stay abreast of peer‐reviewed literature, much less assimilate it at the point of care [2]. This contributes to adoption delays, leading to a widening gap between what is possible at academic research centers and what is practiced in real‐world settings [3]. Consequently, practicing oncologists need new tools to help close this knowledge gap and support adoption of new therapies in an evidence‐based manner so that more patients can benefit from societal investment in research and development [4], [5].
Artificial intelligence (AI) was first introduced in the early 1950s [6], [7] with the goal of replicating the human mind—that is, to perform tasks such as recognition, interpretation, reasoning, and conversing, with the acuity and influence typically attributed to humans. Machine intelligence (a more focused area of AI sometimes also referred to as augmented intelligence) aims to augment human capabilities for which the human mind tends to reach its limits. It excels in areas that humans are generally not very good at, such as assimilating massive quantities of qualitative information to recognize patterns of relevant information [8], [9], [10]. Most famously known for winning against human experts in knowledge and strategy games such as chess [11], Jeopardy! [12], and Go [13], AI is now entering medicine. For example, image recognition, one class of AI, has been applied successfully to imaging‐based clinical diagnoses such as detection of melanoma in dermoscopy [14] or retinopathy in diabetic patients [15]. In cancer, a challenge that is ripe for AI is the augmentation of the human capacity to understand and consistently apply increasingly sophisticated knowledge for clinical decision‐making, and to incorporate increasingly diverse and complex patient data for personalization of care.
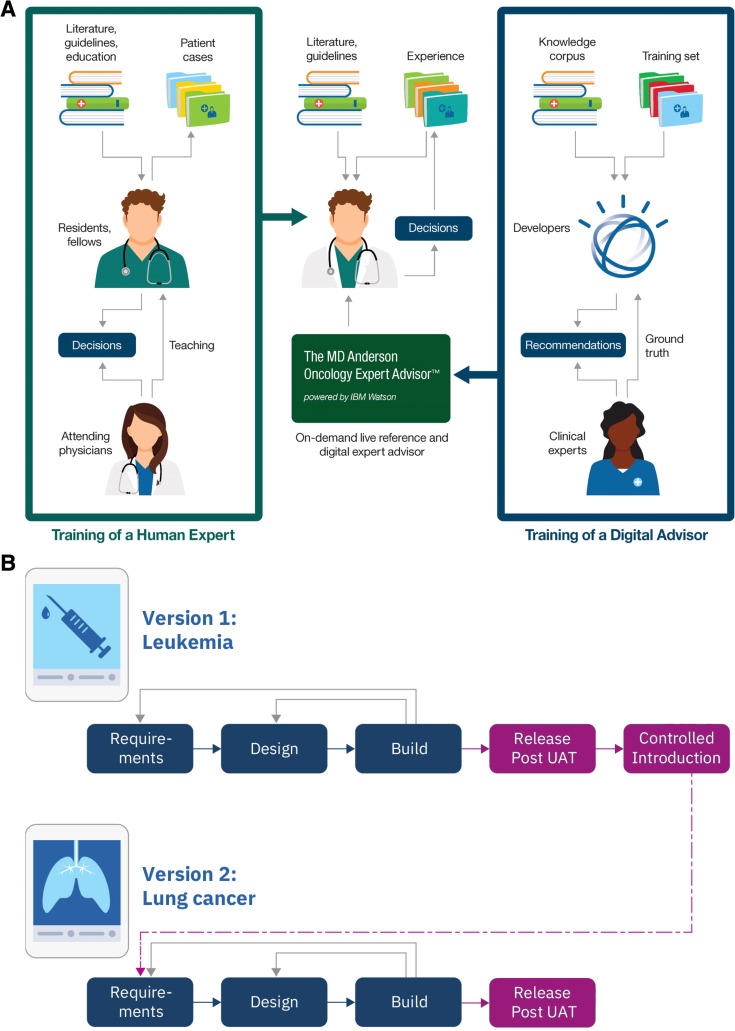
We envisioned an AI‐powered application to augment the knowledge base of practicing oncologists by organizing and vetting the vast literature for relevance to specific patients in real‐time [5], thereby empowering oncologists to consider therapy options based on the latest science for their patients (Fig. 1A). To this end, a multidisciplinary team embarked on an innovation effort to explore approaches to and practicality of developing such an AI application for knowledge democratization.
Figure 1.
Design and development of the Oncology Expert Advisor (OEA). (A): Diagram of the training and intended uses of OEA in relation to the training and responsibilities of a practicing oncologist. (B): Flow chart of the agile design cycle. The design cycles are rapid and iterative to derive continual improvement from lessons learned.
Abbreviation: UAT, user acceptance test.
Materials and Methods
All patient data used in this study were accessed through MD Anderson's electronic medical record system. Patient consent was not required per Institutional Review Board review. For details on training data and knowledge corpus, ground truth generation, and learning module training, as well as methodology for calculating performance accuracy, see supplemental online data.
Results
Conceptualization and Design
As an alternative to referral, practicing oncologists may seek informal advice from colleagues who are recognized experts in an area. This typically happens in an ad hoc fashion, highly dependent on an individual physician's access to and availability of such experts [16]. Here, we conceived an AI application, Oncology Expert Advisor (OEA), that provides practicing oncologists instant access to guidelines and literature, evidence‐based and patient‐specific treatment or clinical trial suggestions, and management advice at points of care, as if they had on‐demand access to the experts (Fig. 1A).
Simulating a consultative exchange, we designed three core functions in OEA: dynamic patient summarization, treatment options recommendation, and management advisory (supplemental online Fig. S1). The Dynamic Patient Summarization module would “read” a patient's medical record and automatically extract relevant attributes pertinent to clinical decisions from both structured (e.g., demographic information or laboratory test results) and unstructured (e.g., transcribed consultation notes or pathology reports) data. This necessitated that OEA be integrated with electronic health records (EHRs) to receive continually refreshed clinical information, rather than relying on manual input of defined parameters. The Treatment Options Recommendation module would search OEA's knowledge corpus to surface both approved and investigational treatment options deemed appropriate for the specific patient with linking of each suggestion to supporting evidence. To maintain currency, OEA's knowledge corpus must therefore be continuously updated with the latest guidelines and literature. Lastly, because treatment choice is only one of many decisions, the Management Advisory module would capture specialists' best practice as advisory for managing patients on a particular therapy.
Dynamic Patient Summarization
In consulting a peer about a patient, physicians communicate with a core patient profile. Such a patient profile typically requires manual searches and synthesis, as it is derived from a composite of structured and unstructured information from multiple sources within the medical record system. Therefore, a foundational capability of OEA is to automatically locate, extract, and analyze patient records to compile such a profile. In addition to demographic information, nine concepts across two categories were identified by clinical experts as key: (a) disease state related: tumor histology, margins of resection, stage at diagnosis, metastasis history, metastatic sites (for solid tumors) or cytogenetics and blast counts (for liquid tumors), and molecular profile; (b) therapy history related: therapy components (chemotherapy, radiotherapy, and surgery), timeline, and response.
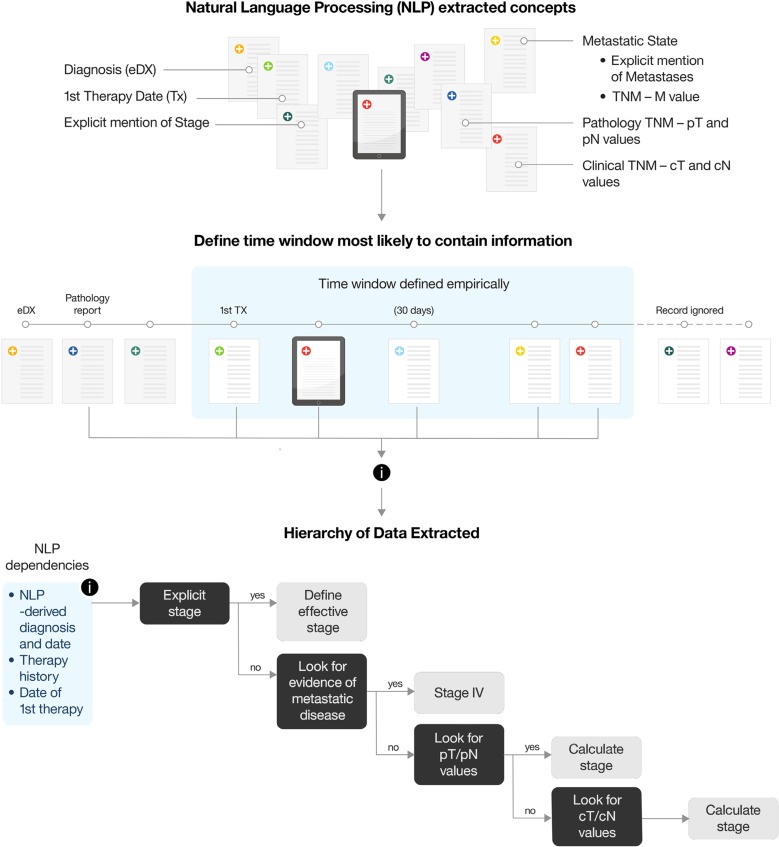
To train learning algorithms to extract each of these concepts from patient records, we had to first identify the best source document or combination of documents for each and then learn their relative weights and underlying relationships. Although intuitive to an experienced clinician, these concepts are built on complex logic layers, as illustrated with the “Stage at Diagnosis” example (Fig. 2). Moreover, although concepts may be common across cancer types, the underlying logic can be cancer type specific. For example, whereas tumor size is monitored to measure response to treatment in lung cancer, percentage of leukemia blast count and peripheral blood count recovery are used to track response in leukemia.
Figure 2.
Determining stage at diagnosis. For effective calculation of Tumor Stage at Diagnosis, several parameters have to be taken into consideration. First, all the relevant Natural Language Processing (NLP) concepts have to be defined in collaboration with subject matter experts (SMEs); each one is measured individually for accuracy. Second, an appropriate window of time in which the information is most likely to appear must be defined, first informed by SMEs and then adjusted empirically as accuracy measurements become available. Third, a layer of medical logic is applied on top of the NLP‐derived attributes, defining the hierarchy of the attributes and the rules on how to best make use of them to effectively derive stage at diagnosis. Tumor Stage at Diagnosis as a concept is then compared against a manually curated Ground Truth, and each one of the components that make up this concept are then adjusted and improved to achieve better accuracy.
Once the logic of a concept was developed, algorithms were coded and trained to interpret clinical documents and determine their relevance within the context of a specific patient. This process turned out to be much more challenging than interpreting published literature, because documentation in medical records often contains private acronyms, sentence fragments, and grammatical errors with unintended or ambiguous meanings [17]. Recognizing that this is the nature of unstructured writings, OEA maintained the links to the original content from where inference of a key concept was made, enabling users to verify OEA's interpretations. This feature could facilitate iterative learning of the algorithms as OEA reads more patient records.
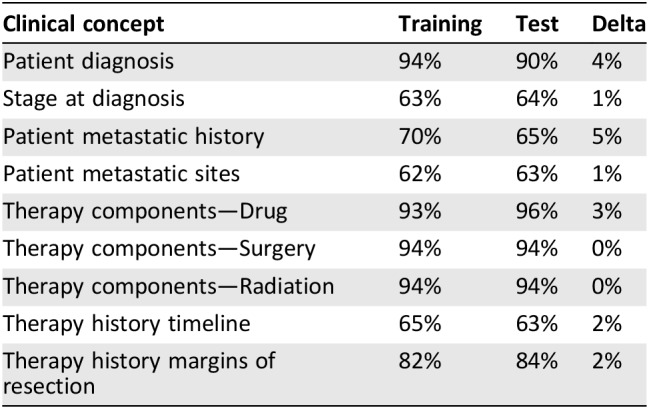
The recall and precision accuracy of the learning models for these nine key clinical concepts improved with iterative training, achieving F1 scores between 60% and 90% (Table 1; supplemental online data). When subsequently tested against a validation sample set, they maintained an F1 score ± 5% of the training value, indicating that the models were relatively stable. Overall, the algorithms were better at correctly extracting non‐time‐dependent clinical concepts (e.g., list of therapies received) than time‐dependent ones (e.g., therapy history timeline). A combination of linguistic and clinical variables contributed to the difference in performance; in particular, the lack of explicit time references or ambiguous or imprecise reporting of events impacted on drawing inferences of chronology, making accurate timeline determination difficult.
Table 1. Assessment by F1 score of natural language processing performance on clinical concepts.
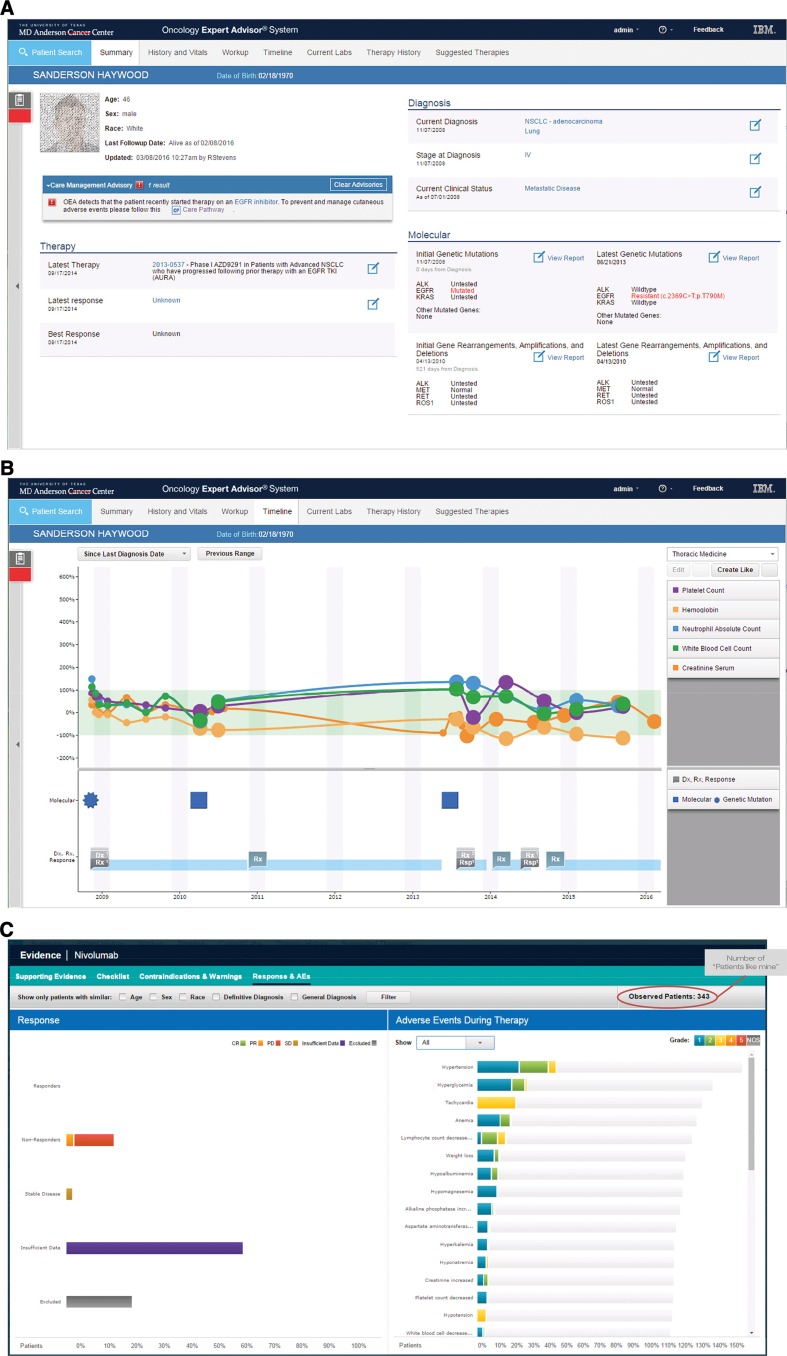
In addition to inferring complex concepts by synthesizing data and interpreting documentation from disparate sources, OEA also organizes and presents this information in an intuitive longitudinal view (Fig. 3A). For example, structured data such as lab results, mutations, and beginning and ending of treatment can be plotted alongside concepts extracted from unstructured documents such as diagnoses, disease progression, or toxicities (Fig. 3B). This creates a harmonized view of patient status and history.
Figure 3.
Demonstration of the Oncology Expert Advisor (OEA) Graphical User Interface. Screenshots from a contrived patient record. (A): Patient summary screen displaying current clinical features and care alerts. Patient information is fictitious and for illustration purposes only. (B): Timeline view of laboratory test results aligned with treatment history and response. (C): Patients like mine population segmentation tool for exploring clinical responses of real patients with similar demographic and diagnostic features.
Approved Therapy Options
A key advantage of consulting an expert compared with researching literature is getting advice that is tailored to specific patient. To simulate this, a core capability of OEA was to vet all treatment options in its knowledge base to surface only those relevant to a patient. Here, to ensure transparency on how OEA arrived at specific suggestions, each option was linked to published literature or a consensus guideline that could be reviewed in real‐time within OEA, facilitating the exercise of the user's own judgment.
In the first OEA solution, which targeted leukemia (inclusive of myelodysplastic syndrome, acute myelogenous leukemia, and acute lymphocytic leukemia), historical cases of patients treated at MD Anderson's Leukemia Center during the prior 2 years were used for training and testing. Outcome (e.g., patient survival) was not used to determine the preferred or correct answer (ground truth), because overall survival is multifactorial and dictated by many variables in addition to treatment choice. Instead, the ground truth for purpose of algorithm training was the actual therapy prescribed by MD Anderson oncologists. Although this approach provided ready “ground truth” for algorithm training, the assumption of only one correct treatment option for each case did not reflect the reality of cancer medicine. Another limitation was the fact that medical advances could render a prior decision suboptimal. Learning from this, we modified the analytic approach to treatment option recommendation in the second OEA solution (which targeted lung cancer). There, we trained a model that expects multiple options being appropriate, using ground truth that was expert curated for each historical case based on current‐day knowledge.
To generate such ground truth, clinical experts first delineated 68 clinical cohorts based on histological, molecular, and other clinical features (supplemental online Table S1). These cohorts spanned non‐small cell lung cancer and small cell lung cancer of all histopathologies, all stages, first diagnosis or recurrent, sensitizing mutations including epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase, and all age brackets. For each cohort, all evidence‐based treatment options were defined. Of note, although programmed death‐ligand 1 (PD‐L1) expression defined a new treatment cohort once anti‐PD‐L1 was approved as a standard of care option by the U.S. Food and Drug Administration (FDA) [18], the lack of PD‐L1 treatment data among retrospective patient cases precluded it from being included, illustrating the limitation of training a model with historical data sets.
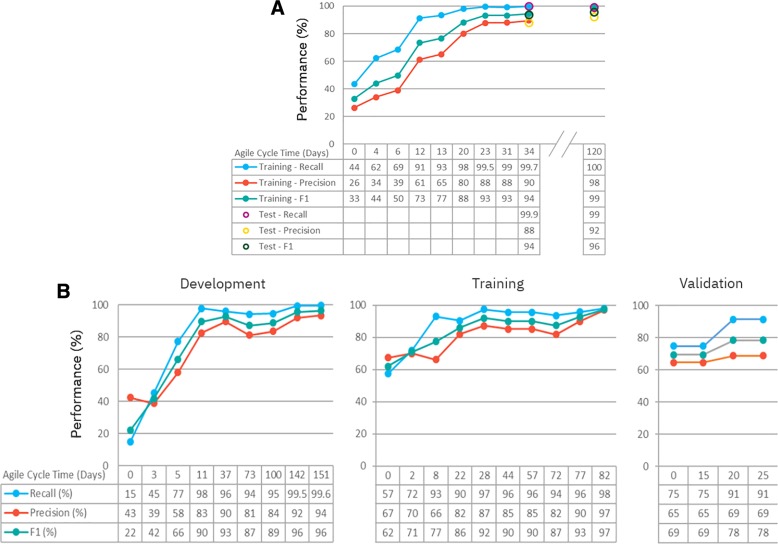
Once ground truth was generated, a retrospective query was performed to capture each encounter in which a new treatment was prescribed for a lung cancer patient during the prior 2‐year period. This resulted in 848 unique cases, each representing a patient profile at a point in time. Of these 848 cases, 585 were randomly selected for model development, 175 for training, and 88 for validation testing. Briefly, a learning model to predict appropriate therapy options was first developed using the 585 cases, then trained iteratively with the 175 training cases. When accuracy improvement plateaued after 10 iterations (Fig. 4A; starting recall of 43.5% and precision of 26.3% to ending recall of >99% and precision of 89.6%), the learning model was then subjected to validation testing in the nonoverlapping 88 cases, achieving a recall 99.9% and precision 88%. It is worth noting that performance of this learning model was not uniform across patient contexts. The model performed better on treatment‐naïve patients (precision from 97.2% to 100%) than on patients having prior therapy (precision from 66.9% to 89.0%). This was due in large part to the challenge of inferring time‐dependent clinical concepts (e.g., distinguishing between adjuvant and systemic use based on proximity to a prior surgical intervention; supplemental online Fig. S2). On the other hand, when new approved therapy options were added (i.e., immunotherapy), the model was retested to show that it maintained its performance (Fig. 4A, recall of 99.1% and precision of 92.2%), suggesting a stable model that can accommodate addition or changes to the approved therapies list.
Figure 4.
Training and evaluation of artificial intelligence modules. (A): Performance of the Oncology Expert Advisor lung approved treatment recommendation model was optimized through a series of design cycles using the training set (n = 175) and then validated on the test set (n = 88). (B): The clinical trial screening model was first run through a series of development iterations using the clinical trials actively recruiting at MD Anderson (n = 16). It was then further trained on an expanded set of trials from ClinicalTrials.gov (n = 165). Finally, the clinical trial screening model was validated on a new set of trials from ClinicalTrials.gov that included non‐lung cancer trials (n = 53).
Investigational Therapy Options
The initial algorithm developed in the leukemia solution was trained to evaluate all clinical trial inclusion and exclusion criteria when determining eligibility. This “matching” strategy proved too stringent as it eliminated many options that clinicians would wish to consider, particularly when the exclusion criteria were modifiable or vague. Learning from the leukemia experience, we modified the strategy in the lung cancer solution, from matching to screening out trials for which the patient was clearly ineligible. In brief, the algorithm screened out options based on a subset of criteria that are considered nonmodifiable, such as diagnosis, histology, mutations, staging, age, and prior therapy, while ignoring modifiable attributes, such as red cell count or renal function status. This approach balances the need to identify as many high‐probability options as appropriate to maximize the chance of qualifying, without overwhelming the care team with too many options that require manual screening.
The investigation trial screening model in Lung Cancer OEA was first built using MD Anderson's internal lung cancer trial protocols (n = 16) curated for eligibility against 3,438 patient/point‐in‐time combinations identified from retrospective query of MD Anderson lung cancer patients over a 5‐year period (Fig 4B, left). A true positive was any patient/point‐in‐time combination that matched the subset of protocol inclusion/exclusion criteria being evaluated. Recognizing that only a small proportion of patients can travel to MD Anderson for clinical trials, and the number of internally open trials was too small as a robust training set, we opted to train this model against a set of 165 lung cancer trials on ClinicalTrials.gov, achieving a recall of 97.9% and precision of 96.9% (Fig. 4B, center). Next, we tested this model on a nonoverlapping set of 53 trials that included non‐lung cancer trials from ClinicalTrials.gov. Not surprisingly, we observed a deterioration of performance (e.g., recall of 74.8% and a precision of 64.6%; Fig. 4B, right), requiring two cycles of iterative training to bring its accuracy back to 91.4% precision and 68.7% recall. This was due to inclusion of clinical trials with data elements that the model was never trained on (not contained in the training set), illustrating the importance of using a training set that is representative of the intended cohort.
Management Advisory
Patient outcomes depend not only on treatment choices but also proper management of therapy [19]. Thus, one of OEA's core functions is to facilitate sharing best practices in the form of expert advisory. Recognizing that much of this aspect of clinical practice is empirical or anecdotal with long time‐lags to publication or consensus guideline, the advisory function was primarily rule based. We first prioritized advisory for adverse events associated with newer classes of therapy (i.e., targeted therapy and immunotherapy) for incorporation into OEA. For agents associated with severe or difficult‐to‐manage adverse reactions, a more complete set of care pathways was defined (supplemental online Table 2) to reflect best practices at MD Anderson. For example, when EGFR inhibitor treatment is suggested, a management framework based on best practice at MD Anderson is surfaced (supplemental online Fig. S3).
Additionally, for each therapy option in OEA's knowledge corpus, known adverse events and contraindications as reported on the FDA label are summarized and viewable by users with a single click. OEA also inferred common adverse events according to Common Terminology Criteria for Adverse Events v4 criteria (Fig. 3C) based on structured information in the medical records. For example, a leukemic patient could be flagged as potentially at risk for tumor lysis syndrome based on laboratory test results (supplemental online Fig. S4). When combined with “patients like mine” analytics using attributes such as age, gender, and ethnicity (Fig. 3C), OEA would enable an oncologist to consider the toxicity profile of a therapy in a particular patient population so that care can be personalized in real‐world practice.
Controlled Introduction
Given the complexity and nature of medicine, we anticipated that there would be accuracy, usability, and utility requirements of an AI application that would only become apparent in actual clinical uses. In other words, an AI‐powered application like OEA requires learning beyond algorithm development and training as described above, not dissimilar to training requirement of physicians after medical school (e.g., residency and fellowship) before independent practice. To model this, we conceptualized Controlled Introduction (CI) as an additional phase in the development cycle for an AI application intended for clinical uses (Fig. 1B), to be conducted, iteratively, in controlled but real‐life clinical environment by clinical users.
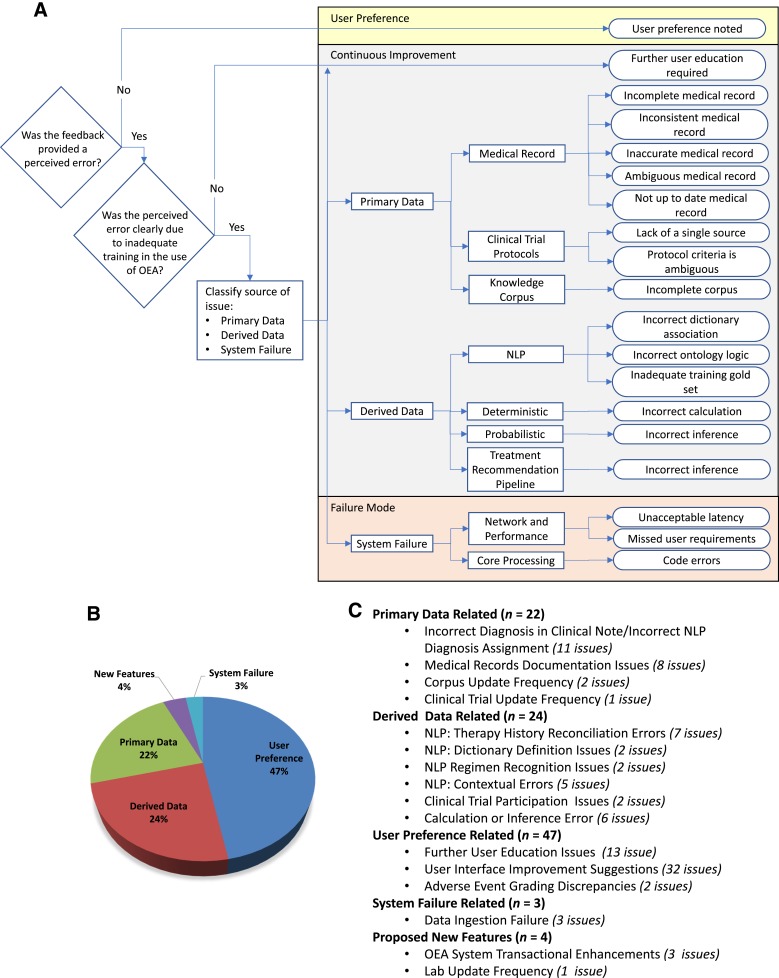
CI begins with functional verification, which is intended to evaluate readiness of the environment for deployment by verifying that the application functions as designed and performs as expected in an actual clinical environment using predefined test scenarios to simulate clinical uses. Then, CI enters clinical testing in which the performance, usability, and relevance of OEA was evaluated by “naïve” users (i.e., those not involved as subject matter experts during development) in real‐life patient cases. In the CI for Leukemia OEA, a total of 49 leukemia clinic team members—comprising 7 faculty, 6 fellows, 14 clinical registered nurses (RNs), 7 physician assistants, 8 research RNs, and 6 patient access staff—participated in evaluating its performance in 352 active patient cases with a diagnosis of acute myeloid leukemia, acute lymphoblastic leukemia, or myelodysplastic syndrome. Clinical users were asked to evaluate OEA and provided feedback on its accuracy and usability, in parallel of routine care of their patients. To capture their feedback, a reporting feature was built within OEA so that evaluators were able to submit comments in real‐time (lowering barriers to feedback). In total, 100 unique observations, issues, or comments were logged during CI. After reproducing each observation or issue, root‐cause analysis was performed to categorize the likely cause (Fig. 5A), evaluate criticality, and identify opportunities for improvement from a people, process, and/or technology perspective. As shown in Figure 5B, more than 85% of issues were related to accuracy, clarity, and/or completeness of the primary medical record (22%) or derived data (24%). Remaining issues included comments reflecting user preference (47%), system feature enhancements (4%), or system failure (3%).
Figure 5.
Root cause analyses of issues identified during controlled introduction. (A): Flow chart of the process for logging errors identified during controlled introduction. (B): Pie chart of the 100 unique, reproducible errors logged during controlled introduction grouped into five root causes: primary data, derived data, user‐preference (primarily relating to user interface/user experience), system performance, and new function features. (C): Detailed summation of subcategories of root causes.
Abbreviations: NLP, Natural Language Processing; OEA, Oncology Expert Advisor.
Additional qualitative evaluation was collected via questionnaires. More than 70% of the responders felt that the Patient Summarization features would be helpful and could save time for general and specialty oncology practices alike, whereas approved therapy option suggestions and the management advisory features would be more useful for general oncology practice (64% and 78%, respectively). Unexpectedly, in more than half of the cases (52%), experts felt that the stringent clinical trial matching feature was eliminating trial options that they would like to consider, leading to new design in the lung OEA solution (as described above). In summary, CI identified issues and revealed context‐specific insights that informed the refinement and improvement of OEA.
Discussion
We showed that an AI application can be developed to (a) locate, search, and extract complex clinical concepts from disparate sources of structured data and unstructured text documents within the medical records, (b) create an integrated patient profile, and (c) organize a chronological view of multidimensional clinical data of the patient. Such summarization is valuable, as it could save time for providers and potentially improve quality of care by automating search, aggregation, and summarization of disparate information. Further, unlike searches and queries using a limited number of parameters, an AI application can be trained to (a) surface treatment options and (b) screen for eligible trials that are tailored to a summarized patient profile, as well as (c) extract data to infer occurrence of adverse events with (d) patients‐like‐mine segmentation analysis. Taken together, an OEA‐like AI application will contribute to a solution that addresses the knowledge gap in oncology practice.
Development of AI for medicine depends on collaboration across industry and academia because it requires expertise from both the clinical and the technical domains. However, it is important that such collaboration be defined as clinical leading, rather than technical leading, as the technical and clinical perspectives might not always agree. In the OEA project, governance was structured such that the MD Anderson clinical team was the lead with IBM's technical team as collaborator. Therefore, any trade‐off between technical and clinical requirements was decided by the clinical team. An example was choosing to use actual patient cases rather than simulated cases for OEA training, even though simulation made large, statistically robust training sets possible, a clear analytic advantage. Furthermore, to assure objectivity in evaluating OEA performance during Controlled Introduction, we engaged a third party independent of both the technical or clinical teams to conduct the technical verification and perform root cause analyses of observations by clinical users (Fig. 5), so that we can be confident of the veracity of OEA's performance within MD Anderson.
However, the major limitation of this study relates to the fact that OEA was not tested outside of MD Anderson Cancer Center; therefore, OEA's performance could not be generalized. We would expect that some functionalities likely require retraining. For example, it is well recognized that significant institution‐specific differences exist in documentation and data source organization. Thus, OEA's dynamic patient summarization module optimized for MD Anderson's system would likely require retraining to adapt to other institutions EHR systems, similar to the example of clinical trial matching (Fig. 4B). On the other hand, modules for recommending approved therapy options or matching to clinical trials based on a constructed summary of a patient's cancer history would be expected to maintain performance across sites. Taking OEA outside of MD Anderson's firewall for testing in another clinical institution was challenging, not only because of a lack of health information technology network infrastructure connecting across institutions but also because of nontechnical barriers, such as investment and opportunity cost, concern over data sharing, and cultural resistance to changes [20].
Development, integration, and implementation of AI are costly, not only because the technology is still maturing and is not yet an off‐the‐shelf commodity but also because it includes opportunity cost for the clinical experts. Time spent training an AI system is time the clinical experts are not applying themselves to either patient care or more traditional forms of research. However, domain expertise is critical to the development of a useful AI application. Therefore, recognizing and supporting clinical experts in such efforts is essential. This is a paradigm change, not dissimilar to the challenge of evolving the academic reward system to acknowledge the value of team science [21]. Indeed, one should equate training of an AI system to training students, residents, and fellows. Additionally, implementing a technology that could disrupt well‐established workflows is challenging for any organization, not to mention the learning and behavioral adaptation required with regard to quality and safety monitoring, security, and compliance. This is particularly difficult for organizations that are still working on or recovering from EHR implementation. Furthermore, a new kind of sustainability model is needed for a health care organization to invest in a digital advisor application for the purpose of improving access and quality of care for patients not being treated by them, and for a network of providers to connect and share their patients' information in order to source and benefit from the collective expertise of the broader oncology community. Such a model will require major realignment of incentives for the organizations that develop and train these advisor applications, for the oncologists who use them to provide evidence‐based care, for the pharmaceutical industry that develops these therapy options, and for the risk‐bearing entities that reimburse these services.
Finally, the application of AI in medicine requires overcoming the natural resistance to change. Historically, the adoption of new technologies in medicine is slow even when there is compelling evidence of clinical benefits or utility [23]. This is in part due to the Hippocratic Oath of “Do No Harm”; many doctors in medicine are reticent of new variables being introduced in their patient care practice [24], particularly when it comes to AI that is being portrayed as a super‐intelligent black box that “knows better.” Therefore, an absolute requirement of AI in medicine is transparency. In OEA, this is through linking every suggestion to supporting literature, so that the human users can accept or reject with their own judgment. Another requirement for adoption is clinical utility, not just technical performance. Therefore, we believe that clinical experts should take the drivers seat in design and development of AI applications for medicine.
Conclusion
It is often said that AI will transform medicine. Indeed, with examples like OEA, we can envision how AI could enable equitable access to the best quality of care no matter where or who the patients are [22]. However, to realize that potential, it will take more than successful design and development of an application like OEA. It will require a new generation of clinical and AI experts who are cross‐trained. It will require us to look beyond AI to consider their integration into a complex system that is needed to deliver care and benefit patients.
See http://www.TheOncologist.com for supplemental material available online.
Acknowledgments
Connectivity to primary medical record data sources in MD Anderson's in‐house EHR system (ClinicStation) was provided by an institutional IT team under deputy Chief Information Officer Keith Perry. We extend special thanks to the Leukemia Center faculty, fellows, and staff under the leadership of Dr. Hagop Kantarjian, chair, for participation in Controlled Introduction. Controlled Introduction execution and root‐cause analyses, as well as overall project management, were provided by PricewaterhouseCooper Healthcare Advisory under Mark A. Mynhier, partner. Editorial assistance (e.g., preparing references and assembling figures and tables) for this manuscript was provided by Jessamine P. Winer‐Jones of IBM Watson Health, supported by IBM Watson Health. This project was supported in part by philanthropic grants to the Department of Genomic Medicine from The Jynwel Charitable Foundation (Hong Kong), The Bosage Family Foundation, and The Ciocca Charitable Foundation, as well as funding to the Lung Cancer Project from the Moonshot Program at MD Anderson Cancer Center.
Author Contributions
Conception/design: Stephen Swisher, Rob High, P. Andrew Futreal, John Heymach, Lynda Chin
Provision of study material or patients: George Simon, Courtney D. DiNardo, Koichi Takahashi, Tina Cascone, Cynthia Powers, Mara B. Antonoff, Daniel Gomez, Quynh Nguyen, Emily Roarty, Sherry Pierce, Jianjun Zhang, Emily Hardeman Barnhill, Kate Lakhani, Kenna Shaw, Brett Smith, Stephen Swisher, John Heymach
Collection and/or assembly of data: Cynthia Powers, Rick Stevens, Joshua Allen, Pat Keane, Fernando Suarez Saiz, Emily Roarty, Sherry Pierce, Emily Hardeman Barnhill, Kate Lakhani, Brett Smith
Data analysis and interpretation: George Simon, Courtney D. DiNardo, Koichi Takahashi, Tina Cascone, Rick Stevens, Joshua Allen, Mara B. Antonoff, Daniel Gomez, Pat Keane, Fernando Suarez Saiz, Quynh Nguyen, Sherry Pierce, Jianjun Zhang, Kenna Shaw, Stephen Swisher, Rob High, P. Andrew Futreal, John Heymach, Lynda Chin
Manuscript writing: Rick Stevens, Fernando Suarez Saiz, Rob High, P. Andrew Futreal, John Heymach, Lynda Chin
Final approval of manuscript: George Simon, Courtney D. DiNardo, Koichi Takahashi, Tina Cascone, Cynthia Powers, Rick Stevens, Joshua Allen, Mara B. Antonoff, Daniel Gomez, Pat Keane, Fernando Suarez Saiz, Quynh Nguyen, Emily Roarty, Sherry Pierce, Jianjun Zhang, Emily Hardeman Barnhill, Kate Lakhani, Kenna Shaw, Brett Smith, Stephen Swisher, Rob High, P. Andrew Futreal, John Heymach, Lynda Chin
Disclosures
Rick Stevens: IBM (E, OI); Joshua Allen: IBM (E, OI); Pat Keane: IBM (E, OI); Fernando Suarez Saiz: IBM (E, OI); Rob High: IBM (E, OI). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Denu RA, Hampton JM, Currey A et al. Influence of patient, physician, and hospital characteristics on the receipt of guideline‐concordant care for inflammatory breast cancer. Cancer Epidemiol 2016;40:7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alper BS, Hand JA, Elliott SG et al. How much effort is needed to keep up with the literature relevant for primary care? J Med Libr Assoc 2004;92:429–437. [PMC free article] [PubMed] [Google Scholar]
- 3.American Society of Clinical Oncology . The state of cancer care in America, 2016: A report by the American Society of Clinical Oncology. J Oncol Pract 2016;12:339–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu P, Artz D, Warner J. Electronic health records (EHRs): Supporting ASCO's vision of cancer care. Am Soc Clin Oncol Educ Book 2014:225–231. [DOI] [PubMed] [Google Scholar]
- 5.Castaneda C, Nalley K, Mannion C et al. Clinical decision support systems for improving diagnostic accuracy and achieving precision medicine. J Clin Bioinforma 2015;5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moor J. The Dartmouth College artificial intelligence conference: The next fifty years. AI Magazine 2006;27:87. [Google Scholar]
- 7.Solomonoff RJ. The time scale of artificial intelligence: Reflections on social effects. Hum Syst Manage 1985;5:149–153. [Google Scholar]
- 8.Flouris AD, Duffy J. Applications of artificial intelligence systems in the analysis of epidemiological data. Eur J Epidemiol 2006;21:167–170. [DOI] [PubMed] [Google Scholar]
- 9.Spangler S, Wilkins AD, Bachman BJ et al. Automated hypothesis generation based on mining scientific literature. In: Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, 24–27 August 2014. New York, NY: ACM; 1877–1886.
- 10.Dayarian A, Romero R, Wang Z et al. Predicting protein phosphorylation from gene expression: Top methods from the IMPROVER Species Translation Challenge. Bioinformatics 2015;31:462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell M, Hoane AJ, Hsu FH. Deep blue. Artificial Intelligence 2002;134:57–83. [Google Scholar]
- 12.Ferrucci D, Levas A, Bagchi S et al. Watson: Beyond Jeopardy! Artificial Intelligence 2013;199:93–105. [Google Scholar]
- 13.Silver D, Huang A, Maddison CJ et al. Mastering the game of Go with deep neural networks and tree search. Nature 2016;529:484–489. [DOI] [PubMed] [Google Scholar]
- 14.Codella N, Cai J, Abedini M et al. Deep learning, sparse coding, and SVM for melanoma recognition in dermoscopy images In: Zhou L, Wang L, Wang Q. et al., eds. Machine Learning in Medical Imaging: 6th International Workshop. Cham, Switzerland: Springer International Publishing, 2015: 118–126. [Google Scholar]
- 15.Gulshan V, Peng L, Coram M et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016;316:2402–2410. [DOI] [PubMed] [Google Scholar]
- 16.Gray SW, Hicks‐Courant K, Cronin A et al. Physicians' attitudes about multiplex tumor genomic testing. J Clin Oncol 2014;32:1317–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feder SL. Data quality in electronic health records research: Quality domains and assessment methods. West J Nurs Res 2018;40:753–766. [DOI] [PubMed] [Google Scholar]
- 18.Chen L, Han X. Anti‐PD‐1/PD‐L1 therapy of human cancer: Past, present, and future. J Clin Invest 2015;125:3384–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segaert S, Van Cutsem E. Clinical signs, pathophysiology and management of skin toxicity during therapy with epidermal growth factor receptor inhibitors. Ann Oncol 2005;16:1425–1433. [DOI] [PubMed] [Google Scholar]
- 20.Poon EG, Jha AK, Christino M et al. Assessing the level of healthcare information technology adoption in the United States: A snapshot. BMC Med Inform Decis Mak 2006;6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Research Council . Enhancing the effectiveness of team science. Washington, DC: National Academies Press, 2015. [PubMed] [Google Scholar]
- 22.Krittanawong C, Zhang H, Wang Z et al. Artificial intelligence in precision cardiovascular medicine. J Am Coll Cardiol 2017;69:2657–2664. [DOI] [PubMed] [Google Scholar]
- 23.Rittenhouse DR, Ramsay PP, Casalino LP et al. Increased health information technology adoption and use among small primary care physician practices over time: A national cohort study. Ann Fam Med 2017;15:56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.24.McGinn T. Putting meaning into meaningful use: A roadmap to successful integration of evidence at the point of care. JMIR Med Inform 2016;4:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]