Abstract
Using assemblies of simple peptides with emergent properties to achieve elaborate functions has attracted increasing attention in recent years. Besides tailoring self-assembly abilities of peptides in vitro, the recent progresses of peptide research are advancing into a new and exciting frontier—that is, rational design of assemblies of peptides (or their derivatives) for biological functions in complex environment, especially in cells or in animals. This minireview highlights some recent developments of peptide assemblies and their applications in biological systems. After introducing the unique merits of peptide assemblies, we discuss the recent progress of designing peptides (or peptide derivatives) for self-assembly with conformational control. Then, we describe biological functions of peptide assemblies, with emphasis on a useful approach—instructed-assembly—for spatiotemporal control of peptide assemblies, in the context of cells (e.g., in the pericellular, intracellular, subcellular, or intercellular space). Finally, we provide a brief perspective to discuss the future promises and challenges of this exciting area of chemistry.
Keywords: peptides, self-assembly, cells, hydrogel, nanostructures
Graphical Abstract
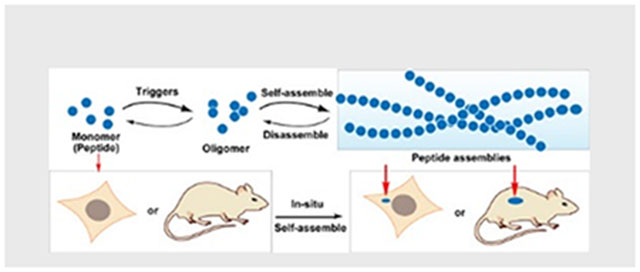
Assemblies of peptides (or their derivatives) for biological functions in complex environment attract increasing attention, especially in cells or in animals. In this minireview, selected examples of peptide assemblies are highlighted, with a focus on instructed-assembly for spatiotemporal control of peptide assemblies in the context of cells.
Graphical Abstract
1. Introduction
Peptides, as a class of bioactive molecules, play essential regulatory roles in cells and organisms. For example, peptide hormones in animals involve in reproductive physiology, energy metabolism, stress, growth, appetite, and cardiac function.[1] Notably, being synthesized in the somas of two neurons and acting as hormones on peripheral targets, oxytocin and vasopressin (Figure 1A) are the two closely related neuropeptides that exert their functions on central and peripheral neurons.[2] Besides studying endogenous peptides in human, researchers have developed various peptides for anticancer, antibacterial, or antiviral applications, and some of them have already entered clinics.[3] While the past successes of peptide chemistry mainly focused on the functions of individual peptides, the assemblies of peptides have attracted increased research attentions.[4]
Figure 1.
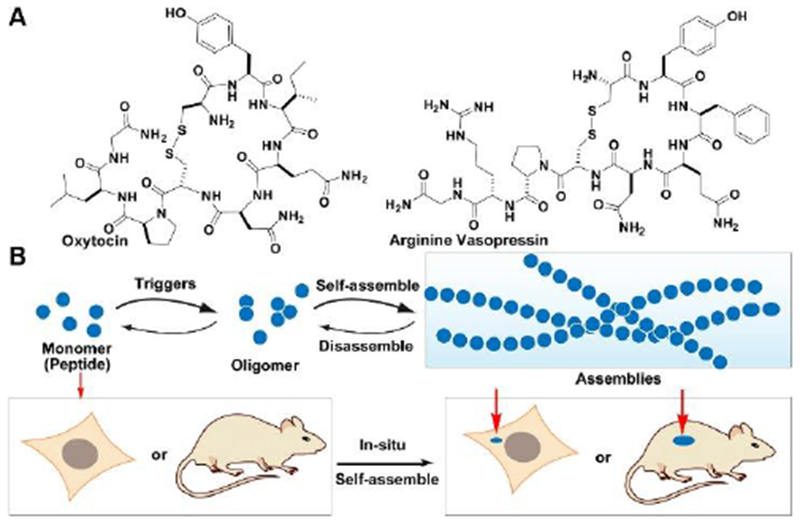
(A) Molecular structures of Oxytocin and arginine vasopressin. (B) Schematic illustration of regulating peptide assemblies via self-assembly and disassembly processes upon suitable triggering in vitro and in vivo.
Using peptides as the build blocks for constructing assemblies confers a number of advantages, including ease of design and tailoring (based on the known structures from proteins), good biocompatibility and degradability, and low immunogenicity, which have facilitated the development of peptide assemblies. Comparing to individual peptides, peptide assemblies have several unique features, such as enabling phase transition (i.e., sol-to-gel)[4c, 4f, 5], providing multivalency,[6] and acting as nanoscale scaffolds.[7] After pioneering works demonstrated these merits, considerable amount of research activities have focused on exploring these advantages, as evidenced by several excellent reviews on peptide assemblies as biomaterials or nanomaterials for various applications.[8]
Another subtle, but key difference between individual peptides and peptide assemblies is the diffusivity: peptide assemblies diffuse much slower than monomeric peptides. Thus, triggering peptide monomers to form peptide assemblies, as a dynamic continuum,[9] provides a unique way to spatiotemporally control the peptide assemblies (Figure 1B). Having recognized this feature of peptide assemblies, we introduced the concept of enzyme instructed self-assembly (EISA), which is a biocatalytic strategy for generating peptide assemblies in aqueous solution through bond cleavage[10] or bond formation[11] in a context-dependent manner.[12] EISA, also being ubiquitous in cells, can be achieved easily so that it has attracted increasing attention after the first report.[10] For example, EISA have found many potential applications, from drug delivery, wound healing, immune modulation to analyte detection.[13]
The ubiquitous roles of enzymes in cells and their expression difference between normal cells and abnormal cells (or between different cell types) or between subcellular locations enable us to start using intracellular enzymes to convert the monomers of peptide in cytoplasm into peptide assemblies in cancer cells (or bacteria) for controlling cell fate.[14] Amplifying the difference between normal and abnormal cells (tissues), using endogenous triggers (e.g., enzymes) to transform peptide monomers to form functional peptide assemblies opens a new direction for developing peptide assemblies in complex environment, such as cells. Since the report of intracellular EISA about a decade ago,[14] we and others have made considerable progresses along the notion of in-situ generation of peptide assemblies (Figure 1B), especially in the applications of these peptide assemblies.[15] These advances warrant a brief reflection on the progresses made and the promises and challenges ahead.
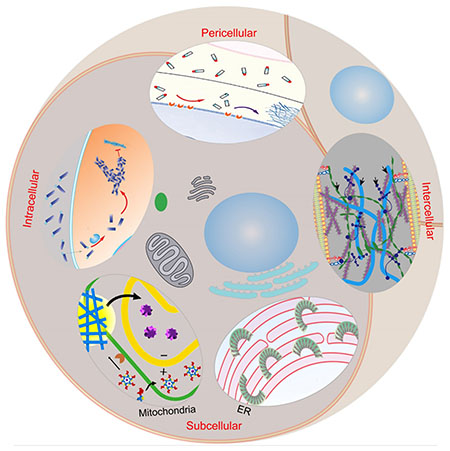
This minireview intends to highlight the recent developments of peptide assemblies and their applications in the context of cells. We start with the less discussed aspects of peptide assemblies—conformational switch. Then, we introduce the concept of instructed-assembly, which emphasizes the initiation and progression of the assembling process. We illustrate its unique merit for spatiotemporal control of peptide assemblies in the context of cells (e.g., in the pericellular, intracellular, subcellular, or intercellular space, also see the Frontispiece Graphic) and the corresponding applications. After that, we provide a brief outlook to discuss the challenges and the future directions. We also like to apologize for the omission of many excellent works due to the space limit.
2. Conformation Switch in Peptide Assemblies
To have sufficient and proper intermolecular interactions for forming the assemblies, peptides need to populate at certain conformations with relative long residence time, but the conformations also can change in the peptide assemblies. The well-established relationship of protein structures and functions has led to several strategies, such as ligand binding, hydrophobic forces, or enzymatic reactions, for switching the conformations of peptides in the assemblies, as shown in the following sections. Such conformation switch underscores the dynamic feature of peptide assemblies, which should be of consideration for peptide assemblies in complex environment.
2.1. Binding switches peptide conformations in the assemblies
A recent report[16] revealed that a heptapeptide from inflammasome, after conjugating with a well-known self-assembly motif, Nap-FF,[17] forms peptide assemblies of which the -helix conformation dominance decreases s upon interacting with ligands (Figure 2). Specifically, molecule 1 consists of KKFKLKL, the segment of ASCPYD,[18] which hardly forms any assemblies in aqueous solution. The attachment of Nap-FF at the N-terminal of KKFKLKL endows self-assembling ability to 1, which largely maintains α-helix conformation. The addition of pyridoxal phosphate (P5P) to the pre-organized nanofibers of 1 results in a hydrogel. Besides dictating the gelation time and the stiffness of resulted hydrogels, P5P decreases the dominance of α-helix conformation of 1. The formation of Schiff-bases and non-covalent interactions between the protonated ɛ-amine groups on the lysine residues of 1 and the phosphate group on P5P are essential (Figure 2) for enhanced assembling, as evidenced by that other small molecules, such as pyridoxal, folinic acid, ATP, and AMP, trigger gelation of 1. The gelation further enables the control of cell proliferation and drug release. This work illustrates conformation change in peptide assemblies upon binding with small bioactive molecules.
Figure 2.
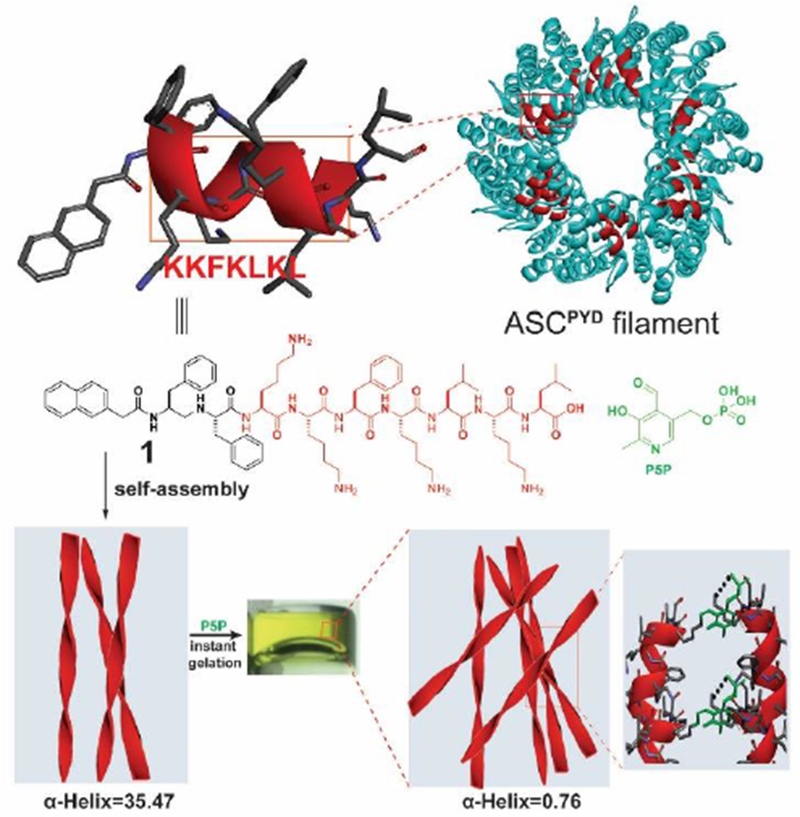
Molecular structure of 1 containing the epitope (KKFKLKL) from the filaments of apoptosis-associated speck-like (ASC) protein and the illustration of hydrogel formed by mixing 1 and P5P to provide covalent and non-covalent interactions, which reduce α-helix composition from 35.47 to 0.76%. Adapted with permission from ref. 16. Copyright 2017, Wiley-VCH.
2.2. Aromatic−aromatic interactions define peptide secondary structures in assemblies
A challenge in designing peptide assemblies is to control the conformations of short peptides. Unlike long peptide sequences that usually maintain their secondary structures as in proteins, isolated short peptides often lose their secondary structures and functions due to the lack of the restriction from proteins. Stabilizing protein structures,[19] aromatic−aromatic interactions are able to enhance intermolecular interactions for defining secondary structures of short peptides.[20] As shown in Figure 3, pentapeptides of RMLRF (2) and IQEVN (3), the segments of a decapeptidic sequence (RMLRFIQEVN) that forms a β-sheet structure at the intermolecular interface of the dimer of protein irisin,[21] hardly form any morphologies or maintain a secondary structure in aqueous solution. Neither does the mixture of 2 and 3. After C-terminal modification of 2 (or 3) with pyrene that provides strong aromatic-aromatic interactions, 2-Py (or 3-Py) forms nanostructures with α-helix like conformation. Notably, mixing 2-Py and 3-Py not only forms uniform nanofibers to entangle to result in a hydrogel, but also makes 2-Py and 3-Py to adopt β-sheet like structures. This facile strategy is applicable for designing assemblies of D-peptide derivatives by simply replacing the L-amino acids with corresponding D-amino acids. This work also agrees with that enhancing intermolecular interactions is a facile way to generate heterotypic peptide assemblies.[22]
Figure 3.
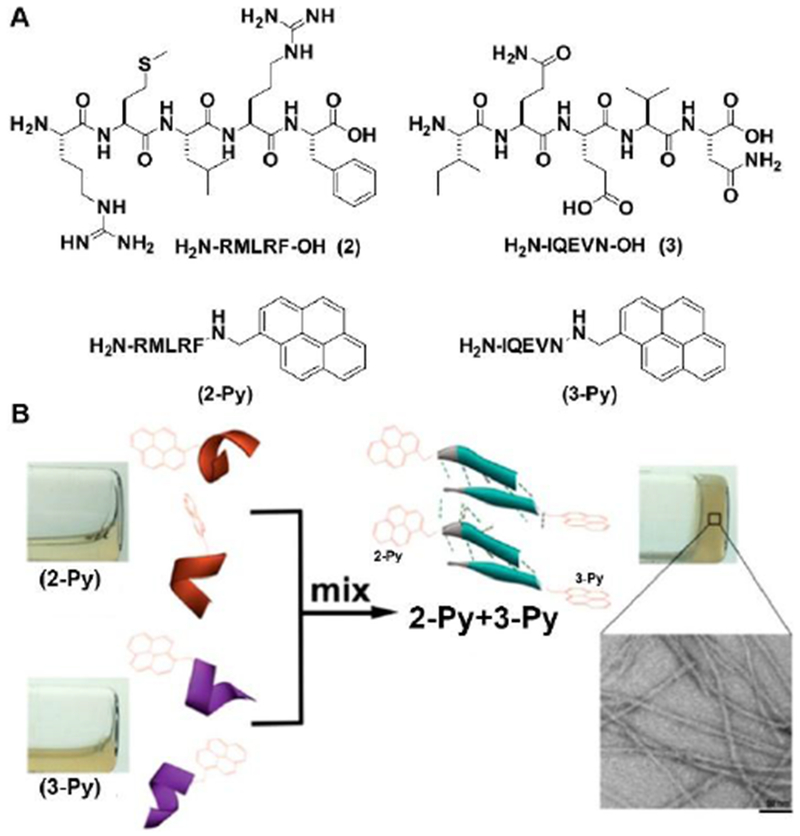
(A) Molecular structure of 2, 3, 2-Py and 3-Py; (B) Aromatic-aromatic interaction enables self-assembly, hydrogelation, and α-helix to β-sheet transition (scale bar = 50 nm). Adapted with permission from ref. 20. Copyright 2016, American Chemical Society.
2.3. Enzymatic selection of peptide secondary structures in assemblies
Although controlling protein conformations and functions by phosphatases and kinases is a common mechanism for signal transduction,[23] transplanting this concept into peptide assemblies to control the conformation of short intrinsically disordered peptides (IDP) is only at its beginning. A recent study reported that enzymatic reaction selects the secondary structures of heterotypic peptide assemblies.[24] Figure 4A shows the molecular structures of two short IDPs, one from protein crystal of PNUTS PP1 binding domain (LREYFY) and another a hypothetical sequence (KLIQFS). C-terminal modification of these two peptides with pyrene results in 4 and 5, which ensures heterotypic assembling. While phosphorylation changes the conformation of 4 from α-helix dominant to disordered structures, dephosphorylation of 4P increases the β-strand conformation. Directly co-assembly of 4 and 5 results in mixed conformations of α-helix and β-strand in the form of aggregates, but enzyme-instructed heterotypic assemblies of 4 and 5 (from 4P and 5) mainly exhibit the β-strand conformation in the form of nanofibers. This study indicates that enzymatic reaction controls the peptide conformations and the superstructures of the formed peptide assemblies. Two recent reports[25] also support this notion. These works on enzyme conversions of peptide conformations may also provide insights for understanding molecular condensates formed by biomacromolecules.[26]
Figure 4.
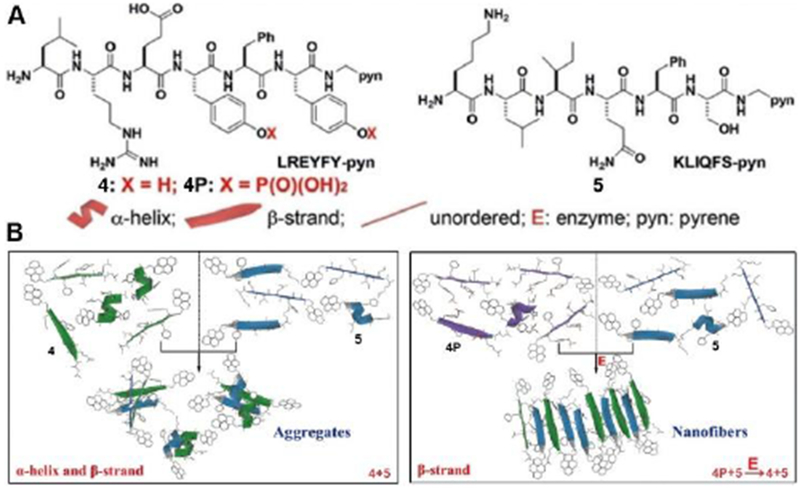
Structures of the peptides and history-dependent conformations of their heterotypic assemblies. Adapted with permission from ref. 24. Copyright 2018, Wiley-VCH.
3. Peptide Assemblies in Cellular Environment and Applications
While most studies have focused on the final structures of the peptide assemblies resulted from self-assembly,[8b, 27] the dynamics of triggering and assembling processes are largely overlooked. Because cells are complex and dynamic, exploring the initiation and progression of peptide assemblies not only matches well with the dynamic nature of cells, but also is unexpectedly fruitful. To emphasize the triggering and the dynamic assembling processes of peptide assemblies, we propose a term—instructed-assembly,[28] which differs from self-assembly. As a thermodynamically equilibrated process in which randomly oriented components (e.g., peptides) spontaneously organize into ordered structures via non-covalent interactions, self-assembly is inherently reversible. Instructed-assembly refers to the formation of ordered superstructures of molecules as the consequence of a triggering event, such as a reaction (especially enzymatic reaction). Like self-assembly, instructed-assembly relies on non-covalent interactions to form large structures from small, simple building blocks (i.e., a “bottom-up” process); unlike self-assembly, instructed-assembly includes an enzymatic reaction (especially enzymatic reaction (ΔG < 0) a multiple step process, can be considered as irreversible. In addition, the resulted assemblies from instructed-assembly highly depend on the path, that is, the history of molecular assembling. Thus, instructed-assembly provides a unique opportunity to explore triggering and assembling processes for designing dynamic peptide assemblies in complex cell environment for unprecedented applications. Among various types of instructed-assembly, EISA in cellular environment has exhibited great potentials, as shown in the following sections.
3.1. Pericellular formation of peptide assemblies
Local control of the complex signaling transduction by enzymatic reaction for regulating diverse cellular functions is ubiquitous in biological systems. Despite its attractiveness, mimicking such phenomenon by synthetic molecules is rare because of the lack of suitable strategies and the complexity of cellular environment. A recent unexpected finding, showing that ectoenzymes-instructed peptide assemblies form nanonets around cancer cells and selectively inhibit the cancer cells,[29] has illustrated a way. As shown in Figure 5, a phosphotyrosine containing D-tripeptide derivative (6P), being dephosphorylated by the overexpressed ectoenzyme (i.e., alkali phosphatase (ALP)), forms a layer of visible hydrogel around cancer cells (HeLa). Such a pericellular hydrogel, consisting of D-peptide assemblies, prevents cellular mass exchange with extracellular environment, entraps secretory proteins,[30] and results in cell apoptosis.[29] Further mechanistic investigation reveals that the assemblies of the D-peptide pleiotropically activate extrinsic death signaling for apoptosis of cancer cells (Figure 5).[31] Such an instructed-assembly process slows the tumor growth in murine models.[31] A relevant study also demonstrated instructed-assembly of D-peptides as a new way for selectively killing undifferentiated iPS cells without harming normal cells.[32]
Figure 5.
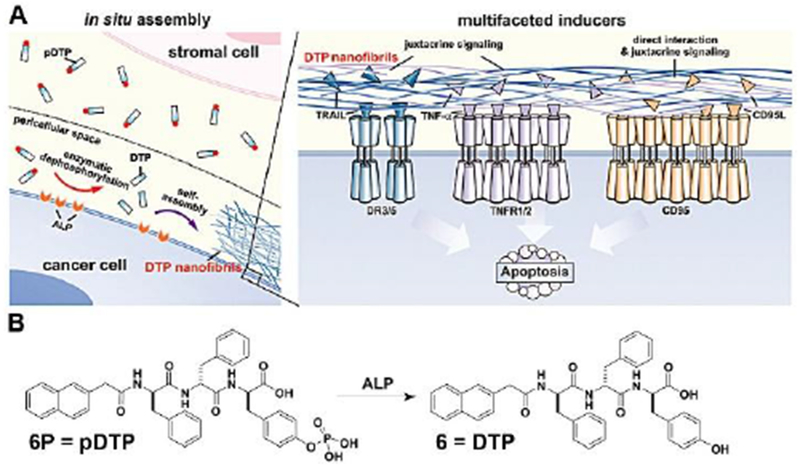
(A) The illustration of the pericellular 6 (DTP) nanofibrils formed by EISA to selectively inhibit cancer cells in co-culture via promiscuously activating cell death signalling. (B) Chemical structures of the precursor 6P (pDTP), the self-assembly tripeptide (DTP), and the dephosphorylation of the precursor catalyzed by ALP. Adapted with permission from ref. 31. Copyright 2017, Nature Publishing Group.
3.2. Intracellular formation of peptide assemblies
Intracellular formation of assemblies of small molecules is more than a scientific curiosity since most reported drug targets are inside cells. Intracellular formation of peptide assemblies, in fact, provides an opportunity to boost cellular uptake and to increase the activity of anticancer therapeutics. For example, a taurine modified D-peptide increases the cellular uptake of D-peptide by 10 times, and results in assemblies inside cytoplasm via esterase-instructed assembly.[33] The formed intracellular peptide assemblies impede actin dynamics (Figure 6A), thus boosting chemotherapy drugs for killing platinum-resistant cancer cells.[34] Besides using enzymes to trigger self-assembly, a recent study revealed that rationally designed nucleopeptide assemblies selectively sequester ATP, disrupt intracellular ATP dynamics, and boost the activity of cancer drug doxorubicin against drug resistant cancer cells.[35] As shown in Figure 6B, 9 forms the assemblies with α-helix dominant secondary structures. A pair of counteracting enzymes are able to modulate the nanostructures formed by the assemblies of 9 and the nucleotides (Figure 6C) by interconverting ATP and ADP. The nucleopeptides, sequestering ATP efficiently in cells, slow down efflux pumps in multidrug resistant cancer cells, thus boosting the efficacy of doxorubicin (Figure 6D). This work illustrates that the intracellular assemblies of nucleopeptides interact with essential biological molecules for controlling cell behavior.
Figure 6.
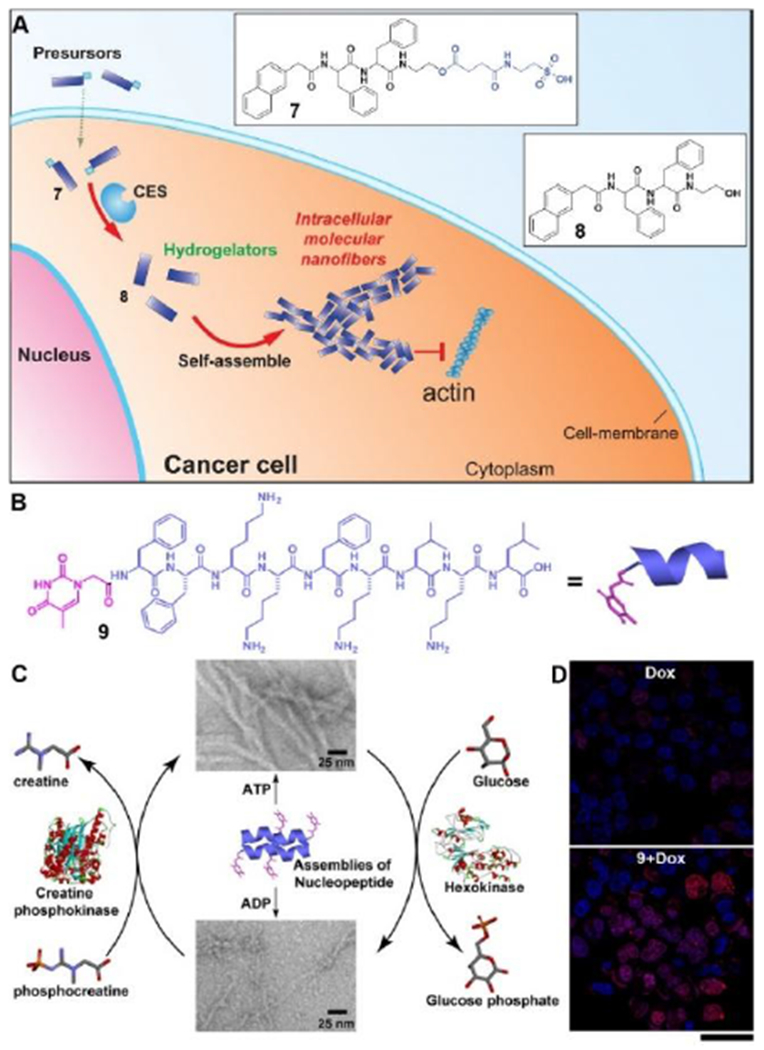
(A) Enzymatic transformation of the precursor (7) as a substrate of carboxylesterase (CES) to the corresponding hydrogelator (8) for intracellular self-assembly. (B) Structure of a nucleopeptide (9) for selective ATP sequestration. (C) Interaction of assemblies of 9 with ATP or ADP and the reversible phase transition of the assemblies controlled by a pair of counteracting enzymes (D) CLSM images show the inhibition of Dox efflux by the assemblies of 9 in MES-SA/dx5 cells at 5 h (changing to fresh medium without Dox after washing three times and further incubation for 5 h). Scale bar is 40 μm. Adapted with permission from ref. 34 and 35. Copyright 2015 and 2018, Wiley-VCH.
Figure 9.
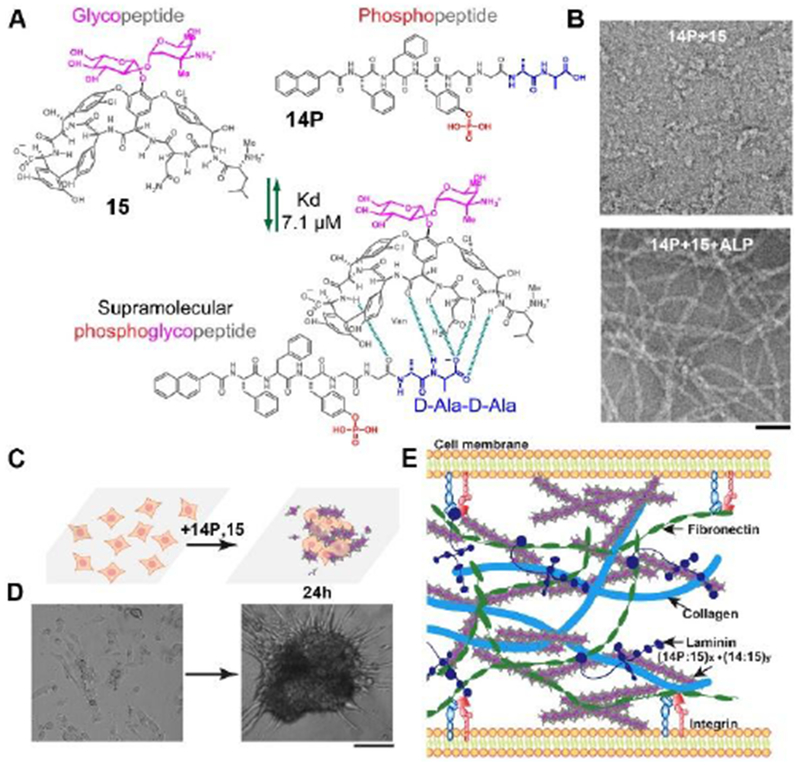
(A) Ligand–receptor interactions (green dash lines, dissociation constant (Kd) is 7.1 μm)) between vancomycin (Van, 15) and the D-Ala-D-Ala containing phosphopeptides (Nap-FFpYGGaa, 14P) result in a sPGP (14P:15). (B) Transmission electron microscopic (TEM) images of 14P:15 (300 μM) before and after being treated by a phosphatase (ALP, 1 UmL−1. 24 h). Scale bar is 50 nm. (C) The illustration of forming 3D spheroids from a 2D cell sheet upon the addition of sPGP and the reversibility of the process. (D) Optical images of time-dependent formation of 3D cell spheroids. Scale bar is 50 μm. (E) Illustration of sPGP assemblies as an ECM mimic to interact with major ECM components. Adapted with permission from ref. 9. Copyright 2017, Wiley-VCH.
3.3. Subcellular (organelle) formation of peptide assemblies
Accumulation of peptide assemblies in subcellular organelles is a promising strategy to improve the efficacy of the assemblies. Mitochondria is an intensively explored organelle due to its crucial cellular functions.[36] A recent report (Figure 7A) combines instructed-assembly with a mitochondria targeting motif (triphenyl phosphinium (TPP)), which modulates the redox potential of mitochondria, to selectively kill cancer cells and minimize acquired drug resistance.[37] TPP containing precursor (10P) first forms redox-modulating assemblies on cancer cell surface upon the dephosphorylation by ALP overexpressed on cancer cells (e.g., Saos2). After entering the cancer cells via endocytosis, escaping from lysosome, and accumulating in mitochondria, these assemblies induce dysfunction of mitochondria to release cytochrome c, thus selectively resulting in the death of the cancer cells. Most importantly, after being stimulated by 10P in multiple rounds, the cancer cells are unable to evolve resistance to 10P. Although the mechanism of endosomal or lysosomal escaping remains to be elucidated, this work validates the spatial control of the assemblies of non-specific cytotoxic agents by instructed-assembly as a molecular process for selectively killing cancer cells without inducing acquired drug resistance, and promises a new way for overcoming the challenging drawback of cancer chemotherapy.
Figure 7.
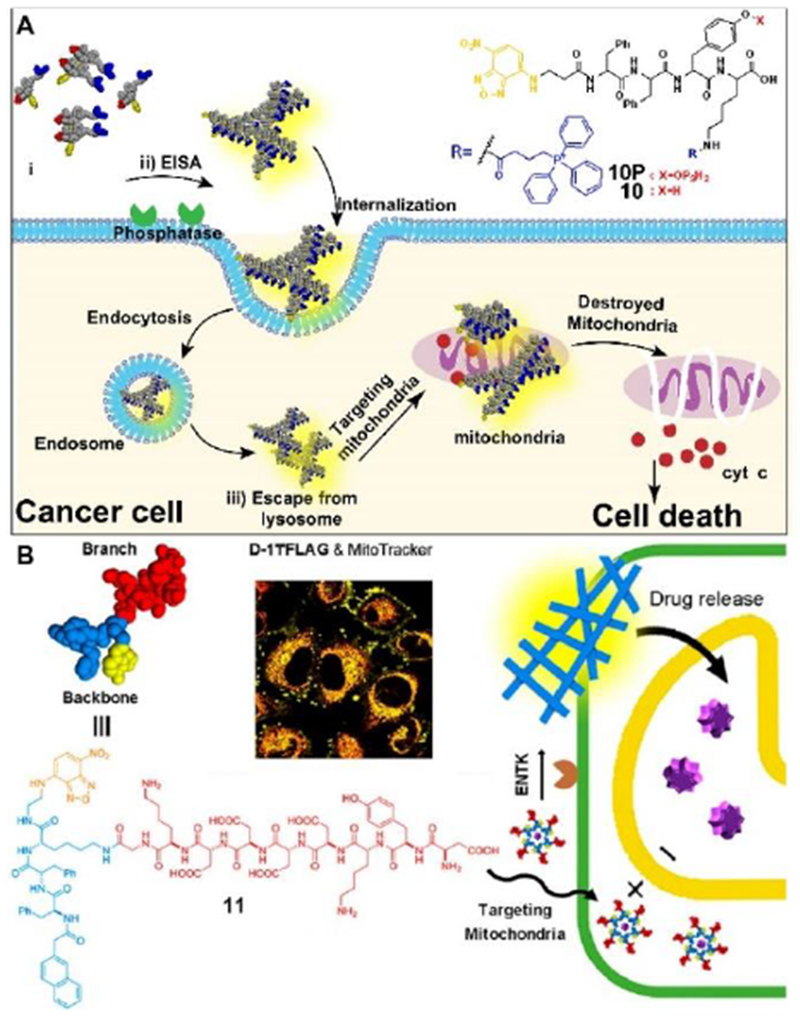
(A) Illustration of EISA for targeting mitochondria and inducing death of cancer cell. (B) Structure of a representative branched peptide and ENTK cleaving the branch to convert micelles to nanofibers on mitochondria. Adapted with permission from ref. 37 and 38. Copyright 2016 and 2018, American Chemical Society.
Unlike the commonly used mitochondria-targeting molecules (lipophilic and cationic), which may cause undesired cytotoxicity, a branched peptide with negative charges is able to target mitochondria via instructed-assembly on mitochondria.[38] 11 consists of a well-established protein tag (i.e., FLAG-tag[39]) and a self-assembling motif,[17] and acts as the substrate of enterokinase (ENTK). After the cells uptake 11 through clathrin-dependent endocytosis, the oligomers of 11 accumulate in mitochondria. ENTK on the mitochondria cleaves the FLAG-tag of 11 to result in the morphology transition from micelles to nanofibers (Figure 7B). In addition, the preformed micelles of 11 are able to deliver cargo (doxorubicin or proteins) into cells, and release them into mitochondria. This work illustrates the use of instructed-assembly for targeting mitochondria, which opens a new route to target subcellular organelles for biomedicine.
Another important organelle is endoplasmic reticulum (ER), which involves biosynthesis, sensing, and signaling. Inducing cancer cell death by excessive endoplasmic reticulum (ER) stress is a promising strategy for the therapeutic intervention of cancer. However, targeting ER for anticancer therapy is less explored due to the complex role of ER in cell signaling.[40] A recent study on instructed-assembly of peptides may lead to a new opportunity for selective targeting ER in cancer therapy.[41] Figure 8A shows the concept that instructed-assembly generates self-limited peptide assemblies to disrupt cell membranes and to target ER for cancer therapy. Serving as a substrate of ALP, 12P undergoes instructed-assembly to form crescent-shaped assemblies with several nanometers in sizes, which interact with lipids and rupture the plasma membrane, as confirmed by live-cell imaging and lactate dehydrogenase (LDH) assays. Further imaging and mechanism studies indicate that the peptide assemblies preferentially accumulate on the ER and induce ER stress, thus activating the caspase signaling cascade for cell death. Differing with the commonly used ER stress inducers,[42] which usually cause undesired side effects, this approach allow selectively targeting ER of cancer cells.
Figure 8.
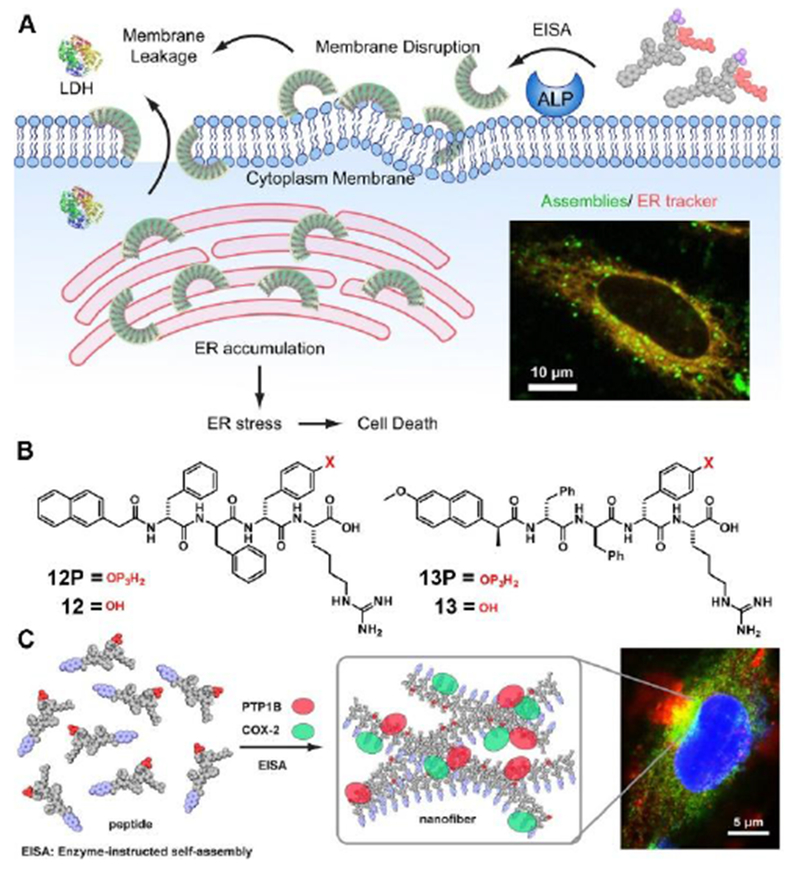
(A) Illustration of EISA Assemblies to Disrupt the Cell Membrane and to Target the ER and Molecular Structure of an EISA Precursor. (B) Molecualr structure of 12P, 12, 13P and 13. (C) Illustration of Instructed Assembly for Intracellular Sequestration of PTP1B and COX-2. Adapted with permission from ref. 41 and 43. Copyright 2018, American Chemical Society.
Another relevant study shows instructed-assembly of short peptides to mimic biomacromolecular condensates[26] for sequester enzymes on ER.[43] Consisting of a short peptide and naproxen (a nonsteroidal anti-inflammatory drug (NSAID) and a ligand of cyclooxygenase-2 (COX-2)) (Figure 8B), 13P acts as a substrate of instructed-assembly. As shown in Figure 8C, partially dephosphorylated by phosphatases, 13P and its corresponding hydrogelator (13) co-assemble to form assemblies that promote the association of COX-2 and PTP1B on the ER in live cells. Besides illustrating the use of supramolecular processes for sequestering enzymes in cells, this work provides alternative insights from supramolecular chemistry perspective for understanding intracellular liquid condensates, that is, biomacromolecular condensates likely are regulated by enzyme reactions as well. These two studies highlight the unique advantage of instructed-assembly, that is, the dynamic features of instructed-assembly are highly important, and exploring them promises useful applications.
3.4. Intercellular formation of peptide assemblies
Cell-cell and cell-extracellular matrix (ECM) interactions play important roles for cell signaling, cell migration, and cell differentiation. Thus, mimicking the microenvironment of cells has been an increasing interest in the last few decades.[5a, 44] Although considerable progresses have been made in developing soft materials for mimicking ECM, the lack of dynamic features and the difficulty in generating ordered structures have limited their applications in tissue engineering. Taken advantages of instructed-assembly and the tight ligand-receptor interactions, a recent study demonstrated the use of in-situ reaction and assembly as a spatiotemporal control to enable cell morphogenesis (i.e., from a 2D cell sheet to 3D cell spheroids).[9] Forming by molecular recognition between a phosphopeptide 14P and its receptor 15 (glycopeptide), supramolecular phosphoglycopeptides (sPGPs) (Figure 9A) form nanoparticles in aqueous solution, which turn into nanofibers upon the addition of enzymes (ALP). Being incubated with cells, the sPGPs undergoes an enzymatic reaction to form dynamic and hierarchical nanofibers (Figure 9B) in-situ on the cell surface and induce the formation of 3D cell spheroids (Figure 9C) from a 2D cell sheet. Detailed mechanism studies indicate that the progress of the instructed-assembly and posttranslational modifications of sPGPs define intercellular interactions (Figure 9D), thus resulting in the formation of 3D cell spheroids. This work not only validates the dynamic continuum of peptide assemblies as an ECM mimic (Figure 9E) to control cell behaviors, but also indicates the importance of dynamics in the design of peptide assemblies for biomaterials.
4. Outlook
Varying their sizes from a few nanometers up to hundreds of micrometers, peptide assemblies can adopt multiple conformations and structures. These features render the research of peptide assemblies as an inexhaustible and exciting subject at the interface of physical and biological sciences. Particularly, the biological relevance and easy access of peptides provide a convenient molecular entry for scientists and engineers from different disciplines to explore fundamental scientific problems and to develop technological innovations. For example, peptide assemblies have attracted interests from several renascent and nascent fields, like nonlinear dynamics,[45] self-replication,[46] orgin of life,[47] supramolecular chemical biology,[48] systems chemistry,[49] drug delivery,[8i, 50] and molecular imaging.[51] Being able to across multiple length scales and to exhibit multiple structures or morphologies, peptide assemblies present a great challenge for reductionist approaches that attempt to formulate structural-activity relationships of peptide assemblies. One notable example is the beta-amyloids (Aβ42). After hundreds of million dollars investment and decade efforts in the research of Aβ42, its relationship with Alzheimer’s disease remains elusive. On the other hand, the instructed-assembly of rather simple peptides, as discussed in this review, is able to achieve relatively important and sophisticated functions in cellular environment. Thus, it likely would be more fruitful by investigating peptide assemblies from a more holistic perspective—that is, directly studying the transformations, functions, and applications of peptide assemblies in complex environment, such as cells. We expect that the future successes in the functions of peptide assemblies, in fact, would stimulate the development of methods and tools for elucidating the atomistic details of peptide assemblies, which in turn would further advance the field.
Cells have relied on non-covalent interactions between molecules to construct high order dynamic continuum for functions for billions of years. As an essence of cellular process, the higher-order assemblies of biomacromolecules in cells are never static, despite reaching steady-states with spatiotemporal controls provided, usually, by enzymatic reactions and intermolecular interactions. Thus, using enzymatic reaction to instruct the assembly of peptides, as a holistic way to mimic the essence of cells, not only provides insights for understanding cellular processes, but also offers a new way to control cell behaviours for developing applications. The examples described in Section 3, undoubtedly, support this notion. Moreover, the knowledge gained from the exploration of peptide assemblies in cellular environment, should also contribute to understanding other molecular assemblies in cell biology, such as lipid rafts[52] and biomacromolecular condensates.[26]
While the assemblies of peptides, forming by instructed-assembly in-situ in complex environment, and their applications are emerging as a promising frontier of peptide research,[13, 15f, 53] several challenges remain: i) Lack of the high-resolution tools for monitoring the dynamics of the peptides assemblies in live cells over large area and extended time; ii) Lack of comprehensive quantitative analysis for correlating the functions and the kinetics of the peptide assemblies, though a recent quantitative analysis of instructed-assembly of peptides in cells started to elucidate the relationship between instructed-assembly and cancer cell death;[54] iii) Lack of guiding principles to combine different triggers to provides more sophisticated applications;[15c, 55] iv) Lack of the understanding of large-scale behaviour emerges from peptide assemblies, although both nanoscale processes and microscale phenomena now are observable. Despite these challenges, we expect the advances of knowledge from different disciplines and the development of tools will gradually address these issues since they are rather generic and similar to the challenges in understanding functions of biomacromolecular assemblies in living organisms. At the same time, exploring peptide assemblies in complex environment and their applications may stimulate the developments for addressing those challenges.
Acknowledgements
This work was partially supported by NIH (CA142746), NSF (DMR-1420382) and W. M. Keck Foundation. ZF thanks the Dean’s fellowship and NIH (F99CA234746).
References
- [1].Siddle K, Hutton J, Oxford University Press, 1991. [Google Scholar]
- [2].a) Ludwig M, Leng G, Nat. Rev. Neurosci. 2006, 7, 126; [DOI] [PubMed] [Google Scholar]; b) Aspé-Sánchez M, Moreno M, Rivera MI, Rossi A, Ewer J, Front. Neurosci. 2016, 9, 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lau JL, Dunn MK, Bioorg. Med. Chem. 2018, 26, 2700–2707. [DOI] [PubMed] [Google Scholar]
- [4].a) Jimenez JL, Guijarro JI, Orlova E, Zurdo J, Dobson CM, Sunde M, Saibil HR, The EMBO journal 1999, 18, 815–821; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Choo DW, Schneider JP, Graciani NR, Kelly JW, Macromolecules 1996, 29, 355–366; [Google Scholar]; c) Schneider JP, Pochan DJ, Ozbas B, Rajagopal K, Pakstis L, Kretsinger J, J. Am. Chem. Soc. 2002, 124, 15030–15037; [DOI] [PubMed] [Google Scholar]; d) Ogihara NL, Weiss MS, Eisenberg D, Degrado WF, Protein Sci. 1997, 6, 80–88; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Kunitake T, Angew. Chem., Int. Ed. 1992, 31, 709–726; [Google Scholar]; f) Zhang S, Holmes T, Lockshin C, Rich A, Proc. Natl. Acad. Sci. U. S. A. 1993, 90, 3334–3338; [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Ghadiri MR, Soares C, Choi C, J. Am. Chem. Soc. 1992, 114, 825–831; [Google Scholar]; h) Gregory DM, Benzinger TL, Burkoth TS, Miller-Auer H, Lynn DG, Meredith SC, Botto RE, Solid State Nucl. Magn. Reson. 1998, 13, 149–166; [DOI] [PubMed] [Google Scholar]; i) Qu Y, Payne SC, Apkarian RP, Conticello VP, J. Am. Chem. Soc. 2000, 122, 5014–5015; [Google Scholar]; j) Pandya MJ, Spooner GM, Sunde M, Thorpe JR, Rodger A, Woolfson DN, Biochemistry 2000, 39, 8728–8734; [DOI] [PubMed] [Google Scholar]; k) Xing B, Yu C-W, Chow K-H, Ho P-L, Fu D, Xu B, J. Am. Chem. Soc. 2002, 124, 14846–14847; [DOI] [PubMed] [Google Scholar]; l) Hartgerink JD, Beniash E, Stupp SI, Science 2001, 294, 1684–1688; [DOI] [PubMed] [Google Scholar]; m) Yu Y-C, Berndt P, Tirrell M, Fields GB, J. Am. Chem. Soc. 1996, 118, 12515–12520. [Google Scholar]
- [5].a) Zhang S, Holmes TC, DiPersio CM, Hynes RO, Su X, Rich A, Biomaterials 1995, 16, 1385–1393; [DOI] [PubMed] [Google Scholar]; b) Pochan DJ, Schneider JP, Kretsinger J, Ozbas B, Rajagopal K, Haines L, J. Am. Chem. Soc. 2003, 125, 11802–11803; [DOI] [PubMed] [Google Scholar]; c) Collier JH, Hu BH, Ruberti JW, Zhang J, Shum P, Thompson DH, Messersmith PB, J. Am. Chem. Soc. 2001, 123, 9463–9464; [DOI] [PubMed] [Google Scholar]; d) Zhang Y, Gu H, Yang Z, Xu B, J. Am. Chem. Soc. 2003, 125, 13680–13681. [DOI] [PubMed] [Google Scholar]
- [6].Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI, Science 2004, 303, 1352–1355. [DOI] [PubMed] [Google Scholar]
- [7].Holmes TC, de Lacalle S, Su X, Liu G, Rich A, Zhang S, Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 6728–6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].a) Boekhoven J, Stupp SI, Adv. Mater. 2014, 26, 1642–1659; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hendricks MP, Sato K, Palmer LC, Stupp SI, Acc. Chem. Res. 2017, 50, 2440–2448; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ulijn RV, Smith AM, Chem. Soc. Rev. 2008, 37, 664–675; [DOI] [PubMed] [Google Scholar]; d) Branco MC, Sigano DM, Schneider JP, Curr. Opin. Chem. Biol. 2011, 15, 427–434; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Gazit E, Chem. Soc. Rev. 2007, 36, 1263–1269; [DOI] [PubMed] [Google Scholar]; f) Adler-Abramovich L, Gazit E, Chem. Soc. Rev. 2014, 43, 6881–6893; [DOI] [PubMed] [Google Scholar]; g) Langer R, Tirrell DA, Nature 2004, 428, 487; [DOI] [PubMed] [Google Scholar]; h) Shigemitsu H, Hamachi I, Acc. Chem. Res. 2017, 50, 740–750; [DOI] [PubMed] [Google Scholar]; i) Cheetham AG, Chakroun RW, Ma W, Cui H, Chem. Soc. Rev. 2017, 46, 6638–6663; [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Cui H, Webber MJ, Stupp SI, Biopolymers 2010, 94, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang H, Shi J, Feng Z, Zhou R, Wang S, Rodal AA, Xu B, Angew. Chem., Int. Ed. 2017, 56, 16297–16301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yang Z, Gu H, Fu D, Gao P, Lam JK, Xu B, Adv. Mater. 2004, 16, 1440–1444. [Google Scholar]
- [11].Toledano S, Williams RJ, Jayawarna V, Ulijn RV, J. Am. Chem. Soc. 2006, 128, 1070–1071. [DOI] [PubMed] [Google Scholar]
- [12].a) Yang Z, Liang G, Xu B, Acc. Chem. Res. 2008, 41, 315–326; [DOI] [PubMed] [Google Scholar]; b) Zhou J, Xu B, Bioconjugate Chem. 2015, 26, 987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Du X, Zhou J, Shi J, Xu B, Chem. Rev. 2015, 115, 13165–13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].a) Yang Z, Xu K, Guo Z, Guo Z, Xu B, Adv. Mater. 2007, 19, 3152–3156; [Google Scholar]; b) Yang Z, Liang G, Guo Z, Guo Z, Xu B, Angew. Chem., Int. Ed. 2007, 46, 8216–8219. [DOI] [PubMed] [Google Scholar]
- [15].a) Pires RA, Abul-Haija YM, Costa DS, Novoa-Carballal R, Reis RL, Ulijn RV, Pashkuleva I, J. Am. Chem. Soc. 2015, 137, 576–579; [DOI] [PubMed] [Google Scholar]; b) Tanaka A, Fukuoka Y, Morimoto Y, Honjo T, Koda D, Goto M, Maruyama T, J. Am. Chem. Soc. 2015, 137, 770–775; [DOI] [PubMed] [Google Scholar]; c) Zhan J, Cai Y, He S, Wang L, Yang Z, Angew. Chem., Int. Ed. 2018, 57, 1813–1816; [DOI] [PubMed] [Google Scholar]; d) Qi GB, Gao YJ, Wang L, Wang H, Adv. Mater. 2018, 1703444; [DOI] [PubMed] [Google Scholar]; e) Wu J, Zheng Z, Chong Y, Li X, Pu L, Tang Q, Yang L, Wang X, Wang F, Liang G, Adv. Mater. 2018, 30, 1805018; [DOI] [PubMed] [Google Scholar]; f) Feng Z, Zhang T, Wang H, Xu B, Chem. Soc. Rev. 2017, 46, 6470–6479; [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Ye D, Liang G, Ma ML, Rao J, Angew. Chem., Int. Ed. 2011, 123, 2323–2327; [Google Scholar]; h) Liang G, Ren H, Rao J, Nat. Chem. 2010, 2, 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang H, Feng Z, Lu A, Jiang Y, Wu H, Xu B, Angew. Chem., Int. Ed. 2017, 129, 7687–7691. [Google Scholar]
- [17].Zhang Y, Kuang Y, Gao Y, Xu B, Langmuir 2010, 27, 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schröder GF, Fitzgerald KA, Wu H, Egelman EH, Cell 2014, 156, 1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Burley S, Petsko GA, Science 1985, 229, 23–28. [DOI] [PubMed] [Google Scholar]
- [20].Li J, Du X, Hashim S, Shy A, Xu B, J. Am. Chem. Soc. 2016, 139, 71–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schumacher MA, Chinnam N, Ohashi T, Shah RS, Erickson HP, J. Biol. Chem. 2013, 288, 33738–33744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yuan D, Du X, Shi J, Zhou N, Zhou J, Xu B, Angew. Chem., Int. Ed. 2015, 54, 5705–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Krebs EG, Fischer EH, Vitam. Horm. 1964, 22, 399–410. [DOI] [PubMed] [Google Scholar]
- [24].Li J, Zhan Z, Du X, Wang J, Hong B, Xu B, Angew. Chem., Int. Ed. 2018, 57, 11716–11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].a) Shi J, Fichman G, Schneider JP, Angew. Chem., Int. Ed. 2018, 130, 11358–11362; [Google Scholar]; b) Liang C, Zheng D, Shi F, Xu T, Yang C, Liu J, Wang L, Yang Z, Nanoscale 2017, 9, 11987–11993. [DOI] [PubMed] [Google Scholar]
- [26].Kato M, McKnight SL, Annu. Rev. Biochem. 2018. [DOI] [PubMed] [Google Scholar]
- [27].Aida T, Meijer E, Stupp S, Science 2012, 335, 813–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].He H, Xu B, Bull. Chem. Soc. Jpn. 2018, 91, 900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kuang Y, Shi J, Li J, Yuan D, Alberti KA, Xu Q, Xu B, Angew. Chem., Int. Ed. 2014, 53, 8104–8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhou R, Kuang Y, Zhou J, Du X, Li J, Shi J, Haburcak R, Xu B, PloS one 2016, 11, e0154126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Du X, Zhou J, Wang H, Shi J, Kuang Y, Zeng W, Yang Z, Xu B, Cell Death Dis. 2017, 8, e2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kuang Y, Miki K, Parr CJ, Hayashi K, Takei I, Li J, Iwasaki M, Nakagawa M, Yoshida Y, Saito H, Cell Chem. Biol. 2017, 24, 685–694. e684. [DOI] [PubMed] [Google Scholar]
- [33].Zhou J, Du X, Li J, Yamagata N, Xu B, J. Am. Chem. Soc. 2015, 137, 10040–10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Li J, Kuang Y, Shi J, Zhou J, Medina JE, Zhou R, Yuan D, Yang C, Wang H, Yang Z, Angew. Chem., Int. Ed. 2015, 54, 13307–13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wang H, Feng Z, Qin Y, Wang J, Xu B, Angew. Chem., Int. Ed. 2018, 57, 4931–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].a) Vyas S, Zaganjor E, Haigis MC, Cell 2016, 166, 555–566; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Green DR, Galluzzi L, Kroemer G, Science 2011, 333, 1109–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang H, Feng Z, Wang Y, Zhou R, Yang Z, Xu B, J. Am. Chem. Soc. 2016, 138, 16046–16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].He H, Wang J, Wang H, Zhou N, Yang D, Green DR, Xu B, J. Am. Chem. Soc. 2018, 140, 1215–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hopp TP, Prickett KS, Price VL, Libby RT, March CJ, Cerretti DP, Urdal DL, Conlon PJ, Nat. Biotechnol. 1988, 6, 1204–1210. [Google Scholar]
- [40].Cubillos-Ruiz JR, Bettigole SE, Glimcher LH, Cell 2017, 168, 692–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Feng Z, Wang H, Wang S, Zhang Q, Zhang X, Rodal AA, Xu B, J. Am. Chem. Soc. 2018, 140, 9566–9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Foufelle F, Fromenty B, Pharmacol. Res. Perspect. 2016, 4, e00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Feng Z, Wang H, Xu B, J. Am. Chem. Soc. 2018, 140, 16433–16437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].a) Powers MJ, Rodriguez RE, Griffith LG, Biotechnol. Bioeng. 1997, 53, 415–426; [DOI] [PubMed] [Google Scholar]; b) Sargeant TD, Rao MS, Koh C-Y, Stupp SI, Biomaterials 2008, 29, 1085–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Epstein IR, Pojman JA, An introduction to nonlinear chemical dynamics: oscillations, waves, patterns, and chaos, Oxford University Press, 1998. [Google Scholar]
- [46].Rubinov B, Wagner N, Rapaport H, Ashkenasy G, Angew. Chem., Int. Ed. 2009, 121, 6811–6814. [DOI] [PubMed] [Google Scholar]
- [47].Mann S, Angew. Chem., Int. Ed. 2013, 52, 155–162. [DOI] [PubMed] [Google Scholar]
- [48].Petkau-Milroy K, Brunsveld L, Org. Biomol. Chem. 2013, 11, 219–232. [DOI] [PubMed] [Google Scholar]
- [49].a) Ashkenasy G, Hermans TM, Otto S, Taylor AF, Chem. Soc. Rev. 2017, 46, 2543–2554; [DOI] [PubMed] [Google Scholar]; b) Ludlow RF, Otto S, Chem. Soc. Rev. 2008, 37, 101–108; [DOI] [PubMed] [Google Scholar]; c) Boekhoven J, Brizard AM, Kowlgi KN, Koper GJ, Eelkema R, van Esch JH, Angew. Chem., Int. Ed. 2010, 122, 4935–4938. [DOI] [PubMed] [Google Scholar]
- [50].a) Zhao F, Ma ML, Xu B, Chem. Soc. Rev. 2009, 38, 883–891; [DOI] [PubMed] [Google Scholar]; b) Gao Y, Kuang Y, Guo Z-F, Guo Z, Krauss IJ, Xu B, J. Am. Chem. Soc. 2009, 131, 13576–13577; [DOI] [PubMed] [Google Scholar]; c) Cheetham AG, Zhang P, Lin Y.-a., Lock LL, Cui H, J. Am. Chem. Soc. 2013, 135, 2907–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].a) Razgulin A, Ma N, Rao J, Chem. Soc. Rev. 2011, 40, 4186–4216; [DOI] [PubMed] [Google Scholar]; b) Takaoka Y, Sakamoto T, Tsukiji S, Narazaki M, Matsuda T, Tochio H, Shirakawa M, Hamachi I, Nat. Chem. 2009, 1, 557; [DOI] [PubMed] [Google Scholar]; c) Mizusawa K, Takaoka Y, Hamachi I, J. Am. Chem. Soc. 2012, 134, 13386–13395. [DOI] [PubMed] [Google Scholar]
- [52].Sezgin E, Levental I, Mayor S, Eggeling C, Nat. Rev. Mol. Cell Biol. 2017, 18, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wang H, Feng Z, Xu B, Chem. Soc. Rev. 2017, 46, 2421–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Li J, Bullara D, Du X, He H, Sofou S, Kevrekidis IG, Epstein IR, Xu B, ACS nano 2018, 12, 3804–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zheng Z, Chen P, Xie M, Wu C, Luo Y, Wang W, Jiang J, Liang G, J. Am. Chem. Soc. 2016, 138, 11128–11131. [DOI] [PubMed] [Google Scholar]


