Abstract
Fucose is a common terminal modification on protein and lipid glycans. Fucose can also be directly linked to protein via an O-linkage to Serine or Threonine residues located within consensus sequences contained in Epidermal Growth Factor-like (EGF) repeats and Thrombospondin Type 1 Repeats (TSRs). In this context, fucose is added exclusively to properly folded EGF repeats and TSRs by Protein O-fucosyltransferases 1 and 2, respectively. In both cases, the O-linked fucose can also be elongated with other sugars. Here we describe the biological importance of these O-fucose glycans and molecular mechanisms by which they affect the function of the proteins they modify. O-Fucosylation of EGF repeats modulates the Notch signaling pathway, while O-fucosylation of TSRs is predicted to influence secretion of targets including several extracellular proteases. Recent data shows O-fucose glycans mediate their effects by participating in both intermolecular and intramolecular interactions.
O-Fucose Biology
Proteins and lipids are commonly modified with glycans containing fucose. The fucose residues typically occur as terminal or core modifications of the glycan, added by Golgi localized fucosyltranferases [1]. These modifications perform many functions including immune modulation, selectin-mediated extravasation of leukocytes, and as blood group antigens. In contrast, fucose can be directly added to proteins via an O-linkage to hydroxyl groups of Serines or Threonines of folded Epidermal Growth Factor-like (EGF) repeats and Thrombospondin Type 1 Repeats (TSR) in the endoplasmic reticulum (ER). ER O-fucosylation of EGF repeats is observed in metazoans [2,3], and O-fucosylation of TSR is observed in metazoans [1,4] as well as Plasmodium falciparum and Toxoplasma gondii [5–9]. Recently, O-fucosylation of nuclear and cytoplasmic proteins was also observed in Arabidopsis [10] and Toxoplasma gondii [11].
EGF repeats and TSRs are small protein modules characterized by three disulfide bonds (Figure 1). Protein O-fucosyltransferases 1 and 2 (POFUT1 and POFUT2) add O-linked fucose to folded EGF repeats and TSRs, respectively [12,13]. These enzymes are highly specific for their substrates [13], due to inherent differences in the three-dimensional shape of EGF repeats and TSRs and complementary differences in the binding pockets of the respective fucosyltransferases [14,15]. Not surprisingly, the cytoplasmic SPINDLY protein O-fucosyltransferase in Arabidopsis, which modifies the disordered domain of the transcriptional repressor DELLA, is unrelated to POFUT1 or 2, but rather is a homolog of the nuclear/cytoplasmic O-GlcNAc transferase (SECRET AGENT in Arabidopsis) [10]. A SPINDLY homolog responsible for O-fucosylation of nuclear pore proteins has also been reported in Toxoplasma gondii [9]. Interestingly, 39 POFUTs are annotated in the Arabidopsis genome [16]. Although none have been demonstrated to have POFUT activity, mutations in some affect growth and reproduction [16].
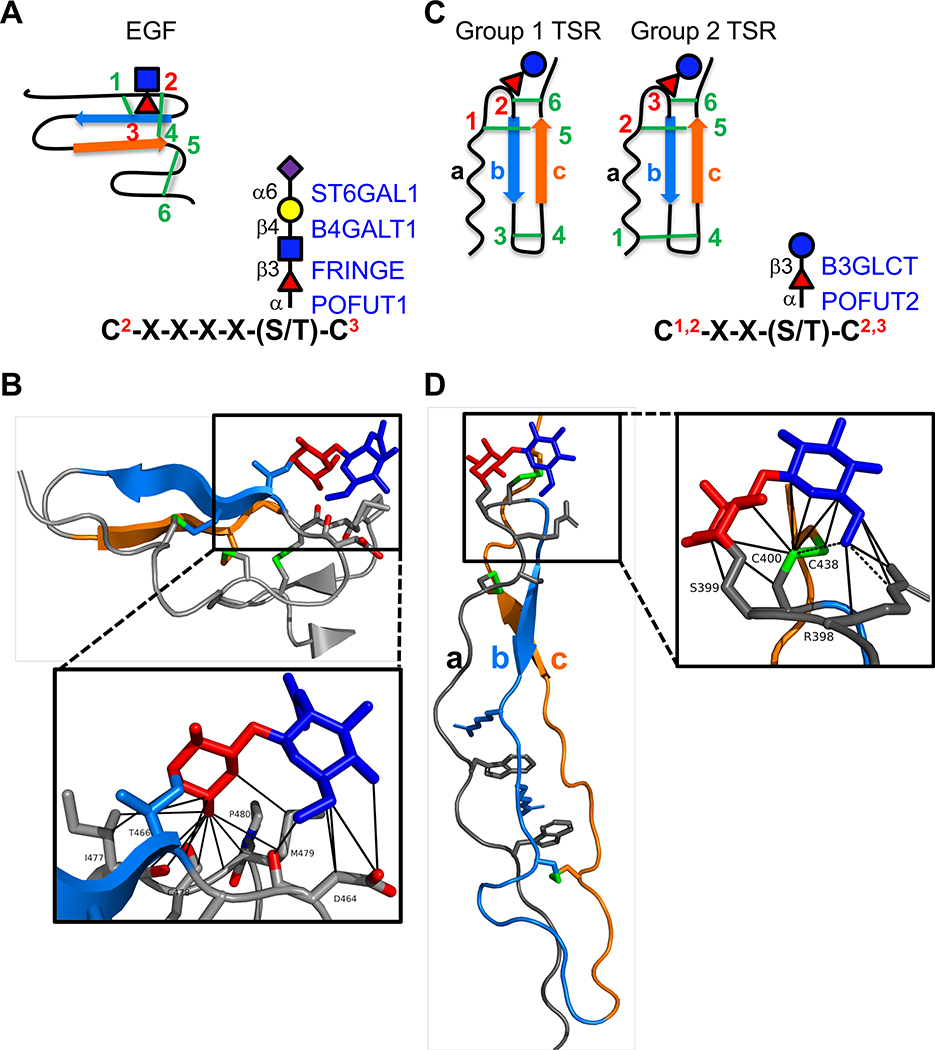
Figure 1. EGF repeats and TSRs are modified by O-fucose glycans A.
(left) Cartoon showing disulfide bonding pattern (green lines) in an EGF repeat. Beta strands are indicated by blue and orange arrows. Site of O-fucosylation and GlcNAc elongation are indicated by red triangle and blue square, respectively. (right) Consensus sequence for POFUT1 modification. C2 and C3 are the second and third conserved cysteine in the EGF repeat. Enzymes responsible for addition of each sugar are indicated in blue on the right with linkages in black on the left. Fucose, red triangle; GlcNAc, blue square; Galactose, yellow circle; Sialic Acid, purple diamond. B. Structure of NOTCH1 EGF12 modified with a GlcNAcβ1–3Fucose disaccharide (from PDB ID 4D0E). Beta strands colored as in A. Fucose in red, GlcNAc in blue, disulfide bonds in green, oxygen atoms highlighted in red. Box shows zoomed in region highlighting interactions of the disaccharide with underlying amino acids identified by MolProbity [73,74] (van der Waals, solid lines). Structures rendered in PyMOL (Version 2.2.2). C. (left) Cartoons showing the two distinct disulfide bonding patterns for TSRs. Beta strands are indicated by blue and orange arrows. Position of O-fucosylation and elongation with glucose are indicated by red triangle and blue circle, respectively. (right) Consensus sequence for POFUT2 modification. The C’s can be C1 and C2 or C2 and C3 depending on whether the TSR is Group 1 or Group 2. Enzymes responsible for addition of each sugar are indicated in blue on the right with linkages in black on the left. Fucose, red triangle; Glucose, blue circle. D. Structure of ADAMTS13 TSR1 modified with Glucoseβ1–3-Fucose disaccharide (from PDB ID 3GHM). The three strands (a, b, and c) of the TSRs are color coded the same in the cartoons (C) and the structure. Fucose in red, glucose in blue, disulfide bonds in green. Box shows zoomed in region highlighting interactions of the disaccharide with underlying amino acids identified by MolProbity [73,74] (H-bonds, dashed lines; van der Waals, solid lines). Structures rendered in PyMOL (Version 2.2.2).
Recent evidence suggests that the O-linked fucose on EGF repeats can participate directly in intermolecular interactions with binding partners. In addition, O-fucose participates in intramolecular interactions with neighboring amino acids of correctly disulfide-bonded EGF repeats and TSRs. These interactions are proposed to stabilize the structure and as a consequence accelerate the overall rate of folding in the ER [17,18]. In this review we will consider structure/functional evidence to support these hypotheses.
Within the ER, POFUT1 adds O-linked fucose to folded EGF repeats containing the consensus sequence C2-X-X-X-X-(S/T)-C3 (Figure 1A) [1,12]. Nearly 100 potential human protein targets, with vastly diverse functions, contain this consensus sequence in the context of an EGF repeat [1]. The Notch receptor family (mammals express four Notch receptors, NOTCH1–4) has more EGF repeats with this consensus sequence than any other protein in databases. Eliminating POFUT1 in mice or flies results in Notch phenotypes [19,20] suggesting that Notch is a major biological target for POFUT1, although O-fucosylation has been reported to affect other POFUT1 targets as well [21,22]. In the Golgi, FRINGE adds a β3-linked GlcNAc to the O-fucose, and this GlcNAcβ1–3Fucose disaccharide can be further extended to a tetrasaccharide by B4GALT1 and ST6GAL1 (Figure 1A) [23]. Three Fringes exist in mammals: LUNATIC, MANIC, and RADICAL FRINGE (LFNG, MFNG, RFNG) [24]. Notably, Fringes are known to modulate Notch function [25–27].
POFUT2 similarly adds O-linked fucose to folded TSRs containing the consensus sequence C1-X-X-(S/T)-C2 or C2-X-X-(S/T)-C3, depending upon the disulfide bonding pattern of the TSR (Figure 1C) [13,14,28]. In contrast to elongation of the O-fucosylated EGF repeat in the Golgi, the O-fucosylated TSR is extended in the ER. There, β3-glucosyltransferase (B3GLCT) adds a terminal glucose to form the characteristic Glucoseβ1–3Fucose disaccharide [29,30]. Based on the presence of the POFUT2 consensus sequences within the context of TSRs, 49 proteins are likely modified by POFUT2 and B3GLCT [1]. Most of these targets are either secreted factors that modulate the function of the extracellular matrix (ECM) or are cell surface proteins that modulate signaling. Nearly half the POFUT2 targets are members of the A-Disintegrin and Metalloproteinase with ThromboSpondin Type-1 motifs (ADAMTS) family of extracellular proteases.
Diseases and phenotypes associated with O-fucosylation defects
In humans, heterozygous mutations in POFUT1 are linked to a rare skin condition, Dowling-Degos Disease [31], and amplification of POFUT1 is associated with several types of cancer [32,33]. Although no human POFUT2 mutations have been reported, knockout of either Pofut1 or Pofut2 is embryonic lethal in mice [19,34]. Loss of Pofut1 causes embryonic lethality with defects in somite, neural tube, heart, and blood vessel development, similar to Notch1 knockout [19]. Similar loss of Notch function is seen in O-fut1 (POFUT1 homolog in flies) mutants in flies [20]. Loss of Pofut2 in mice causes early embryo lethality resulting from abnormalities in gastrulation and axis elongation, similar to Adamts9 mutants [35]. The phenotypic similarities between these POFUT knockouts and knockouts of single target proteins provides strong evidence that O-fucosylation of these targets is essential for their function.
While the O-fucose modification of EGF repeats and TSRs is essential for function, elongation of the O-fucose on these modules has less severe effects. Reduced Notch signaling in Fringe mutants provides evidence that extension of the O-fucose with GlcNAc is not essential for Notch function, but rather is important for modulating Notch signaling. Fringe mutations in flies affects a subset of Notch functions, especially in formation of the wing [25]. Mutations in either mouse or human Lfng/LFNG show skeletal defects consistent with reduced Notch signaling, but are less severe than Pofut1 or Notch1 mutants. In humans, LFNG mutations cause Spondylocostal Dysostosis Type 3, characterized by abnormal development of the bones in the spine and ribs [36]. Similarly, mouse Lfng knockout causes abnormalities in vertebrae and ribs [37,38], but more detailed analysis also shows defects in lungs, T- and B-cell development, reproduction, and vasculature of the retina [39–42]. Knockout of Mfng or Rfng alone result in no major developmental defects [43], but both have been implicated in T- and B-cell development [41] and Mfng in heart development [44]. Both Lfng and Mfng also play roles in cancer [45,46].
Failure to add glucose to O-fucosylated TSR domains causes human Peters Plus Syndrome (PPS) [47]. PPS, caused by loss-of-function mutations in B3GLCT, is characterized by Peters anomaly of the eye, widened craniofacial structures and shortened long bones and digits [48]. The viability of these patients compared to early embryo lethality observed in Pofut2-null mice provides evidence that proteins O-fucosylated by POFUT2 have different requirements for the glucose. As mentioned above, both Pofut2 and Adamts9-null mice are embryonic lethal, but loss of B3GLCT in PPS is not. Since ADAMTS9 is modified with the Glucoseβ1–3Fucose disaccharide [49], this suggests that ADAMTS9 function is more dependent on POFUT2 than B3GLCT.
How does O-fucosylation affect protein function?
O-fucosylation influences intermolecular interaction with binding partners
Recent results suggest that one mechanism by which O-fucose glycans affect protein function is by modulating interactions between binding partners. This is most clearly seen in the case of Notch interactions with its ligands (DLL1 and 4, JAG1 and 2). Twenty of the 36 EGF repeats in the mouse NOTCH1 extracellular domain (ECD) contain the consensus sequence for POFUT1 modification, and mass spectral site mapping revealed that 17 of those sites are modified at high stoichiometry with O-fucose (Figure 2A) [26]. Similar results were seen with Drosophila NOTCH [50]. Thus, O-fucosylation by POFUT1 is a highly efficient process in cells. In contrast to POFUT1, Fringe modifications are site selective (Figure 2A) [26,50]. The basis of this selectivity is unknown, although preliminary results suggests that some EGF repeats contain sequences that block modification by a Fringe [51]. LFNG and MFNG modify similar sites on mouse NOTCH1, while RFNG modifies a subset of these sites (EGF8, 12 and 26) (see [26] for details). Again, the basis for the difference in specificity between the Fringes is unknown.
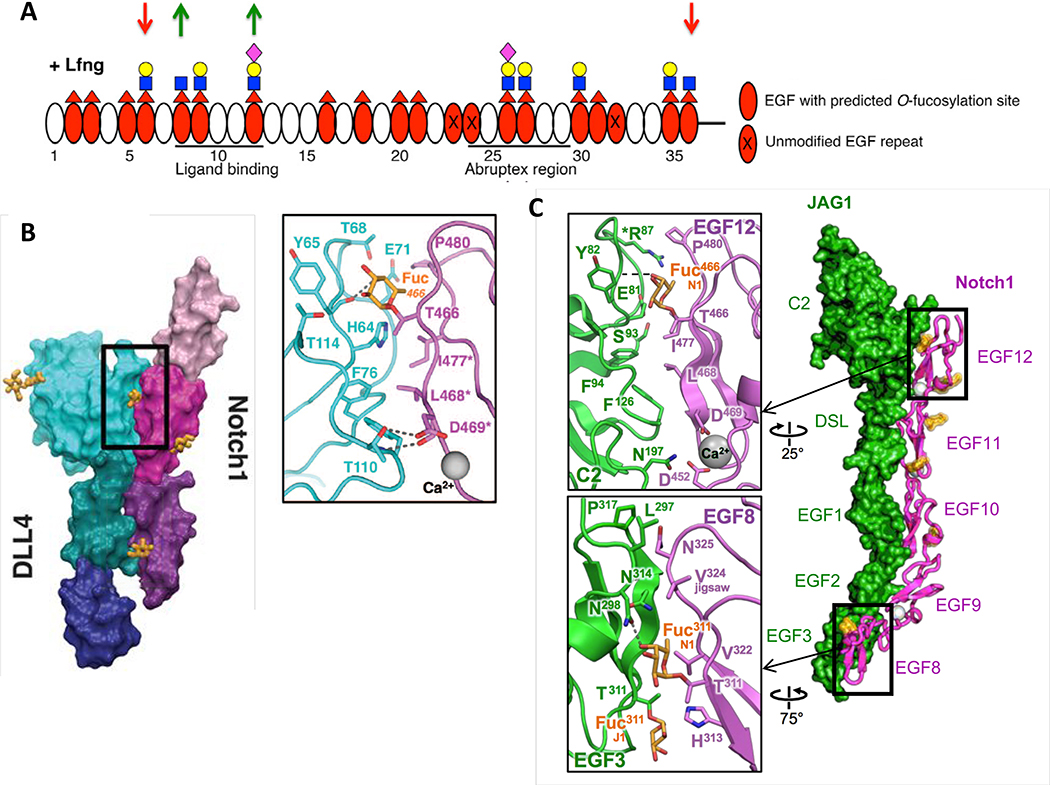
Figure 2. O-Fucose on EGF8 and EGF12 of NOTCH1 is in direct contact with ligands.
A. Domain map of mouse NOTCH1 EGF1–36 showing which EGF repeats are modified by O-fucose and elongated by LFNG (modified from [26]). MFNG elongates similarly, but RFNG only modifies O-fucose on EGF8, 12 and 26. Note that Fringe enzymes were overexpressed in a Fringe-deficient background in these studies. Down red arrows indicate sites where Fringe modification inhibits JAG1-NOTCH1 activation. Green up arrows indicate sites where Fringe modification enhances DLL1-NOTCH1 activation. Fucose, red triangle; GlcNAc, blue square; Galactose, yellow circle; Sialic Acid, purple diamond. B. Co-crystal structure of NOTCH1 EGF11–13 (shades of magenta/purple) and DLL4 (N-terminus to EGF3, shades of blue/green) (modified from [52]). Inset shows direct interaction between O-fucose on NOTCH1 EGF12 with residues in DLL4. C. Co-crystal structure of NOTCH1 EGF8–12 and JAG1 N-EGF3 (modified from [53]). Inset shows direct contacts between O-fucose on EGF8 and EGF12 and residues in JAG1. Note that the structures in B and C were obtained after directed evolution of the ligands toward stronger affinities.
Demonstration that O-fucose directly participates in Notch-ligand interactions was revealed in two recent co-crystal structures, one between a portion of the NOTCH1 ligand-binding domain (EGF11–13) and a portion of DLL4 [52] (Figure 2B), the other between the NOTCH1 ligand-binding domain (EGF8–12) and a portion of JAG1 [53] (Figure 2C). Both structures revealed that the O-fucose on EGF12 of NOTCH1 is in direct contact with backbone and side-chain residues in both ligands. The NOTCH1-JAG1 co-crystal also revealed a direct interaction between the O-fucose on EGF8 of NOTCH1 and the side chain of N298 of JAG1. Elimination of either of these O-fucosylation sites by mutation of the modified Thr to Val reduced the ability of DLL1 or JAG1 to bind to and activate NOTCH1 in cell-based assays, although the mutation at EGF12 had a larger effect on DLL1 than JAG1 [26,53]. Mutation of both sites (EGF8 and 12) resulted in a substantial reduction in NOTCH1 activity, confirming the importance of the O-fucose at these sites for NOTCH1 function. Consistent with these observations, mice homozygous for a knock-in Thr to Ala mutation in the EGF12 O-fucose site display slow growth and defects in T cell development consistent with a hypomorphic Notch phenotype [54].
Fringe modifications of the O-fucose residues on EGF8 and 12 play important roles in enhancing DLL1-mediated NOTCH1 activation. Both sites are modified by Fringes (Figure 2A), and all three Fringes enhance DLL1-mediated NOTCH1 activation [26,27]. Elimination of the O-fucose sites on EGF8 and 12 reduces the ability of the Fringes to enhance DLL1 binding to and activation of NOTCH1 [26]. Thus, Fringe modifications at EGF8 and 12 enhance DLL1-mediated NOTCH1 signaling by enhancing binding. Modeling of the GlcNAc added by Fringe to the O-fucose on EGF12 suggests additional interactions with DLL4, providing a potential molecular explanation for the Fringe effect at this site [52].
In contrast to their effects on DLL1, LFNG and MFNG inhibit JAG1-mediated NOTCH1 activation, while RFNG enhances it [26,27]. Surprisingly, modification of EGF8 and 12 by Fringes enhanced binding between NOTCH1 and JAG1 [26,55]. Since RFNG only modified O-fucose on EGF8, 12 and 26, the additional sites modified by LFNG and MFNG were suspected to be inhibitory (Figure 2A). Mutation of two of the O-fucose sites modified by LFNG and MFNG but not by RFNG, EGF6 and 36, reduced the ability of LFNG or MFNG to inhibit JAG1-mediated NOTCH1 activity (Figure 2A) [26]. The fact that mutations at EGF6 and 36 did not affect NOTCH1-JAG1 binding suggested the basis for this inhibition must be mediated at an event after ligand binding but before proteolytic activation. An intriguing possibility is that Fringe modification at EGF6 affects the establishment or stability of the catch-bond that forms in response to tension generated between JAG1 and NOTCH1 [53]. In addition, Fringe modification at EGF36 could provide stability to the adjacent NOTCH1 Negative Regulatory Domain (NRR), reducing proteolytic activation [56]. These modifications could work together to inhibit JAG1-NOTCH1 activation.
Based on the interactions between O-fucose on Notch EGF repeats and ligands, it is reasonable to propose that the Glucoseβ1–3Fucose disaccharide on TSRs could participate in similar interactions. TSRs are known to bind a number of other proteins including TGFβ and heparan sulfate proteoglycans [57], but to date no one has examined whether the TSR O-fucose glycans affect these interactions.
O-fucosylation generates intramolecular interactions that stabilize folded EGF repeats and TSRs as part of a novel non-canonical ER quality control pathway
The first hint that O-fucose modifications could play a role in quality control of EGF repeat or TSR folding came from demonstration that both POFUT1 and POFUT2 require a folded domain containing the appropriate sequence as a substrate. Consistent with this prediction, consensus sequences are not recognized by the enzymes when located within unfolded domains or linear synthetic peptides [12,13]. Further evidence that these enzymes recognize folded structures came from co-crystal structures of POFUT1 and an EGF repeat [15], and POFUT2 and a TSR [14]. In both cases, the enzymes have a large binding pocket for the respective folded domains, and the enzymes bind these domains in such a way as to orient the hydroxyl group of the Ser/Thr to be modified in the exact position necessary to perform a nucleophilic attack on the anomeric carbon of the fucose of GDP-fucose. These structures explain why the enzymes require both a consensus sequence and a properly folded domain for modification to occur. Thus, unlike classical ER quality control systems that recognize unfolded proteins [58], both POFUT1 and POFUT2 recognize folded domains, modifying them with a fucose after folding. Both enzymes are also localized to the lumen of the ER [28,59], the folding compartment for the secretory pathway. POFUT1 is retained in the ER by a C-terminal KDEL-like ER-retention signal [59]. POFUT2 lacks such a sequence, but appears to be retained in the ER by interaction with other ER resident proteins [17]. B3GLCT is also localized in the ER by a C-terminal ER-retention signal [30], supporting a role for the glucose in quality control. In contrast, the Fringe enzymes are Golgi localized [60,61], consistent with these modifications being modulatory for receptor/ligand interactions.
A direct role for POFUT1 in quality control initially came from studies in flies which showed that knockdown of O-fut1 (Drosophila form of POFUT1) caused reduced cell-surface expression and ER accumulation of NOTCH [62]. Surprisingly, cell-surface expression was partially rescued by an enzymatically inactive form of the enzyme (R240A mutant), suggesting O-fut1 has a fucosyltransferase-independent chaperone activity. Subsequent work has shown that a similar mutation in mouse Pofut1 destabilizes the enzyme [63], and it is not clear whether the R240A mutation has residual enzymatic activity [64], so the dependence of cell-surface expression of Notch proteins on the ability of POFUT1 to transfer fucose is still an open question. More recent results in flies reveal that elimination of O-fut1 causes a temperature-dependent defect in cell-surface expression of NOTCH, consistent with an effect of fucosylation on proper folding of the receptor [65]. Elimination of Pofut1 in mice or in cell lines is reported to reduce cell-surface NOTCH1 in some contexts (e.g. somites [63], HEK293T cells [18], HSCs [66]), but less so in other contexts (e.g. mouse embryonic stem cells [67]), suggesting that the quality control effects of POFUT1 are cell-type specific. This variability has been proposed to be due to differential expression of other chaperones that could also assist in folding and surface expression of Notch proteins, or differences in environmental conditions that could affect protein folding.
A number of cell-based studies demonstrated that knockdown or elimination of POFUT2 in cells blocks secretion of all POFUT2 targets tested to date, including ADAMTS9 [17,35,49,68]. The importance of POFUT2 for secretion of ADAMTS9 provides a potential explanation for the embryonic lethality observed in Pofut2 null mice. Moreover, the marked similarity between Pofut2- and Adamts9-null embryos suggests that ADAMTS9 is the major physiological target for POFUT2 during early embryogenesis [35]. Recent results have also shown that elimination of Pofut2 in Plasmodium falciparum decreased cell-surface expression of Thrombospondin-Related Anonymous Protein (TRAP), a major cell-surface POFUT2 target in those cells, and attenuated infection of mosquito and vertebrate hosts [6]. Eliminating Pofut2 in Toxoplasma gondii lead to defects in stability and localization of Microneme protein 2 (MIC2), a member of the TRAP family, and reduced parasite invasion of the host [7]. However, a similar study reported only a modest effect on MIC2 stability and slight effect on infectivity [8].
Knockdown or deletion of B3GLCT differentially affects secretion of POFUT2 targets [17], suggesting defects in PPS patients result from disruption of a subset of POFUT2 targets that are more sensitive to loss of B3GLCT. Notably, secretion of ADAMTSL2 [17] and ADAMTS17 [68] are significantly impaired by reduction of B3GLCT in secretion assays. Consistent with the prediction that reduced levels of these proteins contribute to abnormalities in PPS patients is the observation that human mutations in ADAMTSL2 and ADAMTS17 cause Geleophysic Dysplasia and Weill-Marchisani Syndromes, which are also characterized by eye abnormalities, short stature, and brachydactyly [69,70]. In contrast, B3GLCT is not required for secretion of ADAMTS13 [17]. Single gene defects in ADAMTS13 result in the clotting disorder Thrombotic Thrombocytopenic Purpura (TTP), but PPS patients do not display clotting defects [48].
The structures of several EGF repeats and TSRs modified with O-fucose glycans (Figure 1) suggest a potential mechanism for how POFUT1 or POFUT2 mediated O-fucosylation ensures efficient secretion of target proteins. Unlike many carbohydrate modifications, the O-fucose glycans in crystal structures of EGF repeats and TSRs are remarkably visible, to the extent that they have been called “surrogate amino acids” because they display thermal mobility (based on B factors) similar to the underlying amino acids [52,55]. The fucose residues lay down on the surface of the EGF or TSR domains, with several contacts with underlying amino acids (Figure 1B, D). These contacts are proposed to stabilize the folded domains. Since the fucose is only added to the domains after they fold, this raises the possibility that addition of fucose drives the domain into an energy well where they are unlikely to move back into a folding equilibrium (Figure 3). Secretion defects in the absence of O-fucose addition would likely result from reentry of the domain into the “folding cycle”, ultimately slowing down the rate of folding. In the context of TSRs, the Glucoseβ1–3Fucose disaccharide interacts directly with the C2-C6 disulfide bond of the TSR, suggesting that the disaccharide protects of the disulfide bond from the surrounding ER folding/unfolding environment (Figure 1D). This may explain how the addition of glucose provides additional stabilization of the TSR. The observation that only a subset of proteins requires B3GLCT for secretion suggests that amino acids located in proximity to the C2-C6 disulfide bond could influence whether the TSR requires the addition of glucose for efficient secretion.
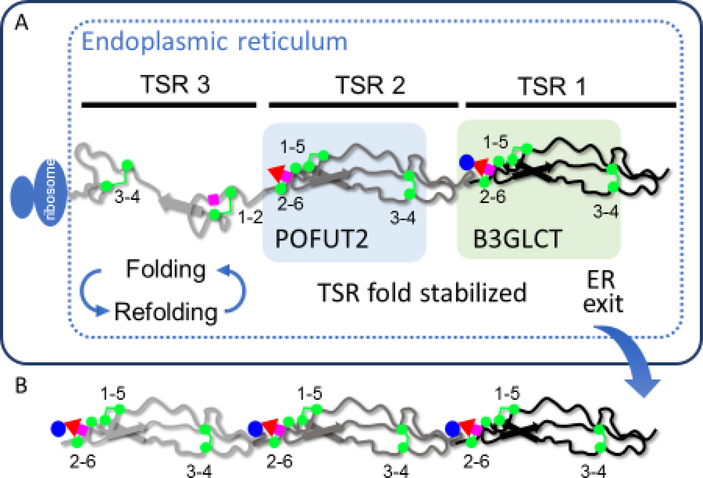
Figure 3. O-Fucosylation stabilizes TSRs by interacting with underlying amino acids.
A. A hypothetical protein with 3 TSRs is being translated and is folding in the ER. TSR3 is in the folding cycle, with some correct and some incorrect disulfide bonds. TSR3 is not modified by O-fucose glycans. TSR2 is fully folded, stabilized by addition of fucose by POFUT2, and is no longer part of a folding cycle. TSR1 is further stabilized by addition of glucose by B3GLCT. Note that the folding pathway for a TSR is not known, so this pathway is hypothetical. B. Once all TSRs with a POFUT2 consensus sequence are modified with Glucoseβ1–3Fucose disaccharide and the protein is fully folded, the native protein exits the ER, transits the secretory system, and is either directed to the cell surface or secreted from the cell depending upon the target protein properties.
Consistent with the prediction that the fucose stabilizes these domains, we observed that O-fucosylated TSRs and EGF repeats unfold significantly slower in reductive unfolding assays compared to unmodified modules [17,18]. Moreover, for TSRs, addition of the glucose has an additional stabilizing effect [17]. Likewise, starting with an unfolded TSR, POFUT2 and GDP-fucose accelerate the in vitro rate of folding, whereas addition of POFUT2 alone has no effect on the rate of folding [17]. Combined, these results provide support for the concept that addition of O-fucose on a properly folded EGF repeat or TSR stabilizes that structure, keeping it from re-entering a folding cycle (Figure 3). This stabilization and acceleration of folding suggests that both POFUT1 and POFUT2 function in novel non-canonical quality control pathways designed for the efficient folding of proteins containing EGF repeats and TSRs, respectively, and provides a likely explanation for the reduced secretion of targets in the absence of these enzymes.
Future directions
While it is clear that O-fucosylation of EGF repeats and TSRs has major effects on the function of proteins, we are just beginning to understand the molecular mechanisms by which the glycans mediate these effects. The data suggesting O-fucose modifications of EGF8 and 12 on NOTCH1 are in direct contact with ligands is compelling, but does not explain what the other 15 O-fucose residues on the NOTCH1 extracellular domain are doing. The stabilizing effects of O-fucose on EGF repeats and importance of POFUT1 for cell-surface expression of Notch receptors may provide part of the explanation for these multiple sites, but the fact that POFUT1 is not required for cell-surface expression in all contexts raises the question of what other mechanisms are used for EGF repeat stabilization. Little is known about how the Glucoseβ1,3Fucose disaccharide on TSRs affects intermolecular interactions. Notably TSRs are often localized within regions of target proteins that are implicated in protein/protein interactions, such as the ancillary domains of the ADAMTS family members [71]. Since defects in folding of EGF repeat or TSR containing proteins would likely lead to ER stress, it is possible that some of the phenotypes observed in Pofut1 or Pofut2-null animals are due to enhancement of Unfolded Protein Response pathways. B3GLCT also appears to stabilize TSRs in an additive fashion and play a role in their folding, but Fringes do not affect cell-surface expression of Notch in cells [26], likely because they are not localized in the ER. Nonetheless, a recent report suggests that cell surface localization of Notch ligands (which also have EGF repeats that are modified by O-fucose) is affected by Fringes in the intestinal epithelium of mice [72]. Answers to these and other questions will help us to better understand how O-fucose glycans affect the function of the proteins they modify.
Highlights.
Protein O-fucosyltransferases 1 and 2 (POFUT1 and POFUT2) are ER-localized and modify EGF repeats and TSRs.
Both POFUT1 and POFUT2 are exquisitely selective for properly folded substrates.
O-Fucose on Notch EGF repeats directly participates in intermolecular interactions with Notch ligands.
O-Fucose glycans on both EGF repeats and TSRs interact with underlying amino acids, stabilizing the folded domains.
Both POFUT1 and POFUT2 are proposed to participate in non-canonical ER quality control pathways for the folding of EGF repeats and TSRs, respectively.
Acknowledgements:
The authors wish to thank Dr. Kelvin Luther for generating the PyMol images and members of the Haltiwanger and Holdener laboratories for helpful comments. Original research was supported by NIH grants GM061126 (RSH), CA123071 (RSH and BCH), HD090156 (RSH and BCH), and HD096030 (RSH and BCH).
Footnotes
Conflict of interest: The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Annotated References:
- 1.Schneider M, Al-Shareffi E, Haltiwanger RS: Biological functions of fucose in mammals. Glycobiology 2017, 27:601–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. *.Varshney S, Stanley P: Multiple Roles for O-Glycans in Notch Signalling. FEBS Lett 2018. 592:3819–3834 Excellent, recent comprehensive review of O-glycans occuring on the Notch receptor [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haltom AR, Jafar-Nejad H: The multiple roles of epidermal growth factor repeat O-glycans in animal development. Glycobiology 2015, 25:1027–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vasudevan D, Haltiwanger RS: Novel roles for O-linked glycans in protein folding. Glycoconj J 2014, 31:417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swearingen KE, Lindner SE, Shi L, Shears MJ, Harupa A, Hopp CS, Vaughan AM, Springer TA, Moritz RL, Kappe SH, et al. : Interrogating the Plasmodium Sporozoite Surface: Identification of Surface-Exposed Proteins and Demonstration of Glycosylation on CSP and TRAP by Mass Spectrometry-Based Proteomics. PLoS Pathog 2016, 12:e1005606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. *.Lopaticki S, Yang ASP, John A, Scott NE, Lingford JP, O’Neill MT, Erickson SM, McKenzie NC, Jennison C, Whitehead LW, et al. : Protein O-fucosylation in Plasmodium falciparum ensures efficient infection of mosquito and vertebrate hosts. Nat Commun 2017, 8:561 Presents evidence for the in vivo importance of POFUT2 in localization and function of the TRAP protein in Plasmodium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. *.Bandini G, Leon DR, Hoppe CM, Zhang Y, Agop-Nersesian C, Shears MJ, Mahal LK, Routier FH, Costello CE, Samuelson J: O-fucosylation of thrombospondin-like repeats is required for processing of microneme protein 2 and for efficient host cell invasion by Toxoplasma gondii tachyzoites. J Biol Chem 2018. Consistent with reference 6, presents evidence for the in vivo importance of POFUT2 in processing and function of the MIC2 protein in Toxoplasma [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. *.Khurana S, Coffey MJ, John A, Uboldi AD, Huynh MH, Stewart RJ, Carruthers V, Tonkin CJ, Goddard-Borger ED, Scott NE: Protein O-fucosyltransferase 2-mediated O-glycosylation of the adhesin MIC2 is dispensable for Toxoplasma gondii tachyzoite infection. J Biol Chem 2018. In contrast to reference 7, shows a less significant effect of elimating POFUT2 in Toxoplasma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gas-Pascual E, Ichikawa HT, Sheikh MO, Serji MI, Deng B, Mandalasi M, Bandini G, Samuelson J, Wells L, West CM: CRISPR/Cas9 and glycomics tools for Toxoplasma glycobiology. J Biol Chem 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zentella R, Sui N, Barnhill B, Hsieh WP, Hu J, Shabanowitz J, Boyce M, Olszewski NE, Zhou P, Hunt DF, et al. : The Arabidopsis O-fucosyltransferase SPINDLY activates nuclear growth repressor DELLA. Nat Chem Biol 2017, 13:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bandini G, Haserick JR, Motari E, Ouologuem DT, Lourido S, Roos DS, Costello CE, Robbins PW, Samuelson J: O-fucosylated glycoproteins form assemblies in close proximity to the nuclear pore complexes of Toxoplasma gondii. Proc Natl Acad Sci U S A 2016, 113:11567–11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Lee GF, Kelley RF, Spellman MW: Identification of a GDP-L-fucose: polypeptide fucosyltransferase and enzymatic addition of O-linked fucose to EGF domains. Glycobiology 1996, 6:837–842. [DOI] [PubMed] [Google Scholar]
- 13.Luo Y, Nita-Lazar A, Haltiwanger RS: Two distinct pathways for O-fucosylation of epidermal growth factor-like or thrombospondin type 1 repeats. J Biol Chem 2006, 281:9385–9392. [DOI] [PubMed] [Google Scholar]
- 14. **.Valero-Gonzalez J, Leonhard-Melief C, Lira-Navarrete E, Jimenez-Oses G, Hernandez-Ruiz C, Pallares MC, Yruela I, Vasudevan D, Lostao A, Corzana F, et al. : A proactive role of water molecules in acceptor recognition by protein O-fucosyltransferase 2. Nat Chem Biol 2016, 12:240–246. Structure of POFUT2 in complex with a TSR showing how it recognizes a folded TSR and orients it for catalysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. **.Li Z, Han K, Pak JE, Satkunarajah M, Zhou D, Rini JM: Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1. Nat Chem Biol 2017, 13:757–763. Structures of POFUT1 in complex with EGF repeats showing how the enzyme recognizes a folded EGF repeat and orients it in the binding pocket for catalysis. [DOI] [PubMed] [Google Scholar]
- 16.Smith DK, Harper JF, Wallace IS: A potential role for protein O-fucosylation during pollenpistil interactions. Plant Signal Behav 2018, 13:e1467687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. **.Vasudevan D, Takeuchi H, Johar SS, Majerus E, Haltiwanger RS: Peters Plus Syndrome Mutations Disrupt a Noncanonical ER Quality-Control Mechanism. Curr Biol 2015, 25:286–295. First description of how POFUT2 and B3GLCT function in a non-canonical ER quality control pathway for folding of TSRs by stabilization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. **.Takeuchi H, Yu H, Hao H, Takeuchi M, Ito A, Li H, Haltiwanger RS: O-Glycosylation modulates the stability of epidermal growth factor-like repeats and thereby regulates Notch trafficking. J Biol Chem 2017, 292:15964–15973. Demontrates how POFUT1 functions in a non-canonical ER quality control pathway for folding of EGF repeats by stabilization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi S, Stanley P: Protein O-fucosyltransferase I is an essential component of Notch signaling pathways. Proc.Natl.Acad.Sci.USA 2003, 100:5234–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okajima T, Irvine KD: Regulation of notch signaling by O-linked fucose. Cell 2002, 111:893–904. [DOI] [PubMed] [Google Scholar]
- 21.Kim ML, Chandrasekharan K, Glass M, Shi S, Stahl MC, Kaspar B, Stanley P, Martin PT: O-fucosylation of muscle agrin determines its ability to cluster acetylcholine receptors. Mol Cell Neurosci 2008, 39:452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serth K, Schuster-Gossler K, Kremmer E, Hansen B, Marohn-Kohn B, Gossler A: O-Fucosylation of DLL3 Is Required for Its Function during Somitogenesis. PLoS One 2015, 10:e0123776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS, et al. : Fringe is a Glycosyltransferase that modifies Notch. Nature 2000, 406:369–375. [DOI] [PubMed] [Google Scholar]
- 24.Johnston SH, Rauskolb C, Wilson R, Prabhakaran B, Irvine KD, Vogt TF: A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway. Development 1997, 124:2245–2254. [DOI] [PubMed] [Google Scholar]
- 25.Panin VM, Papayannopoulos V, Wilson R, Irvine KD: Fringe modulates notch ligand interactions. Nature 1997, 387:908–912. [DOI] [PubMed] [Google Scholar]
- 26. **.Kakuda S, Haltiwanger RS: Deciphering the Fringe-Mediated Notch Code: Identification of Activating and Inhibiting Sites Allowing Discrimination between Ligands. Dev Cell 2017, 40:193–201. Mapped sites of O-fucose and Fringe modification on mouse NOTCH1, and examined which sites are important for Fringe to modulate NOTCH1 function. Note that Fringe enzymes were overexpressed in a Fringe-deficient background in these studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeBon L, Lee TV, Sprinzak D, Jafar-Nejad H, Elowitz MB: Fringe proteins modulate Notch-ligand cis and trans interactions to specify signaling states. Elife 2014, 3:e02950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo Y, Koles K, Vorndam W, Haltiwanger RS, Panin VM: Protein O-fucosyltransferase 2 adds O-fucose to thrombospondin type 1 repeats. J Biol Chem 2006, 281:9393–9399. [DOI] [PubMed] [Google Scholar]
- 29.Kozma K, Keusch JJ, Hegemann B, Luther KB, Klein D, Hess D, Haltiwanger RS, Hofsteenge J: Identification and characterization of a beta1,3-glucosyltransferase that synthesizes the Glc-beta1,3-Fuc disaccharide on thrombospondin type 1 repeats. J Biol Chem 2006, 281:36742–36751. [DOI] [PubMed] [Google Scholar]
- 30.Sato T, Sato M, Kiyohara K, Sogabe M, Shikanai T, Kikuchi N, Togayachi A, Ishida H, Ito H, Kameyama A, et al. : Molecular cloning and characterization of a novel human beta1,3-glucosyltransferase, which is localized at the endoplasmic reticulum and glucosylates O-linked fucosylglycan on thrombospondin type 1 repeat domain. Glycobiology 2006, 16:1194–1206. [DOI] [PubMed] [Google Scholar]
- 31.Li M, Cheng R, Liang J, Yan H, Zhang H, Yang L, Li C, Jiao Q, Lu Z, He J, et al. : Mutations in POFUT1, encoding protein O-fucosyltransferase 1, cause generalized Dowling-Degos disease. Am J Hum Genet 2013, 92:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du Y, Li D, Li N, Su C, Yang C, Lin C, Chen M, Wu R, Li X, Hu G: POFUT1 promotes colorectal cancer development through the activation of Notch1 signaling. Cell Death Dis 2018, 9:995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma L, Dong P, Liu L, Gao Q, Duan M, Zhang S, Chen S, Xue R, Wang X: Overexpression of protein O-fucosyltransferase 1 accelerates hepatocellular carcinoma progression via the Notch signaling pathway. Biochem Biophys Res Commun 2016, 473:503–510. [DOI] [PubMed] [Google Scholar]
- 34.Du J, Takeuchi H, Leonhard-Melief C, Shroyer KR, Dlugosz M, Haltiwanger RS, Holdener BC: O-Fucosylation of thrombospondin type 1 repeats restricts epithelial to mesenchymal transition (EMT) and maintains epiblast pluripotency during mouse gastrulation. Dev Biol 2010, 346:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. *.Benz BA, Nandadasa S, Takeuchi M, Grady RC, Takeuchi H, LoPilato RK, Kakuda S, Somerville RP, Apte SS, Haltiwanger RS, et al. : Genetic and biochemical evidence that gastrulation defects in Pofut2 mutants result from defects in ADAMTS9 secretion. Dev Biol 2016, 416:111–122. Demonstration that early embryonic phenotypes of Pofut2-null mice are likely due to loss of ADAMTS9 function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sparrow DB, Chapman G, Wouters MA, Whittock NV, Ellard S, Fatkin D, Turnpenny PD, Kusumi K, Sillence D, Dunwoodie SL: Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am J Hum Genet 2006, 78:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang N, Gridley T: Defects in somite formation in lunatic fringe-deficient mice. Nature 1998, 394:374–377. [DOI] [PubMed] [Google Scholar]
- 38.Evrard YA, Lun Y, Aulehla A, Gan L, Johnson RL: Lunatic fringe is an essential mediator of somite segmentation and patterning. Nature 1998, 394:377–381. [DOI] [PubMed] [Google Scholar]
- 39.Xu K, Nieuwenhuis E, Cohen BL, Wang W, Canty AJ, Danska JS, Coultas L, Rossant J, Wu MY,Piscione TD, et al. : Lunatic Fringe-mediated Notch signaling is required for lung alveogenesis. Am J Physiol Lung Cell Mol Physiol 2010, 298:L45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hahn KL, Johnson J, Beres BJ, Howard S, Wilson-Rawls J: Lunatic fringe null female mice are infertile due to defects in meiotic maturation. Development 2005, 132:817–828. [DOI] [PubMed] [Google Scholar]
- 41.Song Y, Kumar V, Wei HX, Qiu J, Stanley P: Lunatic, Manic, and Radical Fringe Each Promote T and B Cell Development. J Immunol 2016, 196:232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benedito R, Roca C, Sorensen I, Adams S, Gossler A, Fruttiger M, Adams RH: The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell 2009, 137:1124–1135. [DOI] [PubMed] [Google Scholar]
- 43.Moran JL, Shifley ET, Levorse JM, Mani S, Ostmann K, Perez-Balaguer A, Walker DM, Vogt TF, Cole SE: Manic fringe is not required for embryonic development, and fringe family members do not exhibit redundant functions in the axial skeleton, limb, or hindbrain. Dev Dyn 2009, 238:1803–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D’Amato G, Luxan G, Del Monte-Nieto G, Martinez-Poveda B, Torroja C, Walter W, Bochter MS, Benedito R, Cole S, Martinez F, et al. : Sequential Notch activation regulates ventricular chamber development. Nat Cell Biol 2016, 18:7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang S, Chung WC, Wu G, Egan SE, Miele L, Xu K: Manic Fringe promotes a claudin-low breast cancer phenotype through Notch-mediated PIK3CG induction. Cancer Res 2015, 75:1936–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang S, Chung WC, Xu K: Lunatic Fringe is a potent tumor suppressor in Kras-initiated pancreatic cancer. Oncogene 2016, 35:2485–2495. [DOI] [PubMed] [Google Scholar]
- 47.Lesnik Oberstein SA, Kriek M, White SJ, Kalf ME, Szuhai K, den Dunnen JT, Breuning MH, Hennekam RC: Peters Plus syndrome is caused by mutations in B3GALTL, a putative glycosyltransferase. Am J Hum Genet 2006, 79:562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weh E, Reis LM, Tyler RC, Bick D, Rhead WJ, Wallace S, McGregor TL, Dills SK, Chao MC, Murray JC, et al. : Novel B3GALTL mutations in classic Peters plus syndrome and lack of mutations in a large cohort of patients with similar phenotypes. Clin Genet 2014, 86:142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dubail J, Vasudevan D, Wang LW, Earp SE, Jenkins MW, Haltiwanger RS, Apte SS: Impaired ADAMTS9 secretion: A potential mechanism for eye defects in Peters Plus Syndrome. Sci Rep 2016, 6:33974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harvey BM, Rana NA, Moss H, Leonardi J, Jafar-Nejad H, Haltiwanger RS: Mapping Sites of O-Glycosylation and Fringe Elongation on Drosophila Notch. J Biol Chem 2016, 291:16348–16360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rampal R, Li AS, Moloney DJ, Georgiou SA, Luther KB, Nita-Lazar A, Haltiwanger RS: Lunatic Fringe, Manic Fringe, and Radical Fringe Recognize Similar Specificity Determinants in O-Fucosylated Epidermal Growth Factor-like Repeats. J Biol Chem 2005, 280:4245442463. [DOI] [PubMed] [Google Scholar]
- 52. **.Luca VC, Jude KM, Pierce NW, Nachury MV, Fischer S, Garcia KC: Structural basis for Notch1 engagement of Delta-like 4. Science 2015, 347:847–853. Together with Reference 53, shows that O-fucose on Notch EGF repeats directly participate in ligand interactions. Note that the structures were obtained after directed evolution of the ligand toward stronger affinities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. **.Luca VC, Kim BC, Ge C, Kakuda S, Wu D, Roein-Peikar M, Haltiwanger RS, Zhu C, Ha T, Garcia KC: Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science 2017, 355:1320–1324. Together with Reference 52, shows that O-fucose on Notch EGF repeats directly participate in ligand interactions. Note that the structures were obtained after directed evolution of the ligand toward stronger affinities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ge C, Stanley P: The O-fucose glycan in the ligand-binding domain of Notch1 regulates embryogenesis and T cell development. Proc Natl Acad Sci U S A 2008, 105:1539–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor P, Takeuchi H, Sheppard D, Chillakuri C, Lea SM, Haltiwanger RS, Handford PA: Fringe-mediated extension of O-linked fucose in the ligand-binding region of Notch1 increases binding to mammalian Notch ligands. Proc Natl Acad Sci U S A 2014, 111:7290–7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gordon WR, Zimmerman B, He L, Miles LJ, Huang J, Tiyanont K, McArthur DG, Aster JC, Perrimon N, Loparo JJ, et al. : Mechanical Allostery: Evidence for a Force Requirement in the Proteolytic Activation of Notch. Dev Cell 2015, 33:729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams JC, Tucker RP: The thrombospondin type 1 repeat (TSR) superfamily: diverse proteins with related roles in neuronal development. Develop. Dynamics 2000, 218:280–299. [DOI] [PubMed] [Google Scholar]
- 58.Xu C, Ng DT: Glycosylation-directed quality control of protein folding. Nat Rev Mol Cell Biol 2015, 16:742–752. [DOI] [PubMed] [Google Scholar]
- 59.Luo Y, Haltiwanger RS: O-fucosylation of Notch occurs in the endoplasmic reticulum. J Biol Chem 2005, 280:11289–11294. [DOI] [PubMed] [Google Scholar]
- 60.Munro S, Freeman M: The notch signalling regulator fringe acts in the Golgi apparatus and requires the glycosyltransferase signature motif DXD. Curr.Biol. 2000, 10:813–820. [DOI] [PubMed] [Google Scholar]
- 61.Hicks C, Johnston SH, DiSibio G, Collazo A, Vogt TF, Weinmaster G: Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nature Cell Biology 2000, 2:515–520. [DOI] [PubMed] [Google Scholar]
- 62.Okajima T, Xu A, Lei L, Irvine KD: Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding. Science 2005, 307:1599–1603. [DOI] [PubMed] [Google Scholar]
- 63.Ajima R, Suzuki E, Saga Y: Pofut1 point-mutations that disrupt O-fucosyltransferase activity destabilize the protein and abolish Notch1 signaling during mouse somitogenesis. PLoS One 2017, 12:e0187248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McMillan BJ, Zimmerman B, Egan ED, Lofgren M, Xu X, Hesser A, Blacklow SC: Structure of human POFUT1, its requirement in ligand-independent oncogenic Notch signaling, and functional effects of Dowling-Degos mutations. Glycobiology 2017:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ishio A, Sasamura T, Ayukawa T, Kuroda J, Ishikawa HO, Aoyama N, Matsumoto K, Gushiken T, Okajima T, Yamakawa, et al. : O-fucose monosaccharide of Drosophila Notch has a temperature-sensitive function and cooperates with O-glucose glycan in Notch transport and Notch signaling activation. J Biol Chem 2015, 290:505–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yao D, Huang Y, Huang X, Wang W, Yan Q, Wei L, Xin W, Gerson S, Stanley P, Lowe JB, et al. : Protein O-fucosyltransferase 1 (Pofut1) regulates lymphoid and myeloid homeostasis through modulation of Notch receptor ligand interactions. Blood 2011, 117:5652–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stahl M, Uemura K, Ge C, Shi S, Tashima Y, Stanley P: Roles of Pofut1 and O-fucose in mammalian Notch signaling. J Biol Chem 2008, 283:13638–13651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hubmacher D, Schneider M, Berardinelli SJ, Takeuchi H, Willard B, Reinhardt DP, Haltiwanger RS, Apte SS: Unusual life cycle and impact on microfibril assembly of ADAMTS17, a secreted metalloprotease mutated in genetic eye disease. Sci Rep 2017, 7:41871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morales J, Al-Sharif L, Khalil DS, Shinwari JM, Bavi P, Al-Mahrouqi RA, Al-Rajhi A, Alkuraya FS, Meyer BF, Al Tassan N: Homozygous mutations in ADAMTS10 and ADAMTS17 cause lenticular myopia, ectopia lentis, glaucoma, spherophakia, and short stature. Am J Hum Genet 2009, 85:558–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Le Goff C, Morice-Picard F, Dagoneau N, Wang LW, Perrot C, Crow YJ, Bauer F, Flori E, Prost-Squarcioni C, Krakow D, et al. : ADAMTSL2 mutations in geleophysic dysplasia demonstrate a role for ADAMTS-like proteins in TGF-beta bioavailability regulation. Nat Genet 2008, 40:1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kelwick R, Desanlis I, Wheeler GN, Edwards DR: The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol 2015, 16:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kadur Lakshminarasimha Murthy P, Srinivasan T, Bochter MS, Xi R, Varanko AK, Tung KL, Semerci F, Xu K, Maletic-Savatic M, Cole SE, et al. : Radical and lunatic fringes modulate notch ligands to support mammalian intestinal homeostasis. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd,Snoeyink J, Richardson JS, et al. : MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 2007, 35:W375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC: MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 2010, 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]