Abstract
Culturomics studies the microbial variety of the human microbiome by combining diversified culture conditions, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and 16S rRNA gene identification. This study identifies three putative new bacterial species: Arcanobacterium ihumii sp. nov. strain Marseille-P5647T, Varibaculum vaginae sp. nov. strain Marseille-P5644T and Tessaracoccus timonensis sp. nov. strain Marseille-P5995T, which we describe according to the concept of taxonogenomics.
Keywords: Arcanobacterium ihumii sp. nov., culturomics, Tessaracoccus timonensis sp. nov., vagina, Varibaculum vaginae sp. nov.
Introduction
The vagina is a wet biotope comprising a complex ecosystem colonized by several types of microorganisms [1]. Döderlein bacilli or Lactobacillus are the majority bacteria in the vaginal flora of healthy women of child-bearing age [2]. Many studies have been performed, some of which conclude that this complex ecosystem is dominated by species belonging to the genus Lactobacillus [3]. An imbalance of this flora can lead to vaginosis [4].
Bacterial vaginosis was first described by Schröder in 1921. He noticed dysbiosis characterized by a decrease in Lactobacillus, an increase in the pH of the vaginal mucosa, and a proliferation mainly of Gram-negative anaerobic bacteria such as Gardnerella vaginalis, Atopobium vaginae and Mobiluncus curtisii [5], [6]. The role played by the human vaginal microbiota, in the prevention of many urogenital diseases, such as bacterial vaginosis, sexually transmitted infections and urinary tract infections, is poorly understood [7].
Arcanobacterium spp. have been detected in humans and animal, and some species such as Arcanobacterium haemolyticum and Arcanobacterium pyogenes are occasionally reported as pathogens for humans [8], [9]. To date, there are two valid published bacteria within the genera Varibaculum, previously isolated from human samples [10], [11]. Tessaracoccus species are more frequently isolated from environmental sources and occasionally from animals [12], [13].
Currently, it is important to understand the involvement of bacterial variety in normal physiological functions and exposure to some diseases [14]. The culturomics approach, which permits the isolation of bacteria under diverse culture conditions, is combined with 16S rRNA amplicon sequencing to explore the diversity of the human microbiota [15], [16], [17]. Description of these new species is based on a method combining genotypic and phenotypic characteristics of the bacterium and supplemented by the taxonogenomic strategy previously described [18], [19]. In this study, we describe three new species for the first time, isolated from the vagina.
Isolation and growth conditions
In 2017, as part of a culturomics study of the human microbiome, the three strains Marseille-P5647T, Marseille-P5644T and Marseille-P5995T were isolated from different samples recovered from women living in Dielmo, a West African village. The vaginal swabs were placed directly in liquid medium enriched with sheep's blood and rumen. Initial growth of bacterial cells was obtained after 15 days of pre-incubation in an anaerobic environment. Then the solutions were seeded on 5% sheep's blood agar (bioMérieux, Marcy l’Etoile, France) under anaerobic conditions at 37°C. The colonies were tested using matrix-assisted laser desorption ionization -time of flight mass spectrometry (MALDI-TOF MS; Microflex LT spectrometer; Bruker Daltonics, Bremen, Germany) [20]. Spectra obtained were imported and analysed using the Biotyper 3.0 software against the Bruker database, which was constantly updated from the MEPHI database [14] (Fig. 1). Being potentially new species, no spectra were included in the database and the colonies were not identified using MALDI-TOF.
Fig. 1.
MALDI-TOF MS reference spectra of the three new species described. The reference spectra were generated by comparison of spectra from 12 individual colonies for each species.
Phenotypic characteristics
The strains are bacteria that all grow anaerobically. For each strain, first growth was observed after 15 days incubation at 37°C on Columbia agar medium enriched with 5% sheep's blood (bioMérieux) in an anaerobic atmosphere generated using the GENbag anaer system (bioMérieux). The strain Marseille-P5647 (= CSUR P5647) is Gram-negative. It appears on agar as colonies measuring 1 mm in diameter. Cells are not motile and present no catalase or oxidase activity. The strain Marseille-P5995 (= CSUR P5995) is Gram-negative. Its colonies have a mean size of 0.2 mm. It is a non-motile bacterium with catalase-negative and oxidase-negative activities. Cells of the strain Marseille-P5644 (= CSURP5644) are Gram-positive cocci, non-motile, catalase-positive and oxidase-negative. They form translucent white or grey colonies of 1-mm diameter on Columbia agar medium enriched with 5% sheep's blood at 37°C after 48 hours of anaerobic incubation. the shape of this bacterium was highlighted with a Hitachi TM4000 tabletop scanning electron microscope (Hitachi Group, Krefeld, Germany) (Fig. 2).
Fig. 2.
Scanning electron microscopy (SEM) of the three new species. A colony was collected from agar and immersed into a 2.5% glutaraldehyde fixative solution. Then, a drop of the suspension was directly deposited on a poly-l-lysine-coated microscope slide for 5 minutes and treated with 1% phosphotungstic acid aqueous solution (pH 2.0) for 2 minutes to increase the SEM image contrast. The slide was gently washed in water; air-dried and examined in a tabletop SEM (Hitachi TM4000). Scales and acquisition settings are shown on figures.
Strain identification
To identify these four strains, the 16S rRNA gene of each was amplified using the primer pair fD1 and rP2 (Eurogentec, Angers, France) and sequenced using the Big Dye® Terminator v1.1 Cycle Sequencing Kit and 3500xLGenetic Analyzer capillary sequencer (Thermofisher, Saint-Aubin, France) as previously reported [21]. The 16S rRNA nucleotide sequences were assembled and corrected using CodonCode Aligner software (http://www.codoncode.com). For the strain Marseille-P5647T, the 16S rRNA (Accession number LT993248) gene sequence analyses showed 96.64% sequence similarity with Arcanobacterium phocae strain DSM 10002 (GenBank Accession number NR_117159.1), whereas the 16S rRNA (Accession number LS999997) gene sequence of the strain Marseille-P5644T had 98.22% sequence similarity with Varibaculum cambriense strain CCUG 44998 (GenBank Accession number NR_114873.1). For Marseille-P5995T strain, the analysis of its 16S rRNA gene sequence (Accession number LT996088) showed a sequence similarity of 97.33% with Tessaracoccus oleiagri strain SL014B-20A1 (GenBank Accession number NR_108681.1). These values were lower than the 98.7% 16S rRNA gene sequence threshold recommended by Meier-Kolthoff et al. to delineate a new bacterial species without performing DNA–DNA hybridization [22]. We accordingly propose to classify strains Marseille-P5647T, Marseille-P5644T and Marseille-P5995T as new species within the genus Arcanobacterium, Varibaculum and Tessaracoccus, respectively (Fig. 3).
Fig. 3.
Phylogenetic tree highlighting the position of all the new species relative to their most closely related type strains and validly published. GenBank Accession numbers of 16S rRNA are indicated in parentheses. Sequences were aligned using MUSCLE with default parameters, phylogenetic inferences were obtained using the maximum likelihood method and MEGA 7 software. Numbers at the nodes are percentages of bootstrap values obtained by repeating the analysis 1000 times to generate a majority consensus tree. The scale bar indicates a 5% nucleotide sequence divergence. Helcococcus kunzii DSM 10548 was used as outgroup.
Genome sequencing
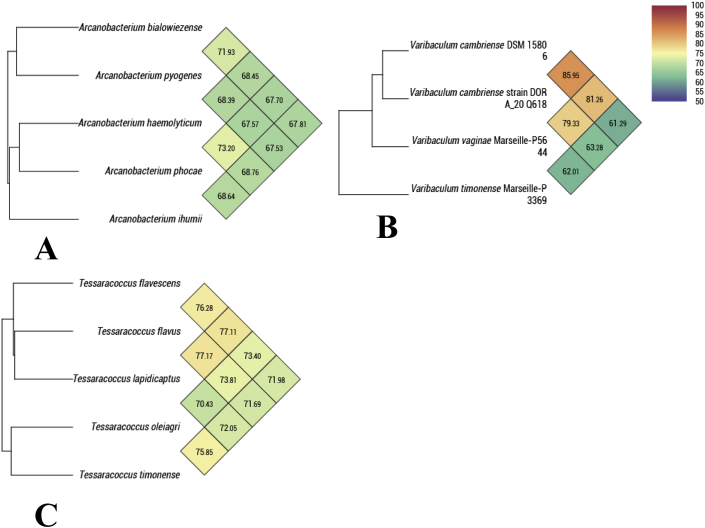
Genomic DNA was extracted using the EZ1 biorobot with the EZ1 DNA tissue kit (Qiagen, Hilden, Germany) and then sequenced on a MiSeq sequencer (Illumina Inc., San Diego, CA, USA) with the Nextera Mate Pair sample prep kit and Nextera XT Paired End (Illumina), as previously described [23]. The assembly was performed using a pipeline containing several softwares (Velvet [24], Spades [25] and Soap Denovo [26]), on trimmed data (MiSeq and Trimmomatic [27] softwares) or untrimmed data (only MiSeq software). GapCloser was used to reduce assembly gaps. Scaffolds <800 bp and scaffolds with a depth value < 25% of the mean depth were removed. The best assembly was selected by using different criteria (number of scaffolds, N50, number of N). The sequencing of these strains gave the sizes and the percentages of G+C content of their respective genomes. The genomes of strains Marseille-P5647T and Marseille-P5644T were, respectively, 2.00 and 2.14 Mb long with a 51.6% and 52.3% G+C content. The genome size of strain Marseille-P5995T was 3.18 Mb long and it had a G + C content of 66.6%. The degree of genomic similarity of these strains with closely related species was calculated using OrthoANI software [28]. When OrthoANI software was used to calculate the similarity between Tessaracoccus timonensis and available genomes within the Tessaracoccus genus (Fig. 4c), we noted that values among closely related species ranged from 77.17%, between Tessaracoccus flavus and Tessaracoccus lapidicaptus, to 70.43%, between Tessaracoccus oleiagri and Tessaracoccus lapidicaptus. When Varibaculum vaginae was compared with its closely related species (Fig. 4b), OrthoANI values ranged from 85.95%, between Varibaculum cambriense strain DSM 15806 and V. cambriense strain DORA_20 Q618, to 61.29%, between V. cambriense strain DSM 15806 and Varibaculum timonense strain Marseille-P3369. Likewise, when OrthoANI computing was performed to estimate the average nucleotide identity between Arcanobacterium ihumii and available genomes within the genus Arcanobacterium (Fig. 4a), we noted that values among closely related species ranged from 73.20%, between Arcanobacterium haemolyticum and Arcanobacterium phocae, to 70.43%, between Arcanobacterium pyogenes and Arcanobacterium ihumii.
Fig. 4.
Heatmaps generated with OrthoANI values calculated using the OAT software for Arcanobacterium ihumii sp. nov. (a), Varibaculum vaginae sp. nov. (b) and Tessaracoccus timonense sp. nov. (c) with other closely related species with standing in nomenclature.
Conclusion
Based on the results from phenotypic characteristics, including MALDI-TOF spectra, a 16S rRNA sequence divergence >1.3% and an OrthoANI value < 95% with the phylogenetically closest species with standing in nomenclature, we formally proposed strains Marseille-P5647T, Marseille-P5644T and Marseille-P5995T, respectively, as being the type strains of Arcanobacterium ihumii sp. nov., Varibaculum vaginae sp. nov. and Tessaracoccus timonensis sp. nov., which are new species in the Bacteria domain.
Description of Arcanobacterium ihumii sp. nov.
Arcanobacterium ihumii (i.hum.i'i. N.L. gen. n. ihumii, based on the acronym IHUMI, the Institut Hospitalo-Universitaire Méditerranée-Infection, where the type strain was isolated). Colonies are 1 mm in diameter. No growth is observed in aerobic and microaerophilic conditions. Growth occurs at 37°C in anaerobic conditions. Cells stain Gram-negative, are non-motile, oxidase-negative and catalase-negative. The G+C content of the genome is 51.6%. The 16S rRNA and genome sequences are deposited in GenBank under Accession numbers LT993248 and UWOS00000000, respectively. The type strain Marseille-P5647 (= CSUR P5647) was isolated from the vaginal flora of a Senegalese woman living in a village.
Description of Varibaculum vaginae sp. nov.
Varibaculum vaginae (va.gi'nae, L. n., from vagina, ‘vagina’; L. gen. n. vaginae, ‘of the vagina’). Varibaculum vaginae strain Marseille-P5644 grows on a 5% sheep's blood enriched Columbia agar medium incubated at 37°C in anaerobic conditions and shows small colonies with a mean diameter of 1 mm. It is a Gram-positive bacterium with catalase-positive and oxidase-negative activities. The 16S rRNA and genome sequences are deposited in GenBank under Accession numbers LS999997 and UWNY00000000, respectively. The genome size is 2 283 357 bp with a G+C content of 52.3%. The type strain Marseille-P5644T (= CSUR P5644) was isolated from the vagina of a woman living in Dielmo, Senegal.
Description of Tessaracoccus timonensis sp. nov.
Tessaracoccus timonensis (ti.mo.nen'sis. L. gen. masc. timonensis, of Timone, the name of the hospital where strain Marseille-P5995T was cultivated). Optimal growth is obtained in anaerobic conditions. No growth is achieved in aerobic and microaerophilic conditions. Cells are Gram-negative and non-motile. Cells are also negative for catalase and oxidase tests. The 16S rRNA and genome sequences are deposited in GenBank under Accession numbers LT996088 and LT996886, respectively. The genome size is 3.18 Mb with a G+C content of 66.6%. The type strain Marseille-P5995T (= CSUR P5995) was isolated from the vagina of a woman living in Dielmo, a Senegalese rural area.
Nucleotide sequence accession numbers
The 16S rRNA gene and genome sequences were deposited in GenBank under the following Accession numbers: LT993248 and UWOS00000000, respectively (Marseille-P5647T); LS999997 and UWNY00000000, respectively (Marseille-P5644T); and LT996088 and LT996886, respectively (Marseille-P5995T)
Deposit in culture collections
Strains Marseille-P5647T, Marseille-P5644T and Marseille-P5995T were deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR) under the following numbers, respectively: CSUR P5647, CSUR P5644 and CSUR P5595.
Acknowledgements
The authors thank Catherine Robert for sequencing the genome, Aurelia Caputo for submitting the genomic sequence to GenBank and Carine Couderc for performing the MALDI-TOF reference spectrum.
Conflicts of interest
None to declare.
Funding sources
This study was supported by the Institut Hospitalo-Universitaire (IHU) Méditerranée Infection, the National Research Agency under the programme Investissements d'avenir, reference ANR-10-IAHU-03.
Ethics and consent
The study and consent procedures were approved by the Senegalese Comité National d’Ethique pour la Recherche en Santé, ethics committee in accordance with the SEN protocol 16/04 under number 00039 as well as by the ethics committee of the Institut Hospitalo-Universitaire Méditerranée Infection, under number 2016-011. The women gave written consent.
References
- 1.Kumar N., Behera B., Sagiri S.S., Pal K., Ray S.S., Roy S. Bacterial vaginosis: etiology and modalities of treatment—a brief note. J Pharm Bioall Sci. 2011;3:496–503. doi: 10.4103/0975-7406.90102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gary Ventolini. Progresses in vaginal microflora physiology and implications for bacterial vaginosis and candidiasis. Womens Health (Lond) 2016;12:283–291. doi: 10.2217/whe.16.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Vos P., Garrity G.M., Jones D., Krieg N.R., Ludwig W., Rainey F.A. The Firmicutes. In: Garrity G.M., editor. Bergey’s manual of systematic bacteriology. Springer; New York, NY: 2009. pp. 465–511. [Google Scholar]
- 4.Fenollar F., Raoult D. Does bacterial vaginosis result from fecal transplantation? J Infect Dis. 2016;214:1784. doi: 10.1093/infdis/jiw472. [DOI] [PubMed] [Google Scholar]
- 5.Pépin J., Deslandes S., Giroux G., Sobéla F., Khonde N., Diakité S. The complex vaginal flora of West African women with bacterial vaginosis. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0025082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shipitsyna E, Roos A, Datcu R, Hallén A, Fredlund H, Jensen JS et al. Composition of the vaginal microbiota in women of reproductive age—sensitive and specific molecular diagnosis of bacterial vaginosis is possible? PLoS ONE 8: e60670. [DOI] [PMC free article] [PubMed]
- 7.Ravela J., Gajera P., Abdob Z., Schneider G.M., Koenig S.S.K., McCulle S.L. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funke G., Ramos C.P., Fernandez-Garayzabal J., Weiss N., Collins M.D. Description of human-derived Centers for Disease Control coryneform group 2 bacteria as Actinomyces bernardiae sp. nov. Int J Syst Bacteriol. 1995;45:57–60. doi: 10.1099/00207713-45-1-57. [DOI] [PubMed] [Google Scholar]
- 9.Hoyles L., Falsen E., Foster G., Rogerson F., Collins M.D. Arcanobacterium hippocoleae sp. nov., from the vagina of a horse. Int J Syst Evol Microbiol. 2002;52:617–619. doi: 10.1099/00207713-52-2-617. [DOI] [PubMed] [Google Scholar]
- 10.Hall V., Collins M.D., Lawson P.A., Hutson R.A., Falsen E., Inganas E. Characterization of some actinomyces-like isolates from human clinical sources: description of Varibaculum cambriensis gen. nov. sp. nov. J Clin Microbiol. 2003;41:640–644. doi: 10.1128/JCM.41.2.640-644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glaeser S.P., Doijad S., Hijazin M., Chakraborty T., Falsen E., Hall V. Varibaculum anthropi sp. nov. represented by three genetically different genomovars isolated from clinical material and emended description of the genus Varibaculum. Syst Appl Microbiol. 2016;39:546–552. doi: 10.1016/j.syapm.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Li G.D., Chen X., Li Q.Y., Xu F.J., Qiu S.M., Jiang Y. Tessaracoccus rhinocerotis sp. nov., isolated from the faeces of Rhinoceros unicornis. Int J Syst Evol Microbiol. 2016;66:922–927. doi: 10.1099/ijsem.0.000812. [DOI] [PubMed] [Google Scholar]
- 13.Thongphrom C., Kim J.H., Bora N., Kim W. Tessaracoccus arenae sp. nov., isolated from sea sand. Int J Syst Evol Microbiol. 2017;67:2008–2013. doi: 10.1099/ijsem.0.001907. [DOI] [PubMed] [Google Scholar]
- 14.Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I. The human microbiome Project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagier J.C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 16.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagier J.C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Ramasamy D., Mishra A.K., Lagier J.C., Padhmanabhan R., Rossi M., Sentausa E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 19.Fournier P.E., Lagier J.C., Dubourg G., Raoult D. From culturomics to taxonomogenomics: a need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe. 2015;36:73–78. doi: 10.1016/j.anaerobe.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 20.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.E., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 21.Lo C.I., Padhmanabhan R., Mediannikov O., Terras J., Robert C., Faye N. High-quality genome sequence and description of Bacillus dielmoensis strain FF4(T) sp. nov. Stand Genom Sci. 2015;10:41. doi: 10.1186/s40793-015-0019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier-Kolthoff J.P., Göker M., Spröer C., Klenk H.P. When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol. 2013;195:413–418. doi: 10.1007/s00203-013-0888-4. [DOI] [PubMed] [Google Scholar]
- 23.Lo C.I., Sankar S.A., Fall B., Ba B.S., Diawara S., Gueye M.W. High-quality draft genome sequence and description of Haemophilus massiliensis sp. nov. Stand Genom Sci. 2016;11:31. doi: 10.1186/s40793-016-0150-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee I., Ouk Kim Y., Park S.C., Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]