Abstract
Objective:
To prospectively validate a previously discovered transcriptomic biomarker consisting of 63 blood leukocyte gene expression (S63) values to discriminate between trauma patients who rapidly recover and those with prolonged hospital stays who would benefit from early biological interventions.
Background:
Many severe trauma patients are successfully resuscitated but have complicated clinical trajectories leading to long-term functional, physical, and cognitive deficiencies. Identifying those trauma patients early would improve treatment plans and resource allocation. Unfortunately, current clinical scores and biomarkers used in trauma clinical trials have typically lacked adequate predictive ability.
Methods:
An independent, prospective, observational cohort study was performed involving 127 trauma subjects. The prospective cohort included patients admitted between October 2013 and August 2016 at 2 United States Level-1 trauma centers. An additional secondary analysis was performed using the Activation of Coagulation and Inflammation in Trauma (ACIT2) database of 26 trauma patients.
Results:
The S63 transcriptomic metric (AUC 0.80) outperformed clinical markers and plasma interleukin-6 for prospectively predicting trauma patients who require intensive care unit stays longer than 5 days with ongoing organ dysfunction. The same metric applied to an existing dataset (ACIT2) was similarly effective (AUC 0.85) at predicting multiorgan failure.
Conclusions:
A single transcriptomic metric of blood leukocyte gene expression can be used in blunt trauma cohorts at 24 hours to distinguish patients who rapidly recover from those with complicated clinical trajectories. The transcriptomic metric has been operationalized on an Food and Drug Administration 510(k)-cleared platform otherwise used for cancer diagnostics. This metric is only modestly improved when combined with clinical markers.
Keywords: chronic critical illness, critical care, injury, multiple organ failure, shock
Advances in the delivery of critical care have reduced early mortality in critically ill trauma patients.1 Unfortunately, approximately 20% of these “trauma survivors” do not rapidly recover, but rather develop chronic critical illness, leading to prolonged hospitalization with increased morbidity, high resource utilization, disposition to a nonhome location, and poor long-term survival.2–5 Improved identification of patients likely to have difficult clinical trajectories would assist caregivers in identifying patients who would benefit from current and novel interventional therapies, and allocate resources more effectively. Clinical trials aimed at immunomodulation in trauma have universally failed in part due to the inability to adequately discriminate between patients who are most likely to have complicated clinical trajectories and those who would rapidly recover.6,7 A reliable metric that could eliminate subjects likely to rapidly recover would potentially enrich populations for those who may benefit from clinical intervention. Such a prognostic metric could identify at-risk patients and allow targeted clinical interventions such as early mobilization, physical therapy, lung-protective ventilation, and high vigilance for organ dysfunction and infection. Such a metric could also be used for clinical trials targeted at high-risk populations.
Progress in high-throughput technologies has encouraged the development of genomic prognostic tools in trauma opening the door for a “precision medicine” approach to the management of these patients.8–12 In sepsis, there have been multiple efforts to identify transcriptomic metrics that can differentiate sepsis from noninfective critical illness in adults10,13,14 and in children.15–18 Retrospective analyses usually of public databases have proven, at least in principle, that such an approach may be prognostic in a critically ill population.19,20 Prior work has shown that a “genomic storm” occurs following severe blunt trauma, indicating a global reprioritization of over 80% of leukocyte cellular functions and pathways.21 This identification of a common genomic change prompted further study into potential prognostic applications of transcriptomic metrics following trauma.
We previously described a transcriptomic metric based on the expression pattern of 63 blood leukocyte genes (S63) from 163 severely injured blunt trauma patients enrolled at 6 institutions between November 2003 and January 2005 and compared with 35 age and gender matched, healthy control subjects.11 The S63 transcriptomic metric at 24hours post-trauma significantly discriminated between trauma patients with uncomplicated versus complicated clinical trajectories over 28 days. We also operationalized this metric on an Food and Drug Administration (FDA) 510(k)-cleared multiplex genomic platform that is currently used for breast cancer prognostics (NanoString PAM50, Prosigna).22 In this report, we conducted a prospective clinical study to directly test this 63 gene metric for its ability to distinguish clinical trajectories and outcomes in an independent cohort of severely injured blunt trauma patients at 2 institutions. We also applied this transcriptomic metric to an additional trauma cohort discoverable in the public domain.
METHODS
Prospective Study Design
A prospective, observational cohort study was conducted between October 2013 and August 2016 at 2 United States Level 1-trauma centers: UFHealth Shands Hospital, Gainesville, Florida, and Harborview Medical Center, Seattle, Washington. The Institutional Review Board of each institution granted approval prior to study initiation, and signed informed consent was obtained from the patient and/or their legally appointed representative. Key aspects of study design are listed here, with additional in-depth methodologic description regarding study sites, subject enrollment, outcomes definitions, and biostatistical analyses provided in S1 Appendix, http://links.lww.com/SLA/B564. The study was prospectively registered with clinicaltrials.gov (NCT 01810328).
Inclusion criteria included patients aged 18-years or older, confirmation of severe blunt traumatic injury with shock defined as systolic blood pressure <90 mm Hg or base deficit of ≥6 meq/L within 60 minutes of arrival. Patients expected to survive less than 48 hours and those with severe traumatic brain injury (TBI; Glasgow Coma Scale <8 and abnormal head computed tomography) were excluded.
Healthy Controls
Thirty-six healthy controls who were age-, race/ethnicity-, and gender-matched to the trauma population were studied. Signed informed consent was obtained prior to blood sampling.
Definition of Outcomes
The primary goal of the prospective trauma study was to validate the S63 transcriptomic metric. The discovery cohort used time to recovery (TTR) to build the predictive model, which was defined as the number of days after injury to resolution of multiple organ dysfunction, without subsequent recurrence (Table 1). Data were discretely divided into 3 groups based on the TTR, as previously described.11 An uncomplicated outcome was defined as a TTR of less than 5 days. In contrast, a complicated outcome was defined as either in-hospital death, a TTR of greater than 14 days, or a TTR of greater than 5 days and discharge to another health care facility with organ dysfunction. Patients with a TTR of between 5 and 14 days were defined as an intermediate outcome. ATTR threshold of 5 or 14 days was selected for clinical reasons. Trauma patients with a TTR <5 days frequently have a complete recovery and are discharged from the ICU for recovery and rehabilitation.5 In contrast, patients who take longer than 14 days to recover generally qualify as having chronic critical illness and demonstrate adverse long-term outcomes.5 For the purpose of predictive modeling, the outcomes were dichotomized so that patients with uncomplicated clinical outcomes were compared with patients with intermediate or complicated outcomes (TTR <5 d vs TTR5 ≥ d), or patients with complicated outcomes were compared with patients with intermediate or uncomplicated outcomes (TTR ≥14 d vs <14 d) (S Table 1, http://links.lww.com/SLA/B564).
TABLE 1.
Parameters for Time to Recovery
| Definition of time to recovery (TTR)* |
| 1st day meeting organ failure recovery criteria in all systems listed below, without any subsequent days with further organ system failure |
| Cardiovascular recovery |
| Mean arterial pressure >60 mm Hg and no inotropic/vasopressor support (dopamine, dobutamine, epinephrine, norepinephrine, phenylephrine, or vasopressin) |
| Hematologic recovery |
| Platelet count >120,000/μL |
| Hepatic recovery |
| Serum bilirubin <3 mg/dL |
| Renal recovery |
| No dialysis and creatinine <1.3 mg/dL |
| Respiratory recovery |
| No mechanical ventilation or PaO2/FiO2 >300 |
Organ injury was derived from a composite of the Denver and Marshall MOF Scores, adding hematologic recovery to the Denver score, and removing the Glasgow Coma Scale (GCS) from the Marshall score. GCS is more subjective and clinician-dependent, and subjects with significant TBI were purposefully excluded from enrollment.
A surviving patient who did not recover by day 28 was assigned a TTR value of 29 days, whereas a patient who died prior to day 28 was assigned a TTR value of 30 days.
Sample Collection and Laboratory Analyses
In the trauma cohort, ethylenediaminetetraacetic acid-anti-coagulated blood samples were collected within 12 hours and at 24 hours following the traumatic insult. Gene expression analysis was conducted on the NanoString nCounter gene expression platform (NanoString Flex) using a custom code set consisting of a 63-gene panel as previously described (S1 Appendix, http://links.lww.com/SLA/B564).11
A single metric was derived from the difference in expression values for the 63 genes (difference from reference; DFR) from age-, race/ethnicity-, and gender-matched healthy subjects using the equation:
where ei is the patient’s expression level for probe set i, Mi is the mean of all controls’ expression of probe set I, and Vi is the variance (squared standard deviation) of all patients’ expression of probe set i. The equation has been modified from the original citation11 in which originally Vi was the control subject and not the trauma patient variance. Division by the patient’s variance is a rescaling that prevents the DFR score from being dominated by genes that are inherently more variable or more highly expressed.9
The 63 genes were also divided into functional groups based on the types of genes and individual transcriptomic metrics were performed from groups of genes encompassing inflammatory and adaptive immunity as well as endothelial activation.
Plasma interleukin (IL)-6 concentrations were determined using the Luminex Magpix (Austin, TX) platform according to the manufacturer’s recommendations. Outcomes were prospectively adjudicated, and the clinical dataset locked before predictive modeling commenced.
Community Analyses
The transcriptomic metric was subsequently evaluated in an additional dataset of blunt trauma patients. This was a secondary analysis of the Activation of Coagulation and Inflammation in Trauma (ACIT2) study (NHS REC: 07/Q0603/29; E-MTAB-5882; https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-5882/), conducted between December 2008 and June 2012, including 6 control subjects and 26 critically ill blunt trauma patients,23 as published in their correction.24
Blood samples were collected on arrival and within 12 hours of injury, and at 24 hours and 72 hours following admission. Transcriptomic analysis was performed on whole blood collected with PAXGene tubes, and gene expression was measured with an Illumina microarray platform. Since the expression of all 63 genes was not available, the transcriptomic metric was constructed using the expression of 58 of the 63 genes (S2 Appendix, http://links.lww.com/SLA/B564). Due to the differing analysis and clinical reporting, outcome variables were dichotomized based on available clinical outcomes used in the original report.
Statistical Analyses
Data are presented as means with standard deviation for continuous variables and are compared using analysis of variance, while those not satisfying normality are presented as medians and quartiles, and are compared using the Kruskal–Wallis test. Categorical variables are presented as frequencies and percentages, and are compared using Fisher exact test. For all multivariate analyses, we report adjusted odds ratios with 95% confidence intervals (95% CI). Area under the receiver operating curve values (AUC) and Hosmer–Lemeshow goodness-of-fit test were used to assess model discrimination and fit. DeLong test was used to compare differences among receiver operating curves. False discovery correction for multiple comparisons was performed using the Benjamini–Hochberg procedure. All analyses were performed using SAS (v.9.4, Cary, NC).
RESULTS
Prospective Validation
A total of 127 subjects were enrolled in the trauma cohort; 46 patients enrolled at the University of Florida Health Shands Hospital and 81 patients at Harborview Medical Center, University of Washington. The overall cohort was predominantly comprised of white (87%) males (69%) with a mean age of 46 years (Table 2). Severity of injury, magnitude of metabolic disturbances, and overall clinical outcomes were similar to that seen in the discovery set.11
TABLE 2.
Trauma Cohort Demographics and Outcomes
| Overall | Uncomplicated | Intermediate | Complicated | P Value | |
|---|---|---|---|---|---|
|
| |||||
| (n = 127) | (n = 34, 26.8%) | (n = 49, 38.6%) | (n = 44, 34.7%) | 3 Groups Overall (K–W or Fisher Exact) | |
| Baseline characteristics | |||||
| Age, meanSD | 46±17 | 41±17 | 44±17 | 51±16 | 0.0383* |
| Male sex (n, %) | 88 (69.3) | 25 (73.5) | 32 (65.3) | 31 (70.5) | 0.7145 |
| Race/ethnicity (n, %) | 0.6433 | ||||
| White | 111 (87.4) | 31 (91.2) | 40 (81.6) | 40 (90.9) | |
| Hispanic | 6 (4.7) | 1 (2.9) | 3 (6.1) | 2 (4.6) | |
| African American | 9 (7.1) | 3 (8.8) | 5 (10.2) | 1 (2.3) | |
| American Indian | 2 (1.6) | 0 (0) | 1 (2.0) | 1 (2.3) | |
| Pacific Islander | 1 (0.8) | 0 (0) | 1 (2.0) | 0 (0) | |
| Asian | 3 (2.4) | 0 (0) | 1 (2.0) | 2 (4.6) | |
| Unknown | 1 (0.8) | 0 (0) | 1 (2.0) | 0 (0) | |
| BMI, median (25th, 75th) | 26.9 (24.5, 30.9) | 26.2 (24.3, 29.8) | 26.9 (24.5, 32.4) | 26.9 (24.5, 32.4) | 0.4969 |
| Number of comorbidities (n, %) | 0.6967 | ||||
| 0 | 45 (35.4) | 12 (35.3) | 16 (32.7) | 17 (38.6) | |
| 1 | 42 (33.1) | 14 (41.2) | 15 (30.6) | 13 (29.6) | |
| ≥2 | 40 (31.5) | 8 (23.5) | 18 (36.7) | 14 (31.8) | |
| APACHE II, median (25th, 75th) | 23 (17, 29) | 18 (13, 26) | 23 (20, 27) | 26 (19.5, 33) | 0.0006* |
| ISS, median (25th, 75th) | 33 (24, 41) | 24 (17, 29) | 34 (27, 43) | 34 (27, 43) | <0.0001 |
| Injury mechanism (n, %) | 0.0498 | ||||
| Fall | 11 (8.6) | 3 (8.8) | 3 (6.1) | 5 (11.4) | |
| Motor vehicle collision | 109 (85.8) | 26 (76.5) | 46 (93.9) | 37 (84.1) | |
| Other | 7 (5.5) | 5 (14.7) | 0 (0) | 2 (4.5) | |
| Total transfusion within 24h, median (25th, 75th) | |||||
| PRBC (units) | 4.0 (1.0, 8.1) | 1.0 (0, 4.7) | 4.0 (1.2, 9.2) | 5.3 (2.3, 9.6) | <0.0001* |
| FFP (units) | 1.4 (0, 3.4) | 0 (0, 1.4) | 1.8 (0, 4.3) | 1.5 (0, 4.7) | <0.0001* |
| Total crystalloid (mL) within 24 h | 8520 (6602, 12,229) | 7558 (6061, 8713) | 8722 (6158, 12,229) | 10419.5 (7395.5, 13,543) | 0.0086* |
| Worst base deficit within 24 h | −7.4 (−11.8, −3.8) | −6.4 (−8.1, −3.7) | −7.0 (−11.4, −1.9) | −10.9 (−13.8, −7) | 0.0008* |
| Highest lactate within 24 h | 4.6 (3.2, 6.2) | 3.9, (2.8, 4.8) | 4.8 (3.4, 5.9) | 5.3 (3.3, 7.1) | 0.0146* |
| Lowest ED SBP (mm Hg) | 85 (68, 99) | 91 (76, 113) | 86 (74, 101) | 74 (63.5, 92.5) | 0.0155* |
| Initial ED SBP (mm Hg) | 118 (98, 137) | 122 (109, 145) | 118 (89, 134) | 110 (98, 134) | 0.1889 |
| ER systolic <90 mm Hg (n, %) | 23 (18.1) | 3 (8.8) | 13 (26.5) | 7 (15.9) | 0.1124 |
| Mechanically ventilated (n, %) | 110 (86.6) | 23 (67.7) | 47 (95.9) | 40 (90.9) | 0.0012 |
| Ventilator-free days (28-d) | 3 (2, 6) | 2, (2, 4) | 4, (2, 6) | 3, (1, 8.5) | 0.0261* |
| MOF (n, %) | 19 (15.0) | 0 (0) | 5 (10.2) | 14 (31.8) | <0.0001* |
| Max. Denver MOF score | 1 (0, 2) | 0 (0, 0) | 1 (0, 2) | 2 (0, 3.5) | <0.0001* |
| Time to recovery (TTR)y (d) | 8 (4, 22) | 3 (2, 4) | 8 (6, 11) | 29 (20, 29) | |
| Cardiovascular recovery | 1 (1, 2) | 1 (1, 1) | 1 (1, 2) | 2 (1, 14) | |
| Hematologic recovery | 3 (1, 6) | 1 (1, 2) | 5 (2, 6) | 5.5 (2.5, 16.5) | |
| Hepatic recovery | 1 (1, 2) | 1 (1, 1) | 1 (1, 1) | 1.5 (1, 3) | |
| Renal recovery | 1 (1, 2) | 1 (1, 1) | 1 (1, 1) | 2 (1, 25) | |
| Respiratory recovery | 4 (1, 11) | 1 (1, 2) | 5 (3, 8) | 14.5 (2, 22) | |
| Infection source (n, %) Pneumonia | 29 (22.8) | 1 (2.9) | 14 (28.6) | 14 (31.8) | 0.0016* |
| Pseudomembranous colitis | 14 (11.0) | 0 (0) | 8 (16.3) | 6 (13.6) | 0.0261* |
| UTI | 9 (7.1) | 0 (0) | 6 (12.2) | 3 (6.8) | 0.0845 |
| Blood stream infection | 3 (2.4) | 0 (0) | 0 (0) | 3 (6.8) | 0.0577 |
| Empyema | 1 (0.8) | 0 (0) | 0 (0) | 1 (2.3) | 0.6142 |
| Other | 5 (3.9) | 1 (2.9) | 0 (0) | 4 (9.1) | 0.0480* |
| Number of nosocomial infections per patient (n, %) | <0.0001* | ||||
| 0 | 80 (63.0) | 32 (94.2) | 25 (51.0) | 23 (52.3) | |
| 1 | 33 (26.0) | 2 (5.9) | 18 (36.7) | 13 (29.6) | |
| ≥2 | 14 (11.0) | 0 (0) | 6 (12.2) | 8 (18.2) | |
| Hospital length of stay (d) | 18 (12, 25) | 12 (5, 17) | 20 (17, 24) | 20 (12.5, 42.5) | <0.0001* |
| ICU length of stay (d) | 8 (4, 15) | 3.5, (2, 6) | 10 (7, 13) | 15 (4, 23.5) | <0.0001* |
| 28-d mortality (n, %) | 6 (4.7) | 0 (0) | 0 (0) | 6 (13.6) | 0.0016* |
| Discharge disposition (n, %) | <0.0001* | ||||
| “Good” disposition | |||||
| Inpatient rehabilitation facility | 22 (17.3) | 5 (14.2) | 11 (22.5) | 6 (13.6) | |
| Home with services | 19 (15.0) | 5 (14.2) | 11 (22.5) | 3 (6.8) | |
| Home | 37 (29.1) | 18 (52.9) | 10 (20.4) | 9 (20.5) | |
| “Poor” disposition | |||||
| SNF | 36 (28.4) | 6 (17.7) | 17 (34.7) | 13 (29.6) | |
| LTAC | 5 (4.0) | 0 (0) | 0 (0) | 5 (11.4) | |
| Death (in-hospital) | 7 (5.5) | 0 (0) | 0 (0) | 7 (15.9) | |
Overall inpatient mortality rate was 5.5% (Table 2). Median ICU and hospital length of stay was eight and 18 days, respectively. Overall, 27% of the trauma cohort was defined as having an uncomplicated recovery, 39% an intermediate recovery, and 35% a complicated recovery.
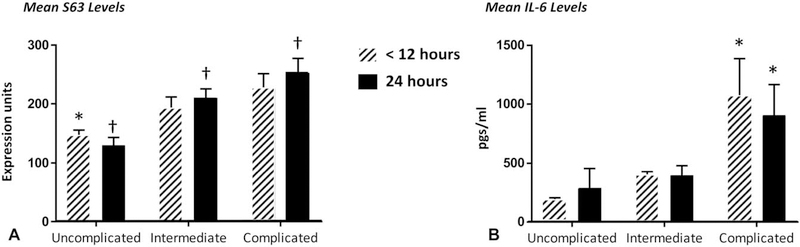
Figure 1 presents the derived transcriptomic metric (Panel A) and plasma IL-6 concentrations (Panel B) obtained at <12 and 24 hours posttrauma in the 3 groups. The magnitude of the disturbances in gene expression and IL-6 concentrations at both <12 and 24 hours increased among the groups of patients based on length of time to recovery. In the uncomplicated clinical group, the transcriptomic metric peaked at <12 hours and declined at 24 hours. In contrast, in patients with either an intermediate or complicated outcome, S63 remained elevated at 24 hours. Correction for false discovery did not alter any significance thresholds.
FIGURE 1.
Mean transcriptomic and plasma IL-6 concentrations in trauma patients with different clinical outcomes. S63 (A) gene expression scores and plasma IL-6 concentrations (B) in patients stratified into uncomplicated, intermediate, and complicated trajectories. S63 (A) scores for the 3 stratified groups were significantly different at 24 hours; †P < 0.05). At <12 h, only the levels of S63 in the uncomplicated group significantly differed from the other 2 groups (*P < 0.05 by ANOVA and Tukey post-hoc comparison). In contrast, IL-6 concentrations at both <12 and 24 h were significantly greater in complicated patients than in the other 2 groups (*P < 0.05).
Table 3 presents the correlations among the 24 hours S63, plasma IL-6 concentrations, and several admission clinical indices. Not unexpectedly, strong positive correlations were seen. The strongest correlations at 24 hours were between S63 and plasma IL-6 concentrations, followed by S63 and clinical indices.
TABLE 3.
Correlations Among Clinical Criteria, Plasma IL-6, and s63
| APACHE II | Denver | Max Denver | IL-6 | S63 DFR | |
|---|---|---|---|---|---|
| ISS | |||||
| Spearman | 0.32451 | −0.1011 | 0.34704 | 0.27484 | 0.38254 |
| P value | 0.0002 | 0.26 | <0.0001 | 0.0022 | <0.0001 |
| APACHE II | |||||
| Spearman | −0.00596 | 0.3863 | 0.32217 | 0.44926 | |
| P value | 0.9472 | <0.0001 | 0.0003 | <0.0001 | |
| Denver | |||||
| Spearman | 0.36375 | 0.26934 | 0.12244 | ||
| P value | <0.0001 | 0.0027 | 0.172 | ||
| Max Denver | |||||
| Spearman | 0.45972 | 0.36996 | |||
| P value | <0.0001 | <0.0001 | |||
| IL-6 | |||||
| Spearman | 0.56546 | ||||
| P value | <0.0001 |
We then examined the ability of these transcriptomic metrics to predict clinical outcomes. The initial selection of the 63 genes was based on a significant (P<0.001) 2-fold difference in expression between subjects with an uncomplicated and complicated clinical outcome.11 Because the discovery cohort used TTR as the clinical outcome, patient outcomes in this validation cohort also used TTR, and outcomes were first dichotomized between groups of subjects with either an uncomplicated or complicated outcome (eliminating the intermediate outcome group). Here, the area under the receiver operating curve (AUC) was 0.81 (P < 0.001), very comparable to the discovery data set.11
However, eliminating the intermediate group negates much of the value of the metric in real-world use. What we really want to know is whether the transcriptomic metric can predict at <12 and 24 hours posttrauma, differences in clinical trajectory among all 3 groups of trauma patients. To achieve this, we dichotomized the patients into 2 groups by combining patients with intermediate outcomes separately with either complicated or uncomplicated outcomes (Table 2).
Using this approach, 3 findings are noteworthy. First, the ability of clinical indices, plasma IL-6, and S63 to discriminate clinical outcomes was markedly better at 24 than <12 hours (Table 4). Second, the S63 transcriptomic metric could better discriminate outcomes than either IL-6 or any of the clinical measures obtained at 24 hours, although in the case of ISS, the improvement was not significant. Using DeLong test, the predictive ability of the S63 metric at 24 hours was significantly better than either plasma IL-6 or APACHE II, but not ISS. Third and finally, the discriminative power of the S63 transcriptomic metric at 24 hours was better at distinguishing patients with a time to recovery <5 days than patients with a time to recovery ≥14 days (Table 4). S63 alone at 24 hours gave an AUC of 0.80 and a 1.014 odds ratio for predicting patients with uncomplicated outcomes compared with patients with either intermediate or complicated outcomes (TTR <5 d vs TTR≥5 d) (Fig. 2A), greater than seen for plasma IL-6, ISS, and APACHE II score. When applied to the prediction of Multiple Organ Dysfunction Syndrome (MODS, defined as Denver multiple organ failure score ≥3), S63 alone had an AUC of 0.78.
TABLE 4.
Area Under Receiver Operating Curve and Odds Ratios for Clinical Indices at 24 h, and Plasma IL-6 and s63 Levels at 12 and 24 h Dichotomizing Trauma Patients Based on Time to Recovery (TTR)
|
Outcome (TTR < 5 d vs ≥ 5 d)
|
12 h
|
24 h
|
||
| Variable | AUC (95% CI) | Odds Ratio (95% CI) | AUC (95% CI) | Odds Ratio (95% CI) |
|
| ||||
| APACHE II | 0.71 (0.60–0.81) | 1.121 (1.053–1.194) | ||
| ISS | 0.77 (0.67–0.87) | 1.092 (1.046–1.141) | ||
| IL-6 | 0.72 (0.62 0.83) | 1.003 (1.001–1.006) | 0.73 (0.64–0.83) | 1.000 (1.000–1.001) |
| S63 Gene DFR | 0.65 (0.54–0.76) | 1.008 (1.002–1.015) | 0.80 (0.70–0.89) | 1.014 (1.007–1.021) |
| Adaptive immunity Gene DFR | 0.62 (0.50–0.73) | 1.013 (1.001–1.025) | 0.77 (0.67–0.87) | 1.027 (1.015–1.039) |
| Endothelial biology Gene DFR | 0.64 (0.53–0.75) | 1.056 (0.999–1.116) | 0.74 (0.64–0.84) | 1.086 (1.015–1.161) |
| Inflammatory responses Gene DFR | 0.73 (0.63–0.82) | 1.127 (1.038–1.223) | 0.70 (0.61 −0.80) | 1.140 (1.022–1.271) |
| S63 DFR + IL-6 | 0.75 (0.65–0.85) | IL-6 1.003 (1.000–1.005) S63 DFR 1.005 (0.998–1.012) |
0.82 (0.73–0.90) | IL-6 1.000 (0.999–1.000) S63 DFR 1.020 (1.011–1.029) |
| S63 DFR + APACHE II + ISS | 0.84 (0.76–0.92) | APACHE II 1.055 (0.981–1.134) ISS 1.062 (1.014–1.113) S63 DFR 1.009 (1.002–1.017) |
||
| S63 DFR + APACHE II + ISS + IL-6 | 0.86 (0.79–0.93) | APACHE II 1.060 (0.979–1.147) ISS 1.057 (1.007–1.110) IL-6 1.000 (0.999–1.000) S63 DFR 1.016 (1.006–1.025) |
||
|
| ||||
|
Outcome (TTR < 14 d vs ≥ 14 d)
|
12 h
|
24 h
|
||
| Variable | AUC (95% CI) | Odds Ratio (95% CI) | AUC (95% CI) | Odds Ratio (95% CI) |
|
| ||||
| APACHE II | 0.66 (0.55–0.76) | 1.094 (1.038–1.152) | ||
| ISS | 0.57 (0.46–0.67) | 1.017 (0.989–1.046) | ||
| IL-6 | 0.68 (0.57–0.78) | 1.001 (1.000–1.002) | 0.68 (0.58–0.78) | 1.000 (1.000–1.001) |
| S63 Gene DFR | 0.59 (0.48–0.70) | 1.003 (1.000–1.006) | 0.66 (0.56–0.76) | 1.005 (1.001–1.008) |
| Adaptive immunity Gene DFR | 0.53 (0.43–0.64) | 1.002 (0.995–1.010) | 0.67 (0.57–0.77) | 1.012 (1.003–1.021) |
| Endothelial biology Gene DFR | 0.58 (0.47–0.68) | 1.011 (0.994–1.028) | 0.65 (0.55–0.75) | 1.033 (1.000–1.068) |
| Inflammatory responses Gene DFR | 0.70 (0.60–0.80) | 1.034 (1.008–1.061) | 0.70 (0.61 −0.80) | 1.053 (1.012–1.095) |
| S63 DFR + IL-6 | 0.68 (0.58–0.79) | IL-6 1.001 (1.000–1.002) S63 DFR 1.001 (0.998–1.005) |
0.69 (0.59–0.79) | IL-6 1.000 (1.000–1.001) S63 DFR 1.005 (1.001–1.008) |
| S63 DFR + APACHE II + ISS | 0.69 (0.59–0.79) | APACHE II 1.076 (1.016–1.140) ISS 0.996 (0.964–1.028) S63 DFR 1.003 (1.000–1.007) |
||
| S63 DFR + APACHE II + ISS + IL-6 | 0.69 (0.59–0.79) | APACHE II 1.061 (0.998–1.128) ISS 0.997 (0.964–1.030) IL-6 1.000 (1.000–1.001) S63 DFR 1.003 (1.000–1.007) |
||
FIGURE 2.
Predictive modeling of clinical outcome. A, Univariate predictive modeling of complicated/intermediate versus uncomplicated outcome for trauma cohort based on values at 24 h. Curves corresponding to ISS, APACHE, IL-6, and S63 models were created with univariate AUC provided in parentheses. B, Multivariate predictive modeling of complicated/intermediate versus uncomplicated outcome for trauma cohort based on values at 24 h. Curves corresponding to multivariate models of S63+IL-6, S63+APACHE+ISS, and S63+APACHE+ISS+IL-6 were created with AUC provided in parentheses. C, S63 Threshold optimization. Here the predictive probability was plotted against the scores for the 2 outcome groups and optimized thresholds identified. D, Predictive modeling of multiple organ dysfunction syndrome versus no multiple organ dysfunction syndrome for community cohort. The community dataset utilized illumina rather than nanostring transcriptomic analysis and reported data on 58 of 63 genes. A curve corresponding to S58 prediction of MODS was created with corresponding AUC in parentheses.
An S63 threshold of 117 U could identify patients who would rapidly recover with a sensitivity of 90% and a specificity of 59% (Fig. 2C). The positive and negative predictive values of the 117 U threshold were 86% and 69%, respectively. Likewise, the corresponding positive and negative likelihood ratios were 2.19 and 0.17, respectively. Meanwhile, a threshold of 212 U yielded a 95% specificity of identifying subjects who would have a TTR ≥5 days, but reduced the sensitivity to 45%. Addition of clinical scores and IL-6 to the S63 measurement only marginally improved the AUC to 0.86 (Fig. 2B).
Community Validation
Raw expression data were obtained for 58 of the 63 genes in 26 critically ill trauma subjects and 6 controls obtained from the available dataset (S2 Appendix, http://links.lww.com/SLA/B564). Presence or absence of MODS was taken from the authors’ own determination. Using 12 and 14 trauma subjects with and without MODS, respectively, the 58 gene metric could discriminate MODS/no MODS with an AUC of 0.85 (Fig. 2D).
Biological Insights
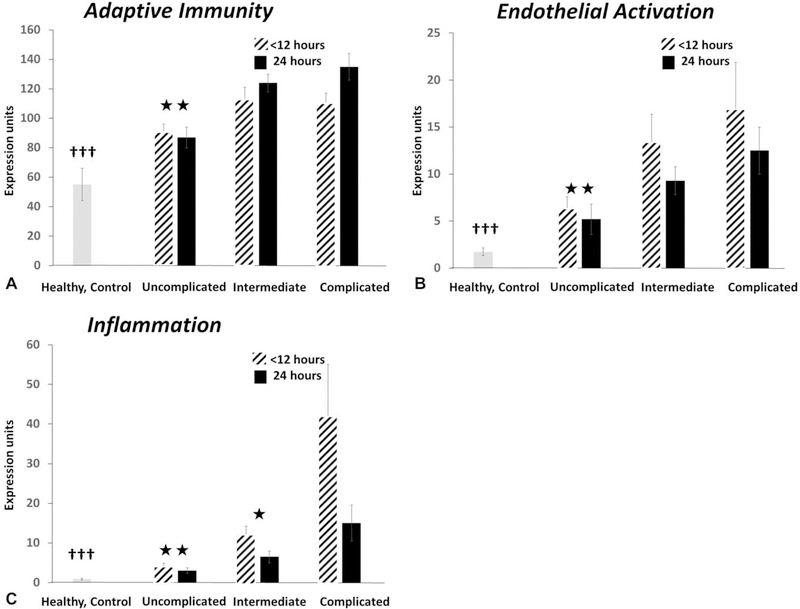
The individual genes used in the S63 metric were originally selected using an unsupervised approach based solely on statistical and fold differences between extremes in clinical outcomes among trauma subjects over 28 days.21 Analysis of the genes comprising the metric revealed that 47 of these 63 genes were involved in the host immune response. It was possible to apportion these genes into 3 functional groups based upon their primary roles in host protective immunity: inflammatory, adaptive immunity and endothelial activation (S Table 2, http://links.lww.com/SLA/B564), and then calculate single transcriptomic metrics for each of these groups. Not surprisingly, the magnitude of the transcriptomic response for all 3 subgroups was less in subjects with uncomplicated outcomes when compared with the other clinical outcomes (Fig. 3). Interestingly, the magnitude of the disturbances in expression of inflammation and endothelial activation genes was significantly higher at <12 than 24hours, whereas disturbances in adaptive immunity gene expression remained elevated or actually increased significantly between 12 and 24 hours in patients with a complicated outcome. Unexpectedly, the predictive ability of these individual metrics was not as strong as the overall S63 metric (Table 4).
FIGURE 3.
Transcriptomic metrics derived from S63 focused on inflammation, adaptive immunity, and endothelial activation. Individual metrics derived from subsets of genes involved in inflammation, endothelial activation, and adaptive immunity (S Table 2, http://links.lww.com/SLA/B564) were calculated in the 3 discrete outcomes. All values from trauma patients at <12 and 24 h were significantly greater than seen in healthy controls (†††). For endothelial activation and inflammation, 24 h metrics were significantly lower than <12 h (P < 0.05 by paired- t test). (*P < 0.05; ** P < 0.01 vs complicated outcome).
DISCUSSION
We report here for the first time both the prospective and subsequent community validation of a transcriptomic expression metric obtained from blood leukocytes 24 hours after severe blunt trauma that could discriminate different clinical outcomes. Importantly, this metric was validated on an FDA 510(k)-cleared instrument currently used for the prediction of breast cancer recurrence based on the expression pattern of 50 different genes.22
In 2011, we reported that severe blunt trauma produced a “genomic storm” in which up to 70% of the blood leukocyte genome changed significantly over a 28-day period.21 More importantly, we identified 2078 genes that were differentially expressed between trauma patients who had a rapid recovery versus those with a sustained complicated outcome. Sixty-three genes were not only statistically different, but had at least a 2-fold difference in gene expression as measured with an Affymetrix GeneChip. The expression of these genes was used to generate a metric based on the sum of the differences of normalized gene expression compared with a cohort of healthy, age- and gender-matched control subjects.11 Here, we have prospectively validated this metric in an additional 127 trauma subjects. This is to our knowledge the first demonstration that a transcriptomic metric has been prospectively validated, and additionally demonstrated to have predictive value when applied to a completely independent public dataset.
Biomarkers that can identify individual patients at risk and who would benefit from an individual therapeutic are the basis for precision medicine. In trauma, the most commonly used biomarkers are clinical scores based on the physiological disturbances observed at the time of admission or at 24 hours. Although these scores have proven to be useful in predicting mortality and are widely used, their precision in predicting other outcomes and demonstrating response to interventions is less well accepted.25 In this report, our transcriptomic metric significantly outperformed the APACHE II measurement, but was only marginally better than ISS. ISS, however, is not a clinically useful metric, as the score is not available for real-time clinical use.
Importantly, mortality to severe blunt trauma without traumatic brain injury has dropped nearly 75% in the past 15 years,5 and the current challenge has focused on reducing infectious and noninfectious complications and increasing early discharge. Protein biomarkers like plasma IL-6, procalcitonin, and C-reactive protein have been promulgated as predictors of outcome,26 but the general consensus is that they too lack sufficient predictive capacity for routine clinical use.25,27 To our knowledge, they have never been applied to discriminating time to recovery until now, and as shown in Table 4, the ability of plasma IL-6 concentrations at 24 hours to predict clinical trajectories was not as good as S63.
Prognostic transcriptomic metrics have become state of the art in the cancer field.28,29 Unique expression patterns of tumor tissue have been associated with both long-term prognostic outcomes, as well as therapeutic responsiveness.30 In fact, several commercially-available, FDA-cleared transcriptomic prognostics are currently available for breast and colorectal cancer (Oncotype DX, PAM50 Prosigna, EndoPredict).22,31–33 Application of this approach to the host response to a variety of inflammatory diseases is also being actively explored. In most cases, blood or blood leukocyte gene expression has been measured. We reported that early changes in blood monocyte gene expression can predict success of lower extremity angioplasty/stenting.34 Others have used transcriptomic metrics in infections and sepsis to discriminate infectious and noninfectious critical illness, and survival.10,13,16,20,35
Importantly, our S63 metric uses the same FDA-cleared analytical platform as the PAM50 Prosigna, but with a unique gene expression set. This metric can be used to identify patients within the heterogeneous nature of trauma who may benefit from a potential therapeutic intervention. Importantly, the platform has a rapid turnaround, less than 24 hours, making it suitable for use in critically ill subjects. Additionally, the threshold can be adjusted (Fig. 2C) depending upon the risk:benefit of the intervention.
In addition to demonstrating a prognostic ability, it is possible, if the genes are properly selected, to use the multiplex transcriptomic markers to better understand the dynamic immunological response to severe trauma. This approach recently identified 3 different “endotypes” in human sepsis.36 Interestingly, earlier analysis of this trauma dataset revealed that the greatest disturbance in genome-wide gene expression occurred within 12 hours and returned to baseline over the subsequent 28 days.21 We see a comparable time course for inflammatory and endothelial activation gene expression changes contained in the S63 metric over the first 24 hours. But the pattern is not the same for genes involved in adaptive immunity (Fig. 3) where the deviation in expression is either constant or increasing over the first 24 hours. Changes in adaptive immune gene expression occur as rapidly as gene expression involved in inflammation, but unlike inflammation which appears to be resolving within the first 24 hours, changes in adaptive immunity at the level of gene expression are maintained. The ability to endotype patients using the same metric makes it potentially useful for exploring therapeutic interventions targeting individual components of the host immune response.
There are a number of limitations to the study that require comment. Although the validation studies were conducted at 2 independent institutions, and the metric was validated with an additional dataset obtained from a prospective, multicenter clinical study, the total number of included patients remains quite small. Second, both the discovery and validation studies were conducted in patients sustaining blunt trauma without traumatic brain injury. Whether this metric will apply to other forms of trauma, such as penetrating trauma or traumatic brain injury will require verification, and therefore limits generalization. Additionally, the discovery and validation cohorts consisted of predominantly Caucasian males; further validation is required to determine if the metric performs well for patients of different gender/race/ethnicity. The genomic response to trauma is dynamic and time-dependent,21 and the data presented here confirms that the transcriptomic metric is more predictive at 24 hours than <12 hours. Whether the metric has value at earlier or later time points will require further validation.
Although mortality to severe blunt trauma has declined dramatically over the past 2 decades, a significant fraction of survivors have a protracted clinical course characterized by secondary infectious and noninfectious complications. Identification of these patients early in their clinical course would provide earlier opportunities for intervention. Here, we prospectively validated an operationalized 63 gene metric (S63) obtained at 24 hours after blunt trauma that can better predict a rapid recovery than either clinical indices or plasma IL-6 concentrations. It is built around a platform already FDA-cleared for genomic analyses. Such a tool would provide the clinician with a rapid test that may assist with clinical management.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the invaluable contributions and efforts of Ricardo Ungaro, M. Cecilia Lopez, and Gabriella Ghita toward the execution of this study.
Supported in part by grants: R01 GM-104481 (L.L.M.), R01 GM-040586 (L.L.M.), R01 GM-113945 (P.A.E.), and P50 GM-111152 (P.A.E., S.C.B., L.L.M.) awarded by the National Institute of General Medical Sciences (NIGMS), and by a postgraduate training grant T32 GM-008721 (R.B.H., J.C.M., J.A.S.) in burns, trauma, and perioperative injury by the NIGMS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors report no conflicts of interest.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.annalsofsurgery.com).
REFERENCES
- 1.DiMaggio C, Ayoung-Chee P, Shinseki M, et al. Traumatic injury in the United States: in-patient epidemiology 2000–2011. Injury. 2016;47:1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kahn JM, Le T, Angus DC, et al. The epidemiology of chronic critical illness in the United States. Crit Care Med. 2015;43:282–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mira JC, Gentile LF, Mathias BJ, et al. Sepsis pathophysiology, chronic critical illness, and persistent inflammation-immunosuppression and catabolism syndrome. Crit Care Med. 2017;45:253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwashyna TJ, Hodgson CL, Pilcher D, et al. Timing of onset and burden of persistent critical illness in Australia and New Zealand: a retrospective, population-based, observational study. Lancet Respir Med. 2016;4:566–573. [DOI] [PubMed] [Google Scholar]
- 5.Mira JC, Cuschieri J, Ozrazgat-Baslanti T, et al. The epidemiology of chronic critical illness after severe traumatic injury at two level-one trauma centers. Crit Care Med. 2017;45:1989–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spruijt NE, Visser T, Leenen LP. A systematic review of randomized controlled trials exploring the effect of immunomodulative interventions on infection, organ failure, and mortality in trauma patients. Crit Care. 2010;14:R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vincent JL. We should abandon randomized controlled trials in the intensive care unit. Crit Care Med. 2010;38(10 suppl):S534–S538. [DOI] [PubMed] [Google Scholar]
- 8.Sweeney TE, Wong HR. Risk stratification and prognosis in sepsis: what have we learned from microarrays? Clin Chest Med. 2016;37:209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warren HS, Elson CM, Hayden DL, et al. A genomic score prognostic of outcome in trauma patients. Mol Med. 2009;15:220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sweeney TE, Shidham A, Wong HR, et al. A comprehensive time-course-based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci Transl Med. 2015;7:287ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuenca AG, Gentile LF, Lopez MC, et al. Development of a genomic metric that can be rapidly used to predict clinical outcome in severely injured trauma patients. Crit Care Med. 2013;41:1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong HR, Cvijanovich NZ, Anas N, et al. Developing a clinically feasible personalized medicine approach to pediatric septic shock. Am J Respir Crit Care Med. 2015;191:309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutherland A, Thomas M, Brandon RA, et al. Development and validation of a novel molecular biomarker diagnostic test for the early detection of sepsis. Crit Care. 2011;15:R149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McHugh L, Seldon TA, Brandon RA, et al. A molecular host response assay to discriminate between sepsis and infection-negative systemic inflammation in critically ill patients: discovery and validation in independent cohorts. PLoS Med. 2015;12:e1001916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raymond SL, Lopez MC, Baker HV, et al. Unique transcriptomic response to sepsis is observed among patients of different age groups. PLoS One. 2017;12:e0184159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sweeney TE, Wynn JL, Cernada M, et al. Validation of the sepsis metascore for diagnosis of neonatal sepsis. J Pediatric Infect Dis Soc. 2017;7:129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong HR, Cvijanovich NZ, Anas N, et al. Pediatric sepsis biomarker risk model-II: redefining the pediatric sepsis biomarker risk model with septic shock phenotype. Crit Care Med. 2016;44:2010–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmerman JJ, Sullivan E, Yager TD, et al. Diagnostic accuracy of a host gene expression signature that discriminates clinical severe sepsis syndrome and infection-negative systemic inflammation among critically ill children. Crit Care Med. 2017;45:e418–e425. [DOI] [PubMed] [Google Scholar]
- 19.Sweeney TE, Khatri P. Benchmarking sepsis gene expression diagnostics using public data. Crit Care Med. 2017;45:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sweeney TE, Perumal TM, Henao R, et al. A community approach to mortality prediction in sepsis via gene expression analysis. Nat Commun. 2018;9:694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao W, Mindrinos MN, Seok J, et al. A genomic storm in critically injured humans. J Exp Med. 2011;208:2581–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallden B, Storhoff J, Nielsen T, et al. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med Genomics. 2015;8:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabrera CP, Manson J, Shepherd JM, et al. Signatures of inflammation and impending multiple organ dysfunction in the hyperacute phase of trauma: a prospective cohort study. PLoS Med. 2017;14:e1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cabrera CP, Manson J, Shepherd JM, et al. Correction: signatures of inflammation and impending multiple organ dysfunction in the hyperacute phase of trauma: a prospective cohort study. PLoS Med. 2018;15:e1002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gentile LF, Cuenca AG, Vanzant EL, et al. Is there value in plasma cytokine measurements in patients with severe trauma and sepsis? Methods. 2013;61:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuschieri J, Bulger E, Schaeffer V, et al. Early elevation in random plasma IL-6 after severe injury is associated with development of organ failure. Shock. 2010;34:346–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oberholzer A, Souza SM, Tschoeke SK, et al. Plasma cytokine measurements augment prognostic scores as indicators of outcome in patients with severe sepsis. Shock. 2005;23:488–493. [PubMed] [Google Scholar]
- 28.Curtit E, Mansi L, Maisonnette-Escot Y, et al. Prognostic and predictive indicators in early-stage breast cancer and the role of genomic profiling: Focus on the Oncotype DX((R)) Breast Recurrence Score Assay. Eur J Surg Oncol. 2017;43:921–930. [DOI] [PubMed] [Google Scholar]
- 29.Sestak I, Buus R, Cuzick J, et al. Comparison of the performance of 6 prognostic signatures for estrogen receptor-positive breast cancer: a secondary analysis of a randomized clinical trial. JAMA Oncol. 2018;4:545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huet S, Tesson B, Jais JP, et al. A gene-expression profiling score for prediction of outcome in patients with follicular lymphoma: a retrospective training and validation analysis in three international cohorts. Lancet Oncol. 2018;19:549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jamieson NB, Maker AV. Gene-expression profiling to predict responsiveness to immunotherapy. Cancer Gene Ther. 2017;24:134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Callari M, Cappelletti V, D’Aiuto F, et al. Subtype-specific metagene-based prediction of outcome after neoadjuvant and adjuvant treatment in breast cancer. Clin Cancer Res. 2016;22:337–345. [DOI] [PubMed] [Google Scholar]
- 33.Geeleher P, Loboda A, Lenkala D, et al. Predicting response to histone deacetylase inhibitors using high-throughput genomics. J Natl Cancer Inst. 2015;107. djv247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeSart K, O’Malley K, Schmit B, et al. Systemic inflammation as a predictor of clinical outcomes after lower extremity angioplasty/stenting. J Vasc Surg. 2016;64:766–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sweeney TE, Wong HR, Khatri P. Robust classification of bacterial and viral infections via integrated host gene expression diagnostics. Sci Transl Med. 2016;8:346ra91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sweeney TE, Azad TD, Donato M, et al. Unsupervised analysis of transcriptomics in bacterial sepsis across multiple datasets reveals three robust clusters. Crit Care Med. 2018;46:915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.