Abstract
Background and study aims Recently, a new Franseen design endoscopic ultrasound-guided fine-needle biopsy (EUS-FNB) needle was developed with the goal of providing more tissue for histology. We compared the tissue adequacy rate and nucleic acid yield of 22G EUS-FNB vs. 22G endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA), in solid gastrointestinal and extra-intestinal lesions.
Patients and methods We conducted a randomized crossover study and recruited 36 patients. We performed three passes for pancreatic lesions and two passes for other lesions, using each needle. We blinded the pathologist to needle assignment. We assessed the diagnostic tissue adequacy rate and compared the total tissue area, diagnostic tissue area, and desmoplastic stroma (DS) area in cases of carcinoma. We also examined the nucleic acid yield of the two needles in pancreatic lesions.
Results The lesions included 20 pancreatic masses (55 %), six gastric subepithelial lesions (17 %), five lymph nodes (14 %) and five other abdominal masses (14 %). Mean ± SD lesion size was 3.8 ± 2.0 cm. The final diagnosis was malignant in 27 lesions (75 %) and benign in nine lesions (25 %). We found EUS-FNB procured significantly more median total tissue area (5.2 mm 2 vs. 1.9 mm 2 , P < 0.001), diagnostic tissue area (2.2 mm 2 vs. 0.9 mm 2 , P = 0.029), and DS area (2 mm 2 vs. 0.1 mm 2 , P = 0.001) in lesions diagnosed as carcinoma (n = 23), as compared to EUS-FNA. In pancreatic lesions, EUS-FNB obtained significantly more nucleic acid than EUS-FNA (median; 4,085 ng vs. 2912 ng, P = 0.02). There was no difference in the cellblock or rapid on-site cytological evaluation (ROSE) diagnostic yield between the needles.
Conclusion The 22G EUS-FNB provides more histological core tissue and adequate nucleic acid yield compared to 22G EUS-FNA. In this study, the diagnostic performance was similar between the needles
Introduction
Endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA is a safe and preferred method for tissue acquisition from solid gastrointestinal and extra-intestinal lesions 1 2 . However, EUS-FNA has certain drawbacks: The yield is predominantly cellular and rarely provide tissue blocks; need to perform multiple passes; and requirement for rapid onsite cytopathology (ROSE) assessment to improve diagnostic yield 3 . In addition, the cellular yield of EUS-FNA limits architecture assessment, performance of immunohistochemistry and molecular analysis 4 5 . Such assessments are essential to establish a diagnosis in neoplasms such as lymphoma and gastrointestinal stromal tumors and they are pivotal for clinical trials evaluating molecular markers for personalized oncological treatment. Multiple techniques of EUS-FNA-guided tissue acquisition have been described, but they have failed to consistently show improved diagnostic yield 6 7 8 .
Newer needles with different tip designs and side fenestration (endoscopic ultrasound-guided fine-needle biopsy [EUS-FNB]) have been developed with the goal of obtaining a core sample for histology. Studies comparing the diagnostic efficacy of EUS-FNA and EUS-FNB have yielded conflicting results 9 10 11 12 13 . A meta-analysis comparing one type of EUS-FNB (ProCore, Cook Endoscopy) to standard EUS-FNA showed no difference in sample adequacy, diagnostic accuracy, or acquisition of core specimen between the two needles 14 .
Recently, an EUS-FNB needle with Franseen geometry (Acquire, Boston Scientific, United States) has been developed to procure tissue specimen for histology 15 16 17 18 . Early evidence assessing diagnostic yield of the Franseen needle in solid pancreatic lesions is promising 19 . However, more evidence of its diagnostic utility in pancreatic and non-pancreatic solid lesions is needed. We performed a randomized trial comparing the outcome of 22G Franseen EUS-FNB (Acquire, Boston Scientific, United States) and 22G standard EUS-FNA (Expect, Boston Scientific, United States) in solid gastrointestinal and extra-intestinal lesions.
Patients and methods
Trial design
We conducted a prospective, randomized, single-blinded crossover trial at Singapore General hospital between April 2017 and November 2017. The institutional review board approved the study. All authors had access to the study data and reviewed and approved the final manuscript (Clinicaltrials.gov, NCT03109639). The study was conducted in accordance with the ethical principles detailed in the Declaration of Helsinki and was consistent with Good Clinical Practices recommendation. We reported our outcome according to the CONSORT recommendation of reporting a randomized trial.
Participants
We enrolled 40 patients who were referred for EUS-guided tissue acquisition. We obtained informed consent from all patients. Inclusions criteria were age older than 18 years, presence of only solid lesions confirmed by endoscopy or radiology, and ability to comply with the procedure and provide informed consent. Patients were excluded if they had active bleeding, coagulopathy (INR > 1.5, platelet count < 50,000), were concurrently taking anticoagulants (e. g., warfarin) and a thienopyridine (e. g., clopidogrel), had an intervening large blood vessel, or had difficulty tolerating the procedure.
Intervention
Procedure
Four experienced endoscopists trained in EUS and EUS-guided tissue acquisition techniques performed the procedures under moderate sedation using midazolam and fentanyl. The endoscopists used the Franseen EUS-FNB in clinical practice and were familiar with the device before participating in the study. They were aware of the type of needle used. We used the curved linear array echoendoscope for the study (GF-UC140P, Olympus, USA; EG-3870UTK, Pentax, Japan; or EG-580UT, Fujifilm, Japan). We identified the lesions using EUS, confirmed the absence of cystic components and then randomized them to the study needles for tissue acquisition.
We advanced the assigned needle into the lesion under EUS guidance, removed the stylet and applied a negative pressure using a 10-cc suction syringe. We accessed the lesions through the same route, whenever possible, using both the needles. We practiced the fanning method and performed three passes for pancreatic lesions and two passes for other lesions using each needle type. All the assigned passes were completed with the initial device before crossing over to the alternate needle ( Fig. 1 ). If there was a technical failure (defined as needle malfunction before we reached a diagnosis), the patient crossed over to the alternative needle.
Fig. 1.
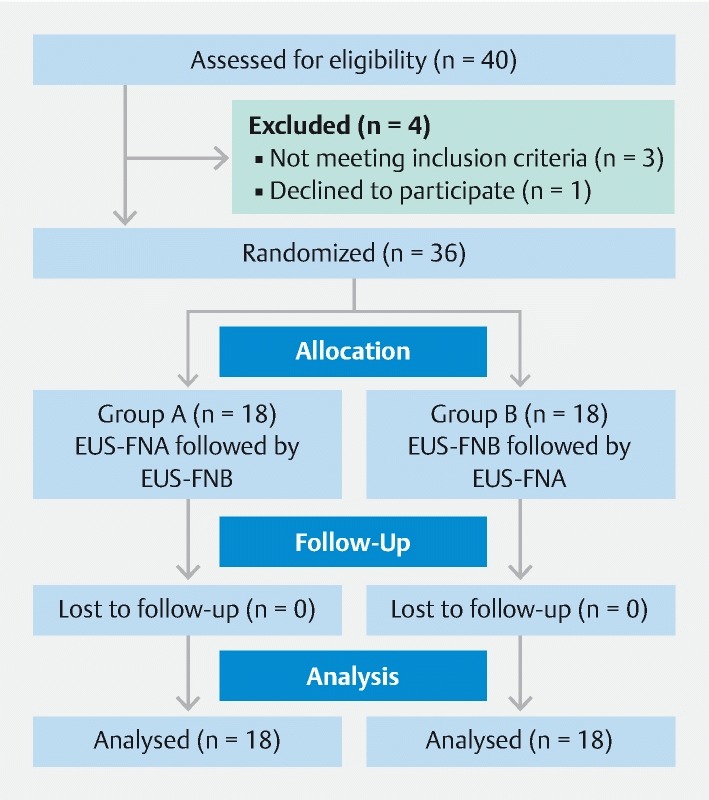
Consort diagram of the study design.
Tissue preparation for pathology
We expressed a portion of the specimen onto a slide using the stylet and prepared air-dried and alcohol-dried smears on-site. We flushed the residual material with normal saline and placed it in formalin solution for cell-block analysis. We collected both the smears and cell-block samples after each needle passes.
The cytotechnician and the pathologist reviewing the smears and slides were blinded to the type of needle used. The slides were reviewed on-site by the cytotechnician after each pass to assess for specimen adequacy. The endoscopist was informed of the outcome of on-site examination only after completion of the assigned number of needle passes to minimize operator bias. We centrifuged and concentrated the cell-block sample and created a tissue clot, fixed the tissue clot in formalin and embedded it with paraffin. We then sectioned and stained it with eosin and hematoxylin for histological assessment. We graded the sample as optimal or suboptimal based on presence of core tissue that enabled adequate evaluation of histological architecture.
We calculated the area of total tissue, diagnostic tissue, and desmoplastic stroma (in carcinoma) using the Philips IntelliSite Pathology solutions image management system. We measured the greatest linear dimension of each tissue core fragments and summed it to estimate the total tissue length. We then identified a representative tissue core fragment and measured the largest diameter. Using these numerical values, we calculated the area of total tissue ( Fig. 2 ). In a similar manner, we estimated the area of diagnostic tissue and desmoplastic stroma. When needed, we performed immunohistochemical or special staining to characterize the lesion better.
Fig. 2.
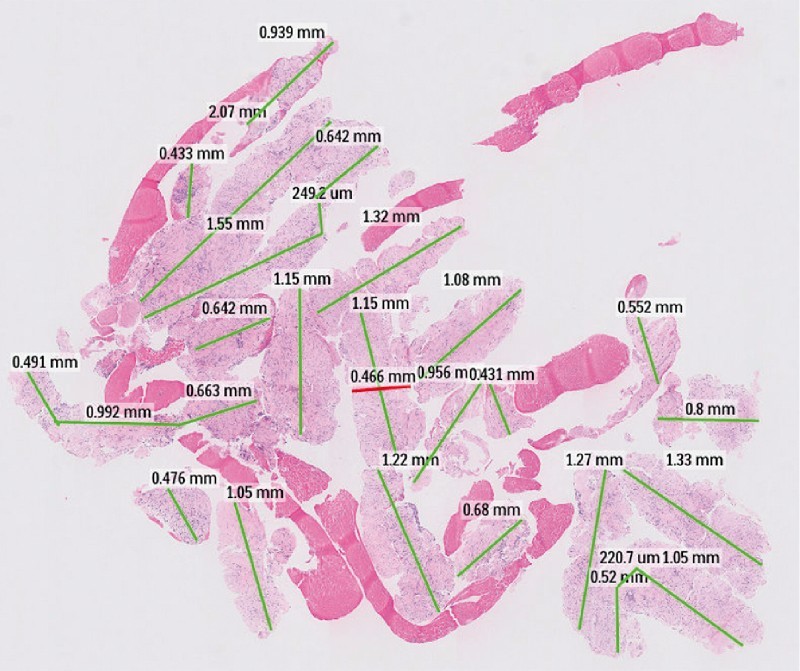
Scanning power photomicrograph of a cell-block specimen. The greatest linear diameter of the tissue core fragments was measured (green line) and summed to obtain the total histological core tissue length. A representative core fragment was identified, and the diameter of it was measured (red line). The area of the total histological core was estimated from these measurements.
Quantification and quality assessment of nucleic acid
We extracted the genomic DNA and RNA from the formalin-fixed paraffin embedded (FFPE) cell blocks from each needle. We estimated the amount of nucleic acid by NanoDrop spectrophotometry (Thermo Scientific, Wilmington, Massachusetts, United States) and further quantified using the Qubit fluorimetry (Life Technologies, Grand Island, New York, United States). We assessed the quality of the DNA and RNA by reading the ratio of absorbance at 260/280 and 260/230 nm. We calibrated the instruments and performed the procedure as per the manufacturer's instructions.
We also explored the possibility of performing molecular studies from EUS-derived tissue samples using Oncomine Comprehensive panel V3 (Life Technologies, Pleasanton, California, United States), an assay that contains 4648 primer pairs designed for hotspots, targeted regions, and gene fusions of 161 known genes relevant to solid tumors. It provides the reagents for library construction and four pools of multiplex polymerase chain reaction primers for preparation of amplicon libraries from formalin-fixed paraffin embedded tumor samples. We randomly selected two solid pancreatic lesions for this analysis and followed optimized protocols.
Outcomes
The primary outcome was to evaluate the diagnostic tissue adequacy rate and compare the area of total tissue, diagnostic tissue, and desmoplastic stroma (in cases with a diagnosis of carcinoma) from the histological core procured using both the needles. Our secondary outcome was to compare the quantity and quality of the DNA and RNA in the samples obtained using the two needles. We defined diagnostic tissue adequacy as presence of histological tissue representative of the sampled lesion and diagnostic yield as the percentage in which a definitive diagnosis could be established from the sampled lesion 20 .
We established the final diagnosis based on: pathology from the surgical specimen; radiological imaging including computed tomography, magnetic resonance imaging, and positron emission tomography scan; or clinical progress. We considered a lesion to be benign if there was spontaneous resolution or interval stability in radiological imaging.
Sample Size
In a preliminary study, the EUS-FNB needle provided diagnostic material for histology in > 95 % of patients 15 . Standard EUS-FNA has a diagnostic histology yield of only 40 % 21 22 . We estimated that a total of 36 patients would be needed to detect a 30 % difference in histological yield between the two needles, with an α of 0.05 and a power of 90 %. We factored in a 10 % dropout rate and enrolled a total of 40 patients.
Randomization and blinding
We randomized the patients to one of two groups (A and B) and recorded the results ( Fig. 1 ). In group A, patients received standard EUS-FNA followed by EUS-FNB. In group B, the assigned passes were performed first using EUS-FNB and then crossed over to EUS-FNA. Randomization was performed in blocks to ensure that the groups were balanced periodically. We used web-based randomization (Sealed Envelope Ltd 2016; https://www.sealedenvelope.com/simple-randomiser/v1 / ) for this purpose. We blinded the cytotechnician and the pathologist to the needle used for specimen collection.
Statistical methods
We expressed continuous variables as the mean ± standard deviation (SD) or median (interquartile range [IQR]). We compared the means using a paired t -test and medians using Wilcoxon ranked sum test. We presented categorical variables in percentage, and the correlation with the different technique was studied using McNemar’s test. Prior to these tests, we performed a linear mixed model with a fixed period and technique effects, and random patient-level intercepts to investigate if there was a significant period effect. We performed statistical analysis using R 3.4.2 software (R Core Team (2017). R: A language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria). A P value < 0.05 was considered significant.
Results
We enrolled 40 patients and excluded four from the study; three had cystic lesions, and one withdrew from the study. The remaining 36 patients were randomized to one of the two study groups. Mean age was 63.5 ± 11.4 years, and the majority were men (56 %, n-20). The lesions sampled included 20 pancreatic masses (55 %), six gastric sub-epithelial (SEL) lesions (17 %), five lymph nodes (14 %) and five other abdominal masses (14%) (2 peri-gastric mass, two retroperitoneal mass, 1 liver metastasis). Mean ± SD size of the lesion was 3.8 ± 2.0 cm. We accessed the lesion through the trans-gastric route in 22 patients (61 %), trans-duodenally in 14 patients (39 %) and trans-esophageally in two patients (5 %). The final diagnosis was malignancy (22 adenocarcinomas, one liposarcoma, one squamous cell carcinoma, one neuroendocrine tumor, two lymphomas) in 27 patients (75 %) and benign lesion (6 spindle cell tumor, 1 splenunculus, 2 inflammatory mass) in 9 (25 %). We did not have any technical difficulty, and the procedure was successful in all patients.
Diagnostic tissue adequacy
We found EUS-FNB obtained histological core tissue more frequently than EUS-FNA (97 % vs. 77 %, P = 0.03). Diagnostic adequacy and yield of the histological tissue was similar between EUS-FNB and EUS-FNA (81 % vs.64 %, P = 0.19). We did not observe any difference in the cell-block diagnostic adequacy and yield between the two needles for pancreatic and non-pancreatic lesions.
When assessing the histology, we found that EUS-FNB provided significantly more median total tissue area than EUS-FNA (5.2 mm 2 vs. 1.9 mm 2 , P < 0.001) in both pancreatic and non-pancreatic lesions ( Table 1 ). Median area of the diagnostic tissue within the histological sample was significantly more with EUS-FNB than with EUS-FNA (2.2 mm 2 vs. 0.9 mm 2 , P = 0.01) in all lesions ( Fig. 3 ). We assessed presence of desmoplastic stroma (DS) in patients with a final diagnosis of carcinoma (64 %, n-23). We found that EUS-FNB provides significantly more DS tissue area than EUS-FNA (2 mm 2 vs. 0.1 mm 2 , P < 0.001) ( Fig. 4 ). When subcategorized, we found EUS-FNB provided significantly more total tissue, diagnostic tissue and desmoplastic stroma in solid pancreatic lesions compared to EUS-FNA. However, in non-pancreatic lesions, EUS-FNB yielded a similar amount of total tissue and diagnostic tissue as EUS-FNA.
Table 1. Comparison of outcomes between EUS-FNB and EUS-FNA.
| Patients (n = 36) | |||
| FNB | FNA | P value | |
| Technical success (%) | 100 % | 100 % | |
| Presence of histology core, n (%) | 35, (97 %) | 28, (77 %) | 0.03 |
| Median total tissue area, mm 2 (IQR) | 5.2 (2.1 – 14.1) | 1.9 (0 .4 – 6.6) | < 0.001 |
| Median diagnostic tissue area, mm 2 (IQR) | 2.2 (0 .5 – 6.1) | 0.9 (0.2 – 3.4) | 0.029 |
| Desmoplastic Fibrosis Assessment (n = 23) | 20 (87 %) | 17 (74 %) | 0.45 |
| Median desmoplastic area, mm 2 (IQR) | 2 (0.4 – 6.9) | 0.1 (0 – 0.5) | 0.001 |
| Conducive for IHC staining, (n = 34) Adverse events, n (%) | 33 (97 %) 0 | 30 (88 %) 0 | 0.35 |
FNB, fine-needle biopsy; FNA, fine-needle aspiration; IQR, interquartile range; IHC, immunohistochemistry
Fig. 3.
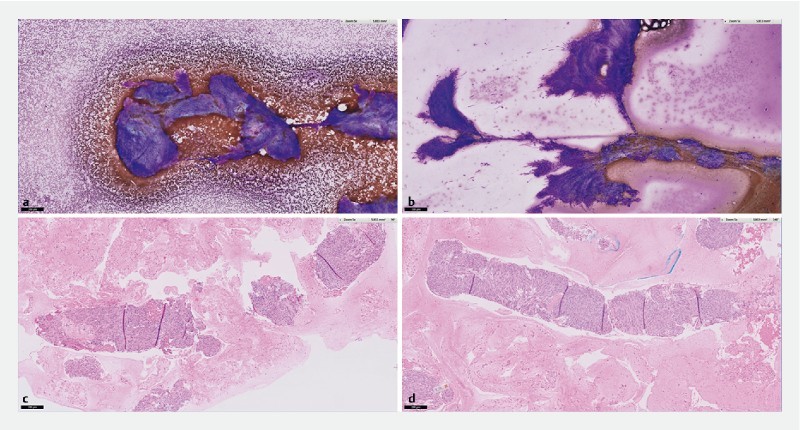
ROSE and cell-block assessment of samples obtained from gastric GIST. a Diff-Quick staining of cytology specimen obtained using Franseen EUS-FNB. b Diff-Quick staining of similar cytology specimen obtained using EUS FNA. c H&E staining of histology obtained using Franseen EUS-FNB. d H&E staining of similar histology obtained using EUS-FNA.
Fig. 4.
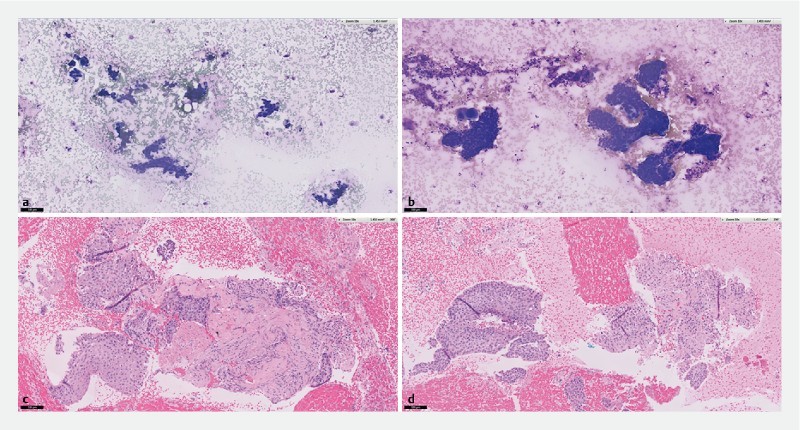
ROSE and cell-block assessment of samples obtained from a lymph node in metastatic squamous cell carcinoma. a Diff-Quick staining of cytology specimen obtained using Franseen EUS-FNB. b Diff-Quick staining of similar cellular yield obtained using EUS FNA. c H&E staining of histology obtained using Franseen EUS-FNB shows malignant cells surrounded by dense desmoplastic stroma. d H&E staining of histology obtained using EUS-FNA shows malignant cells with scanty desmoplastic stroma.
We did not observe any period effect in this crossover trial design ( Table 2 ), and EUS-FNB consistently obtained more histological tissue than EUS-FNA. We evaluated suitability for performing immunohistochemistry (IHC) analysis in 34 lesions where the cell-block had diagnostic tissue and found no significant difference between EUS-FNB and FNA (97 % vs. 88 %).
Table 2. Linear mixed model with a fixed period (Group) and needle (EUS-FNB) effects and random patient-level intercepts for different measurements.
| Estimation | P value | |
| Total histological core area | ||
| Group B | 2.9 (-0.37, 6.26) | 0.08 |
| EUS-FNB | 3.9 (1.94, 5.88) | < 0.001 |
| Total diagnostic tissue area | ||
| Group B | 2.4 (-0.82, 5.53) | 0.141 |
| EUS-FNB | 1.6 (0.17, 3.01) | 0.029 |
| Total desmoplastic stroma area | ||
| Group B | 0.7 (-1.6, 2.92) | 0.549 |
| EUS-FNB | 3.5 (1.6, 5.38) | 0.001 |
EUS-FNB, endoscopic ultrasound-guided fine-needle biopsy
On-site (ROSE) diagnostic yield
We collected smears after each needle pass and assessed for on-site (ROSE) diagnostic yield. We found the overall ROSE diagnostic yield was similar between EUS-FNB (81 %) and EUS-FNA (81 %). In pancreatic lesions, we found the first pass diagnostic yield, as determined by ROSE, was similar between EUS-FNB (80 %) and EUS-FNA (70 %). In non-pancreatic lesions, both EUS-FNB (69 %) and EUS-FNA (69 %) performed suboptimally. We found that additional needle passes using FNB and FNA did not significantly increase the ROSE diagnostic yield in any lesions ( Table 3 ).
Table 3. On-site and cell-block diagnostic rate.
| FNB | FNA | P value | |
| ROSE diagnostic accuracy, n (%) | 29 (81 %) | 29 (81 %) | 1 |
| Pancreatic lesions (n = 20) | |||
|
16 (80 %) | 14 (70 %) | |
|
2 (50 %) | 4 (67 %) | |
|
0 (0 %) | 0 (0 %) | |
| Non-pancreatic lesions (n = 16) | |||
|
11 (69 %) | 11 (69 %) | |
|
0 (0 %) | 0 (0 %) | |
| Cell block diagnostic accuracy, n (%) | 29 (81 %) | 23 (64 %) | 0.19 |
| Pancreatic lesions (n = 20) | |||
|
12 (60 %) | 9 (45 %) | |
|
1 (12 %) | 1 (9 %) | |
|
1 (14 %) | 2 (20 %) | |
| Non-pancreatic lesions (n = 16) | |||
|
13 (81 %) | 10 (63 %) | |
|
2 (40 %) | 1 (14 %) | |
| Combined ROSE and cell block diagnosis, n (%) | 33 (92 %) | 31 (86 %) | 0.71 |
| Pancreatic lesions (n = 20) | 18 (90 %) | 18 (90 %) | |
| Non-pancreatic lesions (n = 16) | 15 (94 %) | 13 (81 %) | |
ROSE, rapid on-site cytological evaluation
When combined with cell-block, the overall diagnostic yield improved to 92 % in EUS-FNB group and 86 % in EUS-FNA group.
Quantification and qualification of nucleic acid
We estimated the volume of DNA and RNA obtained from the pancreatic lesions (n = 20) by the two needles. The lesions included 18 pancreatic adenocarcinomas, one neuroendocrine tumor, and one metastatic renal cell carcinoma. We found that EUS-FNB procured significantly more nucleic acid than EUS-FNA [median; 4,085 (2,804 – 6711) ng vs. 2912 (2,287 – 4,658) ng, P = 0.02). The increased nucleic acid yield corresponds to the increased total tissue yield with EUS-FNB. We noticed the EUS-FNB yielded significantly more RNA [median; 1,634 (1089 – 3,939) ng vs. 1295 (986 – 1,782) ng, P = 0.02] and increased the amount of DNA [median; 2,185 (1,478 – 3,066) ng vs. 1477 (1,151 – 2,522), P = 0.08] when compared to EUS-FNA ( Fig.5 ). We assessed the first-pass nucleic acid yield and found a trend towards increased yield with EUS-FNB compared to EUS-FNA [median;1142 (618 – 2435) vs.1090 (750 – 1341) ng, P = 0.07).
Fig. 5.
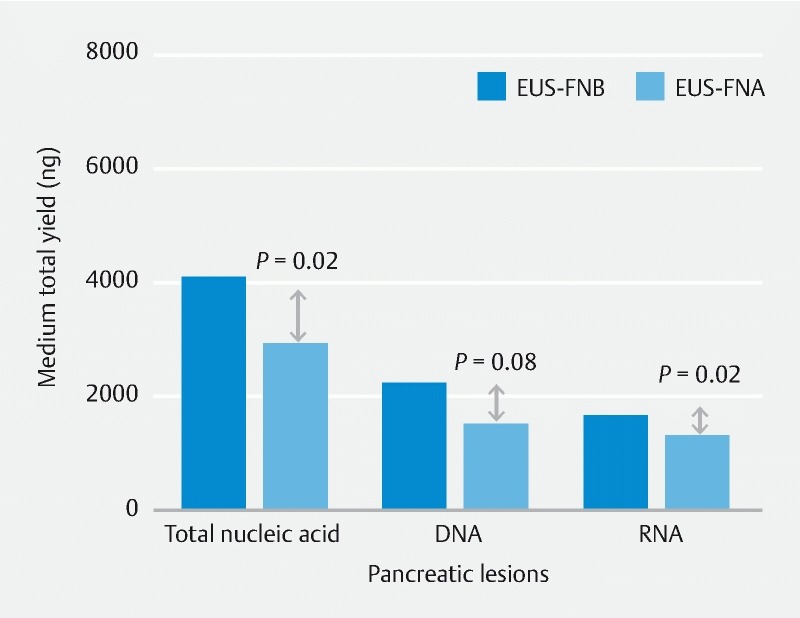
Quantification of nucleic acid obtained by the two needles. EUS-FNB provided significantly more nucleic acid than EUS-FNA. EUS-FNB yielded significantly more RNA and a trend towards an increased amount of DNA.
We analyzed the purity of the DNA and RNA extracted by measuring the ratio of absorbance at 260 /280 nm (optimal threshold range,1.6 – 2.2) and 260 /230 nm (optimal threshold > 1) using Nanodrop spectrophotometer. We found that both EUS-FNB (95 %) and FNA (90 %) needles procured high-quality DNA in pancreatic lesions. There was a trend towards increased higher-quality RNA yield with EUS-FNB (30 %) than with EUS-FNA (10 %). We also evaluated the ability to perform sequencing analysis from the nucleic acid obtained using both needles (n = 2). We found that both needles exceeded the manufacturer recommendation of minimum DNA or RNA concentration (10ng/reaction) needed for tumor profiling using the next-generation sequencing assay (NGS, Oncomine M V3). The NGS results were identical between the two needles.
Complications
There were no complications after EUS-FNB and EUS-FNA in either group.
Discussion
We report the outcome of an RCT comparing Franseen design EUS-FNB versus standard EUS-FNA in solid pancreatic and non-pancreatic lesions. EUS-FNB provided significantly more total histological tissue, diagnostic tissue, and desmoplastic stroma compared to EUS-FNA. However, the diagnostic yield as measured using ROSE and cell block was similar.
The Franseen EUS-FNB needle design has three symmetrical cutting edges. It was postulated that these edges provide stability at puncture, penetrate quickly, and capture maximum tissue with minimal fragmentation. Our study provided some support for this hypothesis and is in agreement with a recent randomized study showing superior histological core and tumor tissue procurements with the 22G Franseen EUS-FNB compared to a standard 22G EUS-FNA needle, in solid pancreatic lesions 19 . It also appears that Franseen EUS-FNB, with its coring ability and tendency to provide a large volume of tissue for cell-block analysis, may overcome the need for ROSE 23 24
In a large retrospective study assessing the utility of Franseen EUS-FNB in solid pancreatic and non-pancreatic lesions, the overall cell-block diagnostic yield (62 %) was found to be significantly lower 25 . This observation was in sharp contrast to the superior cell-block yield (> 90 %) reported in the randomized studies on Franseen EUS-FNB in pancreatic lesions 19 23 . Our results did not support the results of the randomized study. In fact, we found the cell-block diagnostic yield to be lower (81%), and the overall yield improved only with presence of ROSE. One possible cause could be the proportionately larger yield of desmoplastic stroma (10 times higher) than diagnostic tissue (1.5 times higher) compared with EUS-FNA. Other probable contributing factors may include the heterogeneous study cohort, technical difference between the operators, experience of the on-site cytotechnician, cytopathologist expertise and use of specialized imaging software to assess the histology sample.
There are potential issues with obtaining desmoplastic tissue. In pancreatic cancer (PC), a dense, extensive desmoplastic reaction is a typical finding and concern has been raised about its role in drug resistance 26 . Procuring histological samples abundant in desmoplastic stroma may be useful for clinical trials evaluating therapy that targets desmoplastic stroma 27 28 . Similarly, molecular profiling of PC and application of next-generation sequencing may provide an opportunity to advance development of targeted therapies and improve PC treatment outcomes 20 29 30 . Studies evaluating personalized medicine in PC have relied mainly on surgically acquired tissue for performing genetic analysis. Unfortunately, most PC present at an inoperable stage and real-time genetic analysis of advanced cancers is limited. EUS, a safe and minimally invasive technique, is widely used to acquire tissue and establish a diagnosis in such situations. Until now, there was varying evidence on the suitability of EUS-FNA- and EUS-FNB-acquired tissue samples for genomic analysis 31 32 33 34 35 36 37 . Some studies suggest EUS-FNA-acquired cytology samples and liquid cytology specimens (FNA rinse material) have a higher concentration of intact and pure tumor cells and are superior for NGS. Others consider that the cellular material obtained using EUS-FNA is often graded to be insufficient, contaminated, composed of poor-quality DNA 29 38 39 and suboptimal for genetic analysis.
Gleeson et al showed that a cytology slide > 5000 cells, tissue volume > 1.2 cm 2 in FFPE sample and at least 5 ng/uL of recoverable DNA has a higher success rate (> 90 %) at NGS 31 . Our study showed that the Franseen EUS-FNB needle can yield a large amount of tissue and provide an adequate amount of nucleic acids with relatively preserved high-quality DNA and RNA. In the limited samples (n = 2) with NGS results, the quality metrics were all met with high scores, thus providing evidence that FNB-acquired specimens can serve as a tool for genomic studies and a surrogate to surgical specimens, especially in patients not amenable to surgical intervention 40 . Rodriguez et al showed that reliable RNA sequencing (Illumina, Inc, United States) can be performed if the EUS-TA can acquire 100 ng of total RNA 41 . Our study demonstrated that the increased high-quality RNA yield with EUS-FNB may make performance of RNA sequencing feasible. It would provide a platform for measuring gene expression and capturing novel diagnostic and prognostic information about pancreatic cancer.
Our study has certain limitations. It was not possible to blind the endoscopists to the study needles used. We believe by applying a crossover trial design, may have eliminated the operator-related bias. Second, our study is possibly underpowered as the diagnostic histology yield with EUS-FNA (64 %) was higher than the reference rate (40 %) used during power calculation. A type II error cannot be excluded. However, post-hoc analysis demonstrated that the study achieved 90 % power with α of 0.05, when estimated to detect a 1 mm 2 difference in the median total tissue area between the two needles 19 . Third, mean size (3.8 ± 2.0 cm) of the lesions included in the study was larger, which may have contributed to the similar diagnostic efficacy for EUS-FNA and EUS-FNB. Performance of EUS-FNB and EUS-FNA in smaller (< 2 cm) lesions needs to be assessed. Fourth, our secondary outcome was to perform a DNA and RNA quantification in all the lesions that we sampled. Funding limitations prevented us from pursuing more wide-scale NGS. Fifth, we estimated DNA and RNA from FFPE specimens. Formalin fixation and sectioning of the cell block may have degraded and fragmented DNA and RNA, resulting in lower yields. Performing a dedicated additional pass and using RNA later (Ambion, Austin, Texas, United States) or CytoLyt (Hologic Co, Marlborough, Massachusetts, United States) preservative may have increased the RNA and DNA yield further 42 43 . Lastly, we did not use any automated or artificial intelligence software for histological assessment and quantification. Nonetheless, such digital software is not widely implemented, and manual histological assessment is more widespread and applicable at this time point.
Conclusion
In conclusion, our study shows that the Franseen EUS-FNB and EUS-FNA device are equally effective in establishing a diagnosis in all solid gastrointestinal and extra-intestinal lesions. We have demonstrated that the Franseen EUS-FNB device can obtain more histological tissue and has better nucleic acid yield with quality and quantity sufficient for downstream genomics applications like NGS. This is a significant step in EUS-FNB becoming a convenient and safe method for obtaining tumor material for precision genomics.
Acknowledgement
SingHealth-Pitch provided funding for the program.
Footnotes
Competing interests None
References
- 1.Itoi T, Sofuni A, Itokawa F et al. Current status of diagnostic endoscopic ultrasonography in the evaluation of pancreatic mass lesions. Dig Endosc. 2011;23:17–21. doi: 10.1111/j.1443-1661.2011.01132.x. [DOI] [PubMed] [Google Scholar]
- 2.Early D S, Ben-Menachem T, Decker G A et al. Appropriate use of GI endoscopy. Gastrointest Endosc. 2012;75:1127–1131. doi: 10.1016/j.gie.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Lorenzo F, Alberto L et al. Endoscopic ultrasound-guided fine needle aspiration: How to obtain a core biopsy? Endosc Ultrasound. 2014;3:71–81. doi: 10.4103/2303-9027.123011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iglesias-Garcia J, Poley J W, Larghi A et al. Feasibility and yield of a new EUS histology needle: results from a multicenter, pooled, cohort study. Gastrointest Endosc. 2011;73:1189–1196. doi: 10.1016/j.gie.2011.01.053. [DOI] [PubMed] [Google Scholar]
- 5.Jenssen C, Dietrich C F. Endoscopic ultrasound-guided fine needle aspiration biopsy and trucut biopsy in gastroenterology – An overview. Best Pract Res Clin Gastroenterol. 2009;23:743–759. doi: 10.1016/j.bpg.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Rastogi A, Wani S, Gupta N et al. A prospective, single-blind, randomized, controlled trial of EUS-guided FNA with and without a stylet. Gastrointest Endosc. 2011;74:58–64. doi: 10.1016/j.gie.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 7.Puri R, Vilmann P, Săftoiu A et al. Randomized controlled trial of endoscopic ultrasound-guided fine-needle sampling with or without suction for better cytological diagnosis. Scand J Gastroenterol. 2009;44:499–504. doi: 10.1080/00365520802647392. [DOI] [PubMed] [Google Scholar]
- 8.Sachin W. Basic techniques in endoscopic ultrasound-guided fine-needle aspiration: Role of a stylet and suction. Endosc Ultrasound. 2014;3:17–21. doi: 10.4103/2303-9027.123008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bang J Y, Hebert-Magee S, Trevino J et al. Randomized trial comparing the 22-gauge aspiration and 22-gauge biopsy needles for EUS-guided sampling of solid pancreatic mass lesions. Gastrointest Endosc. 2012;76:321–327. doi: 10.1016/j.gie.2012.03.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanbiervliet G, Napoléon B, Saint Paul M C et al. Core needle versus standard needle for endoscopic ultrasound-guided biopsy of solid pancreatic masses: a randomized crossover study. Endoscopy. 2014;46:1063–1070. doi: 10.1055/s-0034-1377559. [DOI] [PubMed] [Google Scholar]
- 11.Săftoiu A, 1, Vilmann P, Guldhammer Skov B et al. Endoscopic ultrasound (EUS)-guided Trucut biopsy adds significant information to EUS-guided fine-needle aspiration in selected patients: a prospective study. Scand J Gastroenterol. 2007;42:117–125. doi: 10.1080/00365520600789800. [DOI] [PubMed] [Google Scholar]
- 12.Tian L, Tang A L, Zhang L et al. Evaluation of 22G fine-needle aspiration (FNA) versus fine-needle biopsy (FNB) for endoscopic ultrasound-guided sampling of pancreatic lesions: a prospective comparison study. Surg Endosc. 2018;32:3533–3539. doi: 10.1007/s00464-018-6075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noh D H, Choi K, Gu S et al. Comparison of 22-gauge standard fine needle versus core biopsy needle for endoscopic ultrasound-guided sampling of suspected pancreatic cancer: a randomized crossover trial. Scand J Gastroenterol. 2018;53:94–99. doi: 10.1080/00365521.2017.1390597. [DOI] [PubMed] [Google Scholar]
- 14.Bang J Y, Hawes R, Varadarajulu S. A meta-analysis comparing ProCore and standard fine-needle aspiration needles for endoscopic ultrasound-guided tissue acquisition. Endoscopy. 2016;48:339–349. doi: 10.1055/s-0034-1393354. [DOI] [PubMed] [Google Scholar]
- 15.Bang J Y, Hebert-Magee S, Hasan M K et al. Endoscopic ultrasonography-guided biopsy using a Franseen needle design: Initial assessment. Dig Endosc. 2017;29:338–346. doi: 10.1111/den.12769. [DOI] [PubMed] [Google Scholar]
- 16.Al-Haddad M, Patel K, Othman M O. Prospective assessment of the performance of a new fine needle biopsy device at 2 large referral centers [abstract] Gastrointest Endosc. 2017;85:AB333. [Google Scholar]
- 17.Mitri R D, Rimbaş M, Attili F et al. Performance of a new needle for endoscopic ultrasound-guided fine-needle biopsy in patients with pancreatic solid lesions: A retrospective multicenter study. Endosc Ultrasound. 2018;7:329–334. doi: 10.4103/eus.eus_33_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukai S, Itoi T, Yamaguchi H et al. A retrospective histological comparison of EUS-guided fine-needle biopsy using a novel Franseen needle and a conventional end-cut type needle. Endosc Ultrasound. 2019;8:50–57. doi: 10.4103/eus.eus_11_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bang J Y, Hebert-Magee S, Navaneethan U et al. EUS-guided fine needle biopsy of pancreatic masses can yield true histology: results of a randomized trial. Gut. 2018;67:2081–2084. doi: 10.1136/gutjnl-2017-315154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wani S, Muthusamy V R, McGrath C M et al. AGA White Paper: Optimizing Endoscopic Ultrasound-Guided Tissue Acquisition and Future Directions. Clin Gastroenterol Hepatol. 2018;16:318–327. doi: 10.1016/j.cgh.2017.10.020. [DOI] [PubMed] [Google Scholar]
- 21.Yang M J, Yim H, Hwang J C et al. Endoscopic ultrasound-guided sampling of solid pancreatic masses: 22-gauge aspiration versus 25-gauge biopsy needles. BMC Gastroenterol. 2015;15:122. doi: 10.1186/s12876-015-0352-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jovani M, Abidi W M, Lee L S. Novel fork-tip needles versus standard needles for EUS-guided tissue acquisition from solid masses of the upper GI tract: a matched cohort study. Scand J Gastroenterol. 2017;52:784–787. doi: 10.1080/00365521.2017.1306879. [DOI] [PubMed] [Google Scholar]
- 23.Bang J Y, Hebert-Magee S, Navaneethan U et al. Randomized trial comparing the Franseen and Fork-tip needles for EUS-guided fine-needle biopsy sampling of solid pancreatic mass lesions. Gastrointest Endosc. 2018;87:1432–1438. doi: 10.1016/j.gie.2017.11.036. [DOI] [PubMed] [Google Scholar]
- 24.Leung Ki E L, Lemaistre A I, Fumex F et al. Macroscopic onsite evaluation using endoscopic ultrasound fine needle biopsy as an alternative to rapid onsite evaluation. Endosc Int Open. 2019;7:E189–E194. doi: 10.1055/a-0770-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdelfatah M M, Grimm I S, Gangarosa L M et al. Cohort study comparing the diagnostic yields of 2 different EUS fine-needle biopsy needles. Gastrointest Endosc. 2018;87:495–500. doi: 10.1016/j.gie.2017.08.033. [DOI] [PubMed] [Google Scholar]
- 26.Whatcott C J, Diep C H, Jiang P et al. Desmoplasia in primary tumors and metastatic lesions of pancreatic cancer. Clin Cancer Res. 2015;21:3561–3568. doi: 10.1158/1078-0432.CCR-14-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bahrami A, Khazaei M, Bagherieh F et al. Targeting stroma in pancreatic cancer: Promises and failures of targeted therapies. J Cell Physiol. 2017;232:2931–2937. doi: 10.1002/jcp.25798. [DOI] [PubMed] [Google Scholar]
- 28.Nicolle R, Blum Y, Marisa L et al. Pancreatic adenocarcinoma therapeutic targets revealed by tumor-stroma cross-talk analyses in patient-derived xenografts. Cell Rep. 2017;21:2458–2470. doi: 10.1016/j.celrep.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chantrill L A, Nagrial A M, Watson C et al. Precision medicine for advanced pancreas cancer: the individualized molecular pancreatic cancer therapy (IMPaCT) trial. Clin Cancer Res. 2015;21:2029–2037. doi: 10.1158/1078-0432.CCR-15-0426. [DOI] [PubMed] [Google Scholar]
- 30.Berry W, Lundy J, Croagh D et al. Reviewing the utility of EUS-FNA to advance precision medicine in pancreatic cancer. Cancers (Basel) 2018:10. doi: 10.3390/cancers10020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gleeson F C, Kipp B R, Voss J S et al. Endoscopic ultrasound fine-needle aspiration cytology mutation profiling using targeted next-generation sequencing: personalized care for rectal cancer. Am J Clin Pathol. 2015;143:879–888. doi: 10.1309/AJCPU3J7FGAYQBRL. [DOI] [PubMed] [Google Scholar]
- 32.Bellevicine C, Vita G D, Malapelle U et al. Applications and limitations of oncogene mutation testing in clinical cytopathology. Semin Diagn Pathol. 2013;30:284–297. doi: 10.1053/j.semdp.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 33.Wei S, Lieberman D, Morrissette J J et al. Using “residual” FNA rinse and body fluid specimens for next-generation sequencing: an institutional experience. Cancer Cytopathol. 2016;124:324–329. doi: 10.1002/cncy.21666. [DOI] [PubMed] [Google Scholar]
- 34.Roy-Chowdhuri S, Stewart J. Preanalytic variables in cytology: lessons learned from next-generation sequencing. Arch Pathol Lab Med. 2016;140:1191–1199. doi: 10.5858/arpa.2016-0117-RA. [DOI] [PubMed] [Google Scholar]
- 35.Roy-Chowdhuri S, Goswami R S, Chen H et al. Factors affecting the success of next-generation sequencing in cytology specimens. Cancer Cytopathol. 2015;123:659–668. doi: 10.1002/cncy.21597. [DOI] [PubMed] [Google Scholar]
- 36.Roy-Chowdhuri S, Chen H, Singh R R et al. Concurrent fine needle aspirations and core needle biopsies: a comparative study of substrates for next-generation sequencing in solid organ malignancies. Mod Pathol. 2017;30:499–508. doi: 10.1038/modpathol.2016.228. [DOI] [PubMed] [Google Scholar]
- 37.Kandel P, Wallace M B. Advanced EUS-Guided tissue acquisition methods for pancreatic cancer. Cancers (Basel) 2018:10. doi: 10.3390/cancers10020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noh D H et al. Factors of endoscopic ultrasound fine-needle aspiration for successful next-generation sequencing in pancreatic cancer. Gastrointest Endosc. 2015;83:AB334. [Google Scholar]
- 39.Navina S, McGrath K, Chennat J et al. Adequacy assessment of endoscopic ultrasound-guided, fine-needle aspirations of pancreatic masses for theranostic studies: optimization of current practices is warranted. Arch Pathol Lab Med. 2014;138:923–928. doi: 10.5858/arpa.2013-0335-OA. [DOI] [PubMed] [Google Scholar]
- 40.Pujan K, Aziza N, Ghassan T et al. Su1326 feasibility of whole exome sequencing and genomic profiling of pancreas tumor tissue obtained with a novel fork-tipped EUS- guided fine needle core biopsy. 2017;85:AB335–AB336. [Google Scholar]
- 41.Rodriguez S A, Impey S D, Pelz C et al. RNA sequencing distinguishes benign from malignant pancreatic lesions sampled by EUS-guided FNA. Gastrointest Endosc. 2016;84:252–258. doi: 10.1016/j.gie.2016.01.042. [DOI] [PubMed] [Google Scholar]
- 42.Lawson M H, Rassl D M, Cummings N M et al. Tissue banking of diagnostic lung cancer biopsies for extraction of high quality RNA. J Thorac Oncol. 2010;5:956–963. doi: 10.1097/JTO.0b013e3181ddbbe9. [DOI] [PubMed] [Google Scholar]
- 43.Dejmek A, Zendehrokh N, Tomaszewska M et al. Preparation of DNA from cytological material. Cancer Cytopathol. 2013;121:344–353. doi: 10.1002/cncy.21276. [DOI] [PubMed] [Google Scholar]


