Introductory Paragraph
Rod-shaped bacteria grow by adding material into their cell wall via the action of two spatially distinct enzymatic systems: The Rod complex moves around the cell circumference, while class A penicillin-binding proteins do not. To understand how the combined action of these two systems defines bacterial dimensions, we examined how each affects the growth and width of Bacillus subtilis, as well as the mechanical anisotropy and orientation of material within their sacculi. Rod width is not determined by MreB, rather it depends on the balance between the systems: The Rod complex reduces diameter, while aPBPs increase it. Increased Rod complex activity correlates with an increased density of directional MreB filaments and a greater fraction of directional PBP2a enzymes. This increased circumferential synthesis increases the relative amount of oriented material within the sacculi, making them more resistant to stretching across their width, thereby reinforcing rod shape. Together, these experiments explain how the combined action of the two main cell wall synthetic systems builds and maintains rods of different widths. Escherichia coli Rod mutants also show the same correlation between width and directional MreB filament density, suggesting this model may be generalizable to bacteria that elongate via the Rod complex.
Introduction
While the length of Bacillus subtilis rods increases with growth rate 1, their width remains constant across different growth conditions 2. How bacteria define and maintain shapes with such precision is not understood, but it must involve controlling the rate and location of glycan insertion into the peptidoglycan (PG) sacculus, the enveloping heteropolymer meshwork that holds cells in shape 3. To understand how bacteria grow in defined shapes, we must understand not only where these enzymes act, but how their activity affects the arrangement of material within the sacculus and its mechanics.
The PG used for elongation is synthesized by two families of penicillin-binding proteins (PBPs): Class A PBPs (aPBPs) both polymerize and cross-link glycans, while class B PBPs (bPBPs) cross-link 4,5 the glycans polymerized by RodA 6. bPBPs and RodA are components of the “Rod complex”, a group of proteins essential for rod shape (Figure 1a). In B. subtilis, the Rod complex contains RodA, the class B transpeptidases PBP2a and/or PbpH, MreC, MreD, RodZ, and filaments of MreB 7,8. MreB polymerizes with other homologs into short, highly curved filaments on the membrane 9,10. To maximize membrane interactions as these curved filaments deform to the membrane, they orient along the direction of highest inward curvature, around the rod width 11. Oriented by MreB filaments, the Rod complex moves directionally around the cell circumference 12–14. This motion is driven by the synthetic activity of RodA/PBP2a, and is believed to reflect the insertion of radially oriented glycans 15.
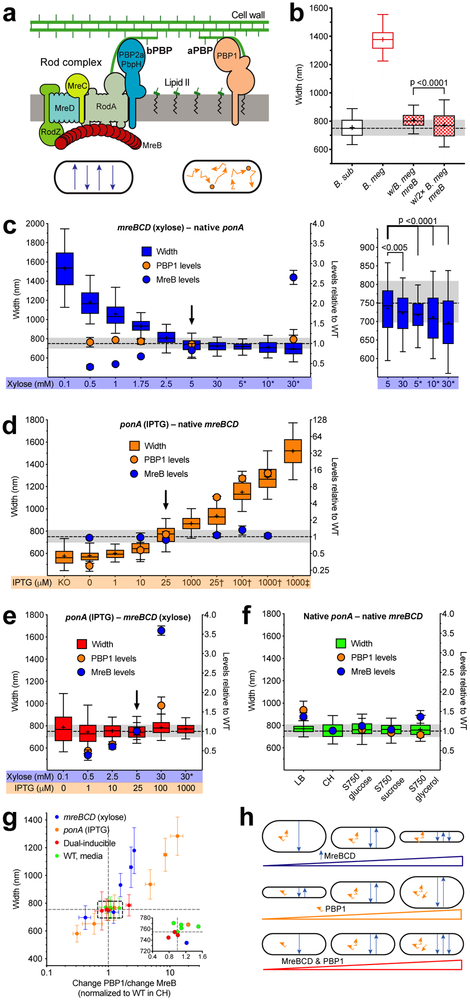
Figure 1 — Rod width depends on the relative levels of widening aPBPs to the thinning Rod system.
Except where indicated, all strains were grown in CH medium. For b–f, the median width of WT B. subtilis grown in CH is depicted by dashed line, grey shading indicates 25–75 percentiles. For details regarding statistics and box plot definitions, see the “Statistics” subheading in “Methods”.
a. Diagram depicting the two peptidoglycan synthesis systems responsible for elongation. Bottom - schematic of each system’s in vivo motions.
b. B. subtilis expressing B. megaterium mreB forms rods close to B. subtilis width. “B. sub” is WT B. subtilis. “B. meg” is B. megaterium. Checkered boxes are bMD465 (amyE::erm Pxyl-mreBCD minCDB. megaterium, ΔmreBCD ΔminCD::spc mreBCD minCDB. megaterium), a B. subtilis strain where the native mreBCD minCD operon was replaced with the same operon from B. megaterium, and an additional B. megaterium mreBCD minCD operon under xylose control at an ectopic locus. “w/B. meg mreB” was grown with 1% glucose to repress ectopic expression. “w/2× B. meg mreB” was grown with 30 mM xylose to overexpress the ectopic B. megaterium mreB operon.
c–f. Titrations of ponA and mreBCD vs. cell width. Strains were grown with the inducer concentrations below each graph. Width plotted on left, mean MreB and PBP1 relative abundances (determined by mass spectrometry, normalized to levels in WT cells grown in CH) on right. Arrowheads are inductions producing WT widths and protein levels. Supplementary Figure 3c shows effects on cell length.
c. Diameter decreases with mreBCD induction.
Inductions of bMD545 (amyE::erm Pxyl-mreBCD, ΔmreBCD::spc), except for those marked * which are bMK355 (amyE::erm Pxyl-mreBCD) containing a xylose-inducible mreBCD in addition to native mreBCD. Right is a zoomed view of highest 5 inductions. Supplementary Figure 1c–d shows MreB levels determined by western blot across the entire range.
d. Cell diameter increases with ponA induction. “KO” is bMK005 (ΔponA::cat). Inductions of bMD598 (yhdG::cat Pspank-ponA, ΔponA::kan), except for the those marked † and ‡, which are under stronger promoters; † is bMD586 (yhdG::cat Phyperspank-ponA, ΔponA::kan), ‡ is bMD554 (yhdG::cat Phyperspank-ponA) which has an inducible ponA in addition to the native copy.
e. Balanced expression of both PG synthetic systems yields normal width across a large range.
Dual inductions of bMD620 (amyE::erm Pxyl-mreBCD, ΔmreBCD::spc, yhdH::cat Pspank-ponA, ΔponA::kan). * indicates bMD622 (amyE::erm Pxyl-mreBCD, yhdG::cat Pspank-ponA, ΔponA::kan) with a xylose-inducible mreBCD in addition to native mreBCD.
f. WT B. subtilis maintains constant width in different media.
g. WT width is maintained within a narrow range of relative PBP1/MreB ratios. Plotted are mean widths (error bars are SD) of cells from c–f against the ratio of fold change in PBP1 to MreB. Inset shows zoomed view of box. Lines indicate mean WT width and PBP1/MreB ratio.
h. Model for how the two PG synthesis systems affect rod width. Top – As circumferentially organized PG synthesis increases (blue arrows), cell diameter decreases. Middle – As non-circumferential synthesis increases (orange squiggles), so does cell diameter. Bottom – As long as non-circumferential and circumferential synthesis is balanced, width remains constant, even across a range of protein levels.
aPBPs also affect rod shape, as B. subtilis cells lacking aPBPs are thinner 16. Single molecule studies have revealed that aPBPs and the Rod system are spatially distinct: Rod complexes move around the cell width, but aPBPs have never been observed to move directionally. Rather, aPBPs display two different interconverting motions: They either 1) diffuse within the membrane, or 2) remain immobile 17. Furthermore, inhibition of aPBP activity has no effect on MreB motion 6,17. Given that Rod complex activity is circumferentially organized while aPBP activity is not, it is not clear how these two PG synthetic machineries work together to create rod-shaped sacculi of defined width.
Current models of rod width have focused on MreB filaments, attributing the altered widths of Escherichia coli MreB mutants to changes in MreB filament curvature, twist, angle, or localization to negative Gaussian curvature 18–23. Not only do these models neglect the contribution of aPBPs, they are A) theoretical, as changes to filament curvature or twist have not been structurally validated, and B) are difficult to reconcile in B. subtilis, which has no detectable negative Gaussian curvature 11 or skew in the angle of filaments relative to the cell body 11,12,14,24.
Rather than focusing on MreB alone, we sought to develop a more thorough understanding of how both circumferential and non-circumferential PG synthesis affects B. subtilis width and growth, as well as the organization and mechanics of its cell wall material. We find that aPBPs and the Rod complex have opposing effects on rod width, and cell diameter depends on their balance. The rate cells expand their sidewalls is largely unaffected by the level of either system, unless both become limiting. As MreBCD expression increases and rods thin, both the density of directionally moving MreB filaments increases as does the fraction of directionally moving enzymes. Increasing Rod complex activity increases the proportion of oriented material within the sacculus, causing the rod to stretch less across its width and more along its length in response to internal turgor. Finally, E. coli Rod mutants show the same correlations between the density of directionally moving MreB filaments and cell width, giving a simple, generalizable model for the how rod-shaped bacteria establish and maintain their cellular dimensions.
Results
The Rod and aPBP systems have opposing effects on cell diameter
The width of rod-shaped bacteria has been attributed to properties encoded within MreB filaments 19,20,23,25. Given that mreBCD genes from other Bacilli can confer rod shape to B. subtilis 26, we reasoned if MreB defined cell width, the mreBCD genes from wider bacteria should produce wider cells. To test this, we replaced genes expressed within the B. subtilis mreB operon (mreBCD, minCD) with those from the 1.7-fold wider Bacillus megaterium. Surprisingly, these cells grew as uniform rods only slightly wider than wild type (WT) B. subtilis. Further overexpression of these B. megaterium genes caused cells to become even thinner (36 nm) (Figure 1b), suggesting that MreB filaments, by themselves, do not encode a specific width.
We next examined how cell width changed as we titrated the levels of the two main B. subtilis PG synthetic systems. We created strains where PBP1 (the major aPBP, encoded by ponA) expression was under IPTG control. As PG synthesis by the Rod system depends upon MreB 17, we created strains where the native mreBCD genes were under xylose control. We grew these strains at different inductions, then measured their widths using fluorescence microscopy (Figure 1c–d, Supplementary Figure 1a–b). For a subset of inductions, we measured the relative protein abundance using proteomic mass spectrometry (Figure 1c–d, Supplementary Data 1) or western blotting (Supplementary Figure 1c–d), normalizing to the levels measured in WT cells.
Varying mreBCD inductions revealed the Rod system has a thinning effect. In the lowest inductions supporting growth, rods were ~2-fold wider than WT, with some cells losing rod shape. As mreBCD induction increased, cells became thinner, reaching WT width when MreB abundance recapitulated WT levels (Figure 1c, Supplementary Figure 1b–d). Inductions above this point resulted in cells becoming thinner than WT, becoming even thinner in mreBCD merodiploids. We verified this thinning using transmission electron microscopy (TEM) (Supplementary Figure 1e). As before, these results demonstrate MreB does not define a given diameter, but rather, as previously hypothesized 11,27, the Rod system acts to reduce cell diameter.
Different ponA inductions revealed aPBPs have a widening effect. With no IPTG, cells contained 0.25 the amount of WT PBP1 and were ~23% thinner, similar to ΔponA 16. As we increased ponA induction cells became wider, reaching WT widths when PBP1 abundance recapitulated WT levels (Figure 1d, Supplementary Figure 1a). Inductions above this point caused cells to become increasingly wider, and by expressing ponA under stronger promoters or in merodiploids we could produce rods almost twice WT diameter.
Inhibition of FtsZ by MciZ 28 verified that both ponA-mediated widening and mreBCD-mediated thinning are independent of the PG synthesis used for cell division (Supplementary Figure 2a). Additionally, both systems showed similar width effects on cells grown in LB media (Supplementary Figure 2b), but required extra magnesium to stabilize rod shape and prevent lysis at low mreBCD inductions 29.
These results demonstrate that the aPBPs and Rod system have opposing effects: The circumferentially moving Rod complex reduces cell diameter, while the non-circumferential aPBPs increase diameter. We hypothesized that a balanced expression of both systems might produce WT diameter rods. We combined the differentially inducible ponA and mreBCD alleles into one “dual-inducible” strain. We found six different pairs of inducer concentrations that produced WT diameter rods (Figure 1e, Supplementary Figure 3a), even though each individual induction resulted in perturbed diameters in the singly-inducible parental strains (Figure 1c–d). Relative quantitation of protein levels revealed that, in induction pairs at or beneath WT levels, cells contained reduced, but relatively balanced amounts of PBP1 relative to MreB (Figure 1e). A similar balance was observed when we measured the widths and protein levels of WT cells grown in different media (Figure 1f, Supplementary Figure 3b). Together, this data suggested cell width depends on the balance between the levels of aPBP and the Rod systems. By plotting the ratio of the [fold change PBP1]/[fold change MreB] for all conditions in the data sets against their width (Figure 1g, Supplementary Data 1), we found that B. subtilis maintains its diameter within ~5% of WT when this ratio was within 0.8 to 1.5; outside of this range, cell diameter diverged. Together, these results indicate that, at the level of PG insertion, cell width is affected by the levels of the two opposing systems inserting material into the sacculus (Figure 1h).
RodA can function outside of the Rod system to widen cells, but only when PBP2a is also in excess
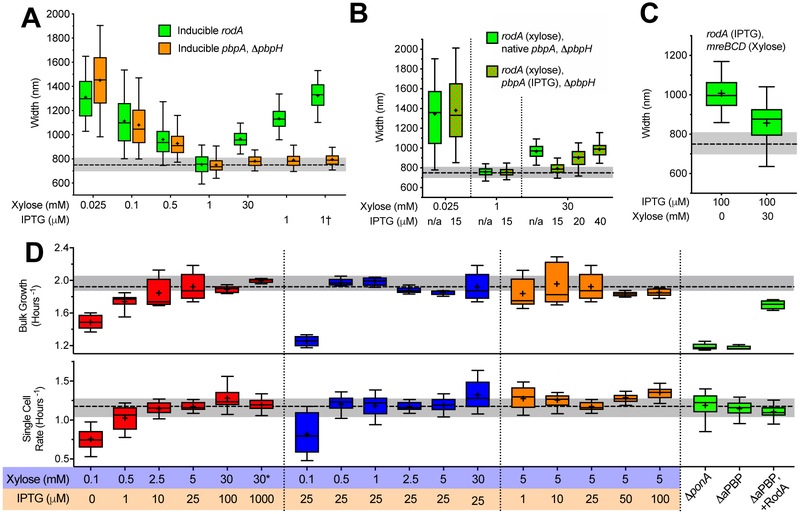
Given that RodA acts with PBP2a (encoded by pbpA) to synthesize PG 17, titrations of rodA and pbpA should show the same thinning effect as mreBCD. However, RodA overexpression restores WT width to thin ΔaPBP cells 6, a widening activity similar to PBP1 overexpression. To investigate this discrepancy, we made strains where either rodA or pbpA (as the only elongation-specific bPBP) were under the control of increasingly strong inducible promoters. As before 30,31, low rodA or pbpA yielded wide cells; and as induction increased, cells gradually thinned to WT width, suggesting both are required for maximal Rod system activity. However, while we expected higher rodA inductions to create thinner cells, once rodA induction exceeded the amount required for WT width, it behaved like PBP1, widening cells at increasing inductions (Figure 2a, Supplementary Figure 4a). In contrast, pbpA inductions beyond this point had a negligible effect on diameter (Figure 2a, Supplementary Figure 4b).
Figure 2 — Effects of RodA/PBP2a on cell width, and how each PG synthetic system affects growth.
All strains were grown in CH medium in the inducer concentrations shown below the graphs. For details regarding statistics and box plot definitions see the “Statistics” subheading in “Methods”.
a–c. Titrations of rodA, pbpA, and mreBCD vs. cell width. The median width of WT B. subtilis grown in CH is depicted by dashed line, grey shading indicates 25–75 percentiles.
a. As rodA or pbpA induction is increased, cell diameter decreases up to a point, beyond which it increases with rising rodA induction, but not for pbpA. Green boxes are bMD592 (rodA::erm Pxyl-rodA), save bMD580 (yhdG::cat Phyperspank-rodA, ΔrodA::kan) and bMD556 (yhdG::cat Phyperspank-rodA – labeled †). Orange boxes are bMD597 (pbpA::erm Pxyl-pbpA, ΔpbpH::spc), save bMD574 (yhdG::cat Phyperspank-pbpA, ΔpbpA::erm, ΔpbpH::spc) and bMD573 (yhdG::cat Phyperspank-pbpA, ΔpbpH::spc – labeled †).
b. Overexpression of rodA increases cell diameter, but only when pbpA expression is also sufficiently high. Green boxes are bMD627 (rodA::erm Pxyl-rodA, ΔpbpH::spc). Green/orange checkered boxes are bMD631 (rodA::erm Pxyl-rodA, yhdG::ble Pspank-pbpA, ΔpbpH::spc).
c. The increase in cell diameter caused by overexpression of rodA is reduced by simultaneous overexpression of mreBCD. Green boxes are bMD583 (yhdG::cat Phyperspank-rodA, ΔrodA::kan, amyE::erm Pxyl-mreBCD).
d. Rates of cell growth measured at the population and single cell level. Rates of growth were measured either by (top) OD600 in a shaking plate reader, or (bottom) by microscopically assaying the rate single cells (grown under a CH agarose pad) increased in perimeter. All measures are bMD620 (amyE::erm Pxyl-mreBCD, ΔmreBCD::spc, yhdG::cat Pspank-ponA, ΔponA::kan), with the exception of: “ΔponA” which is bMK005 (ΔponA::cat), “ΔaPBP” which is bAM268 (ΔpbpF, ΔpbpG, ΔpbpD, ΔponA::kan), and “ΔaPBP+ RodA” which is bAM288 (ΔpbpF, ΔpbpG, ΔpbpD. ΔponA:kan, amyE::spc Phyperspank-rodA-His10), where rodA is induced with 25 μM IPTG. Median growth rate of WT B. subtilis grown in CH is depicted bydashed lines, with shading indicating 25–75 percentiles.
Given the transglycosylase activity within SEDS polymerases is stimulated by their association with transpepdiases 32, we tested if the widening caused by high RodA induction required excess PBP2a. We created two strains with rodA under xylose control; one with native pbpA, and the other with pbpA under IPTG control. As before, when pbpA was under native control, depletion or high inductions of rodA increased diameter (Figure 2b, Supplementary Figure 4c). In contrast, when pbpA was held at the lowest induction required for WT width and rodA simultaneously highly induced, cells remained at WT widths. However, these RodA-overexpressing cells became increasingly wider as we increased pbpA induction (Figure 2b, Supplementary Figure 4d). This demonstrated that 1) excess RodA requires excess PBP2a to increase width, and 2) that native PBP2a levels exceed the amount required for maximal Rod activity, which may explain why bPBPs are observed as mixed populations of directionally moving and diffusive molecules 12,14,17.
These results suggested that, when RodA/PBP2a levels exceed the rest of the rod complex, their activity is no longer oriented by MreB and, like PBP1, widen cells via non-circumferential synthesis. If this hypothesis is correct, wide cells caused by RodA/PBP2a overexpression should thin if MreBCD is also increased, as this should recruit the non-circumferential excess RodA/PBP2a into circumferentially-moving, thinning Rod complexes. Indeed, while strong rodA induction made wide cells, simultaneous overexpression of mreBCD reduced cell diameter (Figure 2c, Supplementary Figure 4e), resulting in a narrowing far more pronounced than when MreBCD was overexpressed in otherwise WT strains (Figure 1c). Thus, the aPBP-like widening activity of RodA/PBP2a occurs once both exceed some level of MreBCD, possibly reflecting a saturation of binding sites within Rod complexes.
Growth rates are maintained across a wide range of enzyme levels, unless both systems become limiting
We next examined how each PG synthetic system affected the rate of cell growth in our dual-induction strain using two assays: 1) single-cell measurements of the rate of surface area change (under agarose pads), and 2) measures of population growth (in shaking plate readers). First, we examined the dual-inducible strain at the induction pairs that produced WT widths (Figure 1e). At the lowest induction pairs, growth was greatly reduced in both assays. Growth increased at each increasing induction pair (Figure 2d) up to the induction pair producing WT PBP1 and MreB protein levels. Thus, growth can be reduced if both PG synthesis systems become limiting.
Next, we assayed growth as we titrated either mreBCD or ponA while holding the other constant. Similar to E. coli MreB studies 33,34, both assays showed no difference in growth across a wide range of mreBCD inductions, except at the lowest induction where cells frequently lost rod shape. Likewise, both assays showed no difference in growth across our ponA induction range. Thus, even though these cells have different geometries, they enlarge their sidewalls at the same rate. We next examined how extremely low levels of aPBPs affected growth. Similar to previous observations 16,35,36, both ΔponA and ΔaPBP (ΔpbpF, ΔpbpG, ΔpbpD, ΔponA::kan) strains showed a marked reduction in bulk growth, and this defect could be rescued by RodA overexpression 6,37. However, single-cell measurements revealed a surprise: Both ΔponA and ΔaPBP cells showed the same single-cell growth rates as WT cells, as did ΔaPBP cells that overexpressed RodA (Figure 2d). Given that the increased rate of lysis of ΔaPBP cells is suppressed by RodA overexpression 6,37, it appears the population growth rate defect of aPBP-deficient cells arises not from a reduction in growth rate, but from an increased frequency of death.
Increased mreBCD induction increases the density of directionally moving MreB filaments and the fraction of directionally moving synthetic enzymes
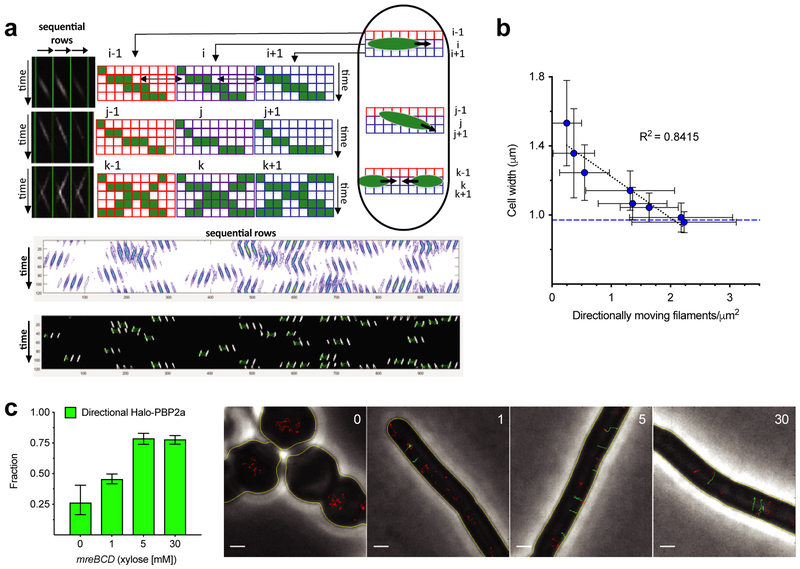
To gain a mechanistic link between the level of each PG synthetic system and cell width, we sought to develop microscopic measures of their activity. While the fraction of stationary aPBPs would be difficult to quantify, Rod complexes exhibit a quantifiable phenotype: As their motion is driven by PG synthesis, their cellular activity can be measured by quantitating the number of directionally moving MreB filaments. Using total internal reflection fluorescence microscopy (TIRFM) data, we developed a method to quantitate the density of directionally moving MreB filaments per surface area of the bacteria. Filaments undergoing directed motion are detected by taking advantage of the temporal correlation occurring between adjacent pixels across the cell width as objects move through them. These objects are then counted over a given time and then normalized to the total cellular surface area (Figure 3a). Simulations demonstrated this method was robust to various parameters, and also more accurate than tracking filaments in the same data, giving values similar to tracking filaments imaged with TIRF-structured illumination microscopy (TIRF-SIM) (Supplementary Figure 5). Using this tool, we examined how the density of directionally moving MreB filaments related to rod width as we titrated the expression of mreB-msfGFPsw, mreCD expressed as the only source. This analysis revealed a well-fit, mostly linear correlation between directional filament density and width (Figure 3b). Thus, an increasing density of directionally moving MreB filaments reduces cell width, possibly by promoting circumferential PG synthesis.
Figure 3 — Increased mreBCD increases directional MreB filament density and the fraction of directional PBP2a molecules.
a. Schematic of our method to quantitate directional MreB filament density. Example data (top) generated from simulated TIRFM movies. First, a kymograph is generated for each row of pixels along the cell midline. These kymographs are lined up side by side to generate a single 2D image, where each column contains a kymograph of each sequential row of pixels in the cell. First, the image is adaptively thresholded, then segmented with contour analysis to extract fluorescent objects (middle). These objects are used to get velocity (slope), time (centroid), and position (row) for each particle. As particles will show similar intensities in adjacent rows, or sometimes move at angles, objects up to two rows apart are grouped based on time, position, and velocity. This yields the final particle count (bottom). See Supplementary Figure 5 for further details and validations.
b. Increasing mreBCD induction correlates with an increasing density of directionally moving MreB filaments. bYS981 (amyE::erm Pxyl-mreB-msfGFPsw mreCD, ΔmreBCD::spc) was grown in different amounts of xylose, imaged with TIRFM, and analyzed as in a. Plotted are mean cell widths (error bars are SD) against the density of directionally moving filaments. Blue dotted line indicates mean width of bYS19, expressing MreB-msfGFPsw at the native locus. (Note that strains that contain fluorescent protein-MreB fusions are wider than WT.)
c. The fraction of directionally moving Halo-PBP2a molecules increases with MreBCD expression. Left – mreBCD was induced in bMK385 (amyE::erm Pxyl-mreBCD, ΔmreBCD::spc, pbpA::cat HaloTag-11aa-pbpA) in different amounts of xylose, and single molecules of JF-549-labeled Halo-PBP2a were imaged by TIRFM. Plotted are the mean (error bars are 95% CI) fraction of labeled PBP2a trajectories over 7 frames in length that moved directionally. Right - Representative montage of Halo-PBP2a trajectories at different levels of mreBCD inductions overlaid on phase images. Directionally moving tracks are green; all other tracks are red. Scale bars are 1 μm. See also Supplementary Movie 1.
We next examined how titrating mreBCD levels affected PBP2a motion. Multiple studies have noted PBP2a molecules exist as a mixed population; some diffuse within the membrane, while others move directionally 12,14,17. We titrated mreBCD induction as we tracked the motions of PBP2a molecules (expressed at the native locus as a HaloTag fusion, sparsely labeled with JF-549 38). At low mreBCD induction, only a small fraction of PBP2a moved directionally. As we increased mreBCD induction, an increasing fraction of PBP2a molecules moved directionally (Figure 3c, Supplementary Movie 1), demonstrating that mreBCD limits the amount of directional PBP2a.
Increased MreBCD:PBP1 correlates with an increased amount of oriented material and structural anisotropy of the cell wall
To explore how an increased density of directional rod complexes reduces rod width, we used polarization microscopy to examine how increasing circumferential synthesis affected the organization of cell wall material. Polarization microscopy reports on both the angle and extent of orientation within optically anisotropic (or birefringent) materials 39, and has been used to assay the orientation of various materials, including plant cell walls 40 and mitotic spindles 41. Polarization microscopy revealed that purified WT sacculi are birefringent, indicating some fraction of the material within them is oriented. Focused at their surface, sacculi showed a predominant slow axis oriented along the rod length (Figure 4a, Supplementary Movie 2). Given that amino acids have a higher refractive index than sugars, this suggests (in agreement with previous models 42) that peptide crosslinks are predominantly oriented along the rod length, and the glycans are oriented around the circumference.
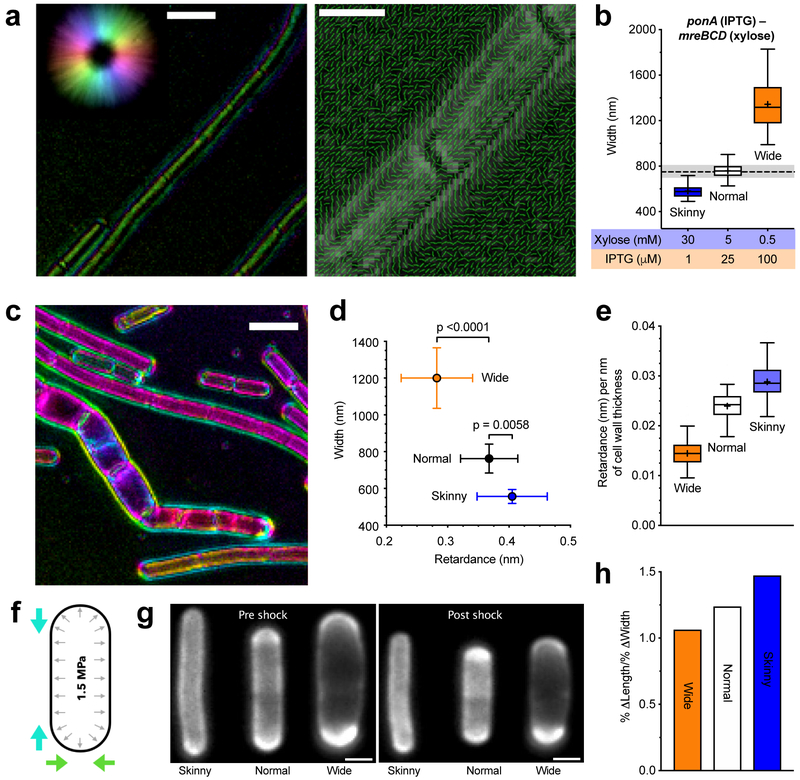
Figure 4 — Increased Rod activity increases both the amount of oriented material within sacculi and their mechanical anisotropy.
For details regarding statistics and box plot definitions, see the “Statistics” subheading in “Methods”.
a. Polarization microscopy reveals oriented material within the cell wall. Retardance is the differential optical path length for light polarized parallel and perpendicular to the axis of molecular alignment; alternatively, it is defined as birefringence (Δn) multiplied by the path length through an anisotropic material. Left – Example LC-PolScope image of purified WT sacculi. Focused at the surface, the wall is seen to be birefringent. Color is the slow axis orientation, intensity corresponds to retardance in that direction (reference, upper left circle). Scale bar is 2 μm. Right - Polarization orientation view of sacculi surface; lines point in predominant orientation of the slow axis. Scale bar is 1 μm. See also Supplementary Movie 2.
b. Inductions used to assay sacculi. bMD620 (amyE::erm Pxyl-mreBCD, ΔmreBCD::spc, yhdG::cat Pspank-ponA, ΔponA::kan) was induced to grow at 3 different widths. Dashed line is the median width of WT B. subtilis, shading indicates 25-75 percentiles.
c. Example LC-PolScope image of bMD620 sacculi induced at different widths. Pairs of purified sacculi (wide and normal, or wide and skinny) were combined and Z-stacks collected in 100 nm steps. Scale bar is 2 μm. See also Supplementary Movie 3.
d–e. The amount of oriented material in the cell wall increases with mreBCD induction, and inversely correlates with width.
d. Mean retardance vs. width of projected Z-stacks of at least 90 different cells for each condition (error bars are SD).
e. Mean retardance normalized to the mean thickness of cell walls in each induction condition (determined with TEM; Supplementary Figure 6a).
f. Schematic of osmotic shock assay of anisotropy. B. subtilis sacculi are normally stretched by high internal turgor (black arrows). Hyperosmotic shocks negate this pressure, allowing observation of how sacculi shrink in length and width (colored arrows).
g. Example FDAA-labeled cells before and after shocks. Scale bars are 1μm.
h. As the relative amount of Rod activity increases, so does the mechanical anisotropy of the sacculus. Percent change in length/percent change in width for each condition following osmotic shock. See also Supplementary Figure 6b–d.
We then examined how the balance between the Rod and aPBP systems affected the relative amount of oriented material in the wall. We grew our dual-induction strain under three different induction conditions to create varied width cells: wide (high ponA, low mreBCD), normal (ponA and mreBCD induced at WT levels), and skinny (low ponA, high mreBCD) (Figure 4b). We purified their sacculi and quantified their total retardance with polarization microscopy (Figure 4c, Supplementary Movie 3). Wide cells had the lowest retardance, skinny cells had the highest retardance, and normal cells were in between (Figure 4d). Normalizing the retardance of each sample to the mean thickness of cell walls from each induction (Supplementary Figure 6a) revealed that, as the relative mreBCD:ponA induction increases, so does the amount of highly ordered material per nanometer of cell wall thickness (Figure 4e). Thus, in agreement with atomic force microscopy studies showing orientated glycans in E. coli require MreB 15, these experiments demonstrate that as Rod system activity increases, so does the amount of oriented material in the wall.
E. coli and B. subtilis sacculi are mechanically anisotropic, stretching more along their length than across their width 43. To test how the ratio of circumferential to non-circumferential synthesis affected this property, we grew our dual-inducer strain at the three mreBCD:ponA inductions described above, labeled their walls with Alexa-488-D-amino carboxamides 4, then assayed their dimensions before and after hyperosmotic shocks (Figure 4f–g). This revealed that increased Rod system activity correlated with an increased mechanical anisotropy of the sacculus: As we increased the expression of MreBCD relative to PBP1, cells shrank less across the rod width, and more along their length (Figure 4h, Supplementary Figure 6b–d). Thus, the Rod system acts to reinforce rod shape against internal turgor pressure by promoting oriented PG synthesis around the rod.
E. coli width mutants show the same correlation between cell width and directional MreB filament density
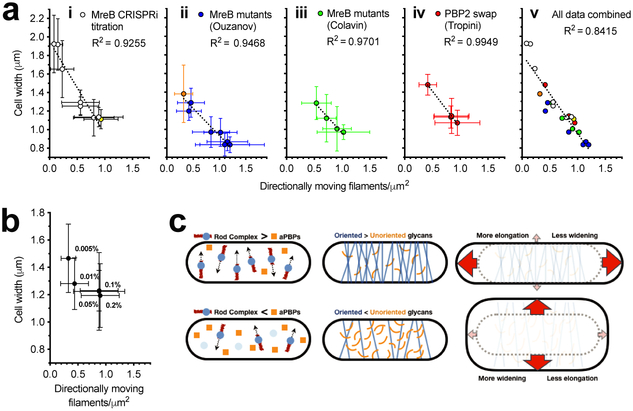
Previous studies have examined how MreB or PBP2 mutations affect the shape of E. coli, hypothesizing their abnormal widths arise from changes in the curvature, twist, or angle of MreB filaments. Our observations suggest an alternative explanation: Abnormal width arises from changes in the amount of Rod system activity. We tested this by measuring the density and velocity of directional GFP-MreB filaments in these same mutant E. coli strains. We first assayed the width of E.coli as we titrated MreB-msfGFPsw expression using CRISPRi against msfGFP 30. As in B. subtilis (Figure 3b), this yielded an inverse correlation between directional filament density and cell width (Figure 5a). Likewise, each group of mutants showed the same relationship: 1) The same trend was observed for the MreB mutants hypothesized to change filament twist 19, and 2) also for the mutants hypothesized to change filament curvature 23. Notably 3), the same correlation was also observed in E. coli where mrdA (encoding PBP2) was replaced with mrdA genes from other species 21 (Figure 5a, Supplementary Figure 7a). TIRF-SIM imaging of these MreB mutants revealed insights into these effects (Supplementary Movie 4): Wide MreB mutants showed either A) longer but fewer filaments, or B) a large fraction of immobile filaments. Conversely, thinner mutants appeared to have more, but shorter filaments. Finally 4), while Colavin et al. hypothesized that RodZ reduces width by changing MreB filament curvature 23, we found that increased RodZ induction increased the density of directional MreB filaments (Figure 5b, Supplementary Movie 5). This suggests that the RodZ dependent decrease in cell width arises not from RodZ changing filament curvature, but rather from RodZ increasing the number of MreB filaments, perhaps via filament nucleation 44. Finally, while a recent study of two MreB mutants noted a correlation between MreB velocity and cell width 45, no such correlation was observed across our expanded set of mutants (Supplementary Figure 7b). Thus, the density of directionally moving Rod complexes correlates with cell width in both B. subtilis and E. coli across multiple genetic perturbations.
Figure 5 — Directional MreB filament density also correlates with cell width of E. coli Rod mutants.
For details regarding statistics see the “Statistics” subheading in “Methods”. Filament densities in a and b were calculated as in Figure 3b.
a. Cell width vs. density of directionally moving MreB filaments in different E. coli strains. (i) AV88 (186::Ptet-dCas9, mreB::mreB-msfGFPsw) allows the tunable expression of MreB-msfGFPsw by expressing various sgRNAs with different matches against msfGFP. Yellow indicates WT expression. (ii) MreB-msfGFPsw mutant strains from Ouzounov et al., 2016. Orange indicates RM478 (ΔrodZ, mreB(S14A)-msfGFPsw) from Morgenstein et al., 2015. (iii) MreB-msfGFPsw mutants believed to change filament curvature from Colavin et al., 2018. (iv) msfGFP-MreB strains from Tropini et al., 2015, where mrdA is replaced with mrdA from other species. (v) All data from (i)–(iv) combined. See also Supplementary Movie 4.
b. Decreased cell width caused by increased RodZ expression correlates with an increased density of directionally moving MreB filaments. KC717 (csrD::kan, mreB::msfGFP-mreB, ProdZ<>(frt araC PBAD)) was grown at different arabinose concentrations (%, indicated on the graph). See also Supplementary Movie 5.
c. Model for how the balance between aPBPs and the Rod system affects cell width. When Rod complex activity is high relative to that of aPBPs (top left), sacculi have more circumferentially oriented material (top center) compared to when aPBP activity is greater (bottom left, bottom center). As the amount of oriented material increases, sacculi become more rigid across their width, but less rigid along their length. Thus, stretched by the internal turgor pressure, sacculi with more Rod activity are better able to maintain their width, and instead, stretch more along their length (top right). In contrast, cells with reduced Rod activity have less circumferentially oriented glycans to reinforce their width, and thus expand more along their width (bottom right).
Discussion
Together, these experiments provide a coarse-grained, mechanistic model explaining how rod-shaped bacteria use the controlled insertion of glycans to define and maintain their dimensions. The shape of bacteria is defined by their cell walls; our data demonstrates the two systems that insert PG into it have opposing roles on its shape. Due to the intrinsic orienting of MreB filaments around the rod width 11, the Rod system inserts material around the rod circumference, reducing its diameter. As the number of MreB filaments increases, so does the fraction of directional enzymes and the amount of oriented material in the wall. In contrast, the aPBPs do not move circumferentially, inserting material that isotropically enlarges the sacculus. Our data indicates that the macroscopic shape of the sacculus arises from the arrangement of the material inserted into it, as this organization determines how it responds to internal turgor pressure: The more material is oriented around the rod circumference by the Rod complex, the less the sacculus stretches across its width, and the more it stretches along its length (Figure 5c). Given that B. subtilis growth is driven by internal pressure 46, the anisotropic stretching caused by MreB patterning could be a key determinant of rod-shaped growth, allowing sacculi to elongate while maintaining their width.
If the balance between the two PG synthetic systems is perturbed, the shape of the sacculus becomes altered, though its rate of expansion remains constant. As both systems utilize the same pool of precursor, the flux through each system depends on their relative levels; reductions in one likely increase the flux through the other. This may explain why cells swell when the Rod system is disrupted 27,31,47, without Rod-mediated thinning, aPBPs add more material uniformly over the cell surface. Likewise, in the absence of widening aPBPs 16, an increased flux through the Rod system would explain why cells become extremely thin. However, if both systems are equivalently reduced, cells grow with normal widths, but at slower rates; as long as the activities are balanced, identical shape arises from the balanced levels of enzymes, but growth is reduced due to their combined activity becoming limiting. This would explain why ponA mutations rescue mreB deletions 48; equally crippling both systems might rebalance the activities so cells retain their normal shape and viability.
Implications for the role of MreB in rod width determination
Given 1) mreBCD from B. megaterium creates normal diameter B. subtilis rods, and 2) B. subtilis diameter depends on mreBCD levels, it appears unlikely that any structural property within MreB filaments defines cell diameter. Rather, MreB simply functions as the orienting component within a rod-thinning system, working in opposition to the widening aPBPs. Indeed, in vitro studies have revealed MreB filaments are curved more than the membrane, allowing them to deform to, and orient around bacteria of any larger width 9,11. While it remains possible that some MreB mutations or interactions with RodZ could alter filament curvature or twist 19,20,23, our data suggests a simpler explanation: These mutations alter MreB’s polymerization dynamics or cellular distribution. Reducing the number of active MreB polymers would reduce Rod activity, making cells wider. Similarly, mutations that alter the filament length distribution or their tendency to bundle would cause Rod activity to become non-uniformly distributed, causing some cellular regions to thin while others widen, as observed for certain MreB mutants 12,27.
The role of aPBPs in growth and survival
These experiments reinforce observations that aPBPs and RodA (when in excess to MreBCD) serve anti-lytic roles: aPBP-deficient cells grow at the same rate as WT, yet have an increased frequency of death, lysing as thin rods without losing shape 6,37. Likewise, aPBP-mediated synthesis increases upon endopeptidase overexpression 49. Given the active state of single aPBP molecules correlates with periods of transient immobility 17,50, their synthesis is likely localized to small regions. Combined, these observations support a model where aPBPs fill gaps in the PG meshwork 17,37. Gaps could arise via damage, hydrolases, or between the imperfectly oriented strands synthesized by the Rod complex 12. If this model is correct, the different spatial activities of these systems might allow the sacculus to maintain integrity at any Rod complex/aPBP ratio: the fewer Rod complexes, the larger the gaps filled by aPBPs.
Methods
Statistics
For all box plots throughout this work, boxes indicate the 25–75 percentiles, whiskers the 5–95 percentiles, the midline indicates median, + indicates mean. P-values are reported in figures and were calculated in GraphPad Prism 7.0 using a two-tailed Mann-Whitney test. Confidence intervals in Figure 3a were calculated using the modified Wald method 51. Means, medians, standard deviations, confidence intervals, and sample sizes for all data points are reported in Supplementary Table 6. Fits to data in Figures 3b and 5a were done using linear regression in GraphPad Prism 7.0. Replicates of experiments are reported in Methods and Supplementary Table 6.
Media and culture conditions
For all experiments, unless otherwise noted, B. subtilis and B. megaterium were grown in casein hydrolysate (CH) medium (where indicated, xylose and/or isopropyl thiogalactoside [IPTG] was added), and E. coli strains were grown in lysogeny broth (LB) medium (where indicated, arabinose or anhydrotetracycline [ATc] was added), at 37°C with rotation. When pre-cultures, grown overnight with rotation at 25°C from single colonies, reached exponential phase the next day (OD600 of 0.4–0.7) they were diluted back into fresh growth medium (and where indicated, with the specified concentrations of inducer) and were grown for at least 3 hours at 37°C with rotation, to an OD600 of ~0.3 to 0.6.
Sacculi purification
Twenty mL cultures were grown in baffled flasks at 37°C with vigorous shaking, to an OD600 of ~0.5, were harvested and centrifuged at 5,000 × g for 5 min at 4°C. Cell pellets were resuspended in 1 mL of ice-cold phosphate buffered saline (PBS), were centrifuged at 6,000 × g for 30 sec, were resuspended in 500 μL of PBS, and were killed by boiling in a water bath for 10 min. Cells were centrifuged at 6,000 × g for 2 min, were resuspended in 500 μL of PBS containing 5% sodium dodecyl sulfate (SDS), and were boiled in a water bath for 25 min; this was repeated once, except boiling was for 15 min. To remove the SDS, the samples were centrifuged at 6,000 × g for 2 min and were resuspended in 500 μL of PBS; this was repeated 5 times. The cells were centrifuged at 6,000 × g for 2 min, were resuspended in 1 mL of 50 mM Tris-HCl, pH 7.5, containing 10 mM sodium chloride, and 2 mg of pronase from Streptomyces griseus (MilliporeSigma, MA), and were incubated at 60°C for 90 min with gentle shaking. To remove the pronase, the samples were boiled twice in PBS/5% SDS, followed by 6 rounds of washes in PBS, exactly as described in the steps above that precede the pronase treatment. The samples were centrifuged at 6,000 × g for 2 min, were resuspended in 1 mL of 25 mM Tris-HCl, pH 8.5, containing 0.5 M sodium chloride, 5 mM magnesium chloride, and 100 U of salt-active nuclease (SAN) (ArticZymes, Norway), and were incubated at 4°C overnight with gentle mixing. To remove the SAN, the samples were boiled twice in PBS/5% SDS, followed by 6 rounds of washes in PBS, exactly as described in the steps above that precede the pronase treatment. The sacculi were centrifuged at 6,000 × g for 2 min, the supernatant was removed, and the pellets were stored at −80°C.
Polarization microscopy
Purified sacculi were resuspended in PBS and placed on ethanol-cleaned No. 1.5 coverslips under a cleaned glass slide. Polarization images were acquired on an inverted LC-PolScope mounted on a Nikon Ti-E equipped with a 60x/1.4NA Plan Apo oil immersion objective and oil condenser with matching NA, and a Hamamatsu Photonics Flash4 camera. Z-stacks in 100 nm steps were taken of each sample using 50 msec acquisitions of 546/12 nm light. All image acquisition, processing, and display, including colored display and line maps were prepared using the OpenPolScope Hardware Kit and plugins for ImageJ/Micro-Manager from OpenPolScope.org. From the previously prepared sacculi, multiple slides were imaged and quantitated, with independent background calibrations. These gave similar results and were combined to yield the final data set.
Calculation of retardance
Z-stacks of the computed total retardance for each slice were exported from OpenPolScope. These stacks were cropped at one frame above and beneath the focal planes of the top and the bottom of the cells, then projected into a single plane using ImageJ. To avoid getting high retardance values from the edge effects arising from the sides of the cell or the septa, we selected the retardance at the middle of the cell, using the average of line scans (5 pixels long and 3 wide) that were drawn down the center of cells, taking care to avoid edges and septa.
Transmission electron microscopy measurements of cell wall thickness and cell diameter
Overnight cultures were diluted into fresh medium and grown to an OD600 of ~0.3 to 0.5. Cells were pelleted by centrifugation at 5,000 × g and fixed by resuspending in 100 mM MOPS buffer pH 7 containing 2% paraformaldayde, 2.5% gluteraldehyde, and 1% dimethyl sulfoxide overnight at 4°C. Cells were centrifuged at 5,000 × g and were washed 3 times with 100 mM MOPS pH 7. The pellet was stained with 2% osmium tetroxide in 100 mM MOPS for 1 hr, washed twice with deionized water, and stained overnight with 2% uranyl acetate. The pellet was washed twice with deionized water and dehydrated by washing once with 50% ethanol, once with 70% ethanol, once with 95% ethanol, and then three times with 100% (v/v) ethanol. Samples were prepared for resin infiltration by washing once with 50% ethanol, 50% propylene oxide (v/v) and then once with 100% propylene oxide. All wash steps were for 5 min. Infiltration of resin was achieved by incubation with 50% Embed 812 (EMS, PA)/50% propylene oxide for 1 hr, followed by incubation with 67% Embed 812/33% propylene oxide for 1 hr, and incubation with 80% Embed 812/20% propylene oxide for 1 hr. Samples were then incubated twice with Embed 812 for 1 hr, followed by an overnight incubation in molds. The molds were baked at 65°C for 18 hr before sectioning.
Serial ultrathin sections (80 nm) were cut with a Diatome diamond knife (EMS, PA) on a Leica Ultracut UCT (Leica Microsystems, Germany) and collected on 200-mesh thin-bar formvar carbon grids. Sections were imaged on an FEI Tecnai transmission electron microscope.
Cell wall thickness measurements were performed using a custom-built MATLAB (Mathworks, MA) script. Image intensity profiles extracted from lines were drawn perpendicular to a user-input line defining the middle of the cell wall. The distance between the two lowest points below a threshold within 40 nm of the middle of the cell wall was measured as the cell wall thickness at ~30 points in each cell. This experiment was conducted once, using multiple cells for the analysis. Cell diameter was measured simultaneously as cell wall thickness using three user-input lines. The average of these three measurements was recorded.
Measurements of Bacillus cell diameters
Cultures grown to an OD600 of ~0.3 to 0.6 were stained with 0.5 μg/mL FM 5–95 (Thermo Fisher Scientific, MA) for 1 min, and were concentrated by centrifugation at 6,000 × g for 30 sec. The cell pellet was resuspended in ~1/20 volume of growth medium, and 3 μL was applied to ethanol-cleaned No. 1.5 coverslips under a 3% agarose pad containing growth medium. Fluorescent cells were imaged with the top surface of the agarose pad exposed to air, in a chamber heated to 37°C. Epifluorescence microscopy was performed using a Nikon Eclipse Ti equipped with a Nikon Plan Apo λ 100×/1.4NA objective and an Andor camera. Cell contours and dimensions were calculated using the Morphometrics software package 52. Each “Width” data point (Figures 1–4) is calculated from at least 79 cells, but most typically hundreds (see Supplementary Table 6), from multiple fields of view across different areas of the agarose pad. Key points in these experiments were repeated multiple times on independent days; including induction conditions for bMD545, bMD598, and bMD620 that resulted in WT width and the extremes of thinning and widening; and PY79 measured in parallel as a width control. All repeat measurements gave similar mean values.
Measurements of single-cell growth rate
Cultures grown to an OD600 below 0.3 were concentrated by centrifugation at 6,000 × g for 30 sec. The cell pellet was resuspended in growth medium, and applied to No. 1.5 glass-bottomed dishes (MatTek Corp., MA). All cells were imaged under a 2% agarose pad containing growth medium, with the top surface exposed to air, in a chamber heated to 37°C. Phase-contrast microscopy was performed using a Nikon Eclipse Ti equipped with a Nikon Plan Apo λ 100×/1.4NA objective and an Andor camera. We used a custom-built package in MATLAB to perform segmentation on phase-contrast time-lapse movies, then calculated the growth rate of the surface area of single B. subtilis chains. Each data point for the single-cell growth rates (Figure 2d) is the result of a single experiment; for each, >50 cells from multiple fields of view across different areas of the agarose pad were imaged and analyzed.
Measurements of bulk growth rate
For cell culture measurements of growth rate, overnight pre-cultures in mid-log growth were diluted in fresh medium and grown for ~3 hr at 37°C to an OD600 of ~0.3 to 0.6. The cultures were diluted back to a calculated OD600 of 0.07 in 100-well microtiter plates (replicated in 3 to 5 wells for each culture), and their growth rates were measured using a Bioscreen-C Automated Growth Curve Analysis System (Growth Curves USA, NJ) plate reader, at 37°C with continuous shaking. Growth rates were calculated from OD600 measurements that were recorded every 5 min for at least 6 hr. This was repeated 6 times for the parental strain PY79, and from 1 to 4 times for all other strains and conditions with PY79 measured in parallel as a growth control.
TIRF microscopy of Halo-PBP2a
Cultures grown to an OD600 below 0.3 were labeled with 10 nM JF549 53, and were concentrated by centrifugation at 6,000 × g for 30 sec. The cell pellet was resuspended in growth medium and applied to ethanol-cleaned No. 1.5 coverslips. All cells were imaged under a 2% agarose pad containing growth medium with the top surface exposed to air, in a chamber heated to 37°C. TIRFM and phase-contrast microscopy were performed using a Nikon Eclipse Ti equipped with a Nikon Plan Apo λ 100×/1.45NA objective and a Hamamatsu ORCA-Flash4.0 V2 sCMOS camera. Fluorescence time-lapse images were collected by continuous acquisition with 300 msec exposures. All data are from a single experiment, where cells were induced at different levels, and tracks from >20 cells were used for analysis of each data point.
Analysis of the density of directionally moving MreB Filaments
Phase images of bacteria were segmented using Morphometrics 52, and the width and length of each cell was calculated. Next, the fluorescence time-lapses were analyzed based on the segmentation mask of the phase image (Supplementary Figure 5c). Filament counting was performed in several steps (Supplementary Figure 5). First, kymographs were generated for each row of pixels along the midline of the cell. Next, the kymographs for each row were placed side by side, converting the TIRF time lapse data into a single 2D image (Supplementary Figure 5d). To identify filaments in the kymograph, closed contours were generated in the 2D image (Supplementary Figure 5e). We only selected contours within a given size range (0.04 μm2 to 0.17 μm2). For these contours, we calculated the total intensity (the sum of the intensities of the pixels in the contour), the centroid, the velocity (calculated from the slope of the major axis line of the contour) (Supplementary Figure 5f), and time (from the centroid). Next, to identify cases where the same MreB filament appears in multiple sequential kymographs, each object in a given kymograph is linked to a corresponding object in the next and previous kymographs based on the above properties of the object (see Supplementary Figure 5f for details). Finally, the counting is verified manually by numbering each filament on the 2D image (Figure 3a). To test the performance of the filament counting, we analyzed simulated data with different filament density, velocity, and orientation settings (Supplementary Figure 5). All of the image analyses were performed using MATLAB. Key points in these experiments – specifically, the density of MreB filaments in E. coli strains from other labs (Figure 5), and the density of MreB filaments in B. subtilis (Figure 3b) – were repeated twice on independent days. All measurements gave similar mean values and were combined into the final data set.
Simulation of directionally moving MreB
The Image Correlation Spectroscopy 6 MATLAB package was used for the simulation of MreB moving around the cell. The following parameters were set for the MreB simulations: velocity, orientation, filament number, and filament length. The default velocity setting is 30 nm/sec and the default orientation is 0, which means MreB moves perpendicular to the central axis. The default filament length is set to 250 nm and each MreB monomer is assumed to be 5 nm. The cell width is set to 1 μm and the cell length is set to 4 μm. The pixel size is 65 nm and the time interval is 1 sec, which is the same as the TIRFM imaging obtained with our Nikon Eclipse Ti equipped with a Hamamatsu ORCA-Flash4.0 V2 sCMOS camera. The particles are randomly distributed on the surface of the cell. Each simulation data point was repeated 5 times for the counting analysis. To compare the correlated motion approach against particle tracking, we counted the number of tracks observed after tracking the simulated data using the Linear Motion LAP tracker in FIJI 2with TrackMate v3.8 54. The threshold for spot size was 0.195 μm and the intensity threshold was 10 counts. The search radius is 0.085 μm, the link radius is 0.085 μm, and the gap size is 1. All traces longer than 6 frames that had moved more than 250 nm were considered directional motions. Each simulation was repeated 5 times. For the simulation of directional moving MreB filaments mixed with diffusive mreB, we used particles of size 5 nm with diffusion coefficient of 0.003 μm2/s, as in Billaudeau et al., 2017 55. For the directional moving mreB, the filament velocity and orientation are set as default, and directional filament density is set to 3.2/μm2 with filaments of 250 nm in length.
TIRF-SIM imaging of E. coli strains
Cells were prepared as described in Media and culture conditions, above. Cells were placed under an LB agarose pad, on a cleaned No.1.5 coverslip, and imaged at 37°C. Live-cell SIM data were acquired as described previously 56 on a Zeiss Axio Observer.Z1 inverted microscope outfitted for structured illumination. An Olympus 100×/1.49NA objective was used instead of the Zeiss 1.45NA objective because the slightly larger NA of the Olympus objective gives higher tolerance for placing the excitation beams inside the TIRF annulus. Data was acquired at 1 sec frame rates, with 20 msec exposures from a 488 nm laser for each rotation. TIRF-SIM images were reconstructed as described previously 56.
TIRF-SIM imaging of B. subtilis strains
Cells were prepared as described in Media and culture conditions, above. Cells were placed under a CH agarose pad in a No. 1.5 glass-bottomed dish (MatTek Corp., MA) for imaging. Images were collected on a DeltaVision OMX SR Blaze system in TIRF mode, using an Edge 5.5 sCMOS camera (PCO AG, Germany) and a 60x objective. 75 msec exposures from a 488 nm diode laser were used for each rotation. Spherical aberration was minimized using immersion oil matching. Raw images were reconstructed using SoftWoRx (GE Healthcare, MA) software.
Particle tracking of JF549-Halo-PBP2a
Particle tracking was performed using the software package FIJI 2 and the TrackMate v3.8 54 plugin. For calculation of particle velocity, the scaling exponent α, and track orientations relative to the midline of the cell, only tracks persisting for 7 frames or longer were used. Particle velocity for each track was calculated from nonlinear least squares fitting using the equation MSD(t) = 4Dt + (vt)2, where MSD is mean squared displacement, t is time interval, D is the diffusion coefficient, and v is speed. The maximum time interval used was 80% of the track length. To filter for directionally moving tracks, we discarded those with a velocity lower than 0.01 nm/sec. Tracks were also excluded if the R2 for log MSD versus log t was less than 0.95, indicating a poor ability to fit the MSD curve.
Osmotic shock experiments
Overnight, exponentially growing cultures (described in Media and culture conditions) were diluted into fresh CH medium, grown at 37°C to an OD600 of 0.1 to 0.2, then stained by growing in 100 μM Alexa Fluor 488-D-Lysine-NH2 for 1 hr. Without washing, cells were then loaded into a CellASIC microfluidic flow cell (MilliporeSigma, MA) pre-conditioned with media and washed in the chamber via channels 6 and 5. Media in channel 6 was replaced with 5 M sodium chloride, and the flow cell was resealed and imaged immediately. After collecting images across the whole chip pre-shock, 5 M sodium chloride was flowed into the chip via channel 6 and imaged immediately. This experiment was repeated in bulk cultures, yielding similar results as in the microfluidics. Each “Width” and “Length” data point (Supplementary Figure 6b) is calculated from at least 145 cells (through >1000; see Supplementary Table 6) from multiple fields of view across different areas of the flow cell.
Alexa Fluor 488-D-Lysine-NH2
Alexa Fluor 488-D-Lysine-NH2 was synthesized as in Lebar et al., 2014 4.Briefly, Boc-D-Lys(Cbz)-OH (Bachem, Switzerland) was reacted with carbonyldiimidazole (CDI) (MilliporeSigma, MA) in dimethylformamide (DMF) for 1.5 hr, then aqueous ammonia was added and stirred for 6 hr to form the carboxamide Boc-D-Lys(Cbz)-NH2. The Cbz protecting group was removed by catalytic hydrogenation (20% Pd(OH)2/C) in methanol. The product, Boc-D-Lys-NH2, was reacted with CDI in DMF for 1.5 hr, then Alexa Fluor 488 carboxylic acid in DMF was added and reacted for 6 hr to yield Boc-D-Lys(Alexa Fluor 488)-NH2. The Boc protecting group was removed by stirring in neat trifluoroacetic acid (TFA) for 30 min. The reaction was stopped by dropwise addition of TFA solution to ice-cold ether. The precipitate was then HPLC-purified to obtain Alexa Fluor 488-D-Lysine-NH2.
CRISPRi titration of MreB expression
We used complementarity-based CRISPR knockdown to titrate the MreB expression level in E. coli. The degree of mreB-msfGFPsw repression is controlled by introducing mismatches between the guide RNA and the target DNA 57. The repression strength can be tuned by modulating spacer complementarity to msfGFP using different numbers of mismatches. To repress msfGFP using CRISPR knockdown, we placed the dCas9 cassette under the control of a Ptet promoter and different plasmids to target msfGFP. We induced dCas9 at a constant high level with 100 ng/ml of ATc and changed the degree of guide RNA complementarity with different plasmids. For pcrRNA plasmid we use four different guide RNAs with 10, 11, 14, and 20 bp of complementarity. For pAV20 plasmid we use four different guide RNAs with 5, 10, 14, and 20 bp of complementarity. The cells were grown and imaged in LB containing 50 μg/ml kanamycin. This experiment was repeated twice, yielding similar means for each data point. All repeats were then combined into the final data set.
Cell lysates
Cell cultures were grown to an OD600 of ~0.3 to 0.6. Cell amounts were normalized by harvesting the equivalent of 1 mL of culture at an OD600 of 0.5 for western blot analysis, or 3 mL for mass spectrometry (MS). Cells were centrifuged at 6,000 × g for 30 sec, washed once in 1 mL of ice-cold 20 mM Tris-HCl, pH 7.5, and 10 mM EDTA (TE), were resuspended in 100 μL of TE, and were killed by boiling in a water bath for 10 min. All samples were frozen at −80°C overnight (or up to 1 week). Thawed samples were digested with 16.7 or 50 μg (for 1 or 3 mL cultures, respectively) of lysozyme (Thermo Fisher Scientific, MA) in the presence of 1 mM phenylmethylsulfonyl fluoride, at 37°C for 15 min.
Western blotting and analysis
Lysozyme-digested cell lysates were resuspended in 1× loading buffer (6× is 50 mM Tris-HCl, pH 6.8, 0.05% bromophenol blue, 30% glycerol, 5% 2-mercaptoethanol, 10% SDS), and denatured at 95°C for 5 min. Proteins were resolved by SDS-PAGE using 10% Bis-Tris gels (Bio-Rad, CA), followed by electrotransfer to 0.2 μm pore size nitrocellulose membranes (Bio-Rad, CA). Western blotting reagents were supplied in a ONE-HOUR Western Advanced Kit (GenScript, NJ), and used according to the manufacturer’s instructions. Briefly, for hybridization against SigA, rabbit anti-SigA polyclonal antibody (gift of R. Losick), was pre-incubated for 1 hour at room temperature with WB-1 Solution (containing anti-rabbit IgG-HRP conjugate), in a 1:100 mixture. Or, for hybridization against MreB, rat anti-MreB polyclonal antibody (gift of R. Carballido) was pre-incubated with anti-rat IgG-HRP (MilliporeSigma, MA) in PBS/0.05% Tween 20, in a 2:1:100 mixture. Membranes were blocked for 5 min in Pretreat Solution A-B and were incubated overnight at 4ºC in WB-2 Solution mixed 333:1, with either the SigA or MreB antibody pre-mixtures. The membranes were washed 3 × 10 min in Wash Solution and were reacted with LumiSensor HRP Substrate for 5 min.
Chemiluminescent bands were digitally imaged with a Sapphire Biomolecular Imager (Azure, CA). Band intensities were quantified using FIJI. MreB signal intensities were normalized by their respective SigA intensity measurements to control for differences in culture density or sample prep.
Peptide labeling, fractionation, mass spectrometry, and analysis
Lysozyme-digested cell lysates were submitted to the Harvard FAS Mass Spectrometry and Proteomics Resource Laboratory (https://proteomics.fas.harvard.edu/). Protein extraction was achieved utilizing a Covaris S220 ultrasonicator (Covaris, MA), under denaturing conditions upon the addition of urea-based Protein Extraction Buffer DF (Covaris, MA), followed by ice-cold methanol/chloroform precipitation. Proteins were digested with trypsin (Promega, WI). Each resulting peptide mixture was labeled with one of a set of up to eleven isotopic tandem mass tags (TMTs) (Thermo Fisher Scientific, MA).
Equal amounts of each TMT-labeled sample were combined and fractionated by electrostatic repulsion-hydrophobic interaction chromatography, on an Agilent 1200 HPLC system (Agilent, CA) using a PolyWAX LP 200 × 2.1 mm, 5 μm, 300Â column (PolyLC, MD). Peptides were separated across a 70 min gradient from 0% of “buffer A” (90% acetonitrile, 0.1% acetic acid) to 75% of “buffer B” (30% acetonitrile, 0.1% formic acid), with 20 fractions collected over time. Each fraction was dried in a SpeedVac (Eppendorf, Germany) and resuspended in 0.1% formic acid before injection to a mass spectrometer.
LC-MS/MS was performed on a Thermo Orbitrap Elite (Thermo Fisher Scientific, MA) mass spectrometer equipped with a Waters nanoACQUITY HPLC pump (Waters Corp., MA). Peptides were separated on a 150 μm inner diameter microcapillary trapping column packed with ~3 cm of C18 Reprosil 5 μm, 100 Å resin (Dr. Maisch GmbH, Germany), followed by an analytical column packed with ~20 cm of Reprosil 1.8 μm, 200 Å resin. Separation was achieved by applying a gradient of 5–27% acetonitrile in 0.1% formic acid, over 90 min at 200 nl/min. Electrospray ionization was achieved by applying a voltage of 2 kV using a home-made electrode junction at the end of the microcapillary column and sprayed from fused-silica PicoTips (New Objective, MA). The Orbitrap instrument was operated in data-dependent mode for the mass spectrometry methods. The mass spectrometry survey scan was performed in the Orbitrap in the range of 410 –1,800 m/z at a resolution of 12 × 104, followed by the selection of the twenty most intense ions for HCD-MS2 fragmentation using a precursor isolation width window of 2 m/z, AGC setting of 50,000, and a maximum ion accumulation of 200 msec. Singly-charged ion species were not subjected to HCD fragmentation. Normalized collision energy was set to 37 V and an activation time of 1 msec. Ions in a 10 ppm m/z window around ions selected for MS2 were excluded from further selection for fragmentation for 60 sec.
Raw data were submitted for analysis in Proteome Discoverer 2.1.0.81 (Thermo Fisher Scientific, MA) software. Assignment of MS/MS spectra was performed using the Sequest HT algorithm by searching the data against a protein sequence database that included all entries from B. subtilis strain PY79 (UniProt proteome ID UP000018540), and other known contaminants such as human keratins and common lab contaminants. Sequest HT searches were performed using a 15 ppm precursor ion tolerance and required each peptide’s N- and C-termini to adhere with trypsin protease specificity, while allowing up to two missed cleavages. TMT tags on peptide N-termini and lysine residues (+229.162932 Da) were set as static modifications while methionine oxidation (+15.99492 Da) was set as a variable modification. An MS2 spectra assignment false discovery rate of 1% on protein level was achieved by applying the target-decoy database search. Filtering was performed using Percolator (64-bit version 58). For quantification, a 0.02 m/z window centered on the theoretical m/z value of each of the TMT reporter ions and the intensity of the signal closest to the theoretical m/z value was recorded. Reporter ion intensities were exported in a results file of the Proteome Discoverer 2.1 search engine in Microsoft Excel format. The total signal intensity across all peptides quantified was summed for each TMT channel, and all intensity values were adjusted to account for potentially uneven TMT-labeling and/or sample handling variance for each labeled channel. For our final relative protein quantitation analysis, all contaminants from the database search were removed from the results, and only the remaining B. subtilis proteins were used to re-normalize all protein abundances.
Code Availability
All particle tracking was done with the Trackmate plugin within FIJI, then analyzed using code available at https://bitbucket.org/garnerlab/hussain-2017-elife. Filament density calculations and filament simulations were done with custom code available at https://bitbucket.org/garnerlab/dion-2018/src/.
Data Availability
All datasets and raw data generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. Raw and proteomic data are available at https://garnerlab.fas.harvard.edu/Dion2019/Raw-MS-data-Dion2019.zip
Supplementary Material
Acknowledgements
We thank Carl Wivagg, Ye Jin Eun, and Leigh Harris, for helpful discussions, Georgia Squyres for TIRF-SIM, Luke Lavis for JF dyes, and Z. Gitai and K.C. Huang for strains. TIRF-SIM was performed at the Advanced Imaging Center at Janelia Research Campus, a facility jointly supported by the Gordon and Betty Moore Foundation and Howard Hughes Medical Institute. This work was funded by National Institutes of Health Grants R01GM114274 to RO, and DP2AI117923 to EG, and support from the Volkswagen Foundation to EG and SVT. Some work was performed at the Center for Nanoscale Systems at Harvard University, supported by NSF ECS-0335765.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Vadia S & Levin PA Growth rate and cell size: a re-examination of the growth law. Curr Opin Microbiol 24, 96–103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharpe ME, Hauser PM, Sharpe RG & Errington J Bacillus subtilis cell cycle as studied by fluorescence microscopy: constancy of cell length at initiation of DNA replication and evidence for active nucleoid partitioning. J Bacteriol 180, 547–555 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vollmer W, Blanot D & de Pedro MA Peptidoglycan structure and architecture. FEMS Microbiol Rev 32, 149–167 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Lebar MD et al. Reconstitution of Peptidoglycan Cross-Linking Leads to Improved Fluorescent Probes of Cell Wall Synthesis. J Am Chem Soc 136, 10874–10877 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banzhaf M et al. Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol Microbiol 85, 179–194 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Meeske AJ et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones LJ, Carballido-López R & Errington J Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104, 913–922 (2001). [DOI] [PubMed] [Google Scholar]
- 8.van den Ent F, Amos L & Löwe J Bacterial ancestry of actin and tubulin. Curr Opin Microbiol 4, 634–638 (2001). [DOI] [PubMed] [Google Scholar]
- 9.van den Ent F, Izoré T, Bharat TA, Johnson CM & Lowe J Bacterial actin MreB forms antiparallel double filaments. Elife 3, e02634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salje J, van den Ent F, de Boer P & Lowe J Direct membrane binding by bacterial actin MreB. Mol Cell 43, 478–487 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussain S et al. MreB filaments align along greatest principal membrane curvature to orient cell wall synthesis. Elife 7, 1239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garner EC et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333, 222–225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Teeffelen S et al. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci USA 108, 15822–15827 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domínguez-Escobar J et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333, 225–228 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Turner RD, Mesnage S, Hobbs JK & Foster SJ Molecular imaging of glycan chains couples cell-wall polysaccharide architecture to bacterial cell morphology. Nat Comms 9, 1263 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McPherson DC & Popham DL Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J Bacteriol 185, 1423–1431 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho H et al. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol 1, 16172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ursell TS et al. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc Natl Acad Sci USA 111, E1025–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouzounov N et al. MreB Orientation Correlates with Cell Diameter in Escherichia coli. Biophys J 111, 1035–1043 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S & Wingreen NS Cell shape can mediate the spatial organization of the bacterial cytoskeleton. Biophys J 104, 541–552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tropini C et al. Principles of Bacterial Cell-Size Determination Revealed by Cell-Wall Synthesis Perturbations. Cell Reports 9, 1520–1527 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi H et al. Deep Phenotypic Mapping of Bacterial Cytoskeletal Mutants Reveals Physiological Robustness to Cell Size. Curr Biol 27, 3419–3429.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Colavin A, Shi H & Huang KC RodZ modulates geometric localization of the bacterial actin MreB to regulate cell shape. Nat Comms 9, 1280 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olshausen PV et al. Superresolution Imaging of Dynamic MreB Filaments in B. subtilis—A Multiple-Motor-Driven Transport? Biophys J 105, 1171–1181 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi H, Bratton BP, Gitai Z & Huang KC How to Build a Bacterial Cell: MreB as the Foreman of E. coli Construction. Cell 172, 1294–1305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schirner K & Errington J Influence of heterologous MreB proteins on cell morphology of Bacillus subtilis. Microbiology (Reading, Engl) 155, 3611–3621 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Harris LK, Dye NA & Theriot JA A Caulobacter MreB mutant with irregular cell shape exhibits compensatory widening to maintain a preferred surface area to volume ratio. Mol Microbiol (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bisson-Filho AW et al. FtsZ filament capping by MciZ, a developmental regulator of bacterial division. Proc Natl Acad Sci USA 112, E2130–E2138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Formstone A & Errington J A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol Microbiol 55, 1646–1657 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Vigouroux A, Oldewurtel E, Cui L, Bikard D & van Teeffelen S Tuning dCas9’s ability to block transcription enables robust, noiseless knockdown of bacterial genes. Molecular Systems Biology 14, e7899 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henriques AO, Glaser P, Piggot PJ & Moran CP Jr. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol Microbiol 28, 235–247 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Fraipont C et al. The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a subcomplex in Escherichia coli. Microbiology (Reading, Engl) 157, 251–259 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Taheri-Araghi S et al. Cell-Size Control and Homeostasis in Bacteria. Current Biology 25, 385–391 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng H et al. Interrogating the Escherichia coli cell cycle by cell dimension perturbations. Proc Natl Acad Sci USA 113, 15000–15005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray T, Popham DL & Setlow P Bacillus subtilis cells lacking penicillin-binding protein 1 require increased levels of divalent cations for growth. (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popham DL & Setlow P Phenotypes of Bacillus subtilis mutants lacking multiple class A high-molecular-weight penicillin-binding proteins. J Bacteriol 178, 2079–2085 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emami K et al. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat Microbiol 2, 16253 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grimm JB et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Meth 12, 244–250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oldenbourg R Polarized light microscopy: principles and practice. Cold Spring Harb Protoc 2013, (2013). [DOI] [PubMed] [Google Scholar]
- 40.Probine MC & Preston RD Cell Growth and the Structure and Mechanical Properties of the Wall in Internodal Cells of Nitella opacaI. Wall Structure and Growth. J Exp Bot 12, 261–282 (1961). [Google Scholar]
- 41.Inouï ï S Polarization Microscopy Current protocols in cell biology 1, (John Wiley & Sons, Inc., 2001). [Google Scholar]
- 42.Verwer RW, Beachey EH, Keck W, Stoub AM & Poldermans JE Oriented fragmentation of Escherichia coli sacculi by sonication. J Bacteriol 141, 327–332 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao X, Jericho M, Pink D & Beveridge T Thickness and elasticity of gram-negative murein sacculi measured by atomic force microscopy. J Bacteriol (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bratton BP, Shaevitz JW, Gitai Z & Morgenstein RM MreB polymers and curvature localization are enhanced by RodZ and predict E. coli’s cylindrical uniformity. - PubMed - NCBI. Nat Comms 9, 12510 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurita K, Shin R, Tabei T & Shiomi D Relation between rotation of MreB actin and cell width of Escherichia coli. Genes Cells (2018). doi: 10.1111/gtc.12667 [DOI] [PubMed] [Google Scholar]
- 46.Rojas ER, Huang KC & Theriot JA Homeostatic Cell Growth Is Accomplished Mechanically through Membrane Tension Inhibition of Cell-Wall Synthesis. Cell Syst 5, 578–590.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leaver M & Errington J Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol Microbiol 57, 1196–1209 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Kawai Y, Daniel RA & Errington J Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol Microbiol 71, 1131–1144 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Lai GC, Cho H & Bernhardt TG The mecillinam resistome reveals a role for peptidoglycan endopeptidases in stimulating cell wall synthesis in Escherichia coli. PLoS Genet 13, e1006934 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee TK, Meng K, Shi H & Huang KC Single-molecule imaging reveals modulation of cell wall synthesis dynamics in live bacterial cells. Nat Comms 7, 13170 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agresti A & Coull BA Approximate Is Better than ‘Exact’ for Interval Estimation of Binomial Proportions. The American Statistician 52, 119 (1998). [Google Scholar]
- 52.Ursell T et al. Rapid, precise quantification of bacterial cellular dimensions across a genomic-scale knockout library. BMC Biology 15, 17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grimm JB et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Meth 12, 244–250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tinevez J-Y et al. TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Billaudeau C et al. Contrasting mechanisms of growth in two model rod-shaped bacteria. Nat Comms 8, 15370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kner P, Chhun BB, Griffis ER, Winoto L & Gustafsson MGL Super-resolution video microscopy of live cells by structured illumination. Nat Meth 6, 339–342 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vigouroux A, Oldewurtel E, Cui L, Bikard D & van Teeffelen S Tuning dCas9’s ability to block transcription enables robust, noiseless knockdown of bacterial genes. Molecular Systems Biology 14, e7899 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kall L, Storey JD & Noble WS Non-parametric estimation of posterior error probabilities associated with peptides identified by tandem mass spectrometry. Bioinformatics 24, i42–i48 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets and raw data generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. Raw and proteomic data are available at https://garnerlab.fas.harvard.edu/Dion2019/Raw-MS-data-Dion2019.zip