Multiple cell types sense fluid flow as an environmental cue. Flow can exert shear force (or stress) on cells and the prevailing model is that biological flow sensing involves the measurement of shear force1, 2. Here, we provide evidence for force-independent flow sensing in the bacterium Pseudomonas aeruginosa. A microfluidic-based transcriptomic approach enabled us to discover an operon of P. aeruginosa that is rapidly and robustly upregulated in response to flow. Using a single-cell reporter of this operon (which we name fro for flow regulated operon), we establish that P. aeruginosa dynamically tunes gene expression to flow intensity through a process we call rheosensing (as rheo- is Greek for flow). We further show that rheosensing occurs in multicellular biofilms, involves signaling through the alternative sigma factor FroR, and does not require known surface sensors. To directly test whether rheosensing measures force, we independently alter the two parameters that contribute to shear stress: shear rate and solution viscosity. Surprisingly, we discover that rheosensing is sensitive to shear rate but not viscosity, indicating that rheosensing is a kinematic (force-independent) form of mechanosensing. Thus, our findings challenge the dominant belief that biological mechanosensing requires the measurement of forces.
Mechanical features shape how organisms interact with their environment such that there are often selective benefits for cells to sense them. While eukaryotic mechanosensing has been studied extensively1, 2, bacterial mechanosensing has been appreciated only recently3, 4. Most studies on mechanosensing have focused on surface sensing, but fluid flow is also an important mechanical cue. Flow is present in many environments where bacteria thrive, such as hosts and associated medical devices. Recent reports have shown that bacteria sense flow to modulate gene expression and signaling5, 6. In Enterohemorrhagic Escherichia coli (EHEC), expression of the LEE virulence factors is induced by flow and host association5, and in Pseudomonas aeruginosa, cyclic-di-GMP levels are induced by flow and surface attachment6. In theory, cells could sense flow by sensing changes in shear rate (the kinematic component of flow) or shear stress (the force-related component of flow). In the best-characterized example of biological flow sensing, mammalian cells use the force-sensitive von Willebrand factor to recruit platelets in response to fluid flow1. By analogy, other cellular systems that sense flow, including the bacterial responses described above, have been interpreted to be triggered by shear force5, 6. However, the conclusion that cells sense flow by measuring shear force has not been directly tested in these systems.
To enable a biophysical characterization of bacterial flow sensing, we focused on the bacterium P. aeruginosa and began with a global assessment of how it changes its transcriptome in response to flow. Specifically, we developed an experimental system that subjects cells to flow in microfluidic channels and monitors global gene expression through RNA-sequencing (Fig. 1A). We discovered a large number of changes in gene expression after 4 hours of exposure to flow (Table S1). To focus on potential direct targets of flow, we repeated our analysis after only 20 minutes of flow exposure (Fig. 1B) (Table S2). A previously-unnamed four-gene operon was the most highly induced operon at this early time point (Fig. 1C). While all four genes in this operon were expressed at relatively low levels (all in the bottom 50% of the genome by expression) before flow exposure, they exhibited strong induction after 20 minutes of flow exposure (approximately ~13 fold, Fig. 1C). Thus, we focused our efforts on this operon as a model for the broader flow response and named its four genes froA through froD for “flow responsive operon.”
Fig. 1: Flow triggers induction of gene expression in Pseudomonas aeruginosa.
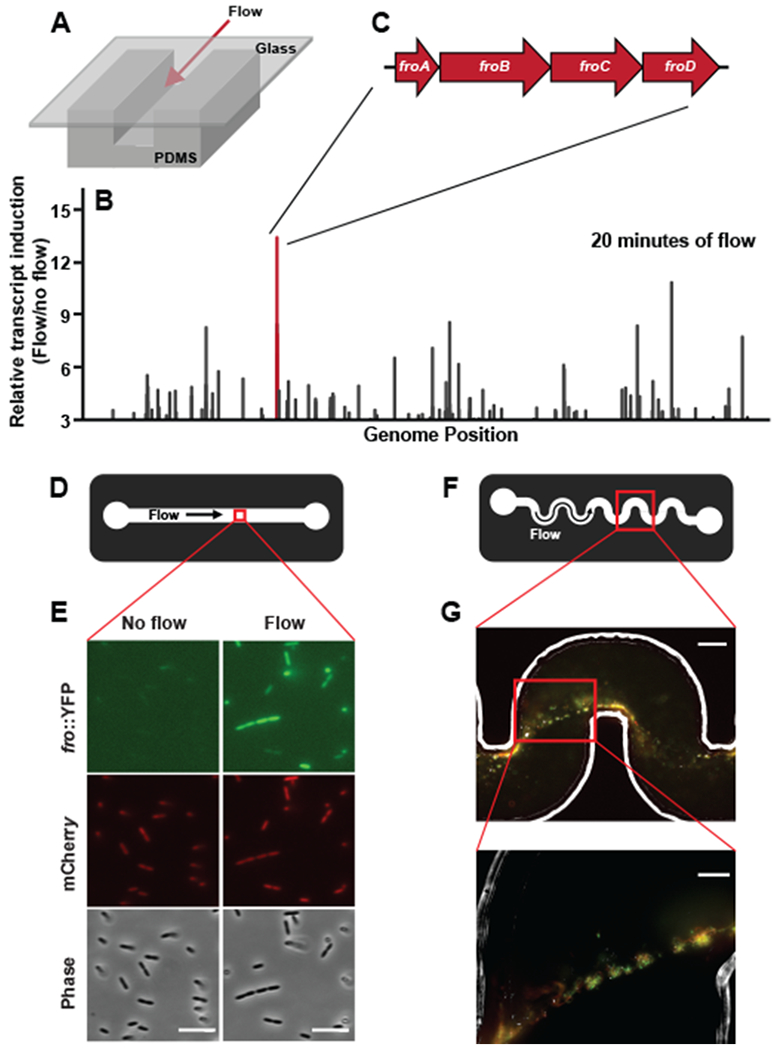
(A) Schematic of microfluidic devices used throughout this study. Channels are custom-fabricated of polydimethylsiloxane (PDMS) and glass. (B) The fold-change in transcript abundance of P. aeruginosa cells subjected to flow for 20 minutes relative to flow-naïve cells. Line length linearly corresponds to fold-change and is plotted as a function of genomic location on the P. aeruginosa chromosome. Only genes induced at least 3-fold are represented, and the raw data used to generate this graph is presented in Supplementary Table 2. The red line corresponds to the fro operon. (D) Schematic depicting the view from above the microchannel used in E. These channels are 50 μm tall by 500 μm wide. (E) Fluorescence and phase images of fro reporter strain in straight microfluidic channels before and after 4 hours of 10 μl/min flow. Images are representative of three independent experiments. Scale bars indicate 5 μm. (F) Schematic of the microchannel used in G. These channels are 90 μm tall by 100 μm wide. (G) A merged image of Phase, YFP, and mCherry from a single optical plane of a representative streamer biofilm projecting off the wall of a microchannel. Images are representative of three independent experiments. Streamers were cultured in 2 μl/min flow for 20 hours. Lower picture is zoomed in on cells that are not directly in contact with the channel surface. Scale bar of top image indicates 50 μm and scale bar of bottom image indicates 20 μm.
In order to probe the P. aeruginosa flow response with single-cell resolution, we engineered a two-color fluorescent reporter strain that reports on fro expression with YFP and uses a constitutively expressed mCherry for normalization (Supplementary Figure 1). In straight microfluidic channels (Fig. 1D), YFP fluorescence increased approximately 6-fold in flow, while mCherry fluorescence remained constant (Fig. 1E, Supplementary Figure 2). These results validate our transcriptional profiling data and demonstrate that individual P. aeruginosa cells induce fro expression in response to flow. We call this form of bacterial environmental sensing rheosensing, as the prefix rheo- is Greek for flow.
P. aeruginosa often exists in biofilms in nature, especially in environments with flow7. Therefore, we examined rheosensing in the context of multicellular communities, focusing on flow-induced biofilm streamers that we generated in microchannels featuring a series of 90-degree bends (Fig. 1F). Biofilms are aggregates of bacteria held together by an extracellular matrix. Biofilm streamers occur under specific conditions of flow in which the biofilm remains attached to the surface at a focal point while a long tendril of cells and matrix extends into the center of the channel7. We detected fro expression throughout cells in biofilm streamers, including in cells significantly removed from the channel surface (Fig. 1G). Therefore, P. aeruginosa cells within a multicellular community are capable of rheosensing.
The single-cell response of the fro reporter enabled us to quantitatively characterize the response of P. aeruginosa to flow. The flow experienced by bacteria on a surface depends on the bulk flow rate and channel geometry. To represent flow intensity in a geometry-independent manner we report the shear rate, which is the rate at which adjacent layers of fluid pass one another. We explored the dynamic range of rheosensing by examining fro expression after cells were subjected to a range of shear rates for 2 hours, fro induction did not occur at low shear rates (8 sec−1), increased in response to intermediate shear rates (40 – 400 sec−1), and plateaued at high shear rates (> 400 sec−1) (Supplementary Figure 3). To formally test the hypothesis that fro induction is modulated by shear rate, we also altered the channel height while maintaining a constant flow rate (the equation in Supplementary Figure 3 shows how channel dimensions relate shear rate and flow rate). Increasing the channel height 10-fold significantly reduced fro induction (Supplementary Figure 3). Together, our results show that fro induction is not binary and is tuned by shear rate. These data also establish that rheosensing is tuned to a physiologically-relevant range of shear rates8, such as those found in average-sized human veins (~100 sec−1) and arteries (~650 sec−1).
Fig. 3: fro induction requires the sigma factor FroR and anti-sigma factor FroI but not known surface sensors.
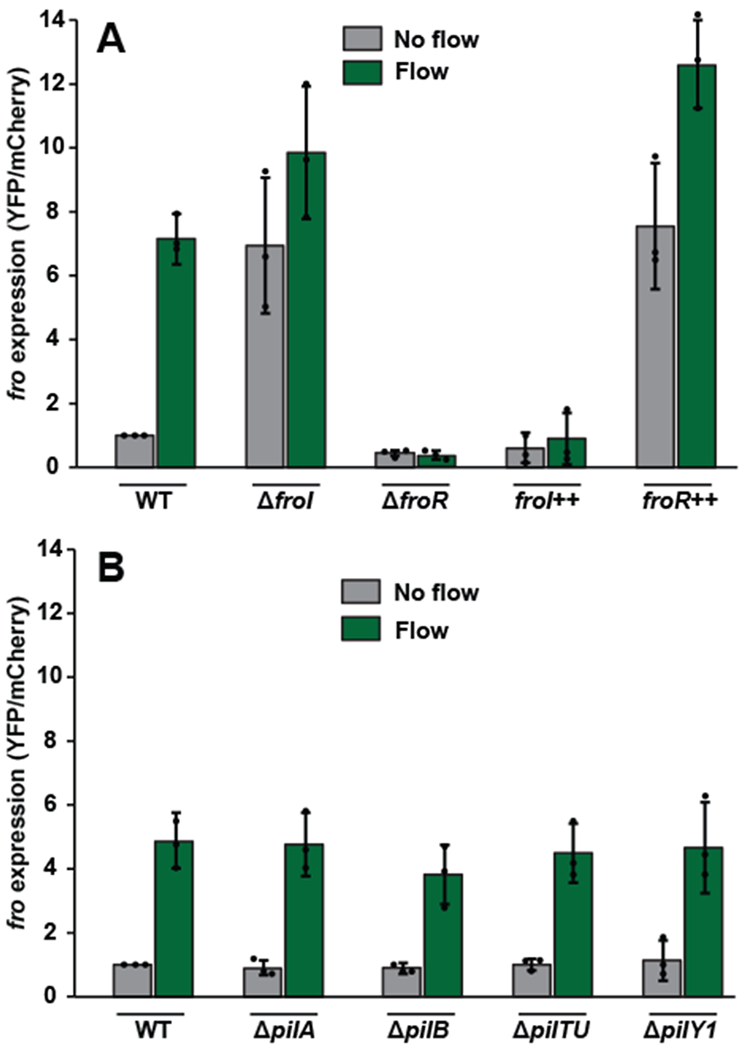
(A) fro expression levels in wild-type cells, ΔfroI mutant cells, ΔfroR mutant cells, froI++ overexpressing cells, and froR++ overexpressing cells either subjected to no flow (gray bars) or 2 hours of flow at a shear rate of 800 sec−1 (green bars). In no flow conditions, wild-type expression is significantly different from ΔfroI (P=0.04), ΔfroR (P=0.008) and froR++ expression (P=0.02), but statistically indistinguishable from froI++ expression (P=0.18), as calculated by a 2-sided T-test. In flow conditions, wild-type expression is significantly different from ΔfroR (P=0.002), froI++ expression (P=0.0005) and froR++ expression (P=0.01), but statistically indistinguishable from ΔfroI (P=0.14), as calculated by a 2-sided T-test. (B) fro expression levels in wild-type cells, ΔpilA mutant cells, ΔpilB mutant cells, ΔpilTU mutant cells, and ΔpilY1 mutant cells either subjected to no flow (gray bars) or 2 hours of flow at a shear rate of 800 sec−1 (green bars). Error bars show SD of three independent replicates and points indicate values for each replicate. In flow conditions, wild-type expression is statistically indistinguishable from ΔpilA (P=0.88), ΔpilB (P=0.28), ΔpilTU (P=0.76), and ΔpilY1 (P=0.95), as calculated by a 2-sided T-test. Values normalized to WT in no flow, which is set to 1 for each replicate. Channels used for these experiments were 50 μm tall by 500 μm wide.
Shear rate could modulate the kinetics of fro induction or the maximal amplitude of fro induction. We thus temporally characterized rheosensing by measuring fro expression over time at a range of shear rates, fro induction began at approximately 45 minutes (Fig. 2A, 2B), which is consistent with the maturation time of the YFP reporter used in this experiment9. Intermediate and high shear rates induced fro expression with different kinetics, as higher shear rate led to more rapid fro induction (Fig. 2B, Supplementary Figure 4). The slope of the fro induction curve demonstrates that induction saturates (Fig. 2C, Supplementary Figure 4), indicating that rheosensing also sets the maximal amplitude of induction. Consistent with the ability of rheosensing to respond to changes in shear rate, cells saturated by exposure to intermediate flow for 2 hours experienced additional fro induction when shifted to higher flow (Fig. 2C). Therefore, we conclude that this type of rheosensing is a tightly controlled sensory modality that fine-tunes the kinetics and amplitude of gene regulation in response to flow.
Fig. 2: Shear rate rapidly and dynamically tunes rheosensing.
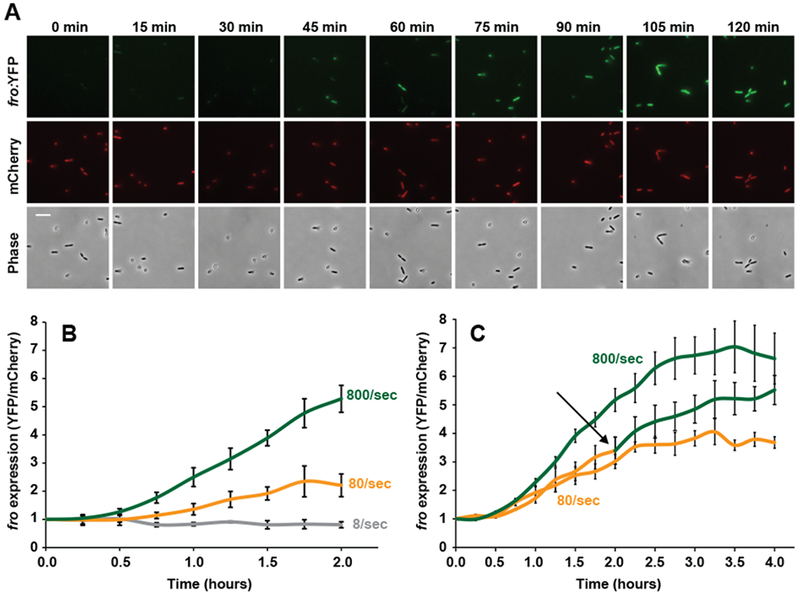
(A) Images of cells exposed to flow at a wall shear rate of 800 sec−1 over 120 min. Top images show the fro reporter (YFP) channel, middle images show the mCherry normalization control channel, and bottom images show the phase contrast channel. Images are representative of three independent experiments. Scale bar indicates 5 μm. (B) fro expression over 2 hours of time in the presence of 8 (gray line), 80 (yellow line), and 800 sec−1 (green line) shear rates. At 2 hours, 8/sec and 80/sec samples are statistically different from each other with P=0.03, calculated by a 2-sided T-test. At 2 hours, 80/sec and 800/sec samples are statistically different from each other with P=0.008, calculated by a 2-sided T-test. (C) fro expression over 4 hours of time in the presence of 80 sec−1 (yellow line), 800 sec−1 (green line), or an upshift from 80 sec−1 to 800 sec−1 (yellow-green line). The black arrow depicts the 2-hour time point where shear rate was increased from 80 sec−1 to 800 sec−1 for the upshifted sample. At 4 hours, the upshifted sample results in fro expression that is statistically different from the 80 sec−1 sample with P=0.03, calculated by a 2-sided T-test. Error bars show SEM of three independent replicates. Each replicate represents quantification from 50 cells, fro expression at time 0 set is set to 1. Channels used for these experiments were 50 μm tall by 500 μm wide.
As rheosensing leads to changes in gene expression, we aimed to discover regulatory factors that control rheosensitive signaling. We focused on two previously uncharacterized genes directly upstream of the fro operon that are predicted to encode an alternative sigma factor and anti-sigma factor10. Deletion of the putative sigma factor eliminated fro induction in flow, while deletion of the putative anti-sigma factor increased fro expression in flow-naïve cells (Fig. 3A, Supplementary Figure 5). Based on these results, we named the gene encoding the sigma factor froR (fro regulator) and the gene encoding the anti-sigma factor froI (fro inhibitor). Overexpression of froR increased fro expression, while overexpression of froI eliminated fro induction (Fig. 3A, Supplementary Figure 5). Together, our results provide evidence for a model where the anti-sigma factor FroI antagonizes the alternative sigma factor FroR to control induction of the fro operon in flow.
Both flow and surfaces exert mechanical forces on cells such that they could use common sensors. To test if rheosensing is related to previously-proposed forms of bacterial mechanosensation, we asked if genes required for surface sensing are required for fro induction. Retraction of the type IV pilus controls surface sensing in P. aeruginosa6, 11–14 and C. crescentus15. However, retraction of the type IV pilus does not control rheosensing, as fro induction is maintained in ΔpilA (lacking the pilus fiber), ΔpilB (lacking pilus extension) and ΔpilTU (lacking pilus retraction) mutant backgrounds (Fig. 3B, Supplementary Figure 6). Similarly, whereas PilY1 is required for surface-activated virulence in P. aeruginosa16, fro induction was still observed in a ΔpilY1 mutant (Fig. 3B, Supplementary Figure 6). We also tested mutants lacking flagella, since the flagellum has been implicated in surface sensing in other bacteria17, 18. Like type IV pili and PilY1, the flagellum is not required for fro induction, as fro induction was also observed in a ΔfliC mutant (Supplementary Figure 7)19. We note that none of the mutants tested dramatically disrupted adhesion (Supplementary Figures 5–7) and that fro induction was normalized on a single-cell basis (Supplementary Figure 1 describes our quantification pipeline). Additional support for the independence of rheosensing from surface sensors came from analysis of our transcriptional profiling, which revealed no statistical overlap between P. aeruginosa genes induced by flow and by surface association (Supplementary Figure 8)16.
To directly test whether surface association alone is sufficient to induce fro expression, we fabricated microfluidic channels with flow-exposed and flow-shielded regions (Fig. 4A). While the bacteria in the flow-shielded regions did not experience flow, they did remain surface-associated for the duration of the experiment. Cells in flow-exposed regions of the channel induced fro expression approximately 9-fold, while cells in flow-shielded regions did not (Fig. 4B, Supplementary Figure 9). As an additional test of whether surface association affects rheosensing we used the chemical MPTMS (3-Mercaptopropyl trimethoxysilane), which increases adhesion between P. aeruginosa and the channel surface20. MPTMS treatment did not affect fro induction (Supplementary Figure 10), suggesting that cells perform rheosensing independently of how they attach to the surface. Together, our data indicate that fro induction is independent of known surface sensors, is not triggered by surface association itself, and is not affected by enhanced surface adhesion.
Fig. 4: Rheosensing is a force-independent sensory modality.
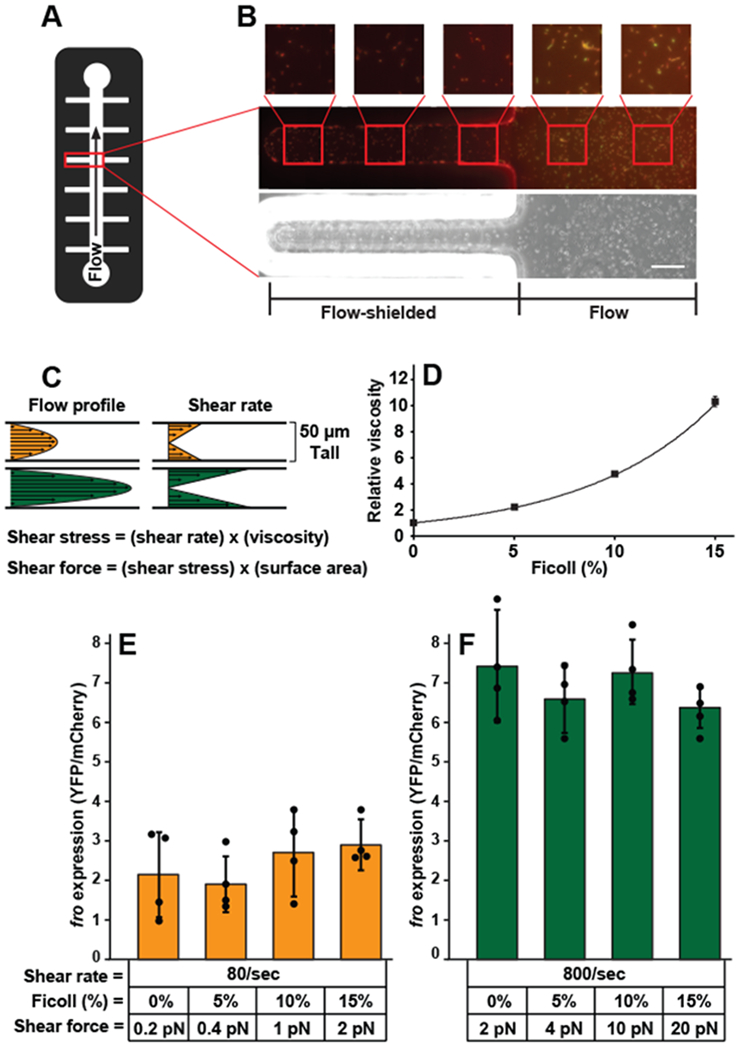
(A) Schematic of the microchannel used in B. (B) Image of fro reporter cells in flow-exposed and flow-shielded regions of the channel after treatment with a shear rate of 800 sec−1 for 4 hours. Phase image is on the bottom and merged YFP/mCherry images are on the top. Smaller square boxes are zoomed in on images depicted by red squares. The scale bar indicates 50 μm. The image is representative of two independent replicates. (C) Schematic showing how the flow profile corresponds to shear rate in a microfluidic device. Equations showing that shear stress is the product of shear rate and fluid viscosity and shear force is the product of shear stress and surface area. (D) Microscopic viscosity of Ficoll solutions as measured with micron-scale beads and optical tweezers. Error bars show SD of three independent replicates. (E, F) Expression of the fro reporter in response to two hours of flow at the defined shear rates and shear forces. Shear forces were calculated by multiplying shear stress by cell surface area, which was estimated at 2.5 μm2. Error bars show SD of four independent replicates and points indicate values for each replicate. fro expression at 80 sec−1 is significantly different from fro expression at 800 sec−1 with treatments of 0% Ficoll (P=0.003), 5% Ficoll (P=0.002), 10% Ficoll (P=0.002), and 15% Ficoll (P=0.006), calculated by a 2-sided T-test. fro expression at 80 sec−1 with no Ficoll is significantly indistinguishable from fro expression at 80 sec−1 with 5% Ficoll (P=0.79), 10% Ficoll (P=0.67), and 15% Ficoll (P=0.37), calculated by a 2-sided T-test. fro expression at 800 sec−1 with no Ficoll is significantly indistinguishable from fro expression at 800 sec−1 with 5% Ficoll (P=0.33), 10% Ficoll (P=0.66), and 15% Ficoll (P=0.10), calculated by a 2-sided T-test. fro expression of cells before flow treatment is set to 1. Channels used for these experiments were 50 μm tall and 500 μm wide.
The independence of rheosensing from previously-proposed forms of bacterial mechanosensing called into question the prevailing model that bacteria sense flow by measuring force. Fluid flow in a microfluidic channel has a kinematic aspect (shear rate, in units of time−1) and a force-related aspect (shear stress, in units of force/area)21. These two aspects are linked by the viscosity of the solution, as shear stress is the product of shear rate and viscosity (Fig. 4C)21. The finding that fro expression is tuned by flow intensity thus enabled us to use changes in viscosity to directly test if P. aeruginosa responds to shear rate or shear force. To modulate viscosity, we used solutions with varying concentrations of the viscous agent Ficoll. These Ficoll solutions act as Newtonian fluids22 and we directly quantified their viscosity at the scale of a bacterial cell using optical tweezers and micron-scale beads (Fig. 4D). Microscopic measurements of viscosity of Ficoll solutions increased exponentially with concentration: 5% Ficoll increased viscosity 2-fold, 10% Ficoll increased viscosity 5-fold, and 15% Ficoll increased viscosity 10-fold (Fig. 4D). If fro expression was triggered by shear force (or stress), we should have observed a linear relationship between viscosity and fro expression when the shear rate is held constant. To our surprise, we found that increasing viscosity up to 10-fold had no effect on fro expression at an intermediate shear rate (80 sec−1, Fig. 4E). To control for the possibility that Ficoll has deleterious effects on bacteria, we confirmed that Ficoll did not affect the full fro induction that occurs at high shear rate (800 sec−1, Fig. 4F). Together, these experiments demonstrate that this form of rheosensing is a force-independent sensory modality.
The observation that fro induction is sensitive to shear rate but not shear force raises the question of whether rheosensing should be considered a form of mechanosensing. Traditionally, the field of mechanics encompasses the study of both motion and force. For example, kinematics is the subfield of mechanics that focuses on motion and deformation while ignoring forces, and the force-independent property of shear rate is considered a fundamental feature of fluid mechanics. Meanwhile, in biological contexts, the term mechanosensing has traditionally been restricted to the study of how cells sense force, potentially leading to premature conclusions about the nature of mechanosensing. The argument over whether kinematic rheosensing should be considered a type of mechanosensing or a distinct process is semantic, but its implications are significant. We suggest that it is more useful to consider rheosensing a form of mechanosensing as it provides proof-of-principle that cells can sense mechanical features of their environment such as flow without measuring force.
One potential benefit of force-independent rheosensing is that such a system robustly measures the speed of flow independent of other fluid properties such as viscosity. Thus, by sensing shear rate instead of shear force, P. aeruginosa could induce fro expression similarly across a wide range of different fluids, such as those found in freshwater streams, medical devices, the blood stream, or lung sputum. Consistently, genomic studies indicate that the froABCD operon and gene encoding the sigma factor FroR are required for colonization of environments that have fluids that vary widely in viscosity, such as the lung23 and gastrointestinal tract24. Furthermore, while the precise physiological role of rheosensing remains to be determined, genomic analysis of flow-induced genes identified a significant number of genes that are also induced during human infection (Supplementary Figure 8)25.
How might bacteria sense flow independently of force? Our finding that fro induction is modulated by shear rate suggests that the bacteria have a mechanism for measuring a rate-dependent biophysical process. Biological processes that are rate-dependent but force-independent include chemical transport and rotational diffusion. For example, flow has previously been shown to impact quorum sensing and this effect is likely dependent on shear rate as higher flow would more rapidly wash away autoinducer. However, rheosensing is induced by flow rather than inhibited by it. Nevertheless, we tested the role of quorum sensing in rheosensing by assaying fro induction in a lasR mutant that eliminates canonical P. aeruginosa quorum sensing. We found that the loss of lasR had no effect on fro induction (Supplementary Figure 11), suggesting that if rheosensing involves chemical transport it does so through a different system than quorum sensing that has not previously been shown to be flow-sensitive.
Another possibility is that P. aeruginosa has a surface-exposed protein that directly senses shear rate. For example, a surface protein with asymmetric domains would be predicted to rotate in a shear-rate-dependent manner, forming a molecular “water-wheel”. While understanding the molecular mechanism of shear rate sensing will require future studies, we do know that rheosensing involves signaling through the ECF-family sigma factor FroR and the anti-sigma factor FroI. As neither froR or froI RNA abundance is regulated by shear flow, post-translational regulation is likely involved in rheosensitive signaling. ECF-family sigma factors and their corresponding anti-sigma factors have traditionally been implicated in sensing extracytoplasmic cues26, such that FroR and FroI are well positioned to link the extracellular input of shear rate to the intracellular output of transcription.
Our discovery that cells can sense flow without sensing shear force suggests that there is value in reevaluating the interpretation of biological responses to flow. Most responses to flow to date have not been thoroughly characterized at the biophysical level. However, a few well-understood examples in mammalian cells involve sensing shear force, such as platelet aggregation induced by the force-sensitive von Willebrand factor1 or ion channel regulation by force-sensitive stereocilia in cochlear hair cells2. Based on analogy to these examples and the intuitive ability to understand how flow can impart a force (or stress), showing that a system is sensitive to flow has often been interpreted as evidence that the system responds to shear force5, 6. Together, our results demonstrate the possibility of kinematic (force-independent) mechanosensing, which challenges the potentially premature conclusion that bacteria sense flow by measuring shear force. Future biophysical studies in both eukaryotes and bacteria will be required to test if rheosensing is sensitive to shear force in other biological systems. It will be particularly interesting to determine if the differences between bacterial and mammalian rheosensing reflect generalizable differences, for example in the need for bacteria to respond to different fluids in contrast to the relatively uniform environments of most mammalian cell types.
Materials and methods
Strains, plasmids and growth conditions
The bacterial strains used in this study are described in Table S3. The primers used are described in Table S4 and the plasmids used are described in Table S5.
P. aeruginosa was grown in liquid LB-Miller (Difco) in a roller drum, and on LB-Miller agar (1.5% Bacto Agar) at 37°C. Antibiotics (Sigma) were used at the following concentrations: carbenicillin 150 μg/ml (liquid) and 300 μg/ml (solid); gentamicin 15 μg/ml (liquid) and 30 μg/ml (solid); tetracycline 100 μg/ml (liquid) and 200 μg/ml solid; and irgasan 25 μg/ml solid.
E. coli was grown in liquid LB-Miller (Difco) in a floor shaker, and on LB-Miller agar (1.5% Bacto Agar) at 37°C. Antibiotics (Sigma) were used at the following concentrations: carbenicillin 50 μg/ml (liquid) and 100 μg/ml (solid); tetracycline 7.5 μg/ml (liquid) and 15 μg/ml solid; and irgasan 25 μg/ml solid.
The fro reporter was generated using the lambda Red recombinase system (27). The fro reporter construct was Gibson-assembled from three PCR products in the following series. First, the 546 bp upstream of the target insertion site amplified from PA14 genomic DNA. Second, a 1903 bp fragment containing a strong bacterial ribosome binding site, a YFP ORF, and an aacC1 ORF flanked by FRT sites amplified from pAS03. Third, the 531 bp downstream of the target insertion site amplified from PA14 genomic DNA. Deletions in the fro reporter background were generated by the lambda Red recombinase system using the aacC1 ORF between the flanking regions of targeted gene of interest.
Constructs targeting the attTn7 phage attachment site were delivered by co-electroporation with pTNS2 (28). Constructs targeting the attB phage attachment site were delivered by conjugation with an S17-1 strain harboring a mini-CTX2 derivative (29).
RNA-sequencing library preparation and data analysis
Total RNA was harvested from cells in fluidic devices by replacing medium with total lysis solution (TLS) (10 mM Tris-HCl, 1 mM EDTA, 0.5 mg/mL lysozyme, 1% SDS) and flowing through the device. TLS was incubated at room temperature for 2 minutes, mixed with sodium citrate pH 5.2 to 0.1 M. The resulting solution was mixed 1:1 with 0.1 M citrate-saturated phenol pH 4.3, incubated at 64°C for 6 minutes, and centrifuged for 10 minutes at 13 k × g at 4°C. The aqueous layer was mixed 1:1 with chloroform, transferred to phase lock tubes (Quanta Bio, Beverly, MA), and centrifuged for 10 minutes at 13 k × g at 4°C. The aqueous layer was precipitated by mixing 1:2 with solution of 30:1 ethanol:3 M sodium acetate pH 5.2, washing with 70% ethanol, and resuspending the resulting pelleting with water. Genomic DNA was removed from nucleic acid preparations using DNA-free DNase (Ambion/Life Technologies, Carlsbad, CA) and purified using ethanol precipitation. The resulting preparations containing RNA was purified of ribosomal RNA using RiboZero (Illumina, San Diego, CA). mRNA libraries were prepared for sequencing using NEBNext Ultra Directional RNA Library Kit (New England Biolabs, Ipswich, MA) with a modified protocol using Sera-Mag SpeedBeads that retains mRNA transcripts as small as 50 bps in length. The resulting mRNA libraries were verified using gel electrophoresis and a Bioanalyzer, multiplexed, and sequenced using an Illumina HiSeq 2500 (Illumina, San Diego, CA) in rapid mode.
The resulting sequence files were processed using the customized Python scripts align_barcode, filter_P7adapter which was written by our lab, aligned using Bowtie 2 (30) and analyzed using the customized scripts tabulateFrequencies and annotateTabulated written by our lab in Python and Perl. The representation of each mRNA transcripts was determined by dividing the number of reads in a particular region by the total number of reads for the library.
Fabrication of microfluidic devices
Microfluidic devices were fabricated using standard soft lithography techniques. Devices were designed in AutoCAD (Autodesk) and masks were printed by CAD/Art Services, Inc. (Bandon, OR). Device molds were produced on silicon wafers (University Wafer) spin-coated with SU-8 photoresist (Microchem). Polydimethylsiloxane (PDMS) chips were plasma-bonded bonded to glass slides at least 24 hours before use.
The devices used to conduct the RNA-seq experiment had twelve parallel channels 400 μm wide × 100 μm high × 5 cm long. All twelve channels shared a single inlet port and a single outlet port. These chips were bonded to Corning 75 mm × 50 mm × 1 mm plain microslides.
The devices used to culture flow-shielded and flow-exposed sub-populations were previously described in (31). The channels were 50 μm high, 500 μm wide in the central channel and 50 μm wide in the crevices. The devices used to culture biofilm streamers were previously described in (7). The channels were 100 μm wide × 90 μm high. Each channel possessed its own inlet and outlet port. All of these chips were bonded to 1.42” × 2.36” #1.5 coverglass (Ted Pella, Inc.).
The devices used in measure fro expression at different shear rates had two parallel channels 500 μm wide × 50 μm high × 1 cm long. Each of the channels possessed its own inlet and outlet port. The devices used in measure fro expression in different mutant backgrounds had five parallel channels 500 μm wide × 50 μm high × 2 cm long. Each of the channels possessed its own inlet and outlet port. These chips were bonded to Fisherbrand 22 mm × 60 mm #1 coverglass.
P. aeruginosa growth in microfluidic devices
In experiments measuring fro expression, cells from mid-log phase cultures were injected directly into the flow chamber inlet with a pipette and allowed to settle for 10 minutes. The flow chamber was fixed on the microscope stage. A plastic, LB-filled, 27G needle-tipped syringe mounted on a syringe pump (KD Scientific Legato 210) was connected to the chamber inlet via tubing (BD Intramedic Polyethylene Tubing .015” ID 0.043” OD). The chamber outlet was connected to a waste container via tubing. The syringe pump was used to generate flow rates of 0.1 to 50 μl/min.
To coat the channel surfaces with MPTMS (3-Mercaptopropyl trimethoxysilane), we used a previously described method (20). Briefly, channels were washed with a 5:1:1 H2O:H2O2:HCl solution for 5 minutes, flushed with H2O, treated with MPTMS for 30 minutes, and flushed with H2O again before adding cells to the channel.
For transcriptional profiling assays, cells from mid-log phase cultures were injected into the flow chamber inlet with a plastic syringe and allowed to settle for 10 minutes. The flow chamber was fixed on the benchtop. A plastic, LB-filled, needle-tipped syringe mounted on a syringe pump was connected to the chamber inlet via tubing (McMaster-Carr Polyethylene Tubing 2 mm ID, 4 mm OD). The chamber outlet was connected to a waste container via tubing. The syringe pump was used to generate a flow rate of 100 μl/min.
Shear rate and shear force calculations
The shear rate at the floor and ceiling of the channel of the rectangular cross-section (where height is << width) is calculated by the equation:
where Q is the flow rate, w is the channel’s width and h is the channel’s height.
Shear stress is calculated as the product of shear rate and viscosity, as shown in Figure 4C. Shear force is calculated as the product of shear stress and the surface area of a cell, which is estimated as 2.5 μm2.
Phase Contrast and Fluorescence Microscopy
Images were obtained with a Nikon Ti-E microscope controlled by NIS Elements (version 3.22.15). The microscope is equipped with a Nikon 10× Plan Fluor Ph1 0.3 NA objective, a Nikon 20× Plan Fluor Ph1 0.45 NA objective, a Nikon 40× Plan Apo Ph2 0.95 NA objective, a Nikon 60× Plan Apo 1.2 NA objective, a Nikon 100× Plan Apo Ph3 1.4 NA objective, a Prior Lumen 200 Pro, and an Andor Clara CCD camera.
Quantification of fro expression
The image analysis pipeline (Supplementary Figure 1) was written in MATLAB (Mathworks, Natick, MA). Cell masks were developed from phase contrast images by a Sobel operator edge-detection algorithm. The YFP and mCherry fluorescence intensity per masked cell was computed. The YFP-to-mCherry ratio of hundreds of individual cells was averaged and expressed as fro expression.
Quantification of Ficoll viscosity
To estimate the microscale viscosity of different Ficoll concentrations, we analyzed the diffusion of optically trapped 500 nm polystyrene beads (32). In brief, a ten second time trace of the bead fluctuation x(t) was recorded at 50 kHz sampling rate. We then computed the positional autocorrelation which yields the autocorrelation time τc = κ/γ. κ = const. is the force sensitivity of the optical trap and depends on the laser power and size of the trapped bead only, hence is constant. γ= 3πDη is the viscous drag coefficient of the bead with diameter D immersed in the Ficoll solution with viscosity η. We then compare the autocorrelation time of identical beads in water to that of different Ficoll concentrations to obtain the Ficoll viscosity ηFicoll / ηWater = τWater / τFicoll relative to water.
Code availability
The custom MATLAB routines used for processing and analyzing the fluorescence microscopy data are freely available from the corresponding author upon request. The custom python and perl scripts used for processing and analyzing the RNA-seq data are freely available from the corresponding author upon request.
Data Availability
The data supporting the findings of the study are available in this article and its Supplementary Information Files. All the RNA-sequencing data used to reach the conclusions of this paper are freely available under NCBI SRA accession number PRJNA530209. Additionally, the raw data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgements
We thank Kevin Kim for assistance in generating flow-shielded and biofilm streamer microfluidic channels. We also thank members of the Gitai lab, Josh Shaevitz, Ned Wingreen, Dan Kearns and Lisa Wiltbank for helpful discussions and comments on the manuscript. This work was supported by grant DP1AI124669 from the National Institutes of Health (to ZG). Additional funding came from the National Science Foundation PHY-1734030 (BPB and MDK), the Glenn Centers for Aging Research (BPB), DFG Award KO5239/1-1 from the German Research Foundation (to MDK) and National Institutes of Health grants K22AI112816 (to AS) and R21AI121828 (BPB and MDK).
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Hansen CE, Qiu Y, McCarty OJT, & Lam WA Platelet mechanotransduction. Annu. Rev. Biomed. Eng 20, 253–275 (2018) [DOI] [PubMed] [Google Scholar]
- 2.Vollrath MA, Kwan KY, & Corey DP The micromachinery of mechanotransduction in hair cells. Annu. Rev. Neurosci 30, 339–65 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Persat A, et al. The mechanical world of bacteria. Cell 5, 988–997 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes KT & Berg HC The bacterium has landed. Science 358, 446–447 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alsharif G, et al. Host attachment and fluid shear are integrated into a mechanical signal regulating virulence in Escherichia coli O157:H7. Proc. Natl. Acad. Sci. USA 17, 5503–5508 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodesney CA, et al. Mechanosensing of shear by Pseudomonas aeruginosa leads to increased levels of the cyclic-di-GMP signal initiating biofilm development. Proc. Natl. Acad. Sci. USA 23, 5906–5911 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drescher K, Shen Y, Bassler BL, & Stone HA Biofilm streamers cause catastrophic disruption of flow with consequences for environmental and medical systems. Proc. Natl. Acad. Sci. USA 11, 4345–4350 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakariassen KS, Orning L, & Turitto VT The impact of blood shear rate on arterial thrombus formation. Future Science OA 4, FSO30 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon A, et al. Single-cell quantification of molecules and rates using open-source microscope-based cytometry. Nature Methods 2, 175–181 (2007) [DOI] [PubMed] [Google Scholar]
- 10.Boechat AL, Kaihami GH, Politi MJ, Lepine F, & Baldini RL A novel role for an ECF sigma factor in fatty acid biosynthesis and membrane fluidity in Pseudomonas aeruginosa. PloS One 12, e84775 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Persat A, Inclan YF, Engel JN, Stone HA, & Gitai Z Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 24, 7563–7568 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee CK, et al. Multigenerational memory and adaptive adhesion in early bacterial biofilm communities. Proc. Natl. Acad. Sci. USA 17, 4471–4476 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inclan YF, et al. A scaffold protein connects type IV pili with the Chp chemosensory system to mediate activation of virulence signaling in Pseudomonas aeruginosa. Molecular Microbiology 4, 590–605 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo Y, et al. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. mBio 1, e02456–14 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellison CK, et al. Obstruction of pilus retraction stimulates bacterial surface sensing. Science 358, 535–538 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siryaporn A, Kuchma SL, O’Toole GA, & Gitai Z Surface attachment induces Pseudomonas aeruginosa virulence. Proc. Natl. Acad. Sci. USA 47, 16860–16865 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hug I, Deshpande S, Sprecher KS, Pfohl T, & Jenal U Second messenger-mediated tactile response by a bacterial rotary motor. Science 358, 531–534 (2017) [DOI] [PubMed] [Google Scholar]
- 18.McCarter L, Hilmen M, & Silverman M Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 3, 345–351 (1988) [DOI] [PubMed] [Google Scholar]
- 19.Brimer CD & Montie TC Cloning and comparison of fliC genes and identification of glycosylation in the flagellin of Pseudomonas aeruginosa a-type strains. Journal of Bacteriology 12, 3209–3217 (1998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siryaporn A, Kim MK, Shen Y, Stone HA, & Gitai Z Colonization, competition, and dispersal of pathogens in fluid flow networks. Current Biology 9, 1201–1207 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S & Springer TA Selectin receptor-ligand bonds: Formation limited by shear rate and dissociation governed by the Bell model. Proc. Natl. Acad. Sci. USA 3, 950–955 (2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez VA, et al. Flagellated bacterial motility in polymer solutions. Proc. Natl. Acad. Sci. USA 50, 17771–17776 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potvin E, et al. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environmental Microbiology 12, 1294–1308 (2003) [DOI] [PubMed] [Google Scholar]
- 24.Skurnik D, et al. A comprehensive analysis of in vitro and in vivo genetic fitness of Pseudomonas aeruginosa using high-throughput sequencing of transposon libraries. PLoS Pathogens 9, e1003582 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cornforth DM, et al. Pseudomonas aeruginosa transcriptome during human infection. Proc. Natl. Acad. Sci. USA 22, E5125–E5134 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helmann JD The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol 46, 47–110 (2002) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of the study are available in this article and its Supplementary Information Files. All the RNA-sequencing data used to reach the conclusions of this paper are freely available under NCBI SRA accession number PRJNA530209. Additionally, the raw data that support the findings of this study are available from the corresponding author upon request.


