Abstract
Listeria monocytogenes is an important zoonotic foodborne pathogen that can tolerate a number of environmental stresses. RsbR, an upstream regulator of the sigma B (SigB) factor, is thought to sense environmental challenges and trigger the SigB pathway. In Bacillus subtilis, two phosphorylation sites in RsbR are involved in activating the SigB pathway and a feedback mechanism, respectively. In this study, the role of RsbR in L. monocytogenes under mild and severe stresses was investigated. Strains with genetic deletion (ΔrsbR), complementation (C-ΔrsbR), and phosphorylation site mutations in the rsbR (RsbR-T175A, RsbR-T209A, and RsbR-T175A-T209A) were constructed to evaluate the roles of these RsbR sequences in listerial growth and survival. SigB was examined at the transcriptional and translational levels. Deletion of rsbR reduced listerial growxth and survival in response to acidic stress. Substitution of the phosphorylation residue RsbR-T175A disabled RsbR complementation, while RsbR-T209A significantly upregulated SigB expression and listerial survival. Our results provide clear evidence that two phosphorylation sites of RsbR are functional in L. monocytogenes under acidic conditions, similar to the situation in B. subtilis.
Keywords: Listeria monocytogenes, RsbR, Sigma B (Sig B) factor, Phosphorylation
1. Introduction
Listeria monocytogenes can cause septicemia, meningitis, and abortion in pregnant women with mortality rates as high as 30% (Varma et al., 2007; Okpo et al., 2015). As a foodborne pathogen, L. monocytogenes encounters acidic environments such as silage, fermented food, stomach, and phagolysosomes within host cells (Nicaogáin and O'Byrne, 2016). Its ability to adapt to relatively harsh environmental conditions is one of the important features enabling its persistence in food processing environments and for establishing infection in the host. L. monocytogenes can maintain its intracellular pH (pHi) homeostasis when exposed to an environmental pH (pHex) of 4.5, and survives well at pHex 3.5 (Cotter et al., 2000). Pre-exposure of L. monocytogenes cells to mild acid stress (pHex 4.5) may induce an acid tolerance response (ATR) that can render them more resistant to fatal acidic stress (O'Byrne and Karatzas, 2008). Sigma B (SigB or σB) is known as a positive regulator of ATR enabling bacteria to cope better with environmental stresses.
SigB was first identified in Bacillus subtilis, and is also present in L. monocytogenes and Staphylococcus aureus (Hecker et al., 2007, 2009; Price, 2011). It plays a pivotal role in resistance to environmental stresses in these bacteria. The SigB operons, consisting of rsbR-rsbS-rsbT-(rsbU)-rsbV-rsbW-sigB-rsbX, are highly conserved between L. monocytogenes and B. subtilis (O'Byrne and Karatzas, 2008), suggesting existence of similar regulatory mechanisms. In B. subtilis, partner switching through serine phosphorylation and dephosphorylation is the main regulatory mechanism of this operon in response to stress (Hecker et al., 2007; Eymann et al., 2011; Hengge, 2011). The operon is composed of the co-antagonist RsbR, antagonist RsbS, and serine-threonine kinase RsbT as the upstream regulators (Delumeau et al., 2006). The phosphorylation of RsbR-RsbT-RsbS could be the initial crucial step to trigger activation of the SigB pathway. There are three important phosphorylation sites in this complex: residues T171 and T205 of RsbRA (formerly RsbR), and residue S59 of RsbS (Chen et al., 2004; Kim et al., 2004a; Liebal et al., 2013). RsbRA-T171 is a prerequisite and related to stress-induced phosphorylation of RsbS-S59, while RsbRA-T205 phosphorylation may function as a second feedback mechanism to limit σB activation (Eymann et al., 2011).
In L. monocytogenes, upstream genes in this operon, rsbT, rsbV, and rsbU, have been proved to play roles in activating σB in response to nutritional and environmental stresses (Chaturongakul and Boor, 2004; Shin et al., 2010; Zhang et al., 2013). Martinez et al. (2010) expressed the RsbR protein of L. monocytogenes in the B. subtilis ΔrsbR mutant, and detected σB induction following nutritional stress. In our previous study (Xia et al., 2016), we showed that RsbX was a negative regulator of L. monocytogenes σB during the recovery period after the primary stress or in the stationary phase, thus affecting its survival under secondary stress. However, the detailed roles of RsbR, RsbT, and RsbS in the resistance of L. monocytogenes to environmental stresses remain unclear. This study was attempted to examine whether RsbR is vital in L. monocytogenes under acid stress, and if the two putative phosphorylation sites are functional.
2. Materials and methods
2.1. Bacterial strains, plasmids, and culture conditions
L. monocytogenes reference strain 10403S, a serotype 1/2a isolate from a human skin lesion, was provided by Dr. Martin Wiedmann (Cornell University, USA) and used as the wild-type strain. The plasmids pMD19-T (TaKaRa, Dalian, China), pERL3 (a gift from Dr. Qin LUO, Huazhong Normal University, Wuhan, China), and pKSV7 (a gift from Dr. M. WIEDMANN) were maintained in Escherichia coli DH5α as the host strain. L. monocytogenes was cultured in brain heart infusion (BHI) medium (Dingguo, Beijing, China), and E. coli was grown in Luria Bertani (LB) broth (Oxoid, Hampshire, UK), all at 37 °C.
2.2. Bioinformatics analysis
Alignment of amino acid sequences was performed with MegAlign using the DNAStar software package (Version 5; Madison, USA). Four RsbR paralogs of B. subtilis, BS-RsbRA (GenBank: P42409), BS-RsbRB (GenBank: O34860), BS-RsbRC (GenBank: O31856), and BS-RsbRD (GenBank: P54504), were used to align with L. monocytogenes RsbR (LM-RsbR; GenBank: CP002002). The three homologous RsbRs in L. monocytogenes were from the genome of 10403S (GenBank: CP002002). The phosphorylation sites of BS-RsbRs were searched in PhosphositePlus (https://www.phosphosite.org), and the phosphorylation sites of LM-RsbR were predicted by DISPHOS (http://www.dabi.temple.edu/disphos).
2.3. Construction of the rsbR deletion mutant
The temperature-sensitive pKSV7 shuttle vector was used for generating the ΔrsbR mutant from the wild-type strain L. monocytogenes 10403S. Gene splicing using the splice-overlap extension PCR (SOE-PCR) procedure was used to construct the homologous arms of rsbR (Table 1), and a homologous recombination strategy was used to construct the ΔrsbR mutant as described by Xia et al. (2016). The ΔsigB mutant kept in our laboratory was constructed using the same approach.
Table 1.
PCR primers used in this study
| Name | Primer sequence (5'→3')a | Purpose |
| rsbR-a | CCGGGATCCATACGGCCTGTTCTCATCATTC | Construction of rsbR null mutant |
| rsbR-b | ACGCGTTGTTTGTAGGTTTCTG | |
| rsbR-c | CAGAAACCTACAAACAACGCGTTACAGCCAGCAGTTGCGATTACAC | |
| rsbR-d | CCCAAGCTTTACAGATTTCTCCTCGTCCAG | |
| rsbR-up | GCGAGTGTACCCATGTCGAAGCAG | Screening of positive clones of rsbR null mutant |
| rsbR-dn | CCCCAATTCCTGTTTAAGTTTTTCCAG | |
| rsbR-W | ACGCGTCGACCAATCCAAGCAAAATAGCTAGGTAGAAA | Complementation of rsbR deletion |
| rsbR-X | CAATCCAAGCAAAATAGCTAGGTAGAAA | |
| rsbR-Y | CTAGCTATTTTGCTTGGATTGATGTATAAAGATTTTGCAAACTT | |
| rsbR-Z | CGAGCTCTCACCCCTCTTTTTCTACT | |
| rsbR175m-F | TGTAATGCCGTTAATTGGAACGATTGACGCAGAAAGAGCCAAG | Construction of mutant rsbR complementary plasmid |
| rsbR175m-R | AGTAAGTTTTCTATGATTAACTTGGCTCTTTCTGCGTCAATCGT | |
| rsbR209m-F | TGATTGATATTACGGGAGTTCCTGTTGTTGATGCAATGGTTGCG | |
| rsbR209m-R | GACGCTTGAATAATATGGTGCGCAACCATTGCATCAACAACAGG | |
| rsbR-RT-F | CGCGGATACGTGGGAAAAG | qRT-PCR |
| rsbR-RT-R | TGCCTGACAGCCGACAAGTCT |
Nucleotides introduced to create restriction sites and point mutations are underlined and in bold, respectively. qRT-PCR: quantitative reverse transcription PCR
2.4. Complementation of the rsbR deletion mutant
Expression of rsbR was complemented based on previous methods (Chen et al., 2011; Xia et al., 2016). SOE-PCR was used with the primer pairs rsbR-W/X and rsbR-Y/Z to fuse the rsbR open reading frame (ORF) with the dlt promoter. The product was ligated to pERL3 and confirmed by sequencing. The recombinant plasmid pERL3-Pdlt-rsbR was electroporated into the L. monocytogenes ΔrsbR strain and transformants were identified by PCR using the primer pair rsbR-Y/Z.
2.5. Site-directed mutagenesis
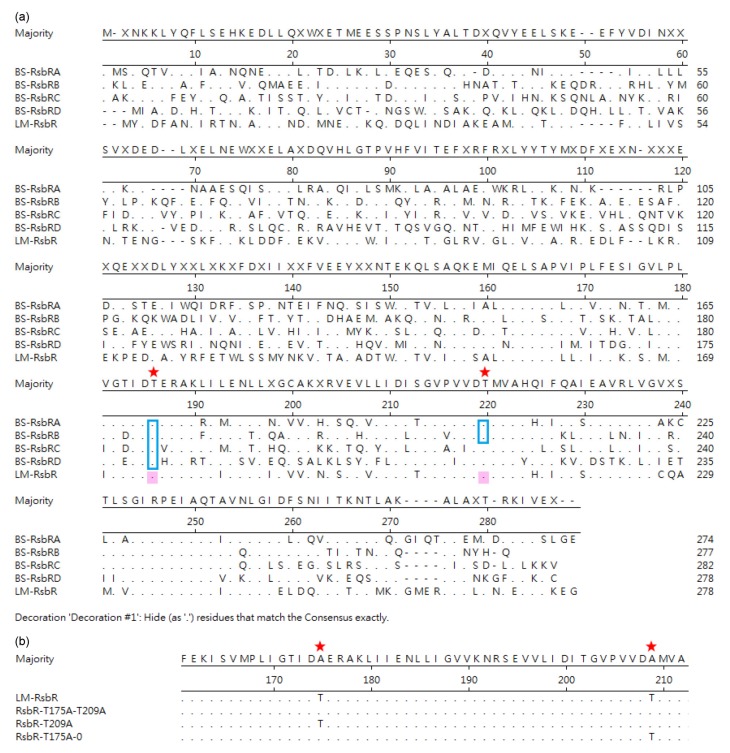
Residues T175 and T209 in RsbR of L. monocytogenes might be the vital phosphorylation sites (Fig. 1) corresponding to residues T171 and T205 of RsbR in B. subtilis. These two sites were chosen for site-directed mutagenesis of three expression plasmids containing single mutations (T175A=M1 or T209A=M2) or double mutations (T175A and T209A=M3). The plasmids were generated from the recombinant vector pERL3-Pdlt-rsbR using the primers listed in Table 1. All three mutated fragments were ligated with pERL3, confirmed by sequencing and electroporated into the L. monocytogenes ΔrsbR strain, as described above.
Fig. 1.
Sequence alignment of RsbR homologs of Bacillus subtilis (BS) and Listeria monocytogenes (LM) (a) and alanine substitutions in the recombinant complementary plasmids (b)
Blue squares indicate the phosphorylation sites in PhosphositePlus (https://www.phosphosite.org); pink shading indicates the phosphorylation sites predicted by DISPHOS (http://www.dabi.temple.edu/disphos); red stars indicate the conservative phosphorylation sites in both BS and LM (Note: for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article)
2.6. Growth and survival assays
All strains (L. monocytogenes 10403S, the ΔrsbR mutant and ΔrsbR complemented (C-ΔrsbR) strains; (Table 2)) were grown to stationary phase with shaking at 37 °C. For the growth assay, the bacteria were adjusted to similar optical density at 620 nm (OD620). Each calibrated strain was inoculated at a 1:20 ratio in BHI at pH 7.4 or 5.0 and then transferred into wells (triplicate wells per strain) on a microplate. The bacterial cultures were incubated at 37 °C for 8 h, and the OD620 was then captured as the end-point measurement to indicate their relative growth potential. For the survival assay, bacterial cells at exponential phase were transferred at a 1:20 ratio in BHI at pH 7.4 and cultured with shaking at 37 °C for 4 h (to early exponential phase, OD620 was approximately 0.3). One milliliter of each culture was centrifuged, resuspended with 1 mL BHI at pH 2.5 and incubated at 37 °C for 30 min. The treated cultures were subjected to immediate dilution in 10 mmol/L phosphate-buffered saline (PBS) at pH 7.4 at the end of 30-min incubation, followed by serial 10-fold dilutions. The diluted samples were plated onto BHI agar for the enumeration of viable bacterial cells. Relative survival was calculated by dividing the colony forming units (CFUs) of the mutants by those of the wild-type strain. The experiments were carried out three times.
Table 2.
Listeria monocytogenes strains
| L. monocytogenes strain | Genotype |
| WT | 10403S |
| ΔrsbR | rsbR null |
| C-ΔrsbR | Pdlt::rsbR |
| C-ΔrsbR-M1 | Pdlt::rsbR-T175A |
| C-ΔrsbR-M2 | Pdlt::rsbR-T209A |
| C-ΔrsbR-M3 | Pdlt::rsbR-T175A-T209A |
Pdlt::rsbR means that the promoter of dlt and open reading frame (ORF) of rsbR are connected together
2.7. Acidic stress for analysis of SigB expression
For transcriptional analysis of sigB after acid stress, all the strains were grown to stationary phase in BHI, diluted 1:20 in 40 mL BHI at pH 7.4 and cultured at 37 °C for 6 h (to late exponential phase, OD620 was approximately 0.45). The cultures were then divided into two parts (each in a different sterile tube) and were centrifuged at 3000 r/min for 5 min, and then resuspended in 20 mL BHI at pH 5.0 (stressed) or BHI at pH 7.4 (as unstressed control) for 1 h. At the end of the treatment, 1 mL of the culture was taken for total RNA isolation, and the rest for extraction of total proteins.
2.8. qRT-PCR and western blotting
Total RNAs from the treatments as described above were prepared using an RNAprep Pure Cell/Bacteria kit (Tiangen Biotech, Beijing, China) and complementary DNAs (cDNAs) were synthesized with ReverTra Ace® reverse transcriptase (Toyobo, Japan). For quantification of sigB and rsbR mRNAs, quantitative reverse transcription PCR (qRT-PCR) was performed as described by Xia et al. (2016). The gyrB gene was used as a reference. Primers for sigB and gyrB were as listed elsewhere (Xia et al., 2016), and primers for rsbR were rsbR-RT-F/R (Table 1). Each treatment was tested in triplicate and the relative expression of each strain was calculated using the 2−ΔΔ C T method (Livak and Schmittgen, 2001), and transcriptional level of sigB was normalized as Shan et al. (2018).
For western blotting, total proteins were extracted from the bacterial cells treated as above following a previous procedure (Xia et al., 2016). The protein concentration was adjusted to the same level for different strains and separated by 12% (0.12 g/mL) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The remaining procedures for western blotting and image capture were followed as described (Zhou et al., 2017), using rabbit polyclonal antibodies against SigB and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Xia et al., 2016) as well as antibodies against RsbR prepared in this study by immunization of rabbits with RsbR protein expressed in E. coli strain BL21. Horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (Sangon, China) was used at 1:2000 (v/v).
2.9. Statistical analysis
All data are expressed as mean±standard deviation (SD) of three independent experiments. Two-tailed Student’s t-tests were employed to extract statistical significance among or between treatments or strains.
3. Results
3.1. Identification of the rsbR deletion mutant and complemented strains
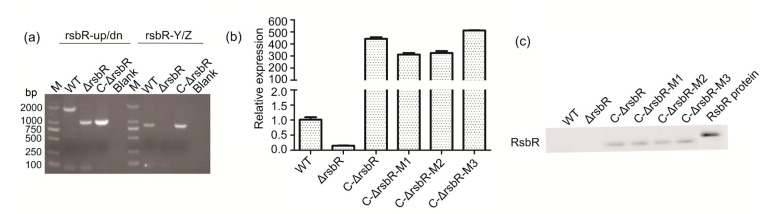
PCR, qRT-PCR, and western blotting were used to confirm successful construction of the rsbR deletion mutant and complemented strain. Fig. 2 shows that the fragments were shorter with the primer pair rsbR-up/dn in ΔrsbR and C-ΔrsbR, suggesting deletion of rsbR in the bacterial genome (Fig. 2a). With the primer pair rsbR-Y/Z, fragments showing the size of the rsbR ORF (687 bp) were present in C-ΔrsbR and the wild-type strain. All strains, including those complemented strains having substitutions of single or double amino acid residues, were confirmed by sequencing. Expression of rsbR was significantly higher in all complemented strains than in their parent strains (Fig. 2b). Western blotting showed visible bands in complemented strains (Fig. 2c). The RsbR band in the wild-type strain was barely visible, possibly because its expression was below a detectable level (the concentration of prokaryotic RsbR protein was 0.27×10−5 mg/mL and we used super sensitive exposure).
Fig. 2.
Identification of Listeria monocytogenes rsbR deletion and complementation strains containing single or double alanine substitutions by PCR (a), qRT-PCR (b), and western blotting (c)
Data are expressed as mean±SD from three independent experiments
3.2. Effects of L. monocytogenes rsbR on growth and survival under acidic conditions
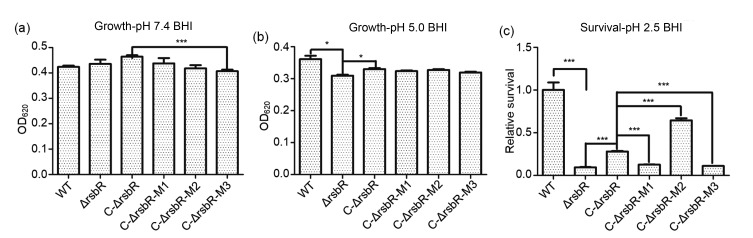
To study whether the rsbR deletion would affect resistance to acidic stress, the ΔrsbR mutant strain and its parent strain 10403S were cultured in BHI at pH 5.0 with pH 7.4 as control. In the control medium, the growth of ΔrsbR and C-ΔrsbR was similar to that of the wild-type strain (P=0.515; Fig. 3a). However, ΔrsbR showed significantly slower growth than its parent strain (P=0.044; Fig. 3b) at acidic pH 5.0 shown as the end-point OD620 at 8 h. Survival of the rsbR deletion mutant was also significantly lower than that of the wild-type strain upon brief exposure at pH 2.5 (Fig. 3c). These effects could be reversed to a significant degree (growth at mild stress, P=0.032; and survival at pH 2.5, P<0.001) by genetic complementation using the strain C-ΔrsbR.
Fig. 3.
Growth of Listeria monocytogenes wild-type (WT) strain and related mutants in BHI medium at pH 7.4 (a) and pH 5.0 (b), and survival in BHI at pH 2.5 (c)
Data are expressed as mean±SD from three independent experiments. * P<0.05, *** P<0.001
3.3. Role of residue T175 contributing to RsbR function in listerial survival under acidic stress
To investigate the key phosphorylated amino acid residues of RsbR, we constructed complement plasmids with a single mutation at T175 or T209, and double mutations at both sites by alanine substitution. In normal and mildly acidic media, there were no significant differences between the growth of the complemented strains with mutations and that of the C-ΔrsbR without alanine substitution (Figs. 3a and 3b). When treated with severe stress at pH 2.5 in BHI, C-ΔrsbR-M1 and C-ΔrsbR-M3 had lower survival (P<0.001), while C-ΔrsbR-M2 showed significantly higher survival (P<0.001) than the non-mutated C-ΔrsbR (Fig. 3c). These findings indicate that T175 (in M1 or M3) is critical for survival.
3.4. Regulaton of RsbR and its residue T175 of SigB expression under stress conditions
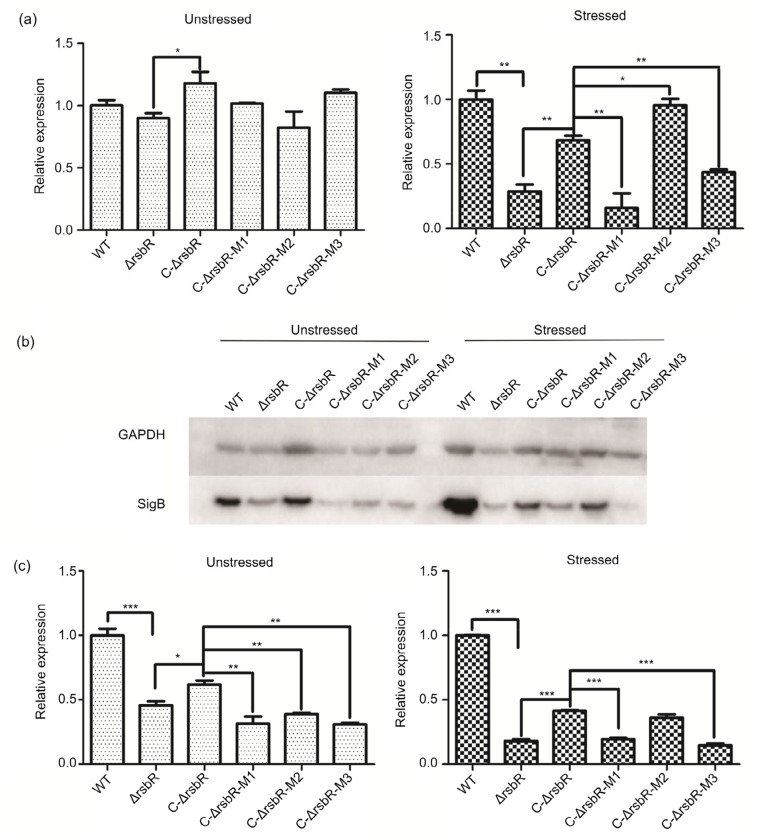
A mild acid stress was introduced to the bacteria (in BHI at pH 5.0) to examine the response of SigB expression to acidic stress. Expression of SigB was 2.0–2.5-fold higher in stressed bacterial cells than in unstressed controls at the mRNA and protein levels. Both qRT-PCR and western blotting revealed that deletion of rsbR led to a significant reduction in SigB expression compared with that in the wild-type strain (P=0.001 and P<0.001 at the mRNA and protein levels, respectively), while complementation (C-ΔrsbR) improved SigB expression significantly (P=0.003 and P<0.001 at the mRNA and protein levels, respectively), though not to the level of the wild-type strain (Fig. 4).
Fig. 4.
Analysis of SigB expression in Listeria monocytogenes wild-type (WT) strain and related mutants in BHI at pH 7.4 (unstressed) or pH 5.0 (stressed) by qRT-PCR (a), western blotting (b), and densitometric quantification of SigB of panels B (c)
Data are expressed as mean±SD from three independent experiments. * P<0.05, ** P<0.01, *** P<0.001
Comparison of the complemented strains with or without mutations of T175 and T209 reveals that C-ΔrsbR-M1 and C-ΔrsbR-M3 down-regulated SigB expression (P<0.01 for sigB mRNA and P<0.001 for SigB protein), while C-ΔrsbR-M2 increased SigB expression (P=0.012 for sigB mRNA and P=0.11 for SigB protein; Fig. 4) compared with that of the non-mutated C-ΔrsbR, suggesting that T175 (M1 or M3) is involved in regulating SigB expression, but not T209 (M2).
4. Discussion
RsbR was identified as a potential receptor to environmental stresses. In B. subtilis, two phosphorylation sites in RsbR behave differently in unstressful and stressful conditions (Kim et al., 2014a, 2004b; Eymann et al., 2011). Until now, there has been no study of the function of this protein in L. monocytogenes. Here we clearly show that RsbR is involved in the growth and survival of L. monocytogenes under acidic stress, and that RsbR-T175 is required to trigger induction of SigB expression in response to acidic stress for improved survival.
It has been reported that deletion of rsbT, rsbV, or rsbU of L. monocytogenes reduces SigB expression under environmental stress (Chaturongakul and Boor, 2004; Shin et al., 2010; Zhang et al., 2013). All these genes are considered essential in the SigB pathway. rsbR, the first gene in the SigB operon, is considered an initial sensory input from environmental stresses. Although the RsbR ortholog in L. monocytogenes has only 49.6% identity to the RsbR in B. subtilis at the amino acid level, the putative key sites for phosphorylation are conserved (T171 and T205 in the RsbR of B. subtilis and T175 and T209 in the RsbR of L. monocytogenes). Therefore, we supposed that LM-RsbR could play a role similar to that of the RsbR in B. subtilis. By genetic deletion, complementation, and site-directed mutagenesis, we proved that RsbR and its residue T175 are involved in listerial survival under acidic stress, most likely via increased expression of the general stress regulator SigB.
In B. subtilis, some of the proteins in the SigB operon (RsbRA, RsbRB, RsbRC, RsbRD, RsbT, and RsbS) form a large molecular mass structure called a stressome and function via a partner switch mechanism in response to environmental stress. The homologous RsbRs that contain the N-and C-terminal sulfate transporter and anti-sigma factor antagonist (STAS) domains and a conserved 13-residue α-helical linker (Marles-Wright et al., 2008) are the major players in this functionality. Each RsbR has two conserved threonine residues in its STAS domain (Pané-Farré et al., 2005). The STAS domain forms the complex core, and receives sensory input from the N-terminal domain (Gaidenko et al., 2012). Chen et al. (2004) suggested that RsbR proteins might serve as redundant co-antagonists necessary for RsbS, as shown by the phenotypes of triple mutants with only one or another RsbR family member. Although the L. monocytogenes genome contains a number of RsbR paralogues, the threonine and serine residues are absent (Pané-Farré et al., 2005). It is also less likely that the RsbR-RsbS-RsbT proteins would form stressomes in L. monocytogenes because these would be physically beyond the holding capacity of the listerial cells. Thus, we speculate that RsbR is the only receptor of environment stress in the L. monocytogenes SigB operon.
RsbRA T171 phosphorylation is thought to be an important prerequisite, but is not the trigger for signaling in B. subtilis. This is supported by the phenotypes of T171A and T171D substitutions and the analysis of the phosphorylation states of the RsbR family in vivo (Kim et al., 2004b). RsbRA T205 is found phosphorylated under strong stresses, and this phosphorylation is considered a feedback mechanism to inhibit signaling under extreme conditions (Eymann et al., 2011). We showed that removal of the phosphorylation residue RsbR-T175A (C-ΔrsbR-M1) could disable the complementation, suggesting that this residue is required to initiate SigB signaling for improved listerial growth or survival. However, dephosphorylation on RsbR-T209A (C-ΔrsbR-M2) seemed to enhance SigB expression and listerial survival, which might indicate a negative regulatory role of T209, similar to that of RsbRA T205 in B. subtilis (Eymann et al., 2011).
In conclusion, this study has provided clear evidence that RsbR T175 is important in triggering increased expression of SigB and listerial survival under acidic stress. It remains to be determined how RsbR residues T175 and T205 coordinate in response to acidic stress, but T175 appeared to function as the major player in this strain with double mutations (C-ΔrsbR-M3).
Footnotes
Project supported by the National Natural Science Foundation of China (Nos. 31702250 and 31600292) and the Open Fund Project by Key Laboratory of Animal Preventive Medicine of Zhejiang Province (No. ZPKLPVM2017KFKT001), China
Contributors: Ke HE and Wei-huan FANG designed the experimentation and data analysis, wrote and edited the manuscript. Ke HE, Yong-ping XIN, Ying SHAN, and Xian ZHANG performed the experimental research and data analysis. Hou-hui SONG contributed to editing of the manuscript. All authors read and approved the final manuscript and, therefore, had full access to all the data in the study and take responsibility for the integrity and security of the data.
Compliance with ethics guidelines: Ke HE, Yong-ping XIN, Ying SHAN, Xian ZHANG, Hou-hui SONG, and Wei-huan FANG declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Chaturongakul S, Boor KJ. Rsbt and RsbV contribute to σB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes . Appl Environ Microbiol. 2004;70(9):5349–5356. doi: 10.1128/AEM.70.9.5349-5356.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen CC, Yudkin MD, Delumeau O. Phosphorylation and RsbX-dependent dephosphorylation of RsbR in the RsbR-RsBs complex of Bacillus subtilis . J Bacteriol. 2004;186(20):6830–6836. doi: 10.1128/JB.186.20.6830-6836.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen JS, Cheng CY, Xia Y, et al. Lmo0036, an ornithine and putrescine carbamoyltransferase in Listeria monocytogenes, participates in arginine deiminase and agmatine deiminase pathways and mediates acid tolerance. Microbiology. 2011;157(11):3150–3161. doi: 10.1099/mic.0.049619-0. [DOI] [PubMed] [Google Scholar]
- 4.Cotter PD, Gahan CGM, Hill C. Analysis of the role of the Listeria monocytogenes F0F1-ATPase operon in the acid tolerance response. Int J Food Microbiol. 2000;60(2-3):137–146. doi: 10.1016/S0168-1605(00)00305-6. [DOI] [PubMed] [Google Scholar]
- 5.Delumeau O, Chen CC, Murray JW, et al. High-molecular-weight complexes of RsbR and paralogues in the environmental signaling pathway of Bacillus subtilis . J Bacteriol. 2006;188(22):7885–7892. doi: 10.1128/JB.00892-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eymann C, Schulz S, Gronau K, et al. In vivo phosphorylation patterns of key stressosome proteins define a second feedback loop that limits activation of Bacillus subtilis σB . Mol Microbiol. 2011;80(3):798–810. doi: 10.1111/j.1365-2958.2011.07609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaidenko TA, Bie XM, Baldwin EP, et al. Two surfaces of a conserved interdomain linker differentially affect output from the RST sensing module of the Bacillus subtilis stressosome . J Bacteriol. 2012;194(15):3913–3921. doi: 10.1128/JB.00583-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hecker M, Pané-Farré J, Völker U. Sigb-dependent general stress response in bacillus subtilis and related Gram-positive bacteria. Annu Rev Microbiol. 2007;61:215–236. doi: 10.1146/annurev.micro.61.080706.093445. [DOI] [PubMed] [Google Scholar]
- 9.Hecker M, Reder A, Fuchs S, et al. Physiological proteomics and stress/starvation responses in Bacillus subtilis and staphylococcus aureus. Res Microbiol. 2009;160(4):245–258. doi: 10.1016/j.resmic.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Kim TJ, Gaidenko TA, Price CW. A multicomponent protein complex mediates environmental stress signaling in Bacillus subtilis . J Mol Biol. 2004;341(1):135–150. doi: 10.1016/j.jmb.2004.05.043. [DOI] [PubMed] [Google Scholar]
- 11.Kim TJ, Gaidenko TA, Price CW. In vivo phosphorylation of partner switching regulators correlates with stress transmission in the environmental signaling pathway of Bacillus subtilis . J Bacteriol. 2004;186(18):6124–6132. doi: 10.1128/JB.186.18.6124-6132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liebal UW, Thomas M, Jon MW, et al. Simulations of stressosome activation emphasize allosteric interactions between RsbR and RsbT. BMC Syst Biol, 7:3. 2013 doi: 10.1186/1752-0509-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔ C T . Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Marles-Wright J, Grant T, Delumeau O, et al. Molecular architecture of the “stressosome,” a signal integration and transduction hub. Science. 2008;322(5898):92–96. doi: 10.1126/science.1159572. [DOI] [PubMed] [Google Scholar]
- 15.Martinez L, Reeves A, Haldenwang W. Stressosomes formed in Bacillus subtilis from the RsbR protein of Listeria monocytogenes allow σB activation following exposure to either physical or nutritional stress. J Bacteriol. 2010;192(23):6279–6286. doi: 10.1128/JB.00467-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.NicAogáin K, O'Byrne CP. The role of stress and stress adaptations in determining the fate of the bacterial pathogen Listeria monocytogenes in the food chain. Front Microbiol, 7:1865. 2016 doi: 10.3389/fmicb.2016.01865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Byrne CP, Karatzas KAG. Chapter 5: the role of sigma B (σB) in the stress adaptations of Listeria monocytogenes: overlaps between stress adaptation and virulence. Adv Appl Microbiol. 2008;65:115–140. doi: 10.1016/S0065-2164(08)00605-9. [DOI] [PubMed] [Google Scholar]
- 18.Okpo E, Leith J, Smith-Palmer A, et al. An outbreak of an unusual strain of Listeria monocytogenes infection in North-East Scotland. J Infect Public Health. 2015;8(6):612–618. doi: 10.1016/j.jiph.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Pané-Farré J, Lewis RJ, Stülke J. The RsbRST stress module in bacteria: a signalling system that may interact with different output modules. J Mol Microbiol Biotechnol. 2005;9:65–76. doi: 10.1159/000088837. [DOI] [PubMed] [Google Scholar]
- 20.Price CW. General stress response in Bacillus subtilis and related Gram-positive bacteria. In: Storz G Hengge R., editor. Bacterial Stress Responses, 2nd Ed. American Society of Microbiology (ASM) Press, Washington, DC; 2011. pp. 301–318. [DOI] [Google Scholar]
- 21.Shan Y, Liu ZQ, Li GW, et al. Nucleocapsid protein from porcine epidemic diarrhea virus isolates can antagonize interferon-λ production by blocking the nuclear factor-κB nuclear translocation. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2018;19(8):570–580. doi: 10.1631/jzus.B1700283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin JH, Brody MS, Price CW. Physical and antibiotic stresses require activation of the RsbU phosphatase to induce the general stress response in Listeria monocytogenes . Microbiology. 2010;156:2660–2669. doi: 10.1099/mic.0.041202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varma JK, Samuel MC, Marcus R, et al. Listeria monocytogenes infection from foods prepared in a commercial establishment: a case-control study of potential sources of sporadic illness in the united states. Clin Inf Dis. 2007;44(4):521–528. doi: 10.1086/509920. [DOI] [PubMed] [Google Scholar]
- 24.Xia Y, Xin YP, Li XL, et al. To modulate survival under secondary stress conditions, Listeria monocytogenes 10403S employs RsbX to downregulate σB activity in the poststress recovery stage or stationary phase. Appl Environ Microbiol. 2016;82(4):1126–1135. doi: 10.1128/AEM.03218-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang ZC, Meng QL, Qiao J, et al. RsbV of Listeria monocytogenes contributes to regulation of environmental stress and virulence. Arch Microbiol. 2013;195(2):113–120. doi: 10.1007/s00203-012-0855-5. [DOI] [PubMed] [Google Scholar]
- 26.Zhou YS, Gu YX, Qi BZ, et al. Porcine circovirus type 2 capsid protein induces unfolded protein response with subsequent activation of apoptosis. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2017;18(4):316–323. doi: 10.1631/jzus.B1600208. [DOI] [PMC free article] [PubMed] [Google Scholar]