Cdc42 enhances the uptake of extracellular vesicles by macrophages during the TLR9-activated immune response.
Abstract
Endosomal Toll-like receptors (TLRs) mediate intracellular innate immunity via the recognition of DNA and RNA sequences. Recent work has reported a role for extracellular vesicles (EVs), known to transfer various nucleic acids, in uptake of TLR-activating molecules, raising speculation about possible roles of EVs in innate immune surveillance. Whether EV-mediated uptake is a general mechanism, however, was unresolved; and the molecular machinery that might be involved was unknown. We show that, when macrophages are stimulated with the TLR9 agonist CpG oligodeoxynucleotides (ODN), the secreted EVs transport ODN into naïve macrophages and induce the release of chemokine TNF-α. In addition, these EVs transfer Cdc42 into recipient cells, resulting in further enhancement of their cellular uptake. Transport of ODN and Cdc42 from TLR9-activated macrophages to naïve cells via EVs exerts synergetic effects in propagation of the intracellular immune response, suggesting a general mechanism of EV-mediated uptake of pathogen-associated molecular patterns.
INTRODUCTION
Intracellular innate immunity has attracted intensive research attention, especially with respect to key mediators such as endosomal Toll-like receptors (TLRs), cyclic guanosine 5′-monophosphate–adenosine 5′-monophosphate synthase, and stimulator of interferon genes in the cytosolic innate immune sensing DNA and RNA from pathogens (1, 2). Human TLRs are an important family of innate immune receptors located either on the cell surface (TLR1, TLR2, TLR4, TLR5, TLR6, and TLR10) or inside cells within the endosomal compartment (TLR3, TLR7, TLR8, and TLR9), which are capable of recognizing pathogen-associated molecular patterns (PAMPs) and danger-associated molecular patterns (DAMPs) (3–5). Intriguingly, a mechanism involving extracellular vesicles (EVs) derived from bacteria has been recently found in the induction of innate immunity activation by facilitating the uptake of lipopolysaccharide (LPS), a classic PAMP for TLR4 on the cellular surfaces (6, 7). With the assistance of EVs, LPS penetrates the cellular membranes and activates various inflammatory caspases in the cytosol (7). Nonetheless, little is known about whether other PAMPs/DAMPs also use similar EV vehicles or what molecular machinery is involved.
EVs play pivotal roles in cell-cell communication during different pathological and physiological processes (8). The passage of EVs between the donor and recipient cells is an important means of transfer for molecules, including proteins, nucleic acids, and lipids, and these specific molecules could, in turn, have an impact on EV cellular uptake (9, 10). A recent study showed that the metastatic potency of cancerous, EV-donating cells could determine the uptake efficiency of the EV-receiving cells (11). This was evidenced by the findings showing that EVs isolated from metastatic site-derived PC-3 cells were more efficiently internalized into the recipient cells, compared with EVs from primary malignant cells (11). EVs secreted from immune cells showed immune-modulatory capacity (12–14). In immune responses, EVs secreted from macrophages upon infection of bacterial antigens induced the activation of dendritic cells and T cells both in vitro and in vivo (14).
Although EVs have been implicated in mediating immune responses, the role of EVs in the cellular internalization of different PAMPs and the activation of intracellular innate immunity warrants further investigation. In this study, we investigated whether EVs transported PAMP/DAMP molecules that affect cell functions upon reception of these EVs. Our findings showed that a TLR9 ligand, CpG oligodeoxynucleotides (ODN), was transported by EVs from infected macrophages into naïve macrophages, suggesting roles for EVs in the mediation of both inter- and intracellular innate immunity with oligo DNA PAMPs as cargos. At the same time, enhanced cellular uptake of EVs from ODN-infected macrophages was observed, compared with EVs from noninfected cells. Moreover, mechanistically, we showed that EVs transported Cdc42, a protein responsible for increased cellular endocytic activity into cells. Particularly, ODN carried by EVs induced the release of tumor necrosis factor–α (TNF-α), which further activated Cdc42 to promote the cellular uptake of EVs. Together, these results may point to a general mechanism whereby EVs facilitate the entry of different PAMPs/DAMPs into recipient cells. These findings not only shed light on the activation of innate immunity but also suggest a previously unidentified regulation strategy for these important biological pathways.
RESULTS
EVs transport ODN into endosomes to interact with TLR9
EVs have been shown to be capable of transporting nucleic acids such as DNA, messenger RNA (mRNA), and microRNA (miRNA) between cells (9, 15). Unmethylated CpG dinucleotide-containing DNA (i.e., ODN) derived from bacteria induces the activation of the endosomal receptor TLR9 (16). Therefore, we hypothesized that EVs may carry ODN from stimulated cells into nonstimulated ones and induce physiological changes in the recipient cells. To test our hypothesis, we added fluorescently labeled fluorescein isothiocyanate (FITC)–ODN into DsRed-marked and TLR9-overexpressing cells, and the progress of EVs release from these cells was monitored using live-cell confocal microscopy. We observed that most EVs released from DsRed-marked cells were labeled with red fluorescence. Some of the EVs were visualized with yellow fluorescence due to the colocalization of FITC-ODN and DsRed protein (Fig. 1A and movie S1).
Fig. 1. EVs transport ODN into recipient cells.
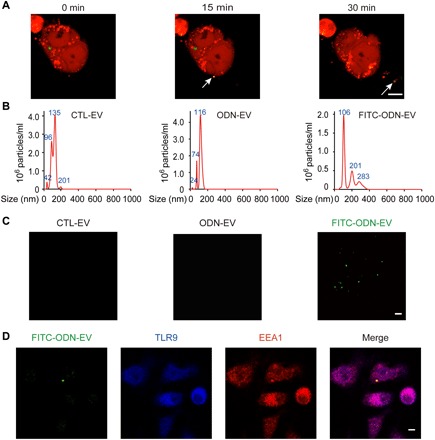
(A) EVs released from FITC-ODN–treated DsRed+ cells (marked by arrows). DsRed+ cells were treated with 0.5 μM FITC-ODN for 12 hours before real-time monitoring by confocal microscopy for 4 hours. Snapshot images taken at 0, 15, and 30 min were shown for the visualization of EV release. Scale bar, 5 μm. (B) Nanoparticle tracking analysis (NTA) displaying the size distribution of particles around the expected size of EVs isolated from the cell culture. EVs were diluted 50 times for NTA analysis. The images are representative of three independent experiments. (C) Immunofluorescence images of EVs isolated from untreated cells (CTL-EV), treated with 0.5 μM ODN (ODN-EV), or treated with 0.5 μM FITC-ODN (FITC-ODN-EV). After 12 hours of the treatment, EVs were isolated from the TLR9-overexpressing human embryonic kidney (HEK) 293 cell culture medium by ultracentrifugation. Then, the isolated EVs were mounted with a fluoromount aqueous mounting medium onto glass slides and observed by the Nikon N-SIM microscopy system. Scale bar, 1 μm. (D) Confocal images showing that FITC-ODN-EV entered DsRed+ cells and colocalized with TLR9 in the endosome. RAW 264.7 macrophage cells were incubated with 0.5 μM FITC-ODN for 12 hours. After the isolated EVs were then added into untreated cells, TLR9 and EEA1 were stained with their respective antibodies, followed by confocal microscopy analysis. Scale bar, 5 μm.
Next, we isolated EVs by the standard ultracentrifugation protocol (fig. S1A) and characterized them with a transmission electron microscope (TEM) and the Western blot. TEM analysis showed that the diameters of our isolated EVs were approximately 150 nm (fig. S1B). Typical protein markers for EVs such as CD63, CD9, and Hsp70 were identified in the Western blot analysis, confirming the identity of those isolated EVs (fig. S1C). The level of β-actin was negligible in EVs compared to that in cells; therefore, it was used as a negative control in our tests (17). Moreover, results from nanoparticle tracking analysis (NTA) using a scattering mode further confirmed the presence of particles around the expected size of EVs in the samples (Fig. 1B). We expected that EVs isolated from FITC-ODN–treated cells would be visible with green fluorescence because of their FITC-ODN cargo. Because of their ultramicroscopic sizes, we observed the isolated EVs using structured illumination microscopy (N-SIM), an established method for visualization of nanoparticles with diameters as small as approximately 100 nm. The results showed that these EVs were indeed visualized with fluorescence at green wavelengths, indicating that FITC-labeled ODN was carried by EVs (Fig. 1C). In contrast, we did not observe any fluorescence from EVs isolated from cells without treatment or treated with nonfluorescent ODN, ruling out the possibility of false positives from background or autofluorescence of EVs (Fig. 1C). In addition, results from high-sensitivity flow cytometry analysis of these EVs showed that FITC fluorescence intensity notably increased by ~20% in EVs derived from FITC-ODN–treated cells, consistent with our observation from N-SIM microscopy imaging (fig. S1D) (18). When the isolated FITC-ODN-EVs were added into cells, the colocalization of TLR9, endosome, and the fluorescent EVs in the recipient cells was observed (Fig. 1D). The ratio for colocalization between FITC-ODN-EVs and TLR9 was 83.17 ± 5.61%. Together, these results indicated that the traffic of EVs between cells led to the transport of ODN into the endosome in recipient cells, which interacted with TLR9.
Cellular uptake of EVs increases upon ODN stimulation
TLR9 is localized in the endosome in response to the inflammatory activation by ODN (19). On the other hand, the endosome is also one of the sources of the EV biogenesis (10). To test whether TLR9 activation correlates with EVs, we first investigated the effect of TLR9 activation on the intercellular EV transfer. A previously reported Cre-LoxP reporter system was used here to visualize the amount of EVs in the cell-cell communication (20). In this reporter system, Cre mRNA from donor cells is loaded into EVs that can be taken up by recipient cells. Upon entry into the recipient cells by endocytosis, the expressed Cre recombinase induces Cre-LoxP recombination, resulting in the excision of the gene encoding the red fluorescent protein and the switch of fluorescent colors from red to green (Fig. 2A) (20). The chosen concentrations of ODN were within the range in which TLR9 activation could be observed, determined by an established secreted embryonic alkaline phosphatase reporter assay (Fig. 2B) (21). The results showed that the ODN challenge increased the number of EVs transferred between cells in a dose-dependent manner, either in short distances when cells were mixed and cultured in the same layer (Fig. 2C) or in long distances when cells were separated by a 400-nm filter in a Transwell chamber, which only allowed EVs to pass (Fig. 2D). Using EVs isolated from the donor (Cre+) cells to stimulate the recipient (reporter+) cells, we observed similar results that the EV transfer increase significantly in the presence of ODN treatment (fig. S2A). In the reporter system, where little TLR9 was expressed, ODN treatment did not significantly increase the transfer of EVs, suggesting that the modulation of EV transfer by ODN was due to the activation of TLR9 (fig. S2, B and C). In addition, when comparing ODN-stimulated with unstimulated cells, neither the percentage of dead cells nor the growth rate of cells was affected, determined by Cell Counting Kit-8 (CCK-8) (fig. S2D) (22).
Fig. 2. ODN enhances EV transfer between cells expressing TLR9.
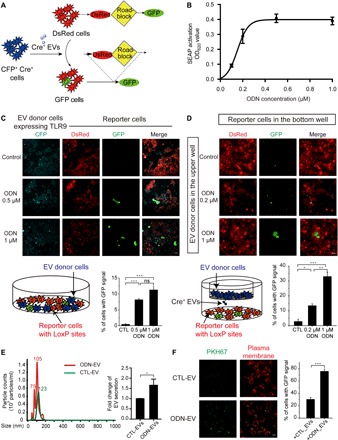
(A) Schematic diagram showing the Cre-LoxP reporter system used to visualize the transfer of Cre+ recombinase [cyan fluorescent protein (CFP), cyan] activity. A red-to-green color switch is induced in reporter+ cells (red) upon the transfer of Cre activity from CFP+ Cre+ cells. (B) Dose-dependent activation of TLR9 by ODN. HEK-Blue hTLR9 cells were treated by different concentrations of ODN (0.2, 0.4, 0.6, 0.8, and 1.0 μM) for 24 hours, and activation was determined by luminescence assay. SEAP, secreted embryonic alkaline phosphatase; OD620, optical density at 620 nm. (C) Images of HEK-293 cultures consisting of a mixture of TLR9-Cre+ (CFP+) and reporter+ (DsRed+) cells. Cells were treated with 0.5 or 1 μM ODN for 24 hours. Graph represents the percentage of reporter+ cells with green fluorescent protein (GFP) signals. Scale bars, 50 μm. ns, not significant. (D) Images of HEK-293 reporter+ cells at the bottom well of a Transwell system and TLR9-Cre+ (cyan) cells in the upper well with or without ODN treatment. TLR9-Cre+ cells were treated with 0.2 or 1 μM ODN for 24 hours and then subcultured in Transwell. Cells were washed twice by centrifugation before the subculture. Graph represents the percentage of reporter+ cells with GFP. Scale bars, 100 μm. (E) NTA displaying the size distribution of EVs isolated from RAW 264.7 cell cultures with (ODN-EV) or without (CTL-EV) the treatment of ODN. The images are representative of three independent experiments. Graph represents the fold change of the numbers between CTL-EV and ODN-EV. (F) Images of RAW 264.7 cells taking up CTL-EV or ODN-EV. CTL-EV and ODN-EV were stained with PKH67 (green), and the plasma membrane of recipient cells was labeled by anti-Glut1 (red). Graph represents the percentage of reporter+ cells or RAW 264.7 cells with green fluorescence signals. Scale bars, 20 μm. Graphs show means ± SEM. [*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant using analysis of variance (ANOVA) one-way test (C and D) or unpaired Student’s t test (E and F)]. Results are representative of at least three independent experiments.
Next, we set out to determine whether the increase of EV traffic between cells was primarily due to the enhancement of the secretion from donor cells or the uptake by recipient cells. With NTA analysis, we quantified the particles around the size of EVs isolated from the culture media of RAW 264.7 cells with or without ODN treatment. The results showed that the number of EVs secreted from cells treated with ODN (ODN-EV) was substantially higher than those of the control-EV (CTL-EV) released from nontreated cells (Fig. 2E). Next, we investigated the cellular uptake of EVs by adding the same amount of PKH67-labeled CTL-EV and ODN-EV into RAW 264.7 cells. Cell plasma membrane was fluorescently labeled with anti–Glut1 to exclude potential false-positive results caused by nonspecific staining of the membrane by PKH67, as shown by Takov et al. (23). We found that the cellular uptake of ODN-EV was also significantly higher compared with CTL-EV, suggesting that ODN-EV was more prone to be internalized by cells (Fig. 2F). In summary, ODN-induced TLR9 activation had an effect on the intercellular traffic of EVs, in both the secretion from donor cells and the uptake by recipient cells.
Cdc42 is enriched in EVs derived from ODN-stimulated cells
Emerging data demonstrated that EV protein composition affected their fusion with recipient cells (24, 25). Thus, we focused on probing the underlying mechanism for the increase of the cellular uptake of EVs through an analysis of their protein cargo. Using liquid chromatography–mass spectrometry (LC-MS) analysis, we identified 1235 different proteins in EVs isolated from RAW 264.7 cell culture supernatant. Among these, in three independent experiments, 42 proteins were found only in ODN-EV and 16 proteins were found only in CTL-EV (Fig. 3A). Moreover, within the proteins identified in both CTL-EV and ODN-EV, 57 proteins significantly increased and five proteins decreased in ODN-EV [1.5-fold cutoff was chosen to show biological significance (26)], compared with CTL-EV (Fig. 3A). Further bioinformatics analyses of gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were performed on 99 proteins, namely, the combination of the 57 proteins that significantly increased and the 42 proteins only present in ODN-EV (fig. S3, A to D). In the KEGG analysis, signaling pathways with P value <0.05, i.e., –log (P value) >1.3, were considered enriched. From our analysis, the most enriched signaling pathway was “endocytosis” (fig. S3A). Since our interest lay in the uptake of EVs into cells, we next investigated those proteins involved in endocytosis. The proteins enriched in endocytosis were further analyzed with their previously reported interaction network summarized in the Search Tool for Recurring Instances of Neighbouring Genes (STRING) database. We found that Cdc42 was the hub of the interaction network of proteins enriched in ODN-EV and endocytosis signaling; therefore, we decided to focus further analysis on this protein (Fig. 3B).
Fig. 3. Proteomic analysis and Western blot analysis of protein cargos of various EVs.
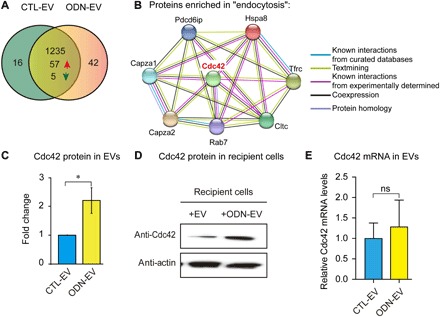
(A) Proteomics analysis of EVs isolated from RAW 264.7 cell cultures in the absence (CTL-EV) or presence of 0.5 μM ODN (ODN-EV) for 12 hours. Liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis was used to identify the proteins. (B) The protein interaction network was generated using the STRING database. Proteins with levels 1.5-fold higher in ODN-EV compared with CTL-EV were first analyzed by KEGG analysis. Then, proteins enriched in “endocytosis” signaling were chosen for the network analysis. (C) Comparison of the level of Cdc42 in CTL-EV and ODN-EV. The relative protein level was quantified by LC-MS analysis. (D) Western blot analysis of Cdc42 protein in cells treated with CTL-EV and ODN-EV. RAW 264.7 cells were treated with CTL-EV or ODN-EV for 12 hours before lysis of cells for Western blot analysis. The images are representative of three repeated experiments. (E) Quantitative reverse transcription polymerase chain reaction analysis of Cdc42 mRNA levels in cells treated with CTL-EV and ODN-EV. Graphs in (C) and (E) show means ± SEM. (*P < 0.05; ns, not significant using unpaired Student’s t test). Results are representative of at least three independent experiments.
Our results showed that the level of Cdc42 was significantly higher in ODN-EV than in CTL-EV with an approximately threefold elevation (Fig. 3C). Further, we carried out biochemical characterizations and found that the level of Cdc42 protein was also markedly higher in cells that received ODN-EV compared with those that received CTL-EV (Fig. 3D). Therefore, we decided to focus on Cdc42 in the following studies. Given that EVs were capable of carrying mRNA to modulate the function of recipient cells, we also investigated the level of Cdc42 mRNA in EVs. Our findings showed that, compared with CTL-EV, ODN-EV carried a slightly higher level of Cdc42 mRNA (~1.2 fold), but the difference is not statistically significant (Fig. 3E). Overall, we found that Cdc42 was present in EVs secreted from macrophages stimulated with the TLR9 agonist ODN and that ODN-EV contained a larger amount of the Cdc42 protein, which was subsequently transferred into the recipient cells.
Cdc42 enhances the cellular uptake of ODN-induced EVs
Distinct endocytic pathways have been suggested in the uptake of EVs, including both clathrin-dependent endocytosis and clathrin-independent pathways such as caveolin-mediated internalization, macropinocytosis, phagocytosis, and lipid raft–mediated uptake (27, 28). Cdc42, a protein belonging to the small guanosine 5′-triphosphate–binding proteins of the Rho family, is associated with the endocytic machinery via a clathrin-independent mechanism in endocytosis (29, 30). We set out to investigate whether Cdc42 was responsible for the internalization of EVs with the Cre-LoxP reporter system. Our findings showed that the overexpression of a constitutively active form of Cdc42, Cdc42-Q61L, in the EV-recipient cells enhanced the cellular internalization of EVs (Fig. 4, A and B). However, the overexpression of the active form of Cdc42 did not significantly increase the number of secreted EVs, characterized by NTA analysis (fig. S4, A and B). The expression of Cdc42-Q61L was confirmed with Western blot analysis (Fig. 4A). Next, we went further to test whether Cdc42 was responsible for the ODN-increased cellular uptake using the same Cre-LoxP cell system. We first knocked down Cdc42 in both donor and recipient cells. Next, we treated donor cells (Cre+ cells) with ODN for 24 hours before they were washed and subcultured with the same amount of recipient cells (reporter+ cells). The depletion of Cdc42 was demonstrated by Western blot analysis (Fig. 4C). The enhancement of ODN-triggered cellular uptake of EVs was abolished in recipient cells where Cdc42 was knocked down, suggesting that Cdc42 was essential in this process (Fig. 4, C and D). GW4869, an established inhibitor of the EV secretion (31), was added to the donor cells in the Cre-LoxP reporter system as a comparison to show the effect on blocking EV transfer in the presence of ODN (Fig. 4D). Furthermore, when Cdc42 was only knocked down in recipient cells, the level of EV uptake was reduced, which is in good agreement with the results showing that the expression of Cdc42-Q61L increased the EV uptake (Fig. 4B and fig. S4C). After being mixed with ODN-pretreated donor cells, the level of EV uptake was lower in Cdc42 knockdown cells compared with the control (fig. S4C). Similarly, when Cdc42 was only knocked down in donor cells before being mixed with recipient cells, the EV uptake was also reduced, possibly because of less transfer of Cdc42 (fig. S4C). On the basis of these findings, we concluded that Cdc42 played an important role in the ODN-increased cellular uptake of EVs.
Fig. 4. Cdc42 enhances the cellular uptake of EVs.

(A) Western blot analysis of the level of Cdc42 in cells expressing Cdc42Q61L. (B) Confocal images of reporter+ cells expressing Cdc42. In the Cre-LoxP reporter system (see Fig. 1A), Cre+ cells were cocultured with reporter+ cells for 48 hours after the transfection of Cdc42Q61L plasmid. Scale bars, 100 μm. (C) Western blot analysis of the level of Cdc42 in reporter+ cells, which were treated with 0, 50, or 100 nM Cdc42 small interfering RNA (siRNA) for 48 hours. (D) Confocal microscopy images of reporter+ cells. Both Cre+ cells and reporter+ cells were treated with 100 nM Cdc42 siRNA. After 6 hours, the medium was replaced with fresh medium. Then, Cre+ cells were treated with 0.5 μM ODN or 20 μM GW4869 for 24 hours before being mixed with reporter+ cells. After 24 hours, cells were observed by confocal microscopy. Scale bars, 50 μm. Graphs in (B) and (D) represent the percentage of reporter+ cells with GFP signals and show means ± SEM. [**P < 0.01 using unpaired Student’s t test (B); *P < 0.05 using ANOVA one-way test (D)]. The percentage of cells with GFP signals was calculated in three different fields of each condition. Images are representative of at least three independent experiments.
EVs transport ODN and induce TNF-α release and Cdc42 elevation
To gain further mechanistic insights, we used FITC-phalloidin to stain actin and examined its morphology to evaluate the activation of Cdc42. The activation of Cdc42 by TNF-α has been implicated in the regulation of endocytosis and induction of actin polymerization (32, 33). Filopodia formation has been proven to be an effective means to monitor the activation of Cdc42 (32, 33). We showed that TNF-α induced the formation of filopodia in RAW 264.7 cells (Fig. 5A), consistent with the previous findings. ODN induced the release of a significant amount of TNF-α in macrophages (fig. S5). The formation of filopodia induced by ODN was suppressed by an inhibitor of TNF-α, indicating that ODN activated Cdc42 through the augmentation of TNF-α release (Fig. 5A).
Fig. 5. The effect of EV and ODN-EV on Cdc42 activity.
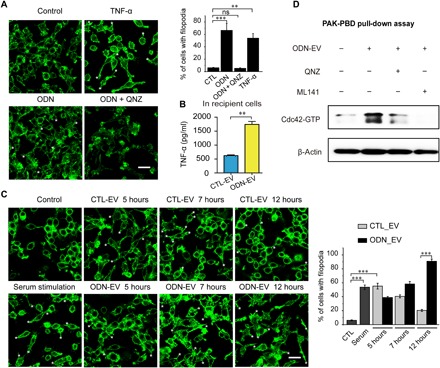
(A) RAW 264.7 cells were serum-starved for 24 hours before actin filament staining with FITC-phalloidin. Cells were treated for 10 min with TNF-α (20 ng/ml) and 24 hours with 0.5 μM CpG ODN after serum starvation. Scale bars, 20 μm. The asterisks (*) highlight the locations of filopodia. Graph represents the percentage of RAW 264.7 cells with filopodia. (B) Enzyme-linked immunosorbent assay (ELISA) was used to determine the concentrations of TNF-α in THP-1 cells, which were treated with 0.5 μM ODN. (C) RAW 264.7 cells were serum-starved for 24 hours before actin filament staining with FITC-phalloidin. Cells were treated with CTL-EV or ODN-EV for the indicated time period after serum starvation. Cells treated with serum for 10 min served as a positive control. Scale bars, 20 μm. The asterisks (*) highlight the locations of filopodia. Graph represents the percentage of RAW 264.7 cells with filopodia. (D) Cdc42 pull-down activation assay showed that the Cdc42 activity was blocked by TNF-α inhibition. QNZ (EVP4593) is a known TNF-α inhibitor. ML141 is a known Cdc42 inhibitor. PAK, human p21 activated kinase; PBD, p21-binding domain. Graphs show means ± SEM. [**P < 0.01; ***P < 0.001; ns, not significant using ANOVA one-way test (A and C) or unpaired Student’s t test (B)]. Results are representative of at least three independent experiments.
Since EVs could deliver ODN into cells, colocalizing with TLR9 within endosomes (Fig. 1D), we investigated whether ODN-EV could induce the release of TNF-α from recipient cells. We observed that the level of TNF-α was significantly higher in the culture medium of macrophages treated with ODN-EV, compared with the medium from cells treated with CTL-EV (Fig. 5B). To rule out the possibility that TNF-α was induced by the impurity from the preparation of EVs, we also isolated EVs by size exclusion chromatography (SEC). Consistent with our findings obtained from EVs isolated by ultracentrifugation, ODN-EV induced a higher level of TNF-α in cells than CTL-EV (fig. S5B). In addition, after being treated with nuclease and protease, ODN-EV was still able to promote the level of TNF-α production (fig. S5C). These results indicate that ODN-EV activated macrophages by ODN enclosed in EVs.
Next, we set out to investigate whether the activation Cdc42 was affected by EVs that transported ODN. We found that both CTL-EV and ODN-EV induced Cdc42 activation in cells at 5 hours, and the maximum amount of activated Cdc42 was observed at 7 hours. At 12 hours, the Cdc42 activation almost disappeared in cells treated with CTL-EV (Fig. 5C). However, in the cells treated with ODN-EV, a significantly larger amount of filopodia was observed, compared with cells treated with CTL-EV (Fig. 5C). The prolonged production of filopodia in cells treated with ODN-EV was possibly due to the production of TNF-α, which accumulated after ODN treatment, exerting an effect on the activity of Cdc42, in addition to the increase of the Cdc42 protein by EV transfer. To prove this concept, we performed Cdc42 pull-down activation assay. The effect of ODN-EV on the activation of Cdc42 was further confirmed (Fig. 5D). Overall, our results suggested that ODN was transported to the endosome of cells by EVs, increasing the activated form of Cdc42 in cells through the enhancement of the TNF-α release.
DISCUSSION
The sensing of PAMPs is a crucial step in innate immune activation (34). Recent studies demonstrated that EVs transport LPS into the cytosol (6). Nonetheless, whether this machinery is a common component of other innate immune receptors remains unclear. In this study, we have shown that EVs secreted from ODN-activated macrophages transport ODN into naïve macrophages, where it interacts with TLR9 and enhances the release of chemokine TNF-α. Furthermore, we found that Cdc42 is transferred from EVs into the recipient cells and further activated by TNF-α, leading to elevated cellular uptake of EVs. Together, our results demonstrate the mechanism that EVs use to regulate their own cellular uptake in macrophages through a feedback loop, which involves amplifying the level of activated Cdc42 protein in the recipient cells (Fig. 6).
Fig. 6. Schematic representation of the proposed mechanism for ODN-stimulated EV uptake via modulation of Cdc42.
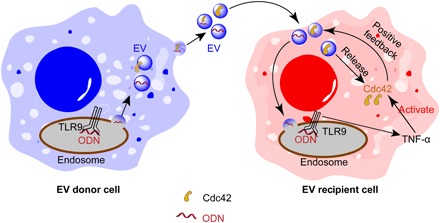
EVs secreted from ODN-activated macrophages carry both Cdc42 and ODN. When EVs internalize into the recipient cells, Cdc42 is transferred from EVs to cells, further increasing the level of Cdc42 in the cells. ODN carried by EVs also induces the release of TNF-α from recipient macrophages, which, in turn, activates Cdc42, forming a positive feedback loop to promote further endocytosis of EVs.
EVs serve as a critical vehicle for intercellular communication in the fusion, internalization, and release of signal molecules to their target cells (9, 13, 35, 36). The influence of EV-carried miRNA was demonstrated in prometastastic inflammatory responses in which TLR7 and TLR8 were involved (36). Our investigation of the cellular uptake of EVs in immune cells provides a better understanding of the general role of EVs in the regulation of intracellular innate immunity. It has been noted that the transfer of EVs was enhanced in pathological states such as cancers and inflammation, compared with most physiological conditions (37). Nonetheless, the underlying mechanism for EV secretion and uptake has not been thoroughly studied. Our effort in deciphering that mechanism by proteomic analysis of EVs led to the finding that Cdc42 is responsible for ODN-mediated EV uptake. Cdc42 activation was previously linked to immune evasion in HIV-1–induced membrane extensions in immature dendritic cells, which facilitated cell-cell exchange of the virus particles between dendritic cells and CD4+ T cell (38). Therefore, we extended our study to characterize the relevance of Cdc42 activation for ODN stimulation. Our investigations demonstrate that the activation of Cdc42 is essential in ODN-triggered EV uptake in macrophages, which may offer insight into the function of Cdc42 activation in various autoimmune diseases.
Our data also show that Cdc42 and ODN are delivered simultaneously to recipient cells to exert a synergetic effect on cellular uptake of EVs. These data are in agreement with a previous finding showing a costimulatory effect of EVs on recipient cells via the transport of PAMPs and major histocompatibility complex molecules from immune antigen-presenting cells (39). ODN has been shown to have great potential in different clinical applications as a vaccine adjuvant (40–42). Our findings show that the level of Cdc42 is enhanced in ODN-derived EVs. Upon entry into recipient cells, Cdc42 was activated by TNF-α, influencing the further uptake of EVs.
In summary, we have deciphered a mechanism by which EVs propagate TLR9-mediated innate immune responses through feedback regulation of their own cellular uptake. Our investigation on the uptake of ODN-carrying EVs could deepen understanding of the role of EVs in TLR-modulated intracellular innate immune responses. Multiple mechanisms have been previously reported for the destinies of the endocytic EVs and their cargos after cell endocytosis (43). Nonetheless, intriguing questions such as the topology of the nucleic acid cargo within the EVs remain unresolved. Thorough characterization of how and when ODN is released from the EVs and consequently interact with TLR9 demands further investigation. Moreover, our findings may facilitate the development of more efficient therapeutic approaches targeting Cdc42 to modulate the uptake of EVs, especially in the context of various inflammatory diseases.
MATERIALS AND METHODS
Purification of EVs
To prepare “EV-depleted medium,” fetal bovine serum (FBS) was centrifuged at 100,000g for 1 hour at 4°C to remove EVs. The centrifuged FBS was then mixed with medium at a ratio of 1:10. For each condition, cells were cultured in the EV-depleted medium at 60 to 70% confluency for 48 hours. Culture medium was then harvested. EV purification from the medium was performed by ultracentrifugation (44) or size exclusion chromatography (SEC).
During ultracentrifugation, the harvested culture medium was first centrifuged at 500g twice for 10 min to eliminate floating cells, at 2000g twice for 15 min to remove cell debris, and centrifuged at 16,500g twice for 20 min. The medium was then filtered through a 0.22-μm filter before an ultracentrifugation step at 100,000g for 1 hour. Then, the supernatant was discarded, followed by a washing step in phosphate-buffered saline (PBS) to eliminate contaminating proteins. EVs were suspended in 100 μl of PBS, and protein content was measured by a bicinchoninic acid (BCA) protein assay kit (Pierce). The number of cells from all the culture conditions was counted.
SEC was performed using commercially available Exo-spin SEC columns (Cell Guidance Systems), following the manufacturer’s protocol. Briefly, the culture medium was centrifuged at 500g twice for 10 min and then at 16,500g for 30 min. The supernatant was transferred to a new centrifuge tube and mixed with 1:2 volume of Exo-spin buffer for 2 hours. Then, the mixture was centrifuged at 16,500g for 1 hour. The supernatant was discarded, while the EV-containing pellet was resuspended in 100 μl of PBS. The sample was further purified using the provided Exo-spin columns. Last, the purified EVs were collected from the eluate.
Nanoparticle tracking analysis
The concentrations and size distribution of EVs were analyzed by NTA using the NanoSight LM14 instrument (Malvern Instruments, UK). EVs were suspended in 100 μl of PBS and diluted 50 times with double-distilled water. Constant flow injection was used, and three videos of 60 s were captured. The camera level was set at level 14 and the detection threshold at 4 or 5. Data were analyzed on the basis of the captured videos by NTA 3.1 software.
PKH67 labeling of EVs
The PKH67 Green Fluorescent Cell Linker Mini Kit for General Cell Membrane Labeling kit (SLBR4806V, Sigma) was used to label EVs. First, EVs from cells were resuspended in 100 μl of diluent C in the kit. PKH67 dye was diluted into a final concentration of 2 μM with diluent C and then mixed with the EVs (~107). After a 5-min incubation at room temperature, the reaction was stopped by adding 500 μl of 5% BSA (bovine serum albumin)/PBS. Labeled EVs were washed with PBS twice by ultracentrifugation and resuspended in PBS.
Immunofluorescence and imaging
Cells were fixed with 4% paraformaldehyde (PFA) for 10 min and permeabilized with 0.1% Triton X-100 for 5 min. The cells were then blocked with 1% BSA/PBS for 45 min and incubated with a primary antibody for 2 hours and a secondary antibody for 1 hour. Washes were done four times with PBS after each step. When desired, the cells were incubated with 4′,6-diamidino-2-phenylindole (1 μg/ml) in PBS for 10 min and washed four times. Last, the cells were photographed with an A1Rsi super-resolution laser scanning confocal microscope (Nikon). Five fields were randomly chosen for images, and three independent experiments were performed. The images were analyzed using ImageJ software (version 2.0.0-rc-54/1.51h).
EVs were fixed by with a fluoromount aqueous mounting medium (Sigma) onto glass slides and then observed by Super Resolution Microscope N-SIM microscopy (Nikon).
Colocalization quantification
The extent of colocalization of two fluorescent labels was measured using the “colocalization” module of Imaris 9.1.2 (Bitplane AG, USA.). The program analyzes stacks of confocal sections obtained in two channels. Green and blue channels were chosen here for analysis. The pixels of the green region (FITC-ODN-EV) were defined as the thresholds in the settings to calculate the percentage of blue area (TLR9) colocalized with the green region. The threshold for all measures was consistent.
Proteomics analysis
Proteins were separated by SDS–polyacrylamide gel electrophoresis, and the gel bands of interest were excised, reduced with 5 mM dithiotreitol, and alkylated with 11 mM iodoacetamide. In-gel digestion was then carried out with sequencing grade modified trypsin in 50 mM ammonium bicarbonate at 37°C overnight. The peptides were extracted twice with 0.1% trifluoroacetic acid (TFA) in 50% acetonitrile aqueous solution. Extracts were centrifuged in a SpeedVac to reduce the volume. Tryptic peptides were dissolved in 20 μl of 0.1% TFA.
For LC–tandem mass spectrometry (LC-MS/MS) analysis, the peptides were separated by a gradient elution at a flow rate of 0.30 μl/min with a Thermo-Dionex UltiMate 3000 HPLC system (Thermo Fisher Scientific), coupled with a Q Exactive mass spectrometer (Thermo Fisher Scientific). The analytical column was a home-made fused silica capillary column (inner diameter, 75 μm; length, 150 mm; Upchurch, Oak Harbor, WA) packed with C-18 resin (300 Å, 5 μm; Varian, Lexington, MA). The mobile phase consisted of 0.1% formic acid, and mobile phase B consisted of 80% acetonitrile and 0.1% formic acid. The Q Exactive mass spectrometer was operated in the data-dependent acquisition mode using Xcalibur 2.1.2 software, and there was a single full-scan mass spectrum in the Orbitrap (300 to 1800 mass/charge ratio, 70,000 resolution), followed by 20 data-dependent MS/MS scans at 27% normalized collision energy [HCD (higher-energy collisional dissociation)].
F-actin staining assay
The cells were fixed with 4% PFA for 10 min and permeabilized by 0.2% Triton X-100 for 5 min at room temperature. F-actin was stained with 100 nM FITC-phalloidin for 30 min in the dark. Actin fragments were imaged with a 100× oil immersion objective on a Nikon A1Rsi super-resolution laser scanning confocal microscope (Nikon).
Enzyme-linked immunosorbent assay
Enzyme-linked immunosorbent assay (ELISA) was performed to measure the level of TNF-α expression. THP-1 cells were pretreated with phorbol 12-myristate 13-acetate (100 ng/ml) before being seeded at 0.5 × 106 per well using a 24-well plate in 0.5 ml of complete cell culture medium with 10% FBS. After 24 hours, cells adhered to the surface of the plate. Fresh medium without FBS was used during the treatment of EVs or ODN-EVs (EV-to-cell ratio was 100:1), or they were left untreated. After 24 hours, the culture medium was collected and levels of TNF-α were determined using the human TNF-α OptEIA ELISA kit (BD Biosciences) according to the manufacturer’s instructions.
Statistical analyses
Data were represented as the means ± SEM. Unpaired Student’s t test was used to analyze data with only two sets. One-way analysis of variance (ANOVA) was performed to determine whether there was a significant difference between more than two datasets, followed by Bonferroni’s post hoc test, using GraphPad Prism 6.0. Group differences at the level of P < 0.05 were considered statistically significant. Asterisk (*) represented P < 0.05; double asterisk (**) represented P < 0.01; triple asterisk (***) represented P < 0.001.
Supplementary Material
Acknowledgments
Funding: This work was funded by the National Key R&D Program of China (no. 2017YFA0505200) and the National Natural Science Foundation of China (nos. 21572114 and 21825702). Y.Z. thanks the China Postdoctoral Science Foundation grant (no. 2017 M620754) for support. Author contributions: H.Y. and Y.Z. conceptualized the project. Y.Z. designed the experiments, analyzed the data, and wrote the manuscript. X.J. performed the experiment of expression of Cdc42-Q61L in cells, observation of EV uptake, and staining of actin filaments. J.L. performed the experiment of quantitative reverse transcription polymerase chain reaction analysis and NTA, Y.G. performed the experiment of EV secretion, and G.S. performed the experiment of knockdown of Cdc42 with small interfering RNA. X.Z. performed the live-cell confocal microscopy analysis. Y.Z. performed all of the other experiments and designed the schematic representation. Competing interests: H.Y. is currently a member at large of the board of directors of the International Society of Extracellular Vesicles (ISEV). The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/7/eaav1564/DC1
General Methods
Fig. S1 Characterization of EVs isolated from cell culture.
Fig. S2. Effect of ODN on EV transfer and cell viability.
Fig. S3. GO bioinformatics analyses of proteins that are more enriched in ODN-EV compared with CTL-EV.
Fig. S4. The quantification of secreted EVs from the Cdc42-overexpressing cells.
Fig. S5. ODN or ODN-EV increases the level of TNF-α in macrophages.
Movie S1. Movie showing real-time EV secretion from FITC-ODN–treated DsRed-marked cells.
REFERENCES AND NOTES
- 1.Chen Q., Sun L., Chen Z. J., Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 17, 1142–1149 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Mackenzie K. J., Carroll P., Martin C.-A., Murina O., Fluteau A., Simpson D. J., Olova N., Sutcliffe H., Rainger J. K., Leitch A., Osborn R. T., Wheeler A. P., Nowotny M., Gilbert N., Chandra T., Reijns M. A. M., Jackson A. P., cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X., Smith C., Yin H., Targeting Toll-like receptors with small molecule agents. Chem. Soc. Rev. 42, 4859–4866 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin H., Flynn A. D., Drugging membrane protein interactions. Annu. Rev. Biomed. Eng. 18, 51–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawai T., Akira S., The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 11, 373–384 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Vanaja S. K., Russo A. J., Behl B., Banerjee I., Yankova M., Deshmukh S. D., Rathinam V. A. K., Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell 165, 1106–1119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi J., Zhao Y., Wang Y., Gao W., Ding J., Li P., Hu L., Shao F., Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Maas S. L. N., Breakefield X. O., Weaver A. M., Extracellular vesicles: Unique intercellular delivery vehicles. Trends Cell Biol. (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tkach M., Thery C., Communication by extracellular vesicles: Where we are and where we need to go. Cell 164, 1226–1232 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Kowal J., Tkach M., Théry C., Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 29, 116–125 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Kim S., Li L., Maliga Z., Yin Q., Wu H., Mitchison T. J., Anticancer flavonoids are mouse-selective STING agonists. ACS Chem. Biol. 8, 1396–1401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mantel P.-Y., Hoang A. N., Goldowitz I., Potashnikova D., Hamza B., Vorobjev I., Ghiran I., Toner M., Irimia D., Ivanov A. R., Barteneva N., Marti M., Malaria-infected erythrocyte-derived microvesicles mediate cellular communication within the parasite population and with the host immune system. Cell Host Microbe 13, 521–534 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schorey J. S., Cheng Y., Singh P. P., Smith V. L., Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep. 16, 24–43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giri P. K., Schorey J. S., Exosomes derived from M. Bovis BCG infected macrophages activate antigen-specific CD4+ and CD8+ T cells in vitro and in vivo. PLOS ONE 3, e2461 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai C. P., Kim E. Y., Badr C. E., Weissleder R., Mempel T. R., Tannous B. A., Breakefield X. O., Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat. Commun. 6, 7029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klinman D. M., Immunotherapeutic uses of CpG oligodeoxynucleotides. Nat. Rev. Immunol. 4, 249–259 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Lotvall J., Hill A. F., Hochberg F., Buzás E. I., Di Vizio D., Gardiner C., Gho Y. S., Kurochkin I. V., Mathivanan S., Quesenberry P., Sahoo S., Tahara H., Wauben M. H., Witwer K. W., Théry C., Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 3, 26913 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian Y., Ma L., Gong M., Su G., Zhu S., Zhang W., Wang S., Li Z., Chen C., Li L., Wu L., Yan X., Protein profiling and sizing of extracellular vesicles from colorectal cancer patients via flow cytometry. ACS Nano 12, 671–680 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Latz E., Schoenemeyer A., Visintin A., Fitzgerald K. A., Monks B. G., Knetter C. F., Lien E., Nilsen N. J., Espevik T., Golenbock D. T., TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 5, 190–198 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Zomer A., Maynard C., Verweij F. J., Kamermans A., Schäfer R., Beerling E., Schiffelers R. M., de Wit E., Berenguer J., Ellenbroek S. I. J., Wurdinger T., Pegtel D. M., van Rheenen J., In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 161, 1046–1057 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng K., Gao M., Godfroy J. I., Brown P. N., Kastelowitz N., Yin H., Specific activation of the TLR1-TLR2 heterodimer by small-molecule agonists. Sci. Adv. 1, e1400139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang M., Weng W., Zhang Q., Wu Y., Ni S., Tan C., Xu M., Sun H., Liu C., Wei P., Du X., The lncRNA NEAT1 activates Wnt/β-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J. Hematol. Oncol. 11, 113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takov K., Yellon D. M., Davidson S. M., Confounding factors in vesicle uptake studies using fluorescent lipophilic membrane dyes. J. Extracell. Vesicles 6, 1388731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrès C., Blanc L., Bette-Bobillo P., André S., Mamoun R., Gabius H.-J., Vidal M., Galectin-5 is bound onto the surface of rat reticulocyte exosomes and modulates vesicle uptake by macrophages. Blood 115, 696–705 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Escrevente C., Keller S., Altevogt P., Costa J., Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer 11, 108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis T. S., Hunt J. B., Aveline L. D., Jonscher K. R., Louie D. F., Yeh J. M., Nahreini T. S., Resing K. A., Ahn N. G., Identification of novel MAP kinase pathway signaling targets by functional proteomics and mass spectrometry. Mol. Cell 6, 1343–1354 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Mulcahy L. A., Pink R. C., Carter D. R., Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 3, 24641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costa Verdera H., Gitz-Francois J. J., Schiffelers R. M., Vader P., Cellular uptake of extracellular vesicles is mediated by clathrin-independent endocytosis and macropinocytosis. J. Control. Release 266, 100–108 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Mayor S., Pagano R. E., Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell Biol. 8, 603–612 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandvig K., Torgersen M. L., Raa H. A., van Deurs B., Clathrin-independent endocytosis: From nonexisting to an extreme degree of complexity. Histochem. Cell Biol. 129, 267–276 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colombo M., Moita C., van Niel G., Kowal J., Vigneron J., Benaroch P., Manel N., Moita L. F., Théry C., Raposo G., Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 126, 5553–5565 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Puls A., Eliopoulos A. G., Nobes C. D., Bridges T., Young L. S., Hall A., Activation of the small GTPase Cdc42 by the inflammatory cytokines TNF(alpha) and IL-1, and by the Epstein-Barr virus transforming protein LMP1. J. Cell Sci. 112 ( Pt 17), 2983–2992 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Sathe M., Muthukrishnan G., Rae J., Disanza A., Thattai M., Scita G., Parton R. G., Mayor S., Small GTPases and BAR domain proteins regulate branched actin polymerisation for clathrin and dynamin-independent endocytosis. Nat. Commun. 9, 1835 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mullard A., Can innate immune system targets turn up the heat on ‘cold’ tumours? Nat. Rev. Drug Discov. 17, 3–5 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Milane L., Singh A., Mattheolabakis G., Suresh M., Amiji M. M., Exosome mediated communication within the tumor microenvironment. J. Control. Release 219, 278–294 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., Lovat F., Fadda P., Mao C., Nuovo G. J., Zanesi N., Crawford M., Ozer G. H., Wernicke D., Alder H., Caligiuri M. A., Nana-Sinkam P., Perrotti D., Croce C. M., MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 109, E2110–E2116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai C. P., Mardini O., Ericsson M., Prabhakar S., Maguire C., Chen J. W., Tannous B. A., Breakefield X. O., Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano 8, 483–494 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikolic D. S., Lehmann M., Felts R., Garcia E., Blanchet F. P., Subramaniam S., Piguet V., HIV-1 activates Cdc42 and induces membrane extensions in immature dendritic cells to facilitate cell-to-cell virus propagation. Blood 118, 4841–4852 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhatnagar S., Shinagawa K., Castellino F. J., Schorey J. S., Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood 110, 3234–3244 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jackson S., Lentino J., Kopp J., Murray L., Ellison W., Rhee M., Shockey G., Akella L., Erby K., Heyward W. L., Janssen R. S., HBV-23 Study Group , Immunogenicity of a two-dose investigational hepatitis B vaccine, HBsAg-1018, using a toll-like receptor 9 agonist adjuvant compared with a licensed hepatitis B vaccine in adults. Vaccine 36, 668–674 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Temizoz B., Kuroda E., Ishii K. J., Vaccine adjuvants as potential cancer immunotherapeutics. Int. Immunol. 28, 329–338 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harshyne L. A., Hooper K. M., Andrews E. G., Nasca B. J., Kenyon L. C., Andrews D. W., Hooper D. C., Glioblastoma exosomes and IGF-1R/AS-ODN are immunogenic stimuli in a translational research immunotherapy paradigm. Cancer Immunol. Immunother. 64, 299–309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Niel G., D’Angelo G., Raposo G., Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213–228 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Thery C., Amigorena S., Raposo G., Clayton A., Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 30, 3.22.1–3.22.29 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/7/eaav1564/DC1
General Methods
Fig. S1 Characterization of EVs isolated from cell culture.
Fig. S2. Effect of ODN on EV transfer and cell viability.
Fig. S3. GO bioinformatics analyses of proteins that are more enriched in ODN-EV compared with CTL-EV.
Fig. S4. The quantification of secreted EVs from the Cdc42-overexpressing cells.
Fig. S5. ODN or ODN-EV increases the level of TNF-α in macrophages.
Movie S1. Movie showing real-time EV secretion from FITC-ODN–treated DsRed-marked cells.


