Abstract
Background
Allele specific real-time PCR and next-generation sequencing (NGS) are widely used to detect somatic mutation in non-small cell lung cancer (NSCLC). Both methods commonly use formalin-fixed paraffin-embedded (FFPE) tissues as diagnostic materials. Real-time PCR has the advantage of being easy to use and more tolerant of variable DNA quality, but has limited multiplex capability. NGS, in contrast, allows simultaneous analysis of many genomic loci while revealing the exact sequence changes; it is, however, more technically demanding and more expensive to employed. A challenge for both platforms is the varied limit of detection (LoD) for target genomic loci, even within the same gene. The variability of detection sensitivity may be problematic if well-known actionable somatic mutations are missed.
Cases
We compared LoDs between real-time PCR and targeted NGS tests for some commonly observed EGFR mutations in NSCLC specimens.
Conclusions
The FDA-approved real-time PCR test was superior to the NGS in detecting low level EGFR exon 19 deletion (near 1% variant allele fraction (VAF)). The cancer hotspot NGS detects low level EGFR c.2369C > T, p.T790M (2–5% VAF) better than the FDA-approved real-time PCR method. We conclude that the real-time PCR and hotspot NGS methods have complementary strengths in accurately determining clinically important EGFR mutations in NSCLC.
Keywords: Non-small cell lung cancer, Epidermal growth factor receptor, Formalin-fixed paraffin-embedded tissue, Fine needle aspiration, Next-generation sequencing, therascreen®, FDA
1. Introduction
Lung cancer is one of the most common tumor types in the U.S; with approximately 80% are NSCLC [1,2]. Approximately 10–35% of NSCLC cases have causative mutations of the epidermal growth factor receptor gene, EGFR [[3], [4], [5]]. While many forms of lung cancer are associated with a poor prognosis, drugs targeting mutated tyrosine kinase receptors are associated with clinical benefit and are widely used in cases positive for EGFR mutations [6]. Appropriate molecular testing methods along with a thorough understanding of these tests’ limitations are therefore relevant in facilitating the timely determination of NSCLC-related gene mutation status. The mutation information obtained can guide therapy choices in NSCLC patients, i.e., the use of tyrosine kinase inhibitors (TKIs) [[7], [8], [9]].
Allele-specific real-time PCR has been widely used in detecting EGFR “hotspot” mutations in cancerous tissues [[10], [11], [12], [13], [14], [15]]. Therascreen® (Qiagen) is a FDA-approved real time PCR in vitro diagnostic (IVD) test that may be used to detect 21 EGFR mutations in exons 18, 19, 20, and 21 against a background of wild type genomic DNA [16]. Advantages of the IVD include: (1) laboratory workflow is straightforward and rapid; (2) suboptimal quality genomic DNA, such as that extracted from FFPE tissues, can be used; and (3) variants with low allele fractions or that may be present in biopsies with low tumor content (<10%) can reasonably be expected to be detected.
In recent years, NGS has been rapidly adopted in molecular diagnostic laboratories to detect gene mutations in cancers [2,17] and can provide results by simultaneously interrogating hundreds of genomic loci. A small amount of input DNA is no longer a limiting factor in targeted library preparation for NGS-based interrogation. Consequently, when performing deep sequencing of targeted NGS panels, mutations may be detected at ~5% VAF. The capability of NGS to detect specific low-level mutations in tumors is important in determining targeted cancer therapy. We present data, based on instructive clinical cases, comparing allele-specific real-time PCR and NGS methods for detection of selected clinically important EGFR mutations.
2. Results
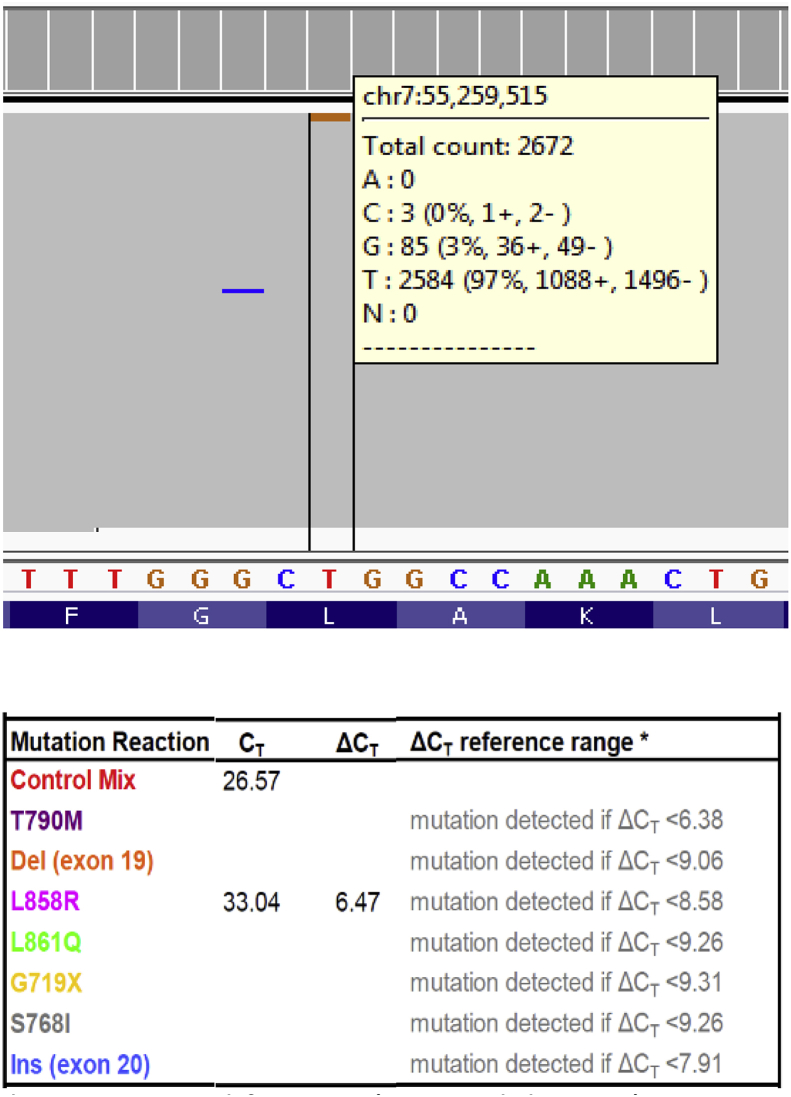
Selected NSCLC specimens with low level EGFR mutations or late therascreen® curves were used in this study. Shown in Fig. 1, the cancer hotspot NGS method revealed an EGFR c.2573T > G (p.Leu858Arg) variant at 3% VAF with a read depth of 2672. The therascreen® test was then performed on the same sample and this EGFR c.2573T > G variant was also detected. This finding indicated that cancer hotspot NGS, similar to the therascreen®, accurately detects a common EGFR mutation at low VAF.
Fig. 1.
The identification of EGFR c.2573T > G (p.Leu858Arg) sequence change at 3% allele fraction by the cancer hotspot NGS (upper panel) and the therascreen® (lower panel) tests.
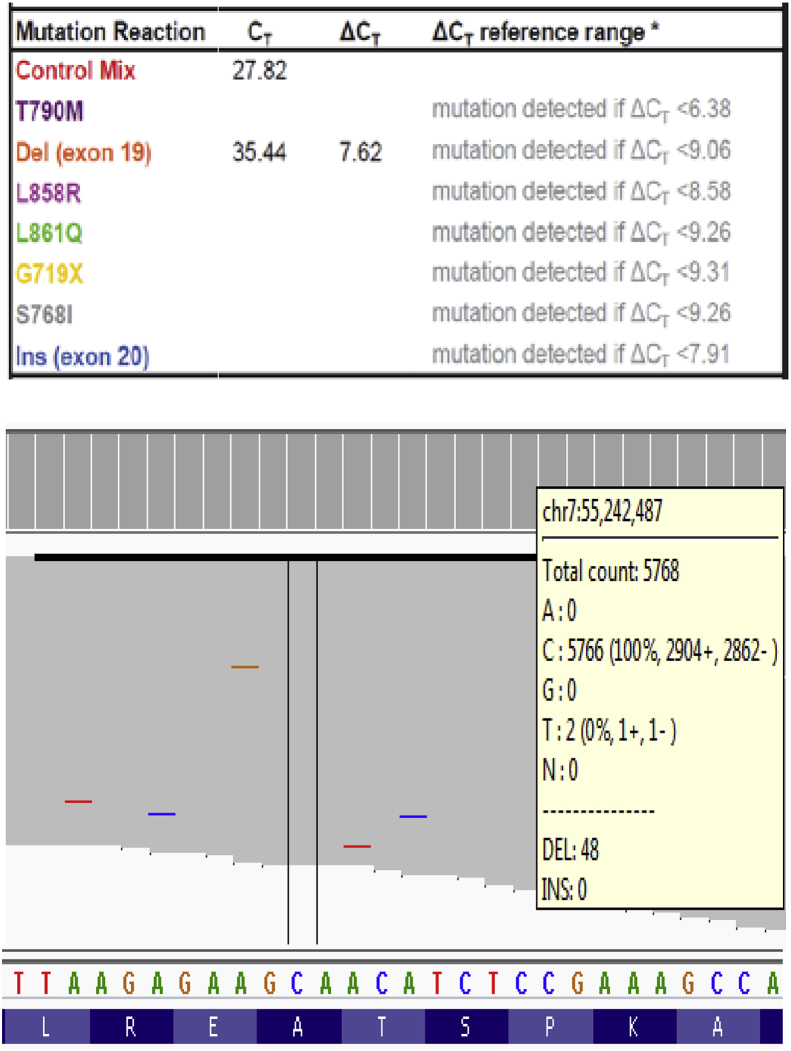
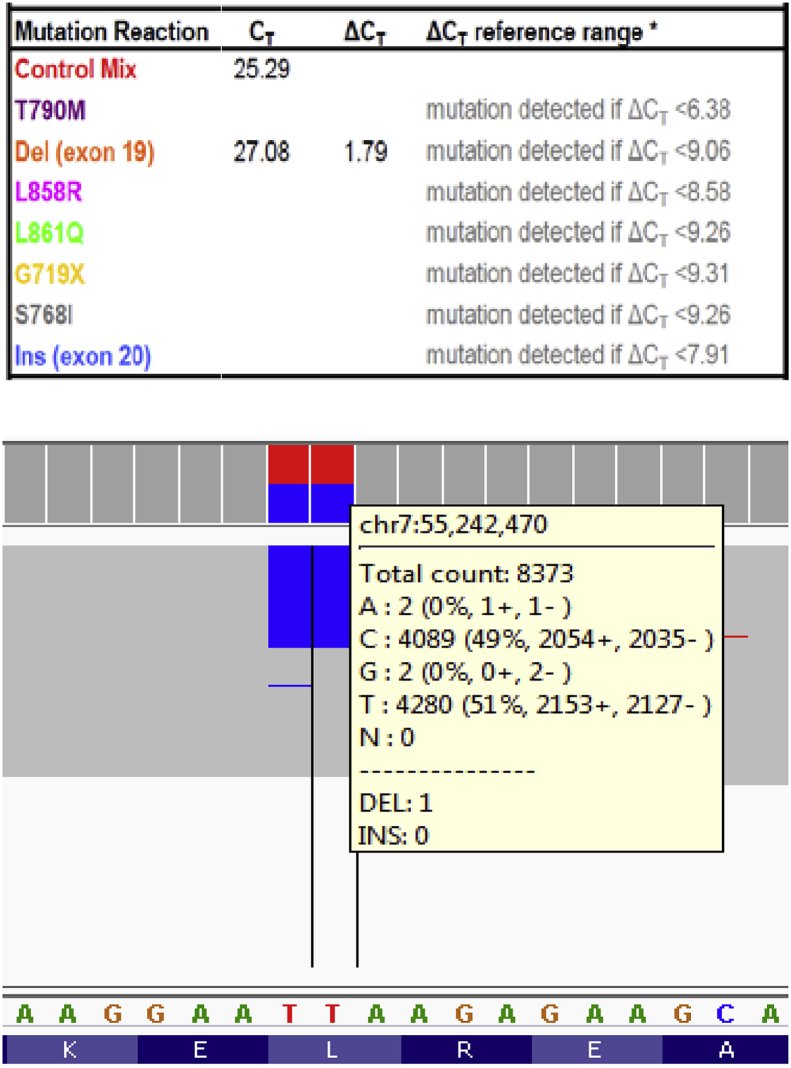
The identification of a low level EGFR exon 19 deletion is shown in Fig. 2. The EGFR exon 19 deletion c.2240_2257delTAAGAGAAGCAACATCTC (p.Leu747_Pro753delinsSer) was accurately determined using the therascreen® test. The observed deltaCt (7.62) is well within the reference range of the assay (<9.06); this test result was therefore confidently reported despite the low allele frequency (see Fig. 2). We also performed the cancer hotspot NGS panel on this sample; this method did not detect the exon 19 deletion variant. Upon manual inspection of the aligned NGS reads, it appeared that this EGFR exon 19 deletion was present at 0.8% VAF (48/5872 reads), which is well below the LoD of cancer hotspot NGS [17].
Fig. 2.
An EGFR exon 19 deletion was accurately scored using the therascreen® assay (upper panel). Manual review of the cancer hotspot NGS data revealed this deletion at 0.8% allele fraction (lower panel).
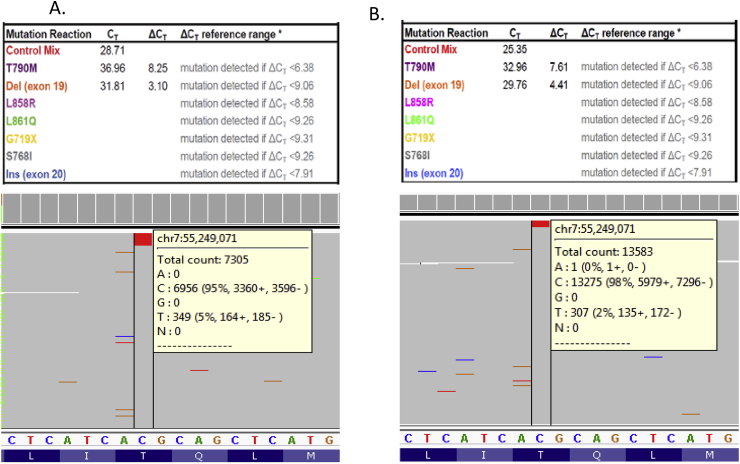
Besides detecting low level EGFR exon 19 deletions, therascreen® detects EGFR T790M variants except when the real-time PCR curves occur much later in the amplification cycles. As shown in Fig. 3A, an EGFR exon 19 deletion and a late curve (Ct = 36.96) associated with EGFR T790M variant were observed with a therascreen® assay. This late curve has a deltaCt value 8.25 that was considered outside the therascreen® T790M reference range (<6.38); as per the therascreen® manual, such a late curve for the T790M variant cannot be accurately called. Thus, we investigated the identity of this therascreen® finding using the cancer hotspot NGS panel [17]. NGS analysis showed the presence of an EGFR exon 19 deletion and the T790M (c.2369C > T) mutation at 19% and 5% VAFs, respectively (Fig. 3A). A similar therascreen® case with a late EGFR T790M amplification curve (Ct = 32.96) and exon 19 deletion are shown in Fig. 3B. This late curve has a deltaCt value 7.61 that is greater than the therascreen® T790M reference cut-off (<6.38). Again, we were able to detect the T790M (c.2369C > T) mutation on the cancer hotspot NGS panel at 2% VAF. These NGS results demonstrate that late therascreen® curves of T790M represent true positive findings.
Fig. 3.
(A) A suspected EGFR T790M variant (Ct = 36.96) was not scored within the therascreen® reference range (upper panel). Alternatively, cancer hotspot NGS accurately called this variant at 5% allele fraction (lower panel). (B) Another example of a late EGFR T790M therascreen® curve (Ct = 32.96) resulted in the deltaCt not within the reference range (upper panel). Cancer hotspot NGS identified this variant at 2% allele fraction (lower panel). Note: the EGFR exon 19 deletions are not shown in the NGS pileup results.
3. Discussion
Therascreen® is a sensitive assay to detect some clinically important EGFR mutations. We show here that therascreen® can accurately identify low level EGFR L858R (3%) and exon 19 deletion (1%) variants (Fig. 1, Fig. 2). It is worth noting that the current cancer hotspot NGS panel was not designed to identify <2% VAF in certain EGFR hotspots, such as exon 19 deletions. In this regard, the therascreen® test has a favorable LoD at the EGFR exon 19 locus. However, the claimable LoDs on the therascreen® user manual vary among the interrogated EGFR mutation hotspots. While the LoD of exon 19 deletions could be as low as 0.81%, the LoDs of exon 20 T790M (c.2369C > T) and exon 18 G719A (c.2156G > C) are reported higher at 17.5% and 32.5% VAFs, respectively.
Since the presence of T790M variant is an indication of TKI resistance, the ability to identify low VAF at this locus is essential in determining drug administration and monitoring therapeutic response. The importance of detecting low level T790M mutations has been stated in the 2018 College of American Pathology molecular testing guideline for selecting NSCLC patients for targeted TKI therapies [18]. The cases described here have late T790M Ct curves that were outside the operating range of the assay, indicating that therascreen® does not detect the T790M mutation when present at low levels and an alternate method should be used to screen for or confirm low level T790M mutation (Fig. 3). In contrast, the in-house developed cancer hotspot NGS assay can detect T790M (c.2369C > T) variant at near 2% VAF.
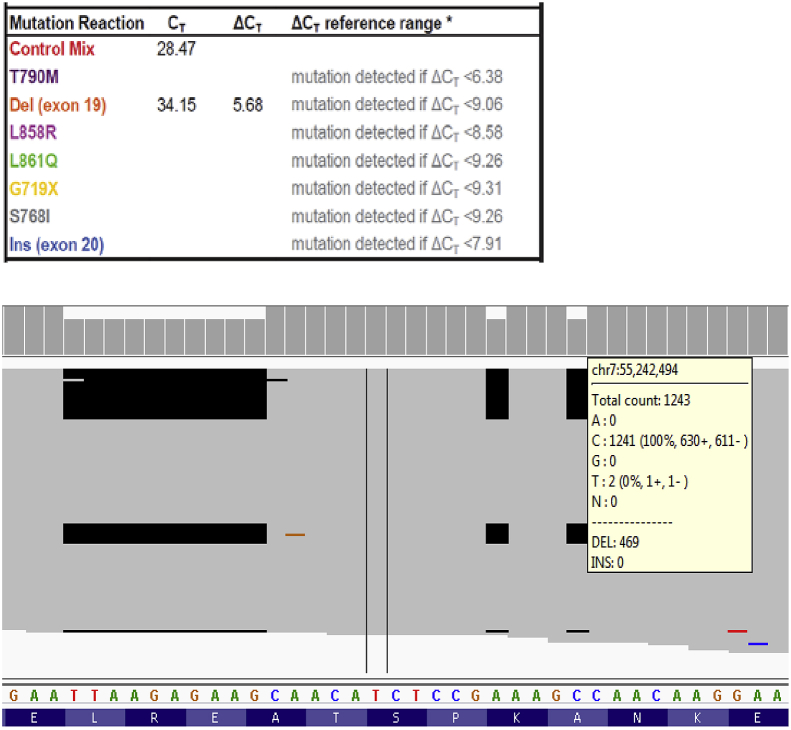
An allele-specific real-time PCR method, such as therascreen®, indirectly reveals a gene's mutation status. It assumes that the identified variants have the same sequence compositions as the interrogating allele-specific primers or probes. This assumption is generally true for most of EGFR mutation hotspots, but there are important exceptions such as the complex cases presented here. As shown in Fig. 4, therascreen® called the EGFR exon 19 deletion without revealing the presence of a 10-base and two single-base deletions. Sanger sequencing was performed and confirmed the NGS result (data not shown). Since the 10-base and the two individual nucleotide deletions occurred on the same sequence read with the same VAFs, these nucleotide changes should be reported as concurrent events and represent a complex exon 19 deletion (c.2239_2264delins14, p.Leu747_Ala755delinsGlnHisLeuArgSer). Of interest, if the sequence changes were not concurrent, each of the three separate deletions would result in a loss-of-function, frame-shift gene product that would not explain the oncogenic character of an EGFR exon 19 variant. Only when all three events occur within the same allele will the deletions create a 12-base, in-frame EGFR exon 19 deletion at this region. For this case, regardless as to whether the exact sequence alteration was determined, there would be no impact on patient care because both test platforms consistently called the EGFR exon 19 deletion and suggest TKI-based therapies for the patient. Although therascreen® did report the sequence changes as an EGFR exon 19 deletion, the complexity of the allele was not identified. By analogy, other EGFR sequence alterations may be inaccurately classified using the therascreen® method (see below).
Fig. 4.
A complex EGFR exon 19 sequence change. The therascreen® result indicated an EGFR exon 19 deletion (upper panel). Cancer hotspot NGS revealed this EGFR exon 19 sequence change consists of a 10-base and two individual single-base deletions (lower panel).
Another example of the importance of accurately revealing the sequence changes of a gene mutation is shown in Fig. 5. This NSCLC specimen has two consecutive missense variants (c.2239_2240delinsCC, p.Leu747Pro, Sanger sequencing confirmed) that should be called as insertion/deletion (indel), but were interpreted by therascreen® test as an EGFR exon 19 deletion event. This difference in variant classification could potentially affect the patient's clinical management; the calling of an EGFR exon 19 deletion indicates that a tumor is likely TKI sensitive, while the calling of a two-base indel results in only one amino acid alteration which is less responsive to TKI treatment [[19], [20], [21]].
Fig. 5.
An unusual EGFR exon 19 sequence change was determined as deletion by therascreen® (upper panel). The cancer hotspot NGS revealed the sequence change involves two consecutive missense variants (lower panel).
In summary, we have shown examples of real-time PCR (therascreen®) and hotspot NGS assays that are able to identify low VAF somatic mutations, despite the LoDs of both platforms varying among different genomic loci. Since it is not cost-effective to test the same sample on two different assay platforms, we suggest those laboratories who routinely run EGFR therascreen® test should have an alternative approach to confirm the findings of late PCR curves. Moreover, for those patients who appear to be resistant to TKI therapy despite having an exon 19 mutation, additional testing should be considered to ensure an accurate classification of the EGFR mutation. For those laboratories that perform cancer hotspot NGS tests, a careful determination of the LoDs during test validation at multiple EGFR loci will reduce false-negative calls. Furthermore, having an alternative method such as therascreen® may be useful when there is limited amount of sample available to confirm the absence of a mutation in EGFR.
4. Methods
4.1. Tumor samples and DNA extraction
Genomic DNA was extracted from tumor specimens collected from NSCLC patients as previously described [17,22,23]. Cytology specimens were briefly kept in PreservCyt solution then were manually extracted according to the Gentra Puregene DNA Extraction Kit (Qiagen, Hilden, Germany) user's manual. DNA from FFPE tissue specimens was extracted using Maxwell® DNA FFPE Kit (Promega, Madison, WI) in an automated fashion. Purified genomic DNA was stored at 4 °C.
4.2. Cancer hotspot panel library preparation, sequencing, and data analysis
Cancer hotspot NGS library preparation was performed as previously described [17]. The sequencing data were aligned to human genome build 19 (HG19) and variants in EGFR were identified using NextGENe Software (Soft Genetics, State College, PA). The Integrative Genomics Viewer (IGV) was used to visually inspect the quality of read alignment and variant calls. A quality score of Q30 was used as filtering criteria to determine the sequence read quality. For a given sample, the minimum coverage requirement of targeted regions was 100X. Variants with VAFs as low as 2% may be identified.
4.3. therascreen®
The EGFR real-time PCR test (therascreen®) was performed as previously described [24]. therascreen® EGFR RGQ PCR Kit (Qiagen) [14] is an FDA approved real-time PCR-based in vitro diagnostic test that was used to cross-check the variant findings from the NGS-based, laboratory-developed AmpliSeq Cancer Hotspot Panel. The PCR test consists of eight separate PCR amplification mixes; amplification occurs in the Rotor-Gene Q MDx instrument. therascreen® can be used to detect common EGFR mutations, e.g., T790M, L858R, L861Q, G719X, S768I, various exon 19 deletions, and exon 20 insertions.
4.4. Sample selection
Fifty-four NSCLC specimens previously analyzed using EGFR therascreen® test were analyzed by the NGS-based study. These specimens included variants identified in late therascreen® PCR cycles, and equivocal variant calls that occurred outside the real-time PCR reference range. Four of the cases with low VAF and late PCR curves are shown in this manuscript. Additionally, two NSCLC specimens with complex and consecutive EGFR sequence changes (Fig. 4, Fig. 5) identified in the NGS test were further interrogated using therascreen® and shown. Since deep sequencing had been performed using the cancer hotspot NGS panel, clinically significant loci were read at >1000 reads. Variants with <5% VAF detected with NGS were evaluated further by manually reviewing the read qualities at each of the EGFR loci.
5. Financial disclosure
All authors have no relevant financial disclosure to report.
Funding
This work was supported by the general research funds in the Department of Laboratory Medicine, Cleveland Clinic.
Acknowledgements
The authors thanks Drs. Daniel Farkas and Marvin Natowicz for the critical reading and instructive modification of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.rmcr.2019.100901.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. Ca - Cancer J. Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. http://www.ncbi.nlm.nih.gov/pubmed/25559415 2015. [DOI] [PubMed] [Google Scholar]
- 2.Doxtader E.E., Cheng Y.W., Zhang Y. Molecular testing of non-small cell lung carcinoma diagnosed by endobronchial ultrasound-guided transbronchial fine-needle aspiration. Arch. Pathol. Lab Med. 2019;143(6):670–676. doi: 10.5858/arpa.2017-0184-RA. http://www.ncbi.nlm.nih.gov/pubmed/29372844 [DOI] [PubMed] [Google Scholar]
- 3.Lynch T.J., Bell D.W., Sordella R., Gurubhagavatula S., Okimoto R.A., Brannigan B.W., Harris P.L., Haserlat S.M., Supko J.G., Haluska F.G., Louis D.N., Christiani D.C., Settleman J., Haber D.A. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. http://www.ncbi.nlm.nih.gov/pubmed/15118073 [DOI] [PubMed] [Google Scholar]
- 4.Paez J.G., Janne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J., Naoki K., Sasaki H., Fujii Y., Eck M.J., Sellers W.R., Johnson B.E., Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. http://www.ncbi.nlm.nih.gov/pubmed/15118125 [DOI] [PubMed] [Google Scholar]
- 5.Pao W., Miller V., Zakowski M., Doherty J., Politi K., Sarkaria I., Singh B., Heelan R., Rusch V., Fulton L., Mardis E., Kupfer D., Wilson R., Kris M., Varmus H. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. U. S. A. 2004;101(36):13306–13311. doi: 10.1073/pnas.0405220101. http://www.ncbi.nlm.nih.gov/pubmed/15329413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lovly C.H., L, Pao W. My Cancer Genome; 2015. EGFR in Non-small Cell Lung Cancer (NSCLC)https://www.mycancergenome.org/content/disease/lung-cancer/egfr/ [Google Scholar]
- 7.Han S.W., Kim T.Y., Hwang P.G., Jeong S., Kim J., Choi I.S., Oh D.Y., Kim J.H., Kim D.W., Chung D.H., Im S.A., Kim Y.T., Lee J.S., Heo D.S., Bang Y.J., Kim N.K. Predictive and prognostic impact of epidermal growth factor receptor mutation in non-small-cell lung cancer patients treated with gefitinib. J. Clin. Oncol. 2005;23(11):2493–2501. doi: 10.1200/JCO.2005.01.388. http://www.ncbi.nlm.nih.gov/pubmed/15710947 [DOI] [PubMed] [Google Scholar]
- 8.Sequist L.V., Besse B., Lynch T.J., Miller V.A., Wong K.K., Gitlitz B., Eaton K., Zacharchuk C., Freyman A., Powell C., Ananthakrishnan R., Quinn S., Soria J.C. Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: results of a phase II trial in patients with advanced non-small-cell lung cancer. J. Clin. Oncol. 2010;28(18):3076–3083. doi: 10.1200/JCO.2009.27.9414. http://www.ncbi.nlm.nih.gov/pubmed/20479403 [DOI] [PubMed] [Google Scholar]
- 9.Keedy V.L., Temin S., Somerfield M.R., Beasley M.B., Johnson D.H., McShane L.M., Milton D.T., Strawn J.R., Wakelee H.A., Giaccone G. American Society of Clinical Oncology provisional clinical opinion: epidermal growth factor receptor (EGFR) Mutation testing for patients with advanced non-small-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J. Clin. Oncol. 2011;29(15):2121–2127. doi: 10.1200/JCO.2010.31.8923. http://www.ncbi.nlm.nih.gov/pubmed/21482992 [DOI] [PubMed] [Google Scholar]
- 10.van Eijk R., Licht J., Schrumpf M., Talebian Yazdi M., Ruano D., Forte G.I., Nederlof P.M., Veselic M., Rabe K.F., Annema J.T., Smit V., Morreau H., van Wezel T. Rapid KRAS, EGFR, BRAF and PIK3CA mutation analysis of fine needle aspirates from non-small-cell lung cancer using allele-specific qPCR. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0017791. http://www.ncbi.nlm.nih.gov/pubmed/21408138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Rios F., Angulo B., Gomez B., Mair D., Martinez R., Conde E., Shieh F., Tsai J., Vaks J., Current R., Lawrence H.J., Gonzalez de Castro D. Comparison of molecular testing methods for the detection of EGFR mutations in formalin-fixed paraffin-embedded tissue specimens of non-small cell lung cancer. J. Clin. Pathol. 2013;66(5):381–385. doi: 10.1136/jclinpath-2012-201240. http://www.ncbi.nlm.nih.gov/pubmed/23386666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong A.T., To R.M., Wong C.L., Chan W.K., Ma E.S. Evaluation of 2 real-time PCR assays for in vitro diagnostic use in the rapid and multiplex detection of EGFR gene mutations in NSCLC. Diagn. Mol. Pathol. 2013;22(3):138–143. doi: 10.1097/PDM.0b013e31827fedcc. http://www.ncbi.nlm.nih.gov/pubmed/23846439 [DOI] [PubMed] [Google Scholar]
- 13.Roma C., Esposito C., Rachiglio A.M., Pasquale R., Iannaccone A., Chicchinelli N., Franco R., Mancini R., Pisconti S., De Luca A., Botti G., Morabito A., Normanno N. Detection of EGFR mutations by TaqMan mutation detection assays powered by competitive allele-specific TaqMan PCR technology. BioMed Res. Int. 2013;2013:385087. doi: 10.1155/2013/385087. http://www.ncbi.nlm.nih.gov/pubmed/24364033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Therascreen® EGFR RGQ PCR Kit Handbook Version 1. Available at https://www.qiagen.com/ch/resources/download.aspx?id=db794cae-999b-4362-aba3-455ebfd807a5&lang=en.
- 15.cobas® EGFR Mutation Test v2. Available at: https://www.accessdata.fda.gov/cdrh_docs/pdf12/P120019S007c.pdf.
- 16.Allegrini S., Antona J., Mezzapelle R., Miglio U., Paganotti A., Veggiani C., Frattini M., Monga G., Balbo P., Boldorini R. Epidermal growth factor receptor gene analysis with a highly sensitive molecular assay in routine cytologic specimens of lung adenocarcinoma. Am. J. Clin. Pathol. 2012;138(3):377–381. doi: 10.1309/AJCPVAGIUC1AHC3Y. http://www.ncbi.nlm.nih.gov/pubmed/22912354 [DOI] [PubMed] [Google Scholar]
- 17.Reynolds J.P., Zhou Y., Jakubowski M.A., Wang Z., Brainard J.A., Klein R.D., Farver C.F., Almeida F.A., Cheng Y.W. Next-generation sequencing of liquid-based cytology non-small cell lung cancer samples. Cancer Cytopathol. 2017;125(3):178–187. doi: 10.1002/cncy.21812. http://www.ncbi.nlm.nih.gov/pubmed/28085233 [DOI] [PubMed] [Google Scholar]
- 18.Lindeman N.I., Cagle P.T., Aisner D.L., Arcila M.E., Beasley M.B., Bernicker E.H., Colasacco C., Dacic S., Hirsch F.R., Kerr K., Kwiatkowski D.J., Ladanyi M., Nowak J.A., Sholl L., Temple-Smolkin R., Solomon B., Souter L.H., Thunnissen E., Tsao M.S., Ventura C.B., Wynes M.W., Yatabe Y. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American pathologists, the international association for the study of lung cancer, and the association for molecular Pathology. Arch. Pathol. Lab Med. 2018;142(3):321–346. doi: 10.5858/arpa.2017-0388-CP. http://www.ncbi.nlm.nih.gov/pubmed/29355391 [DOI] [PubMed] [Google Scholar]
- 19.Walsh K., Wallace W.A., Butler R., Mackean M.J., Harrison D.J., Stirling D., Oniscu A. A cautionary lesson on the use of targeted methods for EGFR mutation analysis: a case report. J. Clin. Pathol. 2014;67(8):734–735. doi: 10.1136/jclinpath-2014-202313. http://www.ncbi.nlm.nih.gov/pubmed/24811487 [DOI] [PubMed] [Google Scholar]
- 20.Wu J.Y., Yu C.J., Chang Y.C., Yang C.H., Shih J.Y., Yang P.C. Effectiveness of tyrosine kinase inhibitors on "uncommon" epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin. Cancer Res. 2011;17(11):3812–3821. doi: 10.1158/1078-0432.CCR-10-3408. http://www.ncbi.nlm.nih.gov/pubmed/21531810 [DOI] [PubMed] [Google Scholar]
- 21.Vallee A., Le Loupp A.G., Denis M.G. Efficiency of the Therascreen(R) RGQ PCR kit for the detection of EGFR mutations in non-small cell lung carcinomas. Clin. Chim. Acta. 2014;429:8–11. doi: 10.1016/j.cca.2013.11.014. http://www.ncbi.nlm.nih.gov/pubmed/24269715 [DOI] [PubMed] [Google Scholar]
- 22.Silvestri G.A., Gonzalez A.V., Jantz M.A., Margolis M.L., Gould M.K., Tanoue L.T., Harris L.J., Detterbeck F.C. Methods for staging non-small cell lung cancer: diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143(5 Suppl) doi: 10.1378/chest.12-2355. http://www.ncbi.nlm.nih.gov/pubmed/23649440 e211S-50S. [DOI] [PubMed] [Google Scholar]
- 23.Rivera M.P., Mehta A.C., Wahidi M.M. Establishing the diagnosis of lung cancer: diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143(5 Suppl) doi: 10.1378/chest.12-2353. http://www.ncbi.nlm.nih.gov/pubmed/23649436 e142S-65S. [DOI] [PubMed] [Google Scholar]
- 24.Reynolds J.P., Tubbs R.R., Minca E.C., MacNamara S., Almeida F.A., Ma P.C., Pennell N.A., Cicenia J.C. EGFR mutational genotyping of liquid based cytology samples obtained via fine needle aspiration (FNA) at endobronchial ultrasound of non-small cell lung cancer (NSCLC) Lung Cancer. 2014;86(2):158–163. doi: 10.1016/j.lungcan.2014.09.003. http://www.ncbi.nlm.nih.gov/pubmed/25263855 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.