Abstract
In developing cerebral cortex, intermediate progenitors (IPs) are transit amplifying cells that specifically express Tbr2 (gene: Eomes), a T‐box transcription factor. IPs are derived from radial glia (RG) progenitors, the neural stem cells of developing cortex. In turn, IPs generate glutamatergic projection neurons (PNs) exclusively. IPs are found in ventricular and subventricular zones, where they differentiate as distinct ventricular IP (vIP) and outer IP (oIP) subtypes. Morphologically, IPs have short processes, resembling filopodia or neurites, that transiently contact other cells, most importantly dividing RG cells to mediate Delta‐Notch signaling. Also, IPs secrete a chemokine, Cxcl12, which guides interneuron and microglia migrations and promotes thalamocortical axon growth. In mice, IPs produce clones of 1–12 PNs, sometimes spanning multiple layers. After mitosis, IP daughter cells undergo asymmetric cell death in the majority of instances. In mice, Tbr2 is necessary for PN differentiation and subtype specification, and to repress IP‐genic transcription factors. Tbr2 directly represses Insm1, an IP‐genic transcription factor gene, as well as Pax6, a key activator of Tbr2 transcription. Without Tbr2, abnormal IPs transiently accumulate in elevated numbers. More broadly, Tbr2 regulates the transcriptome by activating or repressing hundreds of direct target genes. Notably, Tbr2 ‘unlocks’ and activates PN‐specific genes, such as Tbr1, by recruiting Jmjd3, a histone H3K27me3 demethylase that removes repressive epigenetic marks placed by polycomb repressive complex 2. IPs have played an important role in the evolution and gyrification of mammalian cerebral cortex, and TBR2 is essential for human brain development.
Keywords: apoptosis, cortex patterning, cortical development, Eomes, intermediate progenitors, neurogenesis, Tbr2
Introduction
Pioneers such as His and Cajal observed that in most areas of developing vertebrate brain, progenitor cells divide predominantly at the ventricular surface of the ventricular zone (VZ), and fewer cells divide at non‐surface locations (reviewed in Smart, 1973; z, 2006). In developing cerebral cortex, non‐surface divisions are most abundant in the subventricular zone (SVZ) but also occur in the VZ (Boulder Committee, 1970; Smart, 1973).
The significance of non‐surface divisions was unknown for many years. Quantitatively, Smart (1973) reported that non‐surface mitoses account for up to 20% of all divisions in some cortical areas between E12 and E16. Differences in the orientation of the anaphase plate during mitosis were also noted: the plane of division is usually vertical (perpendicular to the ventricular surface) for surface mitoses but horizontal for non‐surface mitoses. Smart (1973) proposed that the non‐surface mitoses produce neurons. Separately, Takahashi et al. (1995) found that non‐surface dividing progenitors accounted for up to 35% of total mitotic activity, and formed a ‘secondary proliferative population’ attributed to gliogenesis.
More recently, analyses of molecular expression patterns provided evidence that SVZ progenitor cells produce projection neurons (PNs). The specific expression of genes such as Svet1/Unc5d and Cux2 in the SVZ and in upper‐layer PNs, linked the SVZ to upper‐layer genesis (Tarabykin et al. 2001; Zimmer et al. 2004).
Definitive characterization of IPs as committed neurogenic progenitors came from studies using time‐lapse microscopy of cortical slice cultures. Multiple groups reported that non‐surface divisions produce cortical PNs during early (Haubensak et al. 2004), middle (Miyata et al. 2004), and late (Noctor et al. 2004) neurogenesis. Haubensak et al. (2004) named the non‐surface dividing cells ‘basal progenitors’ due to their location away from the ventricular (apical) surface of the neuroepithelium. Noctor et al. (2004) called them ‘intermediate progenitors’ for their lineage position, intermediate between radial glia (RG) progenitors and postmitotic PNs.
At about the same time, Tbr2 was demonstrated as a specific marker of IPs (Englund et al. 2005). During the earliest stages of neurogenesis, Tbr2 is also expressed transiently in postmitotic Cajal–Retzius and subplate neurons, but not after embryonic day (E) 12.5 in mice (Englund et al. 2005). Interestingly, Tbr2 is in the same subfamily of T‐box transcription factors as Tbr1, which is specifically expressed in postmitotic cortical PNs (Hevner et al. 2001). Thus, Tbr2 and Tbr1 are expressed sequentially in the cortical PN lineage.
In terms of stem cell biology, IPs can be considered a cortex‐specific form of ‘transit amplifying cells’, defined as ‘a class of cells, with a finite life span, that arise from stem cells, proliferate and then differentiate’ (Tajbakhsh, 2009).
Two IP subtypes in mice
Unbiased single‐cell transcriptome analysis of embryonic day (E) 14.5 mouse VZ/SVZ revealed four cell types: RG cells, two subtypes of IPs, and new postmitotic PNs (Kawaguchi et al. 2008). Although both IP subtypes expressed Tbr2, Afap1, and other general IP markers, one IP subtype was further enriched in neuronal differentiation markers, such as Neurod1 and Mgat5b. Expression patterns of these and other genes, determined by in situ hybridization, showed that the neuronally differentiated IPs were located in the SVZ, whereas less differentiated IPs were mainly in the VZ (Fig. 1).
Figure 1.
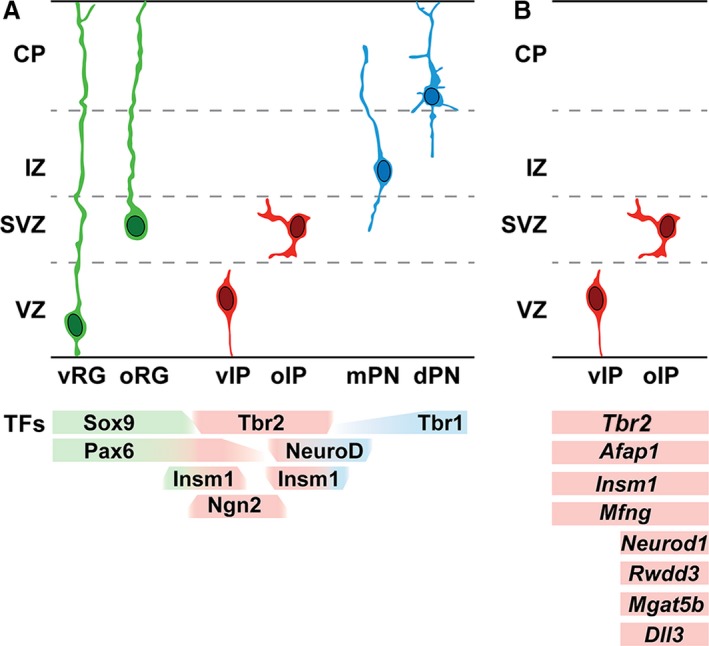
Progenitor cell types and gene expression in the PN lineage of developing mouse cerebral cortex. (A) RG cells (green), IPs (red), and PNs (blue) exhibit distinct morphologies and transcription factor expression (below). Note that Pax6 is expressed in RG cells and some IPs (Englund et al. 2005). (B) IP subtypes are distinguished on the basis of cell morphology and gene expression. Ventricular IPs (vIPs) have short radial morphology, are located in the VZ, and express general markers of IPs, such as Tbr2. Outer IPs (oIPs) have multipolar morphology, are located in the SVZ and express general IP markers, as well as markers of neuronal differentiation (such as Neurod1) and other specialized genes. The gene expression patterns are simplified and represent relative enrichment in the indicated cell types. mPN, migrating PN; dPN, differentiating PN; other abbreviations as in text.
Morphology also distinguishes IP subtypes. Radial bipolar, or ‘pin‐like’, IPs are located in the VZ, and their apical processes initially contact the ventricular surface (Kowalczyk et al. 2009; Ochiai et al. 2009; Borrell et al. 2012). These ventricular IPs (vIPs) correspond to the molecularly less differentiated IP subtype (Kawaguchi et al. 2008) and also to the Tbr2+ subset of ‘short neural precursors’ (Stancik et al. 2010). Indeed, electron microscopy has shown that some short neural precursors contact the ventricle and have apical adherens complexes (Gal et al. 2006). The vIPs usually divide away from the ventricular surface, and rarely at the VZ surface (Gal et al. 2006; Kowalczyk et al. 2009).
Multipolar IPs, located in the SVZ, represent the more neuronally differentiated IP subtype (Kawaguchi et al. 2008) and are designated outer IPs (oIPs) (Fig. 1). These have multiple short processes that extend and retract dynamically in all directions (Noctor et al. 2004; Kowalczyk et al. 2009; Nelson et al. 2013). This mode of oIP activity, called ‘multipolar migration’, results in slow radial or nonradial migration, or no net movement (Tabata & Nakajima, 2003).
Time‐lapse microscopy suggests that vIPs are produced directly from RG cells, then undergo Robo2 signaling‐dependent detachment from the ventricular surface to become oIPs (Borrell et al. 2012). Indeed, vIPs can migrate into the SVZ without mitosis, and convert to multipolar oIP morphology (Tabata et al. 2009). Some vIPs can also generate neurons directly (Gal et al. 2006).
Importantly, RG cells also exhibit two main subtypes distinguished by location in the VZ or SVZ, known as ventricular RG (vRG) and outer RG (oRG) cells (Nowakowski et al. 2016). A specialized type of vRG cell, the truncated RG (tRG), has been observed in developing human cortex (Nowakowski et al. 2016). The tRG maintains apical contact with the ventricular surface, but its basal process does not reach the pial surface.
IPs interact with RG progenitors for Delta‐Notch signaling
Besides migration, another important function of IP processes is to mediate long‐range Delta‐Notch signaling (Fig. 2A). The Delta‐Notch signaling pathway is an evolutionarily conserved system that regulates differentiation of stem cells (Andersson et al. 2011; Pierfelice et al. 2011). Activation of Notch receptors on stem cells is driven by binding of ligands such as Delta, produced by differentiated cells and presented by cell–cell contact.
Figure 2.
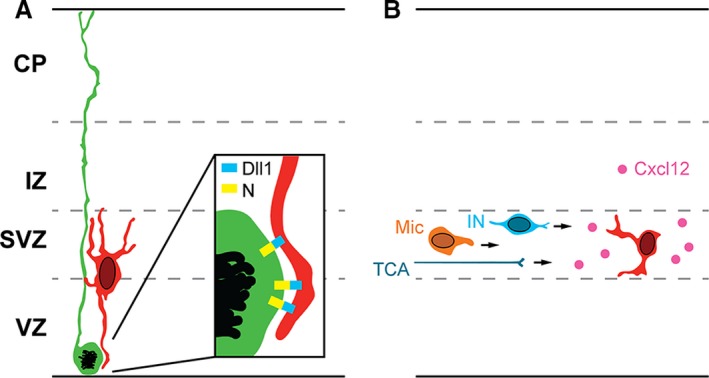
Interactions of IPs with other cell types. (A) IPs express Dll1 on their processes, and activate Notch (N) receptors on RG cells (enlarged in inset). (B) IPs secrete Cxcl12, which guides interneurons (INs) and microglia (Mic) to migrate through the SVZ, and stimulates the growth of thalamocortical axons (TCA).
In developing cortex, IPs produce ligands Delta‐like 1 (Dll1) and Dll3, and RG progenitors express Notch receptors 1–3 (Kawaguchi et al. 2008; Nelson et al. 2013). Activation of Notch instructs RG progenitors to remain as neural stem cells; lack of Notch activation, or pharmacological block, causes RG cells to differentiate rapidly into IPs (Yoon et al. 2008; Nelson et al. 2013). During long‐range signaling, IP processes that contain Dll1 protein extend and contact dividing RG cells (Fig. 2A). Interestingly, similar ‘long‐range’ Delta‐Notch signaling has been reported in Drosophila, where differentiating cells extend filopodia‐like processes that contain Delta (De Joussineau et al. 2003; Rajan et al. 2009; Cohen et al. 2010).
Effective interactions of Delta and Notch require an E3 ubiquitin ligase called mind bomb 1 (Mib1), expressed in the ligand‐producing cell. In cortex, conditional inactivation of Mib1 reduces the efficacy of Dll1 signals from IPs, and causes premature differentiation of RG cells into IPs (Yoon et al. 2008). Thus, IPs are the principal source of active ligands that drive Notch signaling and IPs serve to maintain the balance between RG proliferation and differentiation.
IPs produce glutamatergic projection neurons for all cortical layers
Tbr2+ IPs produce glutamatergic PNs, but not GABAergic interneurons. Glutamatergic PNs can also be produced directly from RG cells, particularly during early stages of cortical neurogenesis, when the cortical preplate is produced (Haubensak et al. 2004; Yoon et al. 2008; Kowalczyk et al. 2009). In layers 2–6, the majority of PNs are produced from IPs (Kowalczyk et al. 2009).
The hypothesis that RG and IP cells produce different types of PNs, such as upper‐layer neurons by IPs, has been tested in a few studies. The possibility that IPs produce upper cortical layers was suggested by the expression patterns of genes such as Svet1/Unc5d and Cux2 (Tarabykin et al. 2001; Zimmer et al. 2004). However, results have shown that proliferating IPs produce PNs for all layers, including the early‐born Cajal–Retzius and subplate neurons of the preplate (Haubensak et al. 2004; Kowalczyk et al. 2009; Vasistha et al. 2015; Mihalas et al. 2016).
The mouse neocortex contains at least 56 molecularly defined subtypes of glutamatergic PNs (Tasic et al. 2018). In a study of five molecularly defined subtypes (Reelin, Tbr1, Ctip2, Satb2, Cux1), IPs were found to produce all of them (Mihalas et al. 2016). Interestingly, genetic lineage tracing in Tbr2‐CreER mice showed that IPs from early stages produced both lower‐ and upper‐layer PNs, whereas IPs from late stages produced upper layers only (Mihalas et al. 2016). These findings supported the ‘progressive restriction’ model of laminar fate specification (Desai & McConnell, 2000). For early IPs to generate upper‐layer PNs, two possibilities were suggested: individual IPs form a mixed population committed to produce different layers, or individual IPs are multipotent for laminar fate, which is determined by contextual factors at the time of neuronal differentiation.
Clonal analysis of IP lineages
Clonal lineage tracing of IPs in Nex/Neurod6‐Cre mice with viral reporter genes indicated that some IPs rapidly differentiate as PNs, whereas other IPs proliferate continuously from one stage to another to produce clones of up to eight PNs (Wu et al. 2005). In another study, larger IP‐derived clones consisting of up to 32 PNs were observed using Tbr2‐Cre mice and electroporated reporters (Vasistha et al. 2015).
To gain higher clonal resolution, we used Tbr2‐CreER mice for mosaic analysis with double markers (MADM) to identify IP daughter cell clones and distinguish daughter clones (Mihalas & Hevner, 2018). With this approach, one IP daughter cell was labeled with green fluorescent protein (GFP), the other with red fluorescent protein (RFP). These studies indicated that most IPs differentiate rapidly to produce just one or two PNs, but a minority of IPs continue proliferating and produce up to 12 neurons, sometimes spanning multiple cortical layers (Mihalas & Hevner, 2018). The results showed that at least some, and possibly all, IPs are indeterminate and multipotent for laminar fate. Rather, laminar fate appears to be determined by local VZ/SVZ factors at the time the new PN is produced.
Cell death is an asymmetric fate choice of many IP daughter cells
In Tbr2‐CreER MADM experiments, the majority of IP‐derived clones (~ 66%) were either GFP+ or RFP+ only, indicating asymmetric cell death. For example, many clones consisted of one GFP+ neuron or one RFP+ neuron (Mihalas & Hevner, 2018). Cell death as a binary fate choice has been reported previously in developing worms and flies (Orgogozo et al. 2002; Arya & White, 2015; Yamaguchi & Miura, 2015).
The high observed rate of IP daughter cell apoptosis accords with previous reports documenting high levels of apoptosis in the SVZ and IZ of embryonic rodent cortex (Blaschke et al. 1996; Thomaidou et al. 1997). In recent preliminary studies, we have observed apoptosis of Tbr2+ IPs by colocalization of Tbr2 with activated caspase‐3 (data not shown), but further studies will be necessary to find the mechanisms that trigger apoptosis. Also, at least some Tbr2+ IPs are phagocytosed by microglia, possibly after apoptosis, or by other mechanisms such as microglial targeting (Cunningham et al. 2013; Arnò et al. 2014). The functions of asymmetric cell death in development are unknown but could potentially include regulation of total neurogenic output, genome quality or PN subtype ratios.
IPs produce Cxcl12 to guide interneurons, microglia, and thalamocortical axons
Interneurons and microglia are important cortical cell types that migrate into the pallium (embryonic cortex) from extracortical progenitor sources. These cells, and thalamocortical axons, are guided into the cortex by IPs which produce a secreted factor, Cxcl12 (Fig. 2B).
Interneurons are produced in the subpallial ganglionic eminences and, beginning around E12.5 in mice, migrate into cortex tangentially through the SVZ and intermediate zone (IZ), and in smaller numbers through the marginal zone (MZ) and subplate (Lim et al. 2018). Interneurons are attracted to the SVZ/IZ by Cxcl12, a C‐X‐C motif chemokine secreted by IPs (Tiveron et al. 2006; Sessa et al. 2010). Interneurons express the Cxcl12 receptors, Cxcr4 and Cxcr7, which activate signaling pathways to regulate interneuron migration (Wang et al. 2011a). Cxcl12 is also produced in the leptomeninges, promoting interneuron migration in the MZ.
Microglia are derived from an extra‐embryonic lineage of erythro‐myeloid progenitors that arise in the yolk sac on E7.5–E9.5 (Casano & Peri, 2015; Gomez Perdiguero et al. 2015; Lopez‐Atalaya et al. 2018). Microglia precursors initially localize in the meninges around E10, and by E14.5 accumulate in the VZ/SVZ, where they ingest cells, including Tbr2+ IPs (Cunningham et al. 2013; Arnò et al. 2014). Like interneurons, microglia are guided by Cxcl12 secreted from IPs, and microglia similarly express the receptors Cxcr4 and Cxcr7 (Arnò et al. 2014). In addition, cortical progenitor cells (IPs and RG cells) produce macrophage migration inhibitory factor (MIF), which stimulates microglia proliferation (Arnò et al. 2014).
Thalamocortical axons enter the cortex from the internal capsule, then grow tangentially through the IZ, along the outer edge of the SVZ. The intracortical growth of thalamic axons has been linked to a growth‐promoting effect of IP‐derived Cxcl12 on thalamic axons that express Cxcr4 (Abe et al. 2015).
By expressing Cxcl12, IPs coordinate PN genesis with thalamocortical innervation, interneuron migration, and microglia proliferation (Fig. 2B).
Tbr2 mutant mouse phenotypes and genetic models
Tbr2 (gene: Eomes) regulates the development of many tissues including trophoblast, mesoderm, T cells, retina, and several brain regions (olfactory bulbs, neocortex, hippocampus, cerebellum, and others). Homozygous inactivation of Tbr2 in all cells from conception causes developmental arrest at the blastocyst (implantation) stage, due to defective trophoblast differentiation and mesoderm movement (Russ et al. 2000).
To study functions of Tbr2 in cerebral cortex development, conditional knockout (cKO) approaches have been used. The first published study used Foxg1‐Cre for Tbr2 cKO in the forebrain (Sessa et al. 2008). That study found that IPs were severely reduced: all cortical layers were thin, and mice died at birth. However, Foxg1‐Cre mice have Foxg1 haploinsufficiency phenotypes, most importantly reduced IPs and PNs (Siegenthaler et al. 2008).
Tbr2 cKO mice have also been generated with other Cre drivers, including Sox1‐Cre (Arnold et al. 2008), Nes11‐Cre (Mihalas et al. 2016), Gfap‐Cre (Arnò et al. 2014), and Emx1‐Cre (Massimino et al. 2018). The Tbr2 cKO mice generated with these other Cre drivers do not die at birth but survive to adulthood, and show other phenotypic and molecular differences from Foxg1‐Cre Tbr2 cKO mice.
Mitotic activity is perturbed in Tbr2 cKO cortex
Given its specific expression in IPs, Tbr2 cKO is expected to alter the differentiation of IPs, and PNs derived from them. In addition, other cortical cell types could potentially be affected by non‐autonomous mechanisms.
In Tbr2 cKO cortex, the abundance of IPs (as detected by non‐surface mitotic activity) and RG cells (surface mitoses) is reported to change, but the magnitude and direction of change differ by genetic model. In Foxg1‐Cre Tbr2 cKO mutants, mitotic divisions were unchanged at the ventricular surface, but were reduced by 75% at non‐surface locations on E15.5 (Sessa et al. 2008). Importantly, Foxg1‐Cre alone reduces IP numbers by 39% on E16.5 (Siegenthaler et al. 2008).
In Sox1‐Cre Tbr2 cKO cortex, surface mitoses were unchanged, whereas non‐surface mitoses were reduced by ~ 25% in E14 Tbr2 cKO cortex (Arnold et al. 2008).
In Nes11‐Cre Tbr2 cKO cortex, dynamic changes in mitotic activity were observed (Mihalas et al. 2016). On E12.5, surface mitoses increased 70%, whereas non‐surface mitoses were not significantly changed. The early increase in surface mitoses might reflect increased RG division or a failure of vIP delamination from the ventricular surface. Since IP delamination is Robo signaling‐dependent (Borrell et al. 2012), and Robo2 is severely reduced in Tbr2 cKO cortex (Elsen et al. 2013), an early failure of IP delamination seems plausible. On E14.5, surface mitoses returned to control levels, whereas non‐surface mitoses were 40% above normal. Furthermore, mitoses occurred ectopically in the IZ and CP of Nes11‐Cre Tbr2 cKO mutants (Fig. 3A). In the absence of Tbr2, the oIPs accumulated due to a defect of differentiation from IPs to PNs.
Figure 3.
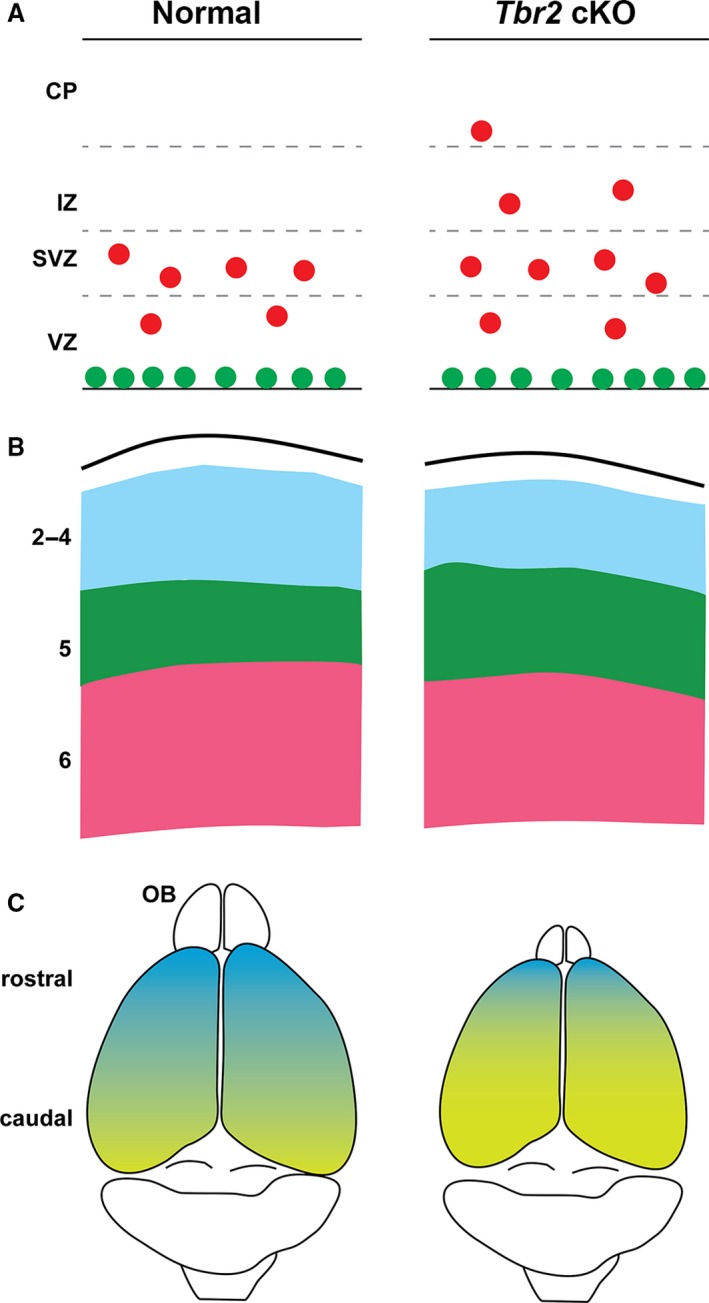
Selected phenotypes of Tbr2 cKO cortex (right panels) compared with normal controls (left panels). (A) On E14.5, non‐surface mitoses (red) are increased in Tbr2 cKO cortex, and occur ectopically in the IZ and CP. The IPs express Pax6 and Insm1 at higher levels than normal, especially outside the VZ. (B) In postnatal Tbr2 cKO cortex, layers 2–4 (blue) are thinner than normal, and layer 5 (green) is thicker than normal. Layer 6 is near normal size. (C) Area patterning is caudalized (yellow) with decreased rostral identity (blue) in Tbr2 cKO cortex. The cortex also has a slightly reduced surface area, and the olfactory bulb (OB) is very small.
Tbr2 regulates the differentiation of cortical layers
In all Tbr2 cKO mice, regardless of the Cre driver used, upper cortical layers (2–4) are the most severely affected. There is a reduced number of upper‐layer neurons (born late in neurogenesis), attributed to accelerated depletion of cortical progenitors (Mihalas et al. 2016). In addition to changes in neuron numbers, upper‐layer gene expression is deficient, as indicated by markers such as Cux1/2 and Satb2 (Arnold et al. 2008; Sessa et al. 2008; Mihalas et al. 2016). In fact, Satb2 is a direct target gene bound by Tbr2, which recruits Jmjd3, an epigenetic factor that removes inhibitory histone marks (Sessa et al. 2017).
Lower layers are also abnormal in Tbr2 cKO cortex. Layer 6 PNs initially show reduced Tbr1 expression on E12.5–E14.5, and the early cortical plate is thin in all models (Sessa et al. 2008; Arnold et al. 2008; Mihalas et al. 2016). However, in contrast to reduced Tbr1, expression of FOG2 (gene: Zfpm2), another marker of layer 6 PNs, is increased in E14.5 Tbr2 cKO cortex (Mihalas et al. 2016). Furthermore, FOG2 was expressed not only in the cortical plate, but also ectopically in the IZ of Tbr2 cKO mutants. The opposing changes in Tbr1 and Zfpm2 expression are directly attributable to Tbr2, which directly activates Tbr1 and directly represses Zfpm2 (Sessa et al. 2017; Elsen et al. 2018).
Why should Tbr2 activate one layer 6 gene but repress another? One possibility is that Tbr2 exerts dual functions to control the timing of layer 6 differentiation, activating some genes to promote differentiation but transiently repressing others to prevent premature layer 6 gene expression. Ultimately, layer 6 differentiates with only slightly reduced thickness in postnatal Tbr2 cKO mice (Arnold et al. 2008; Mihalas et al. 2016).
Layer 5 phenotypes in Tbr2 cKO cortex differ by Cre driver. In Foxg1‐Cre Tbr2 cKO embryos, layer 5 differentiation was reduced, as indicated by Er81/Etv1 and other markers (Sessa et al. 2008). In Sox1‐ or Nes11‐Cre Tbr2 cKO mice, layer 5 differentiated precociously and grew thicker than normal (Arnold et al. 2008; Mihalas et al. 2016). Given the problems with Foxg1‐Cre (Siegenthaler et al. 2008), the layer 5 phenotype observed in this model is an outlier and may be discounted. The phenotypes obtained using other Cre drivers indicate that Tbr2 represses layer 5 differentiation, and layer 5 is thus increased in the absence of Tbr2 (Fig. 3B).
Analysis of epigenetic factors regulated by Tbr2 identified a potential mechanism by which Tbr2 may repress layer 5 differentiation. Tbr2 directly represses Kat6b/querkopf, a histone acetylase gene that is required for layer 5 differentiation (Thomas et al. 2000; Elsen et al. 2018). Increased expression of Kat6b in Tbr2 cKO cortex may bias PNs towards a layer 5 fate (Elsen et al. 2018).
Tbr2 may also regulate general neuronal differentiation. One noteworthy gene repressed by Tbr2 is Zfp423, which encodes a cofactor necessary for neuronal differentiation in response to retinoic acid signaling (Massimino et al. 2018). By repressing Zfp423, Tbr2 may prevent premature neuronal differentiation of IPs, similarly to the mechanism proposed above for Zfpm2/FOG2.
Area patterning is caudalized in Tbr2 cKO mice
During cortical development, Tbr2 is expressed in a gradient such that rostral IPs express higher levels of Tbr2, and caudal IPs express lower levels of Tbr2 (Elsen et al. 2013). In the absence of Tbr2, rostral PN markers such as Bcl6 and Fat3 show decreased expression, whereas caudal PN markers such as Bhlhe22 and Crabp1 show increased and ectopic rostral expression, indicating overall caudalization (Fig. 3C) (Elsen et al. 2013).
The high rostral gradient of Tbr2 mRNA forms in parallel with gradients of Pax6 and Tbr1 expression. Accordingly, Pax6 promotes rostral identity in RG cells (Bishop et al. 2000), and Tbr1 does so in PNs (Bedogni et al. 2010). Thus, the Pax6→Tbr2→Tbr1 cascade transmits rostrocaudal maps from RG cells, which form the ‘protomap’ (Rakic, 1988), to IPs that form an ‘intermediate map’ (Elsen et al. 2013), and then to PNs which form the actual cortical area map in conjunction with innervation patterns. Thus, different sets of genes define rostrocaudal maps at each stage of RG→IP→PN differentiation, and patterning defects arising in RG cells or IPs can be propagated to PNs.
Tbr2 also represses genes associated with non‐cortical cell types, such as Ebf1/2/3 (expressed in striatum) and Th, a dopaminergic enzyme (Kovach et al. 2013; Sessa et al. 2017; Elsen et al. 2018).
Interneurons, microglia, and thalamocortical axons are reduced in Tbr2 cKO cortex
Expression of Cxcl12 by IPs is reduced in Tbr2 cKO cortex, which consequently attracts fewer interneurons (Sessa et al. 2010) and microglia (Arnò et al. 2014). Cxcl12 also stimulates thalamocortical axon growth (Abe et al. 2015). Indeed, Tbr2 cKO mice show disorganized thalamocortical innervation (Arnold et al. 2008).
Tbr2 cKO mice are aggressive and hyperactive
Tbr2 cKO mice generated with Sox1‐Cre are viable and fertile, and were studied behaviorally (Arnold et al. 2008). Aggressiveness, hyperactivity, increased exploratory behavior, and reduced grip strength were salient phenotypes.
Tbr2 directly regulates hundreds of genes, including epigenetic factors
Tbr2 direct target genes (bound and regulated by Tbr2) have been identified by focused analysis of single genes, and by genome‐wide studies of developing rodent neocortex. In one focused study, for example, it was shown that Tbr2 binds and represses Ebf1, a transcription factor gene that is normally expressed in developing striatum (Kovach et al. 2013).
Genome‐wide studies revealed that Tbr2 can activate or repress target genes (Elsen et al. 2013; Sessa et al. 2017). Transcriptome profiling of Tbr2 cKO embryonic cortex has been done on mice produced with Foxg1‐Cre (Sessa et al. 2017) and Nes11‐Cre drivers (Elsen et al. 2013, 2018). Because of problems with Foxg1‐Cre (Siegenthaler et al. 2008), our analyses have used the microarray results from Nes11‐Cre Tbr2 cKO cortex.
To identify direct target genes regulated by Tbr2, differentially expressed genes from transcriptome profiling were compared with Tbr2 binding sites as determined by ChIP (Sessa et al. 2017). Using this ‘chip‐ChIP’ approach, we identified 882 Tbr2‐regulated genes, 502 activated and 380 repressed by Tbr2 (Elsen et al. 2018). Many (18) were epigenetic factor genes, which might have persistent effects on transcription in PNs derived from IPs. Examples include Kdm1a, a histone lysine demethylase gene activated by Tbr2, and Hdac9, a histone deacetylase gene repressed by Tbr2. Remarkably, Tbr2 regulates switching of multiple subunits of the BAF chromatin remodeling complex by activating Smarcd3 (BAF60C), Bcl7a (BAF40A), Bcl11b (BAF110b/Ctip2), and Dpf3 (BAF45c) to drive formation of IP‐ and PN‐specific BAF complex isoforms. BAF subunit switching is critical for neuronal differentiation (Son & Crabtree, 2014). Pax6 and Tbr1 also regulate BAF subunit switching (Elsen et al. 2018).
Importantly, Tbr2 expression in IPs can regulate gene expression in daughter PNs, by two mechanisms: recruiting an epigenetic factor such as Jmjd3 (Sessa et al. 2017), and activating or repressing target genes that encode epigenetic factors. Interestingly, Tbr2/Eomes is itself regulated by epigenetic mechanisms. In IPs, the Tbr2 gene has more activating histone marks (H3K4me3) and fewer repressive marks (H3K27me3) compared with RG cells or PNs (Albert et al. 2017). Thus, IP‐specific Tbr2 expression is controlled in part by active regulation of histone trimethylation marks.
Interestingly, repressive H3K27me3 marks are removed by Jmjd3, a histone demethylase that interacts with Tbr2 to regulate gene expression epigenetically (Sessa et al. 2017). As Tbr2 binds the Tbr2 locus (Elsen et al. 2018), the data suggest that Tbr2 may potentially activate its own transcription in association with Jmjd3. However, the effects of Tbr2 and Jmjd3 on Tbr2 gene expression have not been studied.
Tbr2 represses IP‐genic transcription factor genes Insm1 and Pax6
Along with dysregulation of PN genes, Tbr2 cKO cortex also shows defective repression of genes that promote IP identity. Pax6, an essential activator of Tbr2 gene expression (Quinn et al. 2007; Sansom et al. 2009), is upregulated and expressed ectopically in the IZ and CP of Tbr2 cKO cortex (Mihalas et al. 2016). Insm1, an essential activator of IP genesis (Farkas et al. 2008), is likewise upregulated and ectopically expressed in Tbr2 cKO cortex. Both Pax6 and Insm1 are directly bound and repressed by Tbr2; this feedback is lacking in Tbr2 cKO cortex (Mihalas et al. 2016; Elsen et al. 2018).
The derepression of IP‐genic transcription factor genes explains in part the increased IP numbers and perturbed neuronal differentiation in Tbr2 cKO mice (Mihalas et al. 2016).
Pax6→Tbr2→Tbr1 and the neurogenic transcriptional network
The specific expression of Tbr2 in IPs was initially determined by comparison with Pax6 in RG cells (Götz et al. 1998) and Tbr1 in postmitotic PNs (Hevner et al. 2001). The sequential expression of Pax6→Tbr2→Tbr1 was linked to differentiation from RG→IP→PN (Englund et al. 2005). We further speculated that Pax6→Tbr2→Tbr1 might form a transcription factor cascade (Hevner, 2006). Subsequent research has confirmed this core cascade, and added features of feedback and cross‐regulation with other key transcription factors.
In the transcription factor network around Tbr2 (Fig. 4), Pax6 directly binds and activates Tbr2 (Sansom et al. 2009), and negatively autoregulates its own transcription (Manuel et al. 2007). In turn, Tbr2 directly activates Tbr1, in part by recruiting Jmjd3 for chromatin derepression (Sessa et al. 2017; Elsen et al. 2018). In addition, Tbr2 represses Pax6 in a negative feedback loop (Elsen et al. 2018). The repression of Pax6 by Tbr2 explains why Pax6 protein is increased and ectopically expressed in Tbr2 cKO cortex (Mihalas et al. 2016). Of note, Pax6 mRNA was significantly upregulated in microarray experiments that used Nes11‐Cre, but was not significantly changed in microarray experiments using Foxg1‐Cre for Tbr2 cKO (Sessa et al. 2017). This molecular difference adds to phenotypic differences between Foxg1‐Cre (Sessa et al. 2008) and other Cre drivers for Tbr2 cKO (Arnold et al. 2008; Mihalas et al. 2016). ChIP‐Seq also revealed that Tbr2 binds its own gene, Tbr2/Eomes, presumably for autoregulation, potentially self‐activating in concert with Jmjd3 (Elsen et al. 2018).
Figure 4.
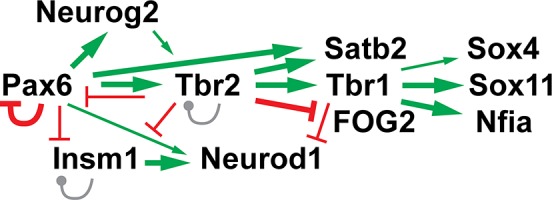
The transcription factor network around Tbr2 in developing neocortex. Green arrows and red bars indicate that the upstream transcription factor directly binds and activates, or represses (respectively) the target transcription factor gene. Gray lines show that Insm1 and Tbr2 bind to their own loci, with unknown effects on autoregulation. Thickness of lines indicates relative strength of the regulatory effects. See text for additional details.
Other key transcription factors linked to the Pax6→Tbr2→Tbr1 cascade include Neurogenin2 (gene: Neurog2), Insm1, and NeuroD (gene: Neurod1) (Fig. 4). The Neurog2 gene is directly activated by Pax6 (Scardigli et al. 2003; Elsen et al. 2018), and Neurogenin2 in turn directly activates Tbr2 (Ochiai et al. 2009; Kovach et al. 2013). Furthermore, Neurogenin2 and Tbr2 binding sites are often adjacent, and they may coordinately regulate gene expression (Sessa et al. 2017). Neurogenin2 also binds and activates Dll1 expression (Castro et al. 2006). Neurod1 is activated by Pax6 and repressed by Tbr1.
Insm1 is a crucial IP‐genic transcription factor (Farkas et al. 2008). The Insm1 gene is directly repressed by Pax6 and Tbr2 (Mihalas et al. 2016; Elsen et al. 2018).
In sum, the Pax6→Tbr2→Tbr1 cascade and other networked transcription factors activate genes required for RG→IP→PN differentiation, and repress genes required for previous stages and alternative pathways of differentiation. This transcription factor cascade is part of a series of overlapping waves of gene expression during PN differentiation (Telley et al. 2016).
TBR2 and IPs are important in evolution, gyrification, and human development
Studies of fetal human, non‐human primate, ferret, and other larger mammals have found that IPs are abundant, and Tbr2 is specifically expressed in IPs, similarly as in small rodents (Fietz et al. 2010; Bakken et al. 2016; Nowakowski et al. 2017). Histologically, the developing cortex of such species, especially primates, displays an expanded SVZ (often divided into inner and outer SVZ by tangentially growing axons) that is rich not only in oIPs but also in abundant oRG cells. In fact, oRG cells were originally characterized in the outer SVZ of humans (Hansen et al. 2010), although they are also present in smaller numbers in small mammals, including mice (Wang et al. 2011b).
In gyrencephalic species, oIPs and oRG cells are most abundant beneath growing gyri, and experimental interference with Tbr2 expression impairs cortical folding (de Juan Romero et al. 2015; Toda et al. 2016). Interestingly, Tbr2 interference in ferrets decreased the abundance of oRG as well as oIP cells, and caused premature neuronal differentiation in the SVZ (Toda et al. 2016), supporting the hypothesis that localized proliferation and migration of oRG and oIP progenitors together drive initial steps in gyrification (Kriegstein et al. 2006; Borrell, 2018).
Outside mammals, Tbr2+ IPs have also been described in the SVZ of cortex equivalent brain structures in diverse vertebrates including lizard, turtle, chicken, dove, and frog (Martínez‐Cerdeño et al. 2016; Nomura et al. 2016; Moreno & González, 2017). Thus, specific features of mammalian neocortex, such as six layers and gyrification, arose much later in evolution than did IPs as a cell type. Evolutionary changes in the balance of direct neurogenesis (from RG cells) and indirect neurogenesis (from IP cells) have been linked to changes in Robo and Dll1 signaling activity, with Robo driving direct neurogenesis and Dll1 driving indirect neurogenesis, i.e. IP genesis (Cárdenas et al. 2018). Interestingly, Robo2 mRNA is highly expressed in IPs, and Tbr2 binds and activates the Robo2 gene (Elsen et al. 2013, 2018; Sessa et al. 2017).
TBR2 is critically important in human brain development. Mutations that perturb the expression of TBR2 cause a severe neurodevelopmental syndrome with microcephaly, severe motor and cognitive delay, hypotonia, polymicrogyria, callosal agenesis, and cerebellar hypoplasia (Baala et al. 2007). As the name implies, polymicrogyria (too many, too small gyri) generally indicates excessive folding, which would seem paradoxical if Tbr2 is necessary to drive gyrification. However, the diagnosis of polymicrogyria in patients with Tbr2 deficiency was based on neuroimaging only, without histologic characterization (Baala et al. 2007). The latter is important because polymicrogyria is used to describe diverse cortical histologies, ranging from unlayered (disorganized) to four‐ or six‐layered, and gyral histology is abnormal in all cases (Juric‐Sekhar & Hevner, 2019). So, the cortical malformation in patients with Tbr2 deficiency can only be described as abnormal gyrification, in the context of overall reduced neurogenesis (microcephaly). Comparisons to animal models must await more detailed studies, such as autopsy neuropathology.
Although much has been learned about IPs and Tbr2 in cortical development, many important questions remain. Why, and by what mechanism, do more than half of IP daughter cells die during normal development? What factors drive the proliferation of some IPs? What factors repress Tbr2 expression upon exit from the cell cycle? At what point in evolution did IPs first become important in brain development? In regenerative medicine, is progression through an IP stage necessary for optimal cortical neuron function, or can neurons produced directly from other sources, such as glial cells, serve equally well? These and other questions continue to stimulate research in this area.
Author contributions
R.F.H. conceived and wrote this review article.
Acknowledgements
This work was supported by the National Institutes of Health [NS092339 and NS085081 to R.F.H.]. The author has no conflicts of interest to declare.
References
- Abe P, Molnár Z, Tzeng YS, et al. (2015) Intermediate progenitors facilitate intracortical progression of thalamocortical axons and interneurons through CXCL12 chemokine signaling. J Neurosci 35, 13053–13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M, Kalebic N, Florio M, et al. (2017) Epigenome profiling and editing of neocortical progenitor cells during development. EMBO J 36, 2642–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson ER, Sandberg R, Lendahl U (2011) Notch signaling: simplicity in design, versatility in function. Development 138, 3593–3612. [DOI] [PubMed] [Google Scholar]
- Arnò B, Grassivaro F, Rossi C, et al. (2014) Neural progenitor cells orchestrate microglia migration and positioning into the developing cortex. Nat Commun 5, 5611. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Huang GJ, Cheung AF, et al. (2008) The T‐box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev 22, 2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya R, White K (2015) Cell death in development: signaling pathways and core mechanisms. Semin Cell Dev Biol 39, 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baala L, Briault S, Etchevers HC, et al. (2007) Homozygous silencing of T‐box transcription factor EOMES leads to microcephaly with polymicrogyria and corpus callosum agenesis. Nat Genet 39, 454–456. [DOI] [PubMed] [Google Scholar]
- Bakken TE, Miller JA, Ding SL, et al. (2016) A comprehensive transcriptional map of primate brain development. Nature 535, 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedogni F, Hodge RD, Elsen GE, et al. (2010) Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci U S A 107, 13129–13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KM, Goudreau G, O'Leary DD (2000) Regulation of area identity in the mammalian neocortex by Emx2 and Pax6. Science 288, 344–349. [DOI] [PubMed] [Google Scholar]
- Blaschke AJ, Staley K, Chun J (1996) Widespread programmed cell death in proliferative and postmitotic regions of the fetal cerebral cortex. Development 122, 1165–1174. [DOI] [PubMed] [Google Scholar]
- Borrell V (2018) How cells fold the cerebral cortex. J Neurosci 38, 776–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell V, Cárdenas A, Ciceri G, et al. (2012) Slit/Robo signaling modulates the proliferation of central nervous system progenitors. Neuron 76, 338–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulder Committee (1970) Embryonic vertebrate central nervous system: Revised terminology. Anat Rec 166, 257–261. [DOI] [PubMed] [Google Scholar]
- Cárdenas A, Villalba A, de Juan Romero C, et al. (2018) Evolution of cortical neurogenesis in amniotes controlled by Robo signaling levels. Cell 174, 590–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casano AM, Peri F (2015) Microglia: multitasking specialists of the brain. Dev Cell 32, 469–477. [DOI] [PubMed] [Google Scholar]
- Castro DS, Skowronska‐Krawczyk D, Armant O, et al. (2006) Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell 11, 831–844. [DOI] [PubMed] [Google Scholar]
- Cohen M, Georgiou M, Stevenson NL, et al. (2010) Dynamic filopodia transmit intermittent Delta‐Notch signaling to drive pattern refinement during lateral inhibition. Dev Cell 19, 78–89. [DOI] [PubMed] [Google Scholar]
- Cunningham CL, Martínez‐Cerdeño V, Noctor SC (2013) Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J Neurosci 33, 4216–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Joussineau C, Soulé J, Martin M, et al. (2003) Delta‐promoted filopodia mediate long‐range lateral inhibition in Drosophila . Nature 426, 555–559. [DOI] [PubMed] [Google Scholar]
- Desai AR, McConnell SK (2000) Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development 127, 2863–2872. [DOI] [PubMed] [Google Scholar]
- Elsen GE, Hodge RD, Bedogni F, et al. (2013) The protomap is propagated to cortical plate neurons through an Eomes‐dependent intermediate map. Proc Natl Acad Sci U S A 110, 4081–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsen GE, Bedogni F, Hodge RD, et al. (2018) The epigenetic factor landscape of developing neocortex is regulated by transcription factors Pax6→Tbr2→Tbr1. Front Neurosci 12, 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, et al. (2005) Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci 25, 247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas LM, Haffner C, Giger T, et al. (2008) Insulinoma‐associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron 60, 40–55. [DOI] [PubMed] [Google Scholar]
- Fietz SA, Kelava I, Vogt J, et al. (2010) OSVZ progenitors of human and ferret neocortex are epithelial‐like and expand by integrin signaling. Nat Neurosci 13, 690–699. [DOI] [PubMed] [Google Scholar]
- Gal JS, Morozov YM, Ayoub AE, et al. (2006) Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci 26, 1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Perdiguero E, Klapproth K, Schulz C, et al. (2015) Tissue‐resident macrophages originate from yolk‐sac‐derived erythro‐myeloid progenitors. Nature 518, 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz M, Stoykova A, Gruss P (1998) Pax6 controls radial glia differentiation in the cerebral cortex. Neuron 21, 1031–1044. [DOI] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PR, et al. (2010) Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, et al. (2004) Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci U S A 101, 3196–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF (2006) From radial glia to pyramidal‐projection neuron: transcription factor cascades in cerebral cortex development. Mol Neurobiol 33, 33–50. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, et al. (2001) Tbr1 regulates differentiation of the preplate and layer 6. Neuron 29, 353–366. [DOI] [PubMed] [Google Scholar]
- de Juan Romero C, Bruder C, Tomasello U, et al. (2015) Discrete domains of gene expression in germinal layers distinguish the development of gyrencephaly. EMBO J 34, 1859–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric‐Sekhar G, Hevner RF (2019) Malformations of cerebral cortex development: molecules and mechanisms. Annu Rev Pathol Mech Dis 14, 291–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi A, Ikawa T, Kasukawa T, et al. (2008) Single‐cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development 135, 3113–3124. [DOI] [PubMed] [Google Scholar]
- Kovach C, Dixit R, Li S, et al. (2013) Neurog2 simultaneously activates and represses alternative gene expression programs in the developing neocortex. Cereb Cortex 23, 1884–1900. [DOI] [PubMed] [Google Scholar]
- Kowalczyk T, Pontious A, Englund C, et al. (2009) Intermediate neuronal progenitors (basal progenitors) produce pyramidal‐projection neurons for all layers of cerebral cortex. Cereb Cortex 19, 2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Noctor S, Martínez‐Cerdeño V (2006) Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci 7, 883–890. [DOI] [PubMed] [Google Scholar]
- Lim L, Mi D, Llorca A, et al. (2018) Development and functional diversification of cortical interneurons. Neuron 100, 294–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Atalaya JP, Askew KE, Sierra A, et al. (2018) Development and maintenance of the brain's immune toolkit: Microglia and non‐parenchymal brain macrophages. Dev Neurobiol 78, 561–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel M, Georgala PA, Carr CB, et al. (2007) Controlled overexpression of Pax6 in vivo negatively autoregulates the Pax6 locus, causing cell‐autonomous defects of late cortical progenitor proliferation with little effect on cortical arealization. Development 134, 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐Cerdeño V, Cunningham CL, Camacho J, et al. (2016) Evolutionary origin of Tbr2‐expressing precursor cells and the subventricular zone in the developing cortex. J Comp Neurol 524, 433–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massimino L, Flores‐Garcia L, Di Stefano B, et al. (2018) TBR2 antagonizes retinoic acid dependent neuronal differentiation by repressing Zfp423 during corticogenesis. Dev Biol 434, 231–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalas AB, Hevner RF (2018) Clonal analysis reveals laminar fate multipotency and daughter cell apoptosis of mouse cortical intermediate progenitors. Development 145, 10.1242/dev.164335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalas AB, Elsen GE, Bedogni F, et al. (2016) Intermediate progenitor cohorts differentially generate cortical layers and require Tbr2 for timely acquisition of neuronal subtype identity. Cell Rep 16, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, et al. (2004) Asymmetric production of surface‐dividing and non‐surface‐dividing cortical progenitor cells. Development 131, 3133–3145. [DOI] [PubMed] [Google Scholar]
- Moreno N, González A (2017) Pattern of neurogenesis and identification of neuronal progenitor subtypes during pallial development in Xenopus laevis . Front Neuroanat 11, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BR, Hodge RD, Bedogni F, et al. (2013) Dynamic interactions between intermediate neurogenic progenitors and radial glia in embryonic mouse neocortex: potential role in Dll1‐Notch signaling. J Neurosci 33, 9122–9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martínez‐Cerdeño V, Ivic L, et al. (2004) Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci 7, 136–144. [DOI] [PubMed] [Google Scholar]
- Nomura T, Ohtaka‐Maruyama C, Yamashita W, et al. (2016) The evolution of basal progenitors in the developing non‐mammalian brain. Development 143, 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Pollen AA, Sandoval‐Espinosa C, et al. (2016) Transformation of the radial glia scaffold demarcates two stages of human cerebral cortex development. Neuron 91, 1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Bhaduri A, Pollen AA, et al. (2017) Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai W, Nakatani S, Takahara T, et al. (2009) Periventricular notch activation and asymmetric Ngn2 and Tbr2 expression in pair‐generated neocortical daughter cells. Mol Cell Neurosci 40, 225–233. [DOI] [PubMed] [Google Scholar]
- Orgogozo V, Schweisguth F, Bellaïche Y (2002) Binary cell death decision regulated by unequal partitioning of Numb at mitosis. Development 129, 4677–4684. [DOI] [PubMed] [Google Scholar]
- Pierfelice T, Alberi L, Gaiano N (2011) Notch in the vertebrate nervous system: an old dog with new tricks. Neuron 69, 840–855. [DOI] [PubMed] [Google Scholar]
- Quinn JC, Molinek M, Martynoga BS, et al. (2007) Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Dev Biol 302, 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan A, Tien AC, Haueter CM, et al. (2009) The Arp2/3 complex and WASp are required for apical trafficking of Delta into microvilli during cell fate specification of sensory organ precursors. Nat Cell Biol 11, 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P (1988) Specification of cerebral cortical areas. Science 241, 170–176. [DOI] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, et al. (2000) Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 404, 95–99. [DOI] [PubMed] [Google Scholar]
- Sansom SN, Griffiths DS, Faedo A, et al. (2009) The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self‐renewal and neurogenesis. PLoS Genet 5, e1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scardigli R, Bäumer N, Gruss P, et al. (2003) Direct and concentration‐dependent regulation of the proneural gene Neurogenin2 by Pax6. Development 130, 3269–3281. [DOI] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, et al. (2008) Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron 60, 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Colasante G, et al. (2010) Tbr2‐positive intermediate (basal) neuronal progenitors safeguard cerebral cortex expansion by controlling amplification of pallial glutamatergic neurons and attraction of subpallial GABAergic interneurons. Genes Dev 24, 1816–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Ciabatti E, Drechsel D, et al. (2017) The Tbr2 molecular network controls cortical neuronal differentiation through complementary genetic and epigenetic pathways. Cereb Cortex 27, 3378–3396. [DOI] [PubMed] [Google Scholar]
- Siegenthaler JA, Tremper‐Wells BA, Miller MW (2008) Foxg1 haploinsufficiency reduces the population of cortical intermediate progenitor cells: effect of increased p21 expression. Cereb Cortex 18, 1865–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart IH (1973) Proliferative characteristics of the ependymal layer during the early development of the mouse neocortex: a pilot study based on recording the number, location and plane of cleavage of mitotic figures. J Anat 116, 67–91. [PMC free article] [PubMed] [Google Scholar]
- Son EY, Crabtree GR (2014) The role of BAF (mSWI/SNF) complexes in mammalian neural development. Am J Med Genet C Semin Med Genet 166C, 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancik EK, Navarro‐Quiroga I, Sellke R, et al. (2010) Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. J Neurosci 30, 7028–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata H, Nakajima K (2003) Multipolar migration: the third mode of radial neuronal migration in the developing cerebral cortex. J Neurosci 23, 9996–10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata H, Kanatani S, Nakajima K (2009) Differences of migratory behavior between direct progeny of apical progenitors and basal progenitors in the developing cerebral cortex. Cereb Cortex 19, 2092–2105. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S (2009) Stem cell: what's in a name? Nature Reports Stem Cells. https://www.nature.com/articles/stemcells.2009.90 [Google Scholar]
- Takahashi T, Nowakowski RS, Caviness VS Jr (1995) Early ontogeny of the secondary proliferative population of the embryonic murine cerebral wall. J Neurosci 15, 6058–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarabykin V, Stoykova A, Usman N, et al. (2001) Cortical upper layer neurons derive from the subventricular zone as indicated by Svet1 gene expression. Development 128, 1983–1993. [DOI] [PubMed] [Google Scholar]
- Tasic B, Yao Z, Graybuck LT, et al. (2018) Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telley L, Govindan S, Prados J, et al. (2016) Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science 351, 1443–1446. [DOI] [PubMed] [Google Scholar]
- Thomaidou D, Mione MC, Cavanagh JF, et al. (1997) Apoptosis and its relation to the cell cycle in the developing cerebral cortex. J Neurosci 17, 1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Voss AK, Chowdhury K, et al. (2000) Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development 127, 2537–2548. [DOI] [PubMed] [Google Scholar]
- Tiveron MC, Rossel M, Moepps B, et al. (2006) Molecular interaction between projection neuron precursors and invading interneurons via stromal‐derived factor 1 (CXCL12)/CXCR4 signaling in the cortical subventricular zone/intermediate zone. J Neurosci 26, 13273–13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Shinmyo Y, Dinh Duong TA, et al. (2016) An essential role of SVZ progenitors in cortical folding in gyrencephalic mammals. Sci Rep 6, 29578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasistha NA, García‐Moreno F, Arora S, et al. (2015) Cortical and clonal contribution of Tbr2 expressing progenitors in the developing mouse brain. Cereb Cortex 25, 3290–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li G, Stanco A, et al. (2011a) CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron 69, 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tsai JW, LaMonica B, et al. (2011b) A new subtype of progenitor cell in the mouse embryonic neocortex. Nat Neurosci 14, 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SX, Goebbels S, Nakamura K, et al. (2005) Pyramidal neurons of upper cortical layers generated by NEX‐positive progenitor cells in the subventricular zone. Proc Natl Acad Sci U S A 102, 17172–17177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Miura M (2015) Programmed cell death in neurodevelopment. Dev Cell 32, 478–490. [DOI] [PubMed] [Google Scholar]
- Yoon KJ, Koo BK, Im SK, et al. (2008) Mind bomb 1‐expressing intermediate progenitors generate notch signaling to maintain radial glial cells. Neuron 58, 519–531. [DOI] [PubMed] [Google Scholar]
- Zimmer C, Tiveron MC, Bodmer R, et al. (2004) Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb Cortex 14, 1408–1420. [DOI] [PubMed] [Google Scholar]


