Abstract
Chronic urogenital schistosomiasis can lead to squamous cell carcinoma of the bladder. The International Agency for Research on Cancer classifies the infection with S. haematobium as a group 1 carcinogen, a definitive cause of cancer. By contrast, hepatointestinal schistosomiasis due to the chronic infection with S. mansoni or S. japonicum associated with liver periportal fibrosis, does not apparently lead to malignancy. The effects of culturing human epithelial cells, HCV29, established from normal urothelium, and H69, established from cholangiocytes, in the presence of S. haematobium or S. mansoni eggs were investigated. Cell growth of cells co-cultured with schistosome eggs was monitored in real time, and gene expression analysis of oncogenesis, epithelial to mesenchymal transition and apoptosis pathways was undertaken. Schistosome eggs promoted proliferation of the urothelial cells but inhibited growth of cholangiocytes. In addition, the tumor suppressor P53 pathway was significantly downregulated when exposed to schistosome eggs, and downregulation of estrogen receptor was predicted in urothelial cells exposed only to S. haematobium eggs. Overall, cell proliferative responses were influenced by both the tissue origin of the epithelial cells and the schistosome species.
Subject terms: Parasite host response, Bladder cancer, Oncogenes, Bladder cancer
Introduction
The blood flukes Schistosoma japonicum and S. mansoni are agents of hepatointestinal schistosomiasis in East Asia, Africa, northeastern South America and the Caribbean, whereas S. haematobium causing urogenital schistosomiasis (UGS) is present through Africa and the Middle East. It is estimated that 4.5 to 70 million disability adjusted life years (DALYs) are lost due to schistosomiasis1, and of >100 million cases of UGS in sub-Saharan Africa, 70 million show hematuria, 18 million major bladder pathology, and 10 million hydronephrosis that would eventually lead to kidney damage2,3. Many of the eggs of S. haematobium become trapped in host tissues, in particular urogenital organs, leading to inflammation and eventually squamous cell carcinoma of the bladder (SCC)4. Accordingly, and based on convincing epidemiological and pathophysiological findings, UGS has been classified as group 1 carcinogen by the International Agency for Research on Cancer5, although the cellular and molecular mechanisms underlying this infection-related carcinogenic process remain unclear. Women with UGS may suffer from female genital schistosomiasis (FGS)6 as consequence of the schistosome egg deposition in the uterus, cervix, vagina and vulva. Moreover, FGS has been associated with female infertility7 and increased susceptibility to HIV8.
Schistosome eggs in the bladder wall release metabolites, presumably to facilitate the egress to the lumen and subsequently to the external environment to propagate the transmission cycle. Mass spectrometric analysis of urine during UGS has revealed estrogen-like metabolites, catechol estrogen quinones (CEQ)-DNA-adducts and novel metabolites derived from 8-oxo-7, 8-dihydro-2′- deoxyguanosine (8-oxodG)9 representing potential bladder carcinogens that may directly damage the DNA, leading to somatic mutations in oncogenes and tumor suppressors10,11. By contrast, S. mansoni dwells in the mesenteric vessels releasing eggs that embolize within the presinusoidal capillary beds of the liver, inducing periportal fibrosis and portal hypertension. Hepatointestinal schistosomiasis does not apparently lead to cell malignant transformation in these organs5.
Epithelial carcinomas are typically classified as conventional and nonconventional carcinomas12; 90% of epithelial carcinomas are of the conventional type and result from either papillary or flat in situ lesions, while nonconventional carcinomas include SCC, adenocarcinoma, and small cell carcinoma. SCC of the bladder is characterized by invasive cells containing desmosomes with keratin formation12. Research of UGS-induced bladder cancer is challenging due to the absence of laboratory animal models that mirror the human disease; in rodent models the vast majority of S. haematobium adult worms reside in the mesenteric veins. Recently, a mouse model was developed by injecting eggs of S. haematobium into the bladder wall of mice provoking egg-associated pathogenesis similar to the human condition13,14. In addition, premalignant lessons associated with epithelial to mesenchymal (EMT)-like profiles occurred following co-administration of nitrosamine in this model15.
In this study, responses of urothelium (HCV29 cells) and bile duct epithelium (H69 cells) to eggs of either S. haematobium or S. mansoni were investigated. Cells were cultured in the presence of schistosome eggs, and cellular proliferation monitored in real time using the xCELLigence system16. Increased proliferation of urothelial cells was evident when exposed to schistosome eggs, in particular to S. haematobium eggs. On the other hand, eggs of both schistosome species induced cell death of cholangiocytes. These phenotypic effects were associated with dysregulation of genes involved in oncogenesis, epithelial-mesenchymal transition and apoptosis pathways. Future studies to decipher cellular and/or molecular mechanisms underlying the association between UGS and bladder cancer will contribute to the discovery of new interventions for this neglected tropical disease-related cancer.
Results
Schistosome eggs promoted growth of urothelial cells but inhibited cholangiocytes
A real-time cell proliferation assay was employed to measure the effect of co-culturing schistosome eggs with two informative human epithelial cell lines. Although we have previously studied human cholangiocyte cells H69 employing the xCELLigence Real Time Cell Assay17, we had not quantified the proliferation of the human urothelial cell line HCV29 using this approach. It is critical to set up a priori the conditions for cell proliferation analysis for every new cell line in the laboratory mediated xCELLigence, even though the same cell line has been already analyzed by others18. Therefore, cell titration experiments were undertaken in order to establish tractable assay parameters, including seeding cell density (Fig. S1). Notably, regardless of the initial number of seeded cells, all the tested conditions showed a delay of at least ~24 hours before the cell index (CI) started to increase, with 20,000 cells per well being the condition that reached a CI of ~3.0 within ~20 hours after it started to increase. Therefore, a seeding cell density of 20,000 cells per well was used in subsequent analyses.
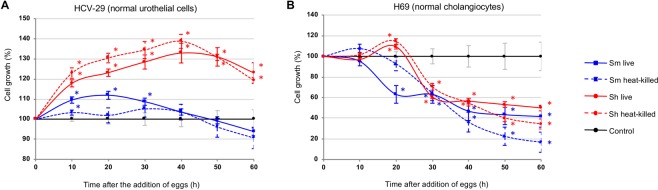
HCV29 cells were seeded in E-plates, and cultured until the CI reached ~1.0. Subsequently, the cells were starved for two to four hours in 1/20 diluted medium before being co-cultured with schistosome eggs. Within ~10 hours following addition of the eggs, proliferation of the cells accelerated, in an egg-concentration dependent manner in comparison to cells cultured without schistosome eggs. Eggs of both S. mansoni and S. haematobium induced cell proliferation, although eggs of S. haematobium induced higher proliferation, i.e. ~30% versus ~10% by eggs of S. haematobium and S. mansoni, respectively (Figs 1A and S2A). Notably, heat-killed eggs showed similar effects to live eggs on HCV29 cell proliferation (Fig. 1A). Similarly, H69 cells were co-cultured in the presence of schistosome eggs. In contrast to HCV29, schistosome eggs (live or dead) inhibited cell proliferation of cholangiocytes (Figs 1B and S2B). Although the S. haematobium eggs briefly induced cell proliferation of H69 cells between ~15 to 20 h post-incubation, eggs of both species overall reduced cell growth by ≥30% (Fig. 1B). The cell proliferation inhibition of H69 cells was evident by 24 h post-incubation for both schistosome species.
Figure 1.
Proliferation of urothelial cells and cholangiocytes in response to schistosome eggs, monitored in real time using the xCELLigence system. Non-contact co-culture of Schistosoma mansoni and S. haematobium eggs with HCV29 cells (Panel A) or H69 cells (Panel B) over time after the addition of the eggs. Cell growth is expressed as percentage of the Normalized Cell Index of cells co-cultured with eggs compared with control cells (control cell growth rate = 100%). All curves represent the averages of at least three technical replicates for the experiment and standard deviations are shown as error bars at each data point. Blue and red asterisks indicate levels of significance (P ≤ 0.01) of growth ratios of cells cultured with S. mansoni and S. haematobium eggs, compared to control cells cultured in the absence of eggs, respectively. Sm live: S. mansoni live eggs; Sm heat-killed: heat-killed S. mansoni eggs; Sh live: S. haematobium live eggs; Sh heat-killed: heat-killed S. haematobium eggs.
To summarize, the effect of the eggs on cell proliferation depended mainly on the cell type, i.e. cholangiocytes or urothelial, rather than on the schistosome species; however, S. haematobium eggs induced more proliferation of HCV29 cells than S. mansoni eggs (Fig. 1A), and a brief increase of H69 cell proliferation before the cell inhibition was evident for both schistosome species. Surprisingly, this proliferative effect was independent of egg viability. However, a cell proliferation increment was also evident when HCV29 cells were exposed to excretory-secretory products (ES) previously collected from either S. mansoni or S. haematobium eggs (Fig. S3), strongly suggesting that the proliferative effect was not due to the presence of a ‘foreign body’ inducing a non-specific effect. Overall, the same trend was evident for both tested conditions, i.e. eggs and ES. When ES products collected from both schistosome egg species were employed in the assay the cell proliferation increased in a concentration-dependent manner. Evidently 10 μg/ml, the highest tested concentration, induced the strongest proliferation, thus the cells started to die sooner. Remarkably, this effect was more pronounced when S. haematobium was employed (Fig. S3). Further studies comparing the compositions of the S. mansoni and S. haematobium ES products may lead to a better understanding of these differences.
Gene expression dysregulation was schistosome species- and contact time- dependent
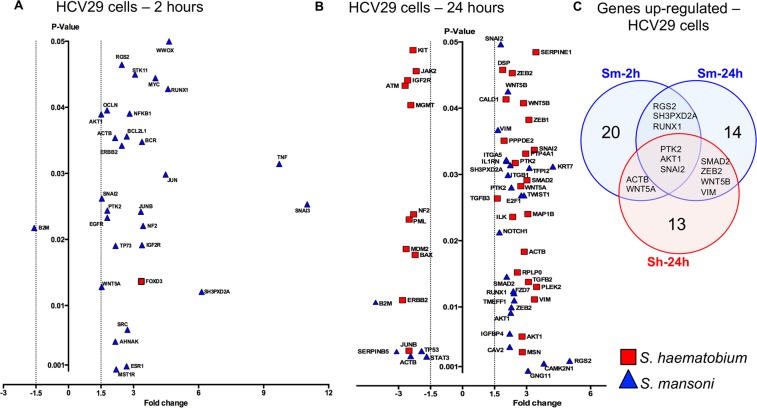
The effect of schistosome eggs on cell proliferation of HCV29 was evident within the first 10 hours after the incubation and persisted beyond 24 hours. Therefore, guided by the xCELLigence findings we decided to analyze the gene expression of cells at two time points post-incubation; cells exposed for 2 hours (‘early time point’) looking for early events in gene expression dysregulation, and cells exposed for 24 hours. Two gene qPCR arrays were employed to detect ‘early’ and ‘late’ molecular events that might be associated with high cell proliferation of the urothelial HCV29 cells co-cultured with schistosome eggs; (1) oncogenes and tumor suppressor gene array, and (2) epithelial to mesenchymal transition (EMT) gene array. The output raw data from these two gene arrays were combined in the final analysis. Whereas urothelial cells rapidly displayed gene dysregulation two hours after the addition of S. mansoni eggs, S. haematobium only induced the upregulation of the transcriptional repressor FOXD3 (Fig. 2A). At 2 hours, the early time point, S. mansoni eggs co-cultured with urothelial cells triggered the upregulation of 28 genes and downregulation of a single gene (B2M - a component of MHC class I molecules.) (Fig. 2A and Table S1). Tumor Necrosis Factor (TNF) and SNAI3 genes were the most upregulated genes in HCV29 cells co-cultured with S. mansoni eggs with fold changes of 9.72 and 11.01, respectively. Notably, the estrogen receptor 1 gene (ESR1) was upregulated 2.68-fold in HCV29 cells co-cultured with S. mansoni eggs at the ‘early’ time point.
Figure 2.
Schistosome eggs induced dysregulation of oncogenes, tumor suppressors and EMT-related genes in HCV29 cells over time. Volcano plots of HCV29 cell gene response to non-contact co-culture with S. mansoni or S. haematobium eggs as indicated, for 2 (Panel A) or 24 (Panel B) hours. Gene expression was measured using qPCR gene arrays designed to assess oncogenesis- and EMT- associated transcripts. Significantly dysregulated (P < 0.05) genes with >±1.5-fold-change, are shown. Panel C. Venn diagram compiled only with the HCV29 cell culture conditions that share differentially expressed genes (DEG) – only upregulated genes were shared among the 3 indicated conditions. (Table S2 includes the complete set of DEG indicated in the Venn diagram).
Urothelial cells exposed to S. mansoni eggs for 24 h (‘late time point’) displayed 29 dysregulated genes (Fig. 2B and Table S1). Interestingly, at this late time point, S. mansoni eggs induced downregulation of p53 tumor suppressor gene by 1.92-fold (Fig. 2B, shown as TP53, and Table S1). The expression of 7 genes dysregulated at the early time point remained altered at the late time point (ACTB, AKT1, B2M, RGS2, RUNX1, SH3PXD2A, SNAI2) (Fig. 2B and Table S1). Although at the early time point a single gene was upregulated, at the late time point 33 genes were dysregulated by S. haematobium eggs (Fig. 2B). Seven genes were dysregulated by both S. mansoni and S. haematobium eggs at the late time point (ACTB, AKT1, SMAD2, SNAI2, VIM, WNT5B, ZEB2) (Fig. 2B and Table S1). It is noteworthy that S. mansoni eggs upregulated 3.39 folds the expression of Insulin Like Growth Factor 2 Receptor (IGF2R) in urothelial cells at both time points examined, the same gene was 2.57-fold downregulated by S. haematobium (Fig. 2B and Table S1). Overall, 64 genes were dysregulated by S. mansoni and S. haematobium eggs when co-cultured with urothelial cells (Fig. 2B and Table S1). Cell culture conditions that displayed overlapped differentially expressed genes are shown in a Venn diagram (Fig. 2C). Surprisingly, relatively few upregulated genes were shared between early and late time points in HCV29 cells exposed to S. mansoni eggs, i.e. RGS2, SH3PXD2A, and RUNX1. Three genes, PTK2, AKT1, and SNAI2 were significantly upregulated in cells exposed to both schistosome species; S. mansoni eggs at both time points and S. haematobium eggs at the late time point. (Fig. 2C, and Table S2). No downregulated genes were shared among the different cell culture conditions for HCV29 cells (Table S2).
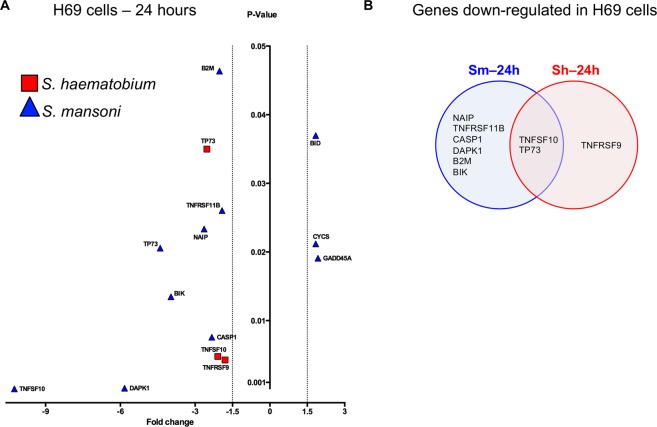
Given schistosome eggs inhibited the cell growth of the H69 human cholangiocyte cell line, an effect that became evident after ~24 hours post-incubation (above), an apoptosis gene array was employed to evaluate the gene expression dysregulation in cells exposed to schistosome eggs for 24 hours. Twelve genes were dysregulated when the cells were co-cultured with S. mansoni and S. haematobium eggs (Fig. 3A). Whereas both TNFSF10 and TP73 genes were downregulated by both schistosome species (Fig. 3B), S. mansoni eggs induced upregulation of three genes, i.e. BID, CYCS and GADD45A, and S. haematobium eggs induced downregulation of TNFRSF9, TNFSF10 and TP73 (Table S3).
Figure 3.
Schistosome eggs induced dysregulation of apoptosis-related genes in H69 cells. Volcano plot of the H69 cell gene response to non-contact co-culture with S. mansoni or S. haematobium eggs as indicated for 24 hours. Gene expression was measured using qPCR gene arrays designed to assess apoptosis-associated transcripts. Significantly dysregulated (P < 0.05) genes with >±1.5-fold-change, are shown. (Table S1 includes the complete data set of gene expression changes.) Panel B. Venn diagram compiled only with the H69 cell culture conditions that share differentially expressed genes (DEG) – only downregulated genes were shared between the two conditions indicated.
Colorectal cancer signaling pathway was significantly dysregulated by S. mansoni but not S. haematobium eggs
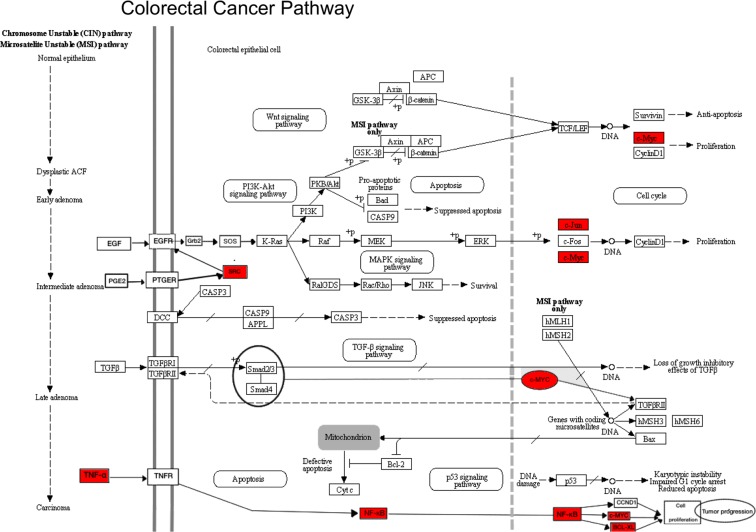
In order to detect significantly altered pathways in cells exposed to schistosome eggs a functional pathway analysis of differentially expressed genes was performed. The functional pathway analysis takes into consideration the differentially expressed genes detected in the gene expression arrays employed herein (see Methods) and determines which biological functions and canonical pathways are significantly associated with these dysregulated genes. At the early time point, i.e. 2 h after incubation of urothelial cells in the presence of S. mansoni eggs, genes associated to colorectal cancer metastasis signaling pathway were upregulated (Fig. 4)19. This pathway was significantly upregulated (with a positive z-score of 7.5 and P = 2.58 × 10−08) showing a cascade of significantly overexpressed genes (Table 1) including TNF, a pro-inflammatory cytokine implicated in colorectal cancers20 upregulated 9.72 fold, the oncogenes MYC and JUN upregulated more than 4 fold, among others (Table 1). Interestingly, urothelial cells co-cultured with S. mansoni eggs or S. haematobium eggs for 24 hours showed upregulation of WNT5B or both WNT5a and WNT5b, respectively (Fig. 2B,C). Notably, upregulation of WNT5a and WNT5b has been implicated in colon rectal cancer via non-canonical WNT signaling pathway21 However, the signaling pathway analysis did not support any significant correlation between the upregulated wnt genes in cells cultured with schistosome eggs and colorectal cancer pathway 24 hours after exposure.
Figure 4.
Significant dysregulation of genes involved in Colorectal Cancer Signaling Pathway in urothelial cells. The pathway was significantly dysregulated at early time point in S. mansoni eggs co-cultured HCV29 cells, and all the perturbed genes were upregulated (genes colored in red). Adapted from map05210 Colorectal cancer, KEEG Database63–65.
Table 1.
Differentially expressed genes associated with Colorectal Cancer Signaling Pathway identified in HCV29 cells exposed to eggs of Schistosoma haematobium (Sh) or S. mansoni (Sm).
| Gene name (ID) | HCV29 cell line* | Gene description | Reference | |
|---|---|---|---|---|
| Sh eggs | Sm eggs | |||
|
TNF (Tumor Necrosis Factor) |
NS | ↑ 9.72 (P = 0.031) 2 h | Pro-inflammatory cytokine implicated in colorectal cancers, associated with tumor growth, proliferation and metastasis | 19, 20, 66 |
|
RUNX1 (Runt Related Transcription Factor 1) |
NS | ↑ 4.59 (P = 0.042) 2 h and 24 h | Transcription factor with tumor suppressor functions associated with growth promotion of skin, oral, breast and ovarian tumor cells. | 67 |
|
MYC (Proto-Oncogene) |
NS | ↑ 4.01 (P = 0.044) 2 h | Transcription factor involved in cell proliferation, inhibition of cell differentiation and apoptosis. Over expressed in over 50% of all cancers, is considered a metastasis marker. | 68, 69 |
|
JUN (Proto-Oncogene, AP-1 Transcription Factor Subunit) |
NS | ↑ 3.34 (P = 0.029) 2 h | Transcription factor involves in cell cycle progression, cell survival, transformation and proliferation. Over expression of JUN has been related to p53 repression. | 70 |
|
NFKB1 (Nuclear Factor Kappa B subunit 1) |
NS | ↑ 2.82 (P = 0.039) 2 h | Transcription factor responsible for the regulation of ~300 genes involved with immune, growth and inflammation processes. Over expressed in many cancer types including colorectal, breast, liver, prostate and kidney. | 71, 72 |
|
SRC (Proto-Oncogene, Non-Receptor Tyrosine Kinase) |
NS | ↑ 2.72 (P = 0.006) 2 h | Proto-oncogene over expressed in colorectal, pancreas and prostate cancer, implicated in tumor progression, cell migration, angiogenesis, and metastasis. | 73, 74 |
|
BCL2 (Apoptosis regulator) |
NS | ↑ 2.69 (P = 0.035) 2 h | Anti-apoptotic regulator involved in several lymphomas. When over expressed, it has been associated with low survival of diffuse large B-cell lymphoma | 75– 77 |
*Fold change and P value, upregulation (↑), downregulation (↓), NS: non-significant. The exposure time of cells co-cultured with indicated schistosome eggs (2 h and/or 24 h) for which the specific gene expression dysregulation was detected is indicated after the Fold change.
Tumor suppressor P53-associated pathway was downregulated by both S. mansoni and S. haematobium eggs
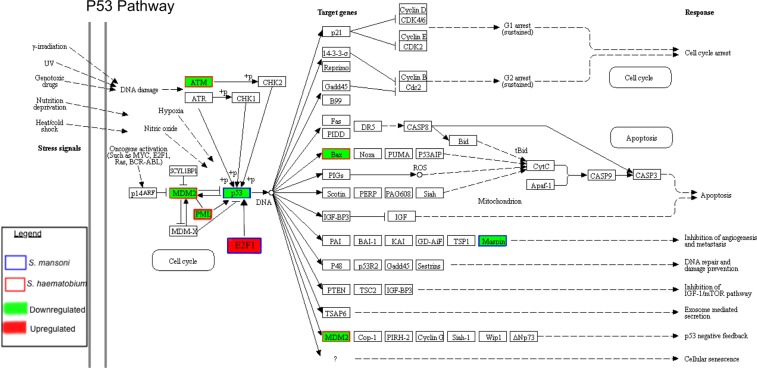
The tumor suppressor P53-associated pathway was significantly downregulated in urothelial cells exposed to both S. mansoni and S. haematobium eggs. Notably, different sets of genes within the pathway were altered by the two schistosome species (Fig. 5 and Table 2), i.e. S. mansoni reduced the expression of P53 (fold change of −1.92, P = 0.0033), E2F1 (fold change of 2.75, P = 0.0268) and SERPINB5 (fold change of −3.09 and P = 0.0032) genes, whereas S. haematobium induced downregulation of ATM (fold change of −2.69, P = 0.0433), MDM2 (fold change of −2.64, P = 0.0187), PML (fold change of −2.50, P = 0.0231) and the BCL2-Associated X (BAX) (fold change of −2.22 and P = 0.0178) (Table 2). Two of these proteins were selected to validate the gene array findings at the protein level, i.e. ELISAs were performed to quantify P53 and BAX in urothelial cells co-cultured in the absence or presence of S. mansoni or S. haematobium eggs for 2 or 24 hours (Fig. S4). In concordance with the gene array findings, no significant changes were observed at 2 hours after egg-exposure for both BAX and P53 proteins among the groups of cells exposed to no eggs, S. mansoni or S. haematobium eggs (Fig. S4). As expected, at 24 hours post-exposure no significant differences were observed between S. mansoni egg-exposed cells (Sm 24 hr) and control (Control 24 hr) for BAX protein; however, a higher level of this protein was quantified in cells exposed to S. haematobium eggs (Sh 24 hr). Similar findings were obtained for P53 (Fig. S4).
Figure 5.
Significant dysregulation of P53 pathway in urothelial cells exposed to either S. haematobium or S. mansoni eggs for 24 hours. P53 pathway highlighting upregulated or downregulated genes in red or green, respectively. Genes affected by S. mansoni or S. haematobium are indicated by blue or red squares, respectively. Adapted from map04115 p53 signaling pathway, KEEG Database63–65.
Table 2.
Differentially expressed genes associated with P53 Signaling Pathway identified in HCV29 cells exposed to either eggs of Schistosoma haematobium (Sh) or S. mansoni (Sm).
| Gene name (ID) | HCV29 cell line* | Gene description | Reference | |
|---|---|---|---|---|
| Sh eggs | Sm eggs | |||
|
SERPINB5 (Serpin Family B, member 5) |
NS | ↓ −3.09 (P = 0.003) 24 h | Inhibitor of cancer cell invasion, metastasis and angiogenesis. Under expression associated with breast, prostate, thyroid and skin tumors. | 78 |
|
ATM (Ataxia Telangiectasia Mutated, Serine/Threonine kinase) |
↓ −2.69 (p = 0.043) 24 h | NS | Mediator in kinase cascade that controls DNA damage response, cell-cycle progression, DNA recombination and apoptosis. Its downregulation inhibits p53 and cell cycle arrest. | 79, 80 |
|
MDM2 (MDM2 Proto-Oncogene) |
↓ −2.64 (P = 0.018) 24 h | NS | Ubiquitin E3 ligase that degrades P53 is under the control of p53 in a regulatory feedback loop (Fig. 4) | 81, 82 |
|
PML (Promyelocytic Leukemia) |
↓ −2.50 (p = 0.023) 24 h | NS | Nuclear protein involved in cell cycle progression, DNA damage response, and apoptosis, i.e. key regulator in the p53 pathway. PML-deficient mice exhibit apoptotic defects. | 83– 85 |
|
BAX (BCL2 Associated X, Apoptosis Regulator) |
↓−2.22 (P = 0.017) 24 h | NS | Major promoter of apoptosis is regulated by p53, it has been involved in tumorigenesis by interfering with cell death. | 86 |
|
P53 (Tumor Protein P53 or TP53 as in Fig. 2) |
NS | ↓ −1.92 (P = 0.003) 24 h | Tumor suppressor, a key transcription factor inhibiting cancer development, being inactivated in most tumors. It responds to cell stress by activating genes responsible for DNA repair, cell cycle arrest, anti-angiogenesis, apoptosis and autophagy | 87, 88 |
|
E2F1 (E2F Transcription Factor 1) |
NS | ↑ 2.75 (P = 0.026) 24 h | Transcription factor that regulates cell cycle progression, involved in either oncogenesis or tumor suppression depending on cellular signals. E2F1 over expression associated with oncogenic transformation in rodent embryonic fibroblasts and tumorigenesis. | 89, 90 |
**Fold change and P value, upregulation (↑), downregulation (↓), NS, non-significant. The exposure time of cells co-cultured with indicated schistosome eggs (2 h and/or 24 h) for which the specific gene expression dysregulation was detected is indicated after the Fold change.
Estrogen receptor and beta-estradiol altered by schistosome eggs
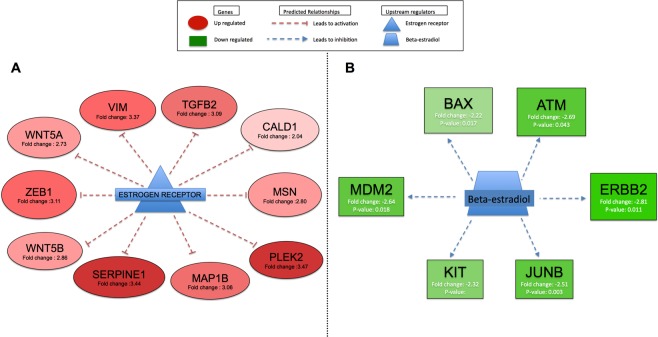
Notably, the estrogen receptor (ESR1) was upregulated 2.68-fold (P = 0.000812) in urothelial cells co-cultured with S. mansoni eggs at the early time point (Fig. 2A), but not with S. haematobium eggs. However, the upstream regulatory analysis (URA) of differentially expressed genes predicted both the estrogen receptor and beta estradiol to be inhibited in urothelial cells co-cultured with S. haematobium eggs for 24 hours, but not with S. mansoni eggs (Fig. 6).
Figure 6.
Upstream regulatory analysis (URA) of differentially expressed genes (DEGs) predicted the estrogen receptor and beta-estradiol (P < 0.05) to be inhibited in HCV29 cells co-culture for 24 h with S. haematobium eggs. Panel A. Significantly upregulated genes are shown in red. These genes, negatively regulated (T-symbol) by the estrogen receptor, were found to be upregulated in HCV29 cells co-cultured with S. haematobium eggs for 24 hours; red dashed lines indicate that these genes were activated. Therefore, the URA analysis significantly predicted the estrogen receptor as an inhibited upstream regulator of this set of genes in this dataset. Panel B. Significantly downregulated genes are shown in green. These genes, positively regulated (arrow) by beta-estradiol, were found to be downregulated in HCV29 cells co-cultured with S. haematobium eggs for 24 hours; blue dashed lines indicate that these genes were inhibited in our dataset. Therefore, the URA analysis significantly predicted beta-estradiol as an inhibited upstream regulator of this set of genes in the dataset.
Discussion
Infectious diseases are associated with >20% of cancers in the developing world22. About a dozen pathogens including human papillomavirus, Epstein-Barr virus and hepatitis B virus are well known agents of cancer-associated infections. Infections with several parasitic flatworms are also associated with carcinogenesis5,22. The International Agency for Research on Cancer (IARC) categorizes infection with the food-borne trematodes Opisthorchis viverrini and Clonorchis sinensis, both liver flukes, and the blood fluke Schistosoma haematobium as group 1 biological carcinogens5. More recently we have shown that the chronic infection with the liver fluke Opisthorchis felineus might also be associated with liver cancer23. In addition to parasitism directly damaging development, health and prosperity of infected populations, chronic infection with these liver and blood flukes leads to cholangiocarcinoma (CCA) (bile duct cancer) and squamous cell carcinoma (SCC) of the urinary bladder, respectively5. By contrast, chronic infection with phylogenetic relatives, including trematodes of the phylum Platyhelminthes which are major pathogens, has not been proven to be carcinogenic. These noteworthy differences suggest that either helminth-specific metabolites contribute to tumorigenesis and/or that certain tissues or organs directly or indirectly exposed to the parasite and/or parasite-derived metabolites are particularly susceptible to infection-induced malignancy24. Consequently, aiming to compare/contrast the effect of eggs of a well-established carcinogenic species of schistosome, i.e. S. haematobium, with eggs of a non-carcinogenic schistosome, i.e. S. mansoni, established epithelial cells derived from two discrete tissues, HCV29, from human urothelium, and the H69, from cholangiocytes, were cultured in the presence of eggs of S. haematobium or S. mansoni. Remarkably, epithelial cells from urinary and biliary tract displayed differential responses to S. haematobium and S. mansoni. The proliferative response and associated changes in signaling pathways responses of these cells appear to have been influenced by both the tissue origin of the epithelial cells and the schistosome species.
Both S. haematobium and S. mansoni eggs stimulated cellular growth in HCV29 urothelial cells when compared to control cells in the absence of schistosome eggs. Moreover, genes strongly associated with cell migration (PTK2 and VIM)25, proliferation (AKT1)26, inhibition of apoptosis (SNAI2)27, and carcinogenesis mediated by the TGFβ signaling pathway (SMAD2 and ZEB2)28 were upregulated in cells exposed to both species. Therefore, it is tempting to speculate that the carcinogenicity of the chronic infection with these parasites not only depends on the schistosome species but also the host tissue exposed to antigens and excretory-secretory products from the eggs. More studies, including in vitro cell co-culture experiments with informative cell lines derived from different tissues will contribute to the resolution of this issue. Several cancer-inducing parasites stimulate cellular proliferation when co-cultured with mammalian cells. Excretory-secretory products (ES) from the liver flukes, O. viverrini and C. sinensis promoted growth of mouse fibroblast NIH-3T3 and human kidney embryonic epithelial cells HEK29329,30. S. haematobium egg antigens induced cell proliferation, apoptosis inhibition and altered cell migration in vitro31. In addition, cells pretreated with S. haematobium egg antigens induced tumor development when injected in immunodeficient mice32. Moreover, intravesical administration of S. haematobium total antigen into CD-1 mice induces a high incidence of urothelial dysplasia33. Other reports reveal that human urothelial and bovine endothelial cells exhibit increased proliferation following exposure to soluble egg antigen of S. haematobium34. S. mansoni also promotes proliferation of some cell lineages as granulomas are a rich source of cytokines and growth factors and stimulates tissue proliferation and fibroplasia in mammalian tissue35 and total antigen from S. mansoni egg stimulates fibroblasts in vitro36. S. mansoni soluble egg antigens induce proliferation of H69 cells and murine cholangiocytes37. However, herein we showed that when each of the species of schistosome eggs were co-cultured with the H69 cholangiocyte cell line a consistent decrease in cellular proliferation, even using heat-inactivated schistosome eggs, was observed.
Schistosome soluble egg antigens stimulate cellular proliferation of murine splenocytes whereas higher concentrations induce apoptosis in a dose-dependent fashion38. Antigens from both S. japonicum and S. mansoni negatively regulate hepatic stellate and CD4 + ve T cells39,40. Some of the disparities among these reports may relate to methods to prepare egg-secreted proteins (ESP) or soluble egg antigens (SEA). Egg-secreted proteins differ significantly when compared to soluble egg antigen in terms of protein sizes and the presence of proteases that are found in ESP but not in SEA41. In addition, we cannot completely rule out the effect observed in the current study was due to a foreign body co-cultured with the cells. However, we speculate that if that was the case the same unspecific effect should have been observed on both cell lines regardless the schistosome species. In this regard, the findings showed (1) species-specific effects on the same cell line, e.g. even though both schistosome eggs stimulated cell proliferation of HCV29, S. haematobium eggs induced more proliferation than S. mansoni eggs, and (2) differences on effect intensities between live and heat-killed eggs, e.g. even though similar effects were induced by heat-killed or live S. mansoni eggs, the latter clearly had a stronger effect on both stimulating HCV29 cells and inhibiting H69 cells. By contrast, S. haematobium heat-killed eggs slightly induced a stronger effect on the cells compared to live eggs. These differences induced by the two schistosomes on different cells presumably would not have been detected if the effects were due to just the presence of a foreign inert body. Last, there was increased proliferation when the cells were cultured in the presence of ES only collected from the eggs, i.e. no eggs were used in the experiment. This strongly suggested the effect was not due to the presence of an inert body in the experimental setting.
The Ingenuity Pathway Analysis (IPA) identified the colorectal metastasis-signaling pathway significantly dysregulated in urothelial HCV29 cells co-cultured with S. mansoni eggs, but not with S. haematobium eggs after being co-cultured for 2 hours. In contrast, exposure of the urothelial cells to S. haematobium eggs for 24 hours induced upregulation of WNT5a and WNT5b. Although not significantly associated to Colorectal cancer pathway in our analysis, these genes have been involved in carcinogenesis, including colorectal cancer via non-canonical WNT signaling pathway21. On the other hand, S. mansoni eggs induced upregulation of these same genes (WNT5a and WNT5b) at both time points. A link has been proposed between infection with S. mansoni and colorectal cancers42. IARC classifies infection with S. haematobium as a group 1 carcinogen, whereas infection with S. mansoni is included in group 3, indicating there is insufficient evidence to determine its carcinogenicity5. Nonetheless, schistosomal colitis is commonly associated with earlier onset of multicentric colorectal cancer, and notably mucinous adenocarcinoma often presents at an advanced stage in regions endemic for infection with S. mansoni43. Moreover, a higher incidence of altered p53 expression with schistosomal colitis-associated colorectal cancer suggests an association between schistosomiasis and alterations in p53 activation43. Intestinal schistosomiasis caused by S. japonicum exhibits a geographic distribution that overlaps with populations affected by early onset of colon cancer in China44. In addition, several genes were upregulated in HCV29 cells at the early time point when exposed to S. mansoni eggs. Interestingly, only β-2 microglobulin (B2M) was downregulated 1.5 folds and more than 3 folds 2- and 24-hours post-exposure, respectively. B2M belongs to the major histocompatibility complex (MHC) class I45. Because B2M is not only markedly downregulated in colorectal cancer tissue compared with normal mucosa46, but also plays a prominent role in antigen presentation, it is tempting to speculate that its downregulation might impede host clearance of the fluke infection.
Surprisingly, significantly dysregulated pathways were not identified in HCV29 cells co-cultured with S. haematobium eggs at the early time point (2 hours); FOXD3 was the only gene to be upregulated. This transcription factor is a member of the forkhead family with a key role in progenitor cell pluripotency and differentiation, and regulation of cellular transition from naïve to primed pluripotency47. FOXD3 is upregulated in renal cancer and endometrial tumors and downregulated in cancers of the colon and cervix48. Remarkably, by 24 hours post-exposure not only were many more genes were upregulated in HCV29 cells exposed to S. haematobium eggs, but also the number of dysregulated genes outnumbered those dysregulated by S. mansoni eggs. It is tempting to speculate that this is related to the oncogenic nature of S. haematobium infection, although further studies are needed to clarify this point. Several genes dysregulated by either or both species of schistosomes have been linked to inflammation and schistosomiasis-associated disease but have not been associated with cancer, at least in a direct manner, e.g. Tumor Necrosis Factor (TNF) or Nuclear Factor Kappa B subunit 1 (NFK-B). Notably, elevated TNF levels can occur during acute schistosomiasis and severe pathology is associated with elevated circulating levels of TNF and TNF receptors (TNFRs). Moreover, TNF might be involved in maintaining S. mansoni adult viability in the portal system49, and would be required by the parasite for egg-laying and egg-excretion from the host50. Consistent with this hypothesis, here TNF was upregulated >9 folds by eggs of S. mansoni but not S. haematobium.
Estradiol-like metabolites have been characterized both in S. haematobium parasites and in urine during urogenital schistosomiasis9. We have recently predicted pathways and enzymes that are involved in the production of these metabolites and emphasized their potential effects on the dysregulation of the tumor suppressor gene p53 expression during urogenital schistosomiasis (reviewed in51). Mutations of p53 in the context of UGS-induced bladder cancers from Angola along with sialylated glycans have been suggested as surrogate biomarkers of bladder carcinogenesis associated with S. haematobium infection highlighting links between infection and cancer development52. Eggs of S. haematobium express the sialyl-Lewis sLea and sLex antigens in mimicry of human leukocytes glycosylation, which may play a role in the colonization and disease dissemination. Tumor suppressors including P53 and genes involved in the P53 pathway and metalloprotease inhibitors were downregulated when exposed to schistosome eggs. In bladder cancers, recurrent mutations occur in >30 other genes involved in cell proliferation, differentiation, genetic stability, and specifically cell-cycle regulation, chromatin regulation, and kinase signaling pathways53. The downregulation of p53 is characteristic of cancers in general and SCC of the bladder in particular. In this regard, we found the tumor suppressor P53-associated pathway significantly downregulated in urothelial cells exposed to both S. mansoni and S. haematobium eggs. The P53 signaling pathway is responsible for hundreds of downstream transcriptional targets involved in the inhibition of cell growth, induction of apoptosis and regulation of metabolism in response to oncogene activation and other signals54. In a mouse model of infection-related bladder cancer, S. haematobium egg-induced bladder urothelial abnormalities were dependent on p53 in a host sex dependent manner10,14. We decided to validate the dysregulation of P53 and BAX proteins performing ELISA assays, key genes detected in our dataset involved in carcinogenesis. The protein quantification for BAX and P53 concurred with the gene array data 2 hours after exposure. In addition, as in the gene array data BAX protein was not dysregulated in S. mansoni egg-exposed cells at 24 hours post-exposure. However, BAX and P53 proteins were over expressed in S. haematobium egg-exposed cells and P53 was not down-regulated in S. mansoni egg-exposed cells. Among confounders that might explain the unexpected differences, different batches of eggs and cells employed in the two experiments, which were separated widely in time. In addition, recent studies highlight the lack of correlation between mRNA and protein levels for many genes, including those involved in carcinogenesis. This would reflect the complexity of the gene expression regulation at several levels including post-transcriptional regulation of mRNA and post-translational modifications of proteins55,56. The latter could also explain the inconsistencies between the qPCR gene array signals for the bax and p53 genes and concentrations of BAX and P53 in cells exposed to schistosome eggs. Future studies involving high throughput transcriptomic and proteomic analyses from cells exposed to eggs and excretory-secretory products from eggs may clarify these contradictory findings.
Upstream regulatory analysis (URA) of differentially expressed genes (DEGs) predicted that the exposure of urothelial cells to eggs of S. haematobium but not S. mansoni downregulated expression of the estrogen receptor. This phenomenon was previously reported in other experimental models showing that an estradiol-like molecule antagonist to estradiol represses the transcriptional activity of the estrogen receptor (ER)57. We have reviewed how an estrogen-DNA adduct mediated pathway may be involved in the pathogenesis of the squamous cell carcinoma of the bladder associated with urogenital schistosomiasis58,59. Extracts of S. haematobium eggs induced tumor-like phenotypes in cultured cells and, in addition, estrogen-derived, reactive metabolites occurred in this pathogen and in sera during UGS including catechol estrogen quinones (CEQ) and CEQ-DNA-adducts9.
To conclude, H69 and HCV29 cells displayed differential responses to eggs of S. haematobium and S. mansoni, supporting the hypothesis that the carcinogenic nature of infection with S. haematobium is related to its interaction with the specialized epithelium of the human bladder. Proliferative responses of these cells were influenced by both the tissue origin of the epithelial cells and the schistosome species. Focused studies of the molecular mechanisms underlying the association between UGS and SSC of the bladder are expected to enhance prospects to manage this neglected tropical disease-related cancer.
Materials and Methods
Ethics statement
Male LVG hamsters were purchased from Charles River (Wilmington, MA) and maintained at the Biomedical Research Institute’s (BRI), Rockville, MD, animal facility accredited by the American Association for Accreditation of Laboratory Animal Care (AAALAC; #000779). BRI is a USDA registered animal facility (51-R-0050), and has an Animal Welfare Assurance on file with the National Institutes of Health, Office of Laboratory Animal Welfare (OLAW), A3080-01. Maintenance of hamsters exposed to Egyptian strain of S. haematobium cercariae and harvesting of tissues were approved by the BRI Institutional Animal Care and Use Committee. All procedures employed were consistent with the Guide for the Care and Use of Laboratory Animals.
Mice infected with the NMRI (Puerto Rican) strain of Schistosoma mansoni were supplied by BRI and maintained at The George Washington University (GW) following protocols approved by the GW Institutional Animal Case and Use Committee.
Schistosome eggs and excretory-secretory products (ES) from eggs
Isolation of eggs from liver of experimentally-infected rodents was performed as previously described60. In brief, 3–5 dissected livers from S. mansoni-infected mice, and 2–3 livers from S. haematobium-infected Syrian golden hamsters were briefly washed in 70% ethanol solution, chopped finely with a sterile scalpel blade and transferred to a 50 ml conical tube containing 45 ml phosphate-buffered saline (1X PBS), 2% antibiotic-antimycotic solution (Life Technologies) and 0.5% clostridial collagenase (Sigma). The minced livers were incubated with gentle shaking overnight at 37 °C, centrifuged at 400 g for 5 min, and washed with 1X PBS 3 times. After the last wash, the pellet was resuspended in 25 ml 1X PBS, passed successively through sterile 250 μm and 150 μm sieves, and the filtrate centrifuged at 400 g for 5 min. The liver mixture filtrate mixture was centrifuged at 400 g for 5 min, the supernatant discarded and the pellet resuspended in 3 ml of 1X PBS, applied to a column of Percoll, prepared by mixing 8 ml of Percoll (GE Healthcare Bio-Science AB) with 32 ml of 0.25 M sucrose, and centrifuged at 800 g for 10 min. Liver cells and debris that remained on the top of the Percoll were removed with a Pasteur pipette. The schistosome eggs, which pelleted tightly at the bottom of the tube, were washed three times with 1X PBS to remove residual host tissues. Further purification was achieved by resuspending the eggs in 0.5 ml of 1X PBS and a second Percoll column, prepared by mixing 2.5 ml of Percoll with 7.5 ml of 0.25 M sucrose. The eggs were pelleted, washed as before, counted and cultured in DMEM, 10% FBS and 2% antibiotic-antimycotic at 37 °C, 5% CO2. Egg viability was evaluated by hatching an aliquot of a few hundred eggs in water under light. When heat-killed eggs were included as controls, eggs were transferred to a 1.5 ml tube and incubated for 10 min at 80 °C in a heat-block. For excretory-secretory products (ES), eggs were washed 3 times in 1X PBS, resuspended in DMEM, 2% antibiotic-antimycotic solution and protease inhibitor cocktail (Amresco), and incubated in 2 ml of serum-free media in 12-well plates overnight at 37 °C, 5% CO2. The supernatant (i.e., ES) was collected, clarified by centrifugation at ~14,000 rpm, 4 °C, 30 min, transferred to a new tube where additional protease inhibitor cocktail was added and stored at −80 °C. Protein concentrations of ES was determined using the bicinchoninic acid assay (BCA kit, Pierce, Rockford, IL).
Human epithelial cell lines
The human cholangiocyte H69 cell line is a SV40-transformed human bile duct epithelial cell line derived from a normal liver61. The H69 cells were used between passages 10 and 30 and grown in H69 medium that included 43.82% DMEM medium (Dulbecco’s Modified Eagle’s Medium), 43.82% DMEM/Ham’s F12, 10% fetal bovine serum (FBS), supplemented with 1% adenine, 0.1% insulin, 0.1% epinephrine, 0.1% T3-T, 0.03% epidermal growth factor (EGF), 0.03% of a 4% hydrocortisone dilution in DMEM and 1% penicillin/streptomycin (PS). The human HCV29 cell line is derived from non-malignant urothelium of the ureter62. The HCV29 cells, used between passages of 10 to 30, were maintained in DMEM supplemented with 10% FBS and 1% PS (complete DMEM). Passages of both cell lines were undertaken when the confluence reached 90% in a 75 cm2 flask in a 1/10 dilution using trypsin to dislocate cells. Both cell lines tested negative for mycoplasma (not shown).
Real time assessment of cell proliferation by xCELLigence system
Cellular proliferation of H69 and HCV29 cells was assessed using the xCELLigence DP system (ACEA Biosciences, San Diego, CA, USA), designed to monitor events in real time by measuring electrical impedance across inter-digitated microelectrodes integrated on the bottom of tissue culture E-plates, see http://www.aceabio.com/main.aspx16. H69 cells were seeded at 5,000 cells/well in E-plates in H69 medium, cultured for one day, rinsed in 1X PBS and cultured in 1/20 diluted H69 medium in 50% DMEM and 50% DMEM/Ham’s F12, containing 1% of PS, i.e. 0.5% FBS final concentration, as described17. For HCV29 cells a cell number titration experiment was first performed (Fig. S1), being 20,000 cells/well the selected number of cells for subsequent experiments. HCV29 cells were seeded in E-plates in complete DMEM medium, cultured for one day, rinsed with 1X PBS, and cultured in 1/20 diluted DMEM media. Both cell lines were fasted in 1/20 diluted medium for 4–6 h after which the cells were co-cultured with viable or heat-killed (below) schistosome eggs. The eggs were placed in a tissue culture insert transwell where the base of the insert is a polyethylene terephthalate (PET) membrane with a median pore size of 0.4 μm (ACEA catalog no. 06465382001). The transwell carrying the eggs were inserted into tissue culture plates were the cells were growing. Consequently, cells cultured on plastic in the tissue culture wells were exposed to the excretory-secretory products (ES) diffusing from the insert chamber carrying the eggs. For experiments where HCV29 cells were cultured in the presence of ES previously collected from S. mansoni or S. haematobium eggs as described above, the cells were fasted in 100 μl of 1/20 diluted medium for 4–6 hours in the xCELLigence platform after which 100 μl of 2X ES of the desire final concentration, i.e. no ES (control), 10, 2 or 0.4 μg/ml, was added to the well. The cellular growth was monitored in real time for at least 60 hours. The normalized cell index (CI), obtained by dividing the CI value at each time point by the CI at the time of addition of the eggs was ascertained with the assistance of the RTCA Software 1.2 (ACEA). Normalized CI values were imported into Microsoft Excel for analysis where the cellular growth of cells exposed to the eggs was expressed as the percentage of the normalized CI of cells cultured with indicated eggs compared to (i.e. divided by) that of control cells cultured in 1/20 diluted media. (Control cells growth rate = 100%.) Statistical significance among the groups was assessed using Analysis of Variance (ANOVA) and Student’s t-test. P values of ≤0.05 were considered significant.
Epithelial cells co-cultured with schistosome eggs
H69 and HCV29 cells were seeded into wells of 6-well plates at a cell density ranging from 2.5 × 105 and 3 × 105 cells/well in 2 ml media and cultured until 80–90% confluence (usually 12–24 h) before being co-cultured with schistosome eggs. Before the addition of the eggs, the cells were starved for at least four hours in 1/20 dilution with 1% PS of their respective media. Thereafter, wells containing starved cells were fitted with Corning 6 Well Transwell Inserts, 0.4 μm PET Membrane, TC Treated, and ~2.5 × 103 eggs of S. mansoni or S. haematobium were added to the Transwell. The Transwell ensured exposure of the cells to the excretory-secretory products (ES) of the eggs. Transwells were removed after two hours (‘early time point’) or 24 hours (‘late time point’) at which time cells were harvested for RNA isolation and gene expression analysis, or protein quantification. Cells not exposed to eggs but otherwise cultured similarly served as the control.
ELISA-based quantification of selected human proteins
Human urothelial HCV29 cells exposed to schistosome eggs or control cells exposed to no eggs for 2 or 24 hours in 6 -well plates in triplicates were harvested for protein isolation and quantification of selected human proteins using commercially available ELISA kits (Abcam). Based on the qPCR gene array findings three human proteins were measured by ELISA, i.e. BAX (https://www.abcam.com/human-bax-elisa-kit-ab199080.html), MASPIN (SERPIN B5) (https://www.abcam.com/human-maspin-elisa-kitab218269.html), and P53 (https://www.abcam.com/human-p53-elisa-kit-ab171571.html). At the indicated time points the cell supernatant was removed and the cells washed twice with 1x PBS. The cells were exposed to 20μl of protease inhibitor cocktail (Calbiochem 539137), collected using a cell scraper and transferred to a 1.5 ml tube. One ml of 1xPBS was used to rinse the well and transferred to the corresponding tube that was kept on ice all the time. Thereafter, the cells were pelleted at 400 × g, for 10 min at 4 °C, and the cell pellet resuspended in 1 ml of 1xPBS. The cells were centrifugated as above and the pellet resuspended in 60μl of CEB PTR buffer (ELISA Abcam kit). The protein concentration for each sample was determined using the bicinchoninic acid assay (BCA kit, Pierce, Rockford, IL), and the input total protein concentration was 20 ng/ml for each reaction. The ELISA assays were performed following the manufacturer’s instructions. In brief, a colorimetric reaction became apparent after the addition of the TMB substrate to the protein lysate mixed with the protein-specific antibodies in a ‘sandwich’ format in a 96-well plate. Standard curves for each target protein were included in the assay as positive control and as reference for the quantification. Optical densities at 450 nm were determined using a spectrophotometer plate reader, and the raw readouts were exported and analysed using Graphpad Prism software. Three biological replicates with three technical replicates per biological repeat were carried out. Levels of MASPIN were below the level of detection of the kit.
RNA isolation and gene arrays analysis
Total RNA was isolated from cells using the miRNeasy Mini kit (Qiagen) following the manufacturer’s instruction. Briefly, 700 μl of QIAzol Lysis Reagent was added to each cell pellet and homogenized using a sterile micropestle (Kontes, Vineland NJ). Chloroform, 140 μl, was added, contents mixed, and clarified at 12,000 × g, 15 min at 4 °C. The upper aqueous phase was transferred to a new tube containing 100% ethanol; subsequent washes through a spin column and on-column DNase treatment were performed. RNA was eluted in ~35 μl of RNAse-free water, and concentration, purity and integrity evaluated using spectrophotometry (Nanodrop 1000) and Agilent 2100 Bioanalyzer. The RNA preps were stored at −80 °C until processed for cDNA synthesis using 300 to 500 ng of total RNA and qPCR following the PrimerPCR Assay protocol (Bio-Rad). The qPCR experiments were performed using a Bio-Rad iCycler iQ5 with an initial activation step of 95 °C for 10 min followed by 40 cycles of 95 °C for 10 sec and 60 °C for 1 min. A melting curve analysis from 55 °C to 95 °C and 0.5 °C temperature increment every 30 sec was included at the end of the run. Specific gene arrays were selected; (1) Oncogenes and tumor suppressor genes (SAB Target List), http://www.bio-rad.com/en-uk/prime-pcr-assays/predesigned-plate/sybr-green-oncogenes-tumor-suppressor-genes-sab-target-list-h96; (2) Epithelial to mesenchymal transition (EMT) (SAB Target List), http://www.bio-rad.com/en-uk/prime-pcr-assays/predesigned-plate/sybr-green-epithelial-mesenchymal-transition-emt-sab-target-list-h96; and (3) Apoptosis (SAB Target List), http://www.bio-rad.com/en-uk/prime-pcr-assays/predesigned-plate/sybr-green-apoptosis-sab-target-list-h96. These predesigned arrays allow the detection of 84 genes along with controls that include DNA contamination control, positive PCR control, RNA quality control, and reverse transcription control. Ct values were exported and analyzed using Prime PCR Analysis software (http://www.bio-rad.com/en-us/sku/genestudy-1-0-030-1023-primepcr-analysis-software), and relative quantitation performed using the 2−ΔΔCt method employing a panel of three housekeeping genes: glyceraldehyde-3-phosphate dehydrogenase, hypoxanthine phosphoribosyltransferase 1, and TATA-box binding protein. Control groups (cells exposed to media alone) were used as calibrator samples. Three biological replicates were performed. Venn diagrams were performed using the online free tool http://bioinformatics.psb.ugent.be/webtools/Venn/.
Gene pathway analysis
Functional pathway and upstream regulatory analysis (URA) of differentially expressed genes (DEGs) was performed with the assistance of Ingenuity Pathway Analysis (IPA QIAGEN Redwood City, www.qiagen.com/ingenuity) software. In brief, the dataset from the real time PCR analysis was imported into the IPA to define the genes which were significantly up and downregulated and P-values were set up equal or lower than 0.05 as the significant threshold. IPA includes Benjamini-Hochberg analysis of multiple testing correction based on the Fisher’s exact test p-value to calculate false discovery rate between multiple comparisons. Additionally, IPA uses Right-Tailed Fisher’s Exact Test to determine which biological functions and canonical pathways are significantly associated with the genes of interest compared with the whole Ingenuity knowledge base (http://www.ingenuity.com/science/knowledge-base). In addition, to determine predictions about upstream processes, IPA provides a z-score, where z-scores >2 or <−2 are considered significant. This analysis considers the directional effect of one molecule or process on another, and the direction of this change in the dataset, i.e. it represents the statistical measure of the correlation between the direction of the change (up- or down- regulation) and gene expression (https://chhe.research.ncsu.edu/wordpress/wp-content/uploads/2015/10/IPA-Data-Analysis-training-slides-2016_04.pdf). The IPA analysis also enables the identification of biological networks, global functions and functional pathways for a particular dataset. The analysis estimates the significance of the genes in the network, the other genes with which it interacts, and how the products of the genes directly or indirectly act on each other, including those not involved in the analysis (determining up stream regulators). The networks are ranked depending on their number of significantly differentially expressed genes and associated relevant diseases. Genes in the network are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). The intensity of the node color is related to the fold change of the differentially expressed genes, i.e. upregulated or downregulated genes indicated in red or green, respectively. All edges are supported by at least one published report and/or canonical definition in the Ingenuity Pathways Knowledge Base.
Statistical analysis
For gene expression studies, fold change values of significantly DEGs (P ≤ 0.05) from the four analyzed gene arrays (above) using Prime PCR Analysis were exported to GraphPad Prism 6.02, pooled and plotted in a volcano plot. The genes with P-value ≤ 0.05 and a significant fold-change >1.5 or <−1.5 for upregulated and downregulated genes, respectively, were included in subsequent analyses.
Supplementary information
Acknowledgements
Rafael Nacif-Pimenta was supported by PhD fellowships from Coordenação de Aperfeiçoamento de Pessoal de Nivel Superior (CAPES) and the Fundação de Amparo a Pesquisas de Minas Gerais (FAPEMIG). We thank Dra. Mónica C. Botelho, Department of Infectious Diseases, R&D Unit, INSA-National Health Institute Dr. Ricardo Jorge, Porto, Portugal & IPATIMUP, Institute of Pathology and Molecular Immunology of the University of Porto, Portugal for the HCV29 cell line and informative discussion, and Christian Owusu from the Wellcome Sanger Institute for expert technical assistance. Hamsters and mice infected with Schistosoma haematobium and S. mansoni, respectively, were provided by the NIAID Schistosomiasis Resource Center for distribution through BEI Resources, NIH-NIAID Contract HHSN272201000005I. We acknowledge support from the GW SPARC program (IAM) and award R01CA164719 from the National Cancer Institute (NCI), NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of CAPES, FAPEMIG, NCI or the NIH.
Author Contributions
Conceived and designed the study: G.R. Resources: G.R., V.H.M., M.H.H. and P.J.B. Performed the experiments: R.N.P., G.R., I.M., K.I. and S.K. Data analysis: R.N.P., A.d.S.O. and G.R. Analysis and interpretation of the overall data: R.N.P., A.d.S.O., I.M., S.K., V.H.M., P.M.Z.C., J.M.C.d.C., M.H.H., P.J.B. and G.R. Supervision: G.R. and P.J.B. Writing: R.N.P., A.d.S.O., I.M., S.K., V.H.M., P.M.Z.C., J.M.C.d.C., M.H.H., P.J.B. and G.R.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Paul J. Brindley, Email: pbrindley@gwu.edu
Gabriel Rinaldi, Email: gr10@sanger.ac.uk.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-46917-y.
References
- 1.King CH, Dangerfield-Cha M. The unacknowledged impact of chronic schistosomiasis. Chronic Illn. 2008;4:65–79. doi: 10.1177/1742395307084407. [DOI] [PubMed] [Google Scholar]
- 2.King CH. Parasites and poverty: the case of schistosomiasis. Acta Trop. 2010;113:95–104. doi: 10.1016/j.actatropica.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gryseels B, Polman K, Clerinx J, Kestens L. Human schistosomiasis. Lancet. 2006;368:1106–1118. doi: 10.1016/S0140-6736(06)69440-3. [DOI] [PubMed] [Google Scholar]
- 4.Hodder SL, et al. Predisposition to urinary tract epithelial metaplasia in Schistosoma haematobium infection. Am J Trop Med Hyg. 2000;63:133–138. doi: 10.4269/ajtmh.2000.63.133. [DOI] [PubMed] [Google Scholar]
- 5.Humans IWGotEoCRt. Biological agents. Volume 100 B. A review of human carcinogens. IARC Monogr Eval Carcinog Risks Hum. 2012;100:1–441. [PMC free article] [PubMed] [Google Scholar]
- 6.Hotez PJ, Fenwick A, Kjetland EF. Africa’s 32 cents solution for HIV/AIDS. PLoS Negl Trop Dis. 2009;3:e430. doi: 10.1371/journal.pntd.0000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santos J, et al. Urinary estrogen metabolites and self-reported infertility in women infected with Schistosoma haematobium. PLoS One. 2014;9:e96774. doi: 10.1371/journal.pone.0096774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldmeier H, Krantz I, Poggensee G. Female genital schistosomiasis as a risk-factor for the transmission of HIV. Int J STD AIDS. 1994;5:368–372. doi: 10.1177/095646249400500517. [DOI] [PubMed] [Google Scholar]
- 9.Gouveia MJ, et al. Estrogen-like metabolites and DNA-adducts in urogenital schistosomiasis-associated bladder cancer. Cancer Lett. 2015;359:226–232. doi: 10.1016/j.canlet.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 10.Honeycutt J, Hammam O, Fu CL, Hsieh MH. Controversies and challenges in research on urogenital schistosomiasis-associated bladder cancer. Trends Parasitol. 2014;30:324–332. doi: 10.1016/j.pt.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kjetland EF, et al. Association between genital schistosomiasis and HIV in rural Zimbabwean women. Aids. 2006;20:593–600. doi: 10.1097/01.aids.0000210614.45212.0a. [DOI] [PubMed] [Google Scholar]
- 12.Hansel DE, et al. A contemporary update on pathology standards for bladder cancer: transurethral resection and radical cystectomy specimens. Eur Urol. 2013;63:321–332. doi: 10.1016/j.eururo.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Fu CL, Odegaard JI, Herbert DR, Hsieh MH. A novel mouse model of Schistosoma haematobium egg-induced immunopathology. PLoS Pathog. 2012;8:e1002605. doi: 10.1371/journal.ppat.1002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honeycutt J, Hammam O, Hsieh MH. Schistosoma haematobium egg-induced bladder urothelial abnormalities dependent on p53 are modulated by host sex. Exp Parasitol. 2015;158:55–60. doi: 10.1016/j.exppara.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chala B, et al. Development of Urinary Bladder Pre-Neoplasia by Schistosoma haematobium Eggs and Chemical Carcinogen in Mice. Korean J Parasitol. 2017;55:21–29. doi: 10.3347/kjp.2017.55.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ke N, Wang X, Xu X, Abassi YA. The xCELLigence system for real-time and label-free monitoring of cell viability. Methods Mol Biol. 2011;740:33–43. doi: 10.1007/978-1-61779-108-6_6. [DOI] [PubMed] [Google Scholar]
- 17.Matchimakul P, et al. Apoptosis of cholangiocytes modulated by thioredoxin of carcinogenic liver fluke. Int J Biochem Cell Biol. 2015;65:72–80. doi: 10.1016/j.biocel.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Memon AA, et al. Soluble HER3 predicts survival in bladder cancer patients. Oncol Lett. 2018;15:1783–1788. doi: 10.3892/ol.2017.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci. 2013;14:16365–16385. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popivanova BK, et al. Blocking TNF-alpha in mice reduces colorectal carcinogenesis associated with chronic colitis. J Clin Invest. 2008;118:560–570. doi: 10.1172/JCI32453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schatoff EM, Leach BI, Dow LE. Wnt Signaling and Colorectal Cancer. Curr Colorectal Cancer Rep. 2017;13:101–110. doi: 10.1007/s11888-017-0354-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Martel C, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 23.Gouveia MJ, et al. Infection with Opisthorchis felineus induces intraepithelial neoplasia of the biliary tract in a rodent model. Carcinogenesis. 2017;38:929–937. doi: 10.1093/carcin/bgx042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brindley PJ, Loukas A. Helminth infection-induced malignancy. PLoS Pathog. 2017;13:e1006393. doi: 10.1371/journal.ppat.1006393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sulzmaier FJ, Jean C, Schlaepfer DD. FAK in cancer: mechanistic findings and clinical applications. Nat Rev Cancer. 2014;14:598–610. doi: 10.1038/nrc3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar A, Rajendran V, Sethumadhavan R, Purohit R. AKT kinase pathway: a leading target in cancer research. ScientificWorldJournal. 2013;2013:756134. doi: 10.1155/2013/756134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganesan R, Mallets E, Gomez-Cambronero J. The transcription factors Slug (SNAI2) and Snail (SNAI1) regulate phospholipase D (PLD) promoter in opposite ways towards cancer cell invasion. Mol Oncol. 2016;10:663–676. doi: 10.1016/j.molonc.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conidi A, et al. Few Smad proteins and many Smad-interacting proteins yield multiple functions and action modes in TGFbeta/BMP signaling in vivo. Cytokine Growth Factor Rev. 2011;22:287–300. doi: 10.1016/j.cytogfr.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Smout MJ, et al. A granulin-like growth factor secreted by the carcinogenic liver fluke, Opisthorchis viverrini, promotes proliferation of host cells. PLoS Pathog. 2009;5:e1000611. doi: 10.1371/journal.ppat.1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sripa B, et al. Liver fluke induces cholangiocarcinoma. PLoS Med. 2007;4:e201. doi: 10.1371/journal.pmed.0040201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Botelho M, et al. Schistosoma haematobium total antigen induces increased proliferation, migration and invasion, and decreases apoptosis of normal epithelial cells. Int J Parasitol. 2009;39:1083–1091. doi: 10.1016/j.ijpara.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 32.Botelho M, et al. Tumourigenic effect of Schistosoma haematobium total antigen in mammalian cells. Int J Exp Pathol. 2009;90:448–453. doi: 10.1111/j.1365-2613.2009.00650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Botelho MC, Oliveira PA, Lopes C, Correia da Costa JM, Machado JC. Urothelial dysplasia and inflammation induced by Schistosoma haematobium total antigen instillation in mice normal urothelium. Urol Oncol. 29. 2009;6:809–814. doi: 10.1016/j.urolonc.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 34.El-Awady MK, et al. Schistosoma hematobium soluble egg antigens induce proliferation of urothelial and endothelial cells. World J Urol. 2001;19:263–266. doi: 10.1007/s003450100217. [DOI] [PubMed] [Google Scholar]
- 35.Boloukhere M, Baldo-Correa E, Borojevic R. Experimental schistosomiasis mansoni: characterization of connective tissue cells in hepatic periovular granulomas. J Submicrosc Cytol Pathol. 1993;25:505–517. [PubMed] [Google Scholar]
- 36.Wyler DJ, Tracy JW. Direct and indirect effects of soluble extracts of Schistosoma mansoni eggs on fibroblast proliferation in vitro. Infect Immun. 1982;38:103–108. doi: 10.1128/iai.38.1.103-108.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereira TA, et al. Schistosome-induced cholangiocyte proliferation and osteopontin secretion correlate with fibrosis and portal hypertension in human and murine schistosomiasis mansoni. Clin Sci (Lond) 2015;129:875–883. doi: 10.1042/CS20150117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rumbley CA, Sugaya H, Zekavat SA, Perrin PJ, Phillips SM. Elimination of lymphocytes, but not eosinophils, by Fas-mediated apoptosis in murine schistosomiasis. Am J Trop Med Hyg. 2001;65:442–449. doi: 10.4269/ajtmh.2001.65.442. [DOI] [PubMed] [Google Scholar]
- 39.Anthony B, Mathieson W, de Castro-Borges W, Allen J. Schistosoma mansoni: egg-induced downregulation of hepatic stellate cell activation and fibrogenesis. Exp Parasitol. 2010;124:409–420. doi: 10.1016/j.exppara.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 40.Lundy SK, Boros DL. Fas ligand-expressing B-1a lymphocytes mediate CD4(+)-T-cell apoptosis during schistosomal infection: induction by interleukin 4 (IL-4) and IL-10. Infect Immun. 2002;70:812–819. doi: 10.1128/IAI.70.2.812-819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashton PD, Harrop R, Shah B, Wilson RA. The schistosome egg: development and secretions. Parasitology. 2001;122:329–338. doi: 10.1017/S0031182001007351. [DOI] [PubMed] [Google Scholar]
- 42.Al-Mashat F, et al. Rectal cancer associated with schistosomiasis: report of two cases and review of the literature. Ann Saudi Med. 2001;21:65–67. doi: 10.5144/0256-4947.2001.65. [DOI] [PubMed] [Google Scholar]
- 43.Madbouly KM, et al. Colorectal cancer in a population with endemic Schistosoma mansoni: is this an at-risk population? Int J Colorectal Dis. 2007;22:175–181. doi: 10.1007/s00384-006-0144-3. [DOI] [PubMed] [Google Scholar]
- 44.Qiu DC, Hubbard AE, Zhong B, Zhang Y, Spear RC. A matched, case-control study of the association between Schistosoma japonicum and liver and colon cancers, in rural China. Ann Trop Med Parasitol. 2005;99:47–52. doi: 10.1179/136485905X19883. [DOI] [PubMed] [Google Scholar]
- 45.Gussow D, et al. The human beta 2-microglobulin gene. Primary structure and definition of the transcriptional unit. J Immunol. 1987;139:3132–3138. [PubMed] [Google Scholar]
- 46.Bianchini M, et al. Comparative study of gene expression by cDNA microarray in human colorectal cancer tissues and normal mucosa. Int J Oncol. 2006;29:83–94. [PubMed] [Google Scholar]
- 47.Plank-Bazinet JL, Mundell NA. The paradox of Foxd3: how does it function in pluripotency and differentiation of embryonic stem cells? Stem Cell Investig. 2016;3:73. doi: 10.21037/sci.2016.09.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li D, et al. FOXD3 is a novel tumor suppressor that affects growth, invasion, metastasis and angiogenesis of neuroblastoma. Oncotarget. 2013;4:2021–2044. doi: 10.18632/oncotarget.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davies SJ, et al. Involvement of TNF in limiting liver pathology and promoting parasite survival during schistosome infection. Int J Parasitol. 2004;34:27–36. doi: 10.1016/j.ijpara.2003.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amiri P, et al. Tumour necrosis factor alpha restores granulomas and induces parasite egg-laying in schistosome-infected SCID mice. Nature. 1992;356:604–607. doi: 10.1038/356604a0. [DOI] [PubMed] [Google Scholar]
- 51.Vale N, et al. The role of estradiol metabolism in urogenital schistosomiasis-induced bladder cancer. Tumour Biol. 2017;39:1010428317692247. doi: 10.1177/1010428317692247. [DOI] [PubMed] [Google Scholar]
- 52.Santos J, et al. P53 and cancer-associated sialylated glycans are surrogate markers of cancerization of the bladder associated with Schistosoma haematobium infection. PLoS Negl Trop Dis. 2014;8:e3329. doi: 10.1371/journal.pntd.0003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cancer Genome Atlas Research, N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y, Beyer A, Aebersold R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 57.Botelho MC, et al. Inactivation of estrogen receptor by Schistosoma haematobium total antigen in bladder urothelial cells. Oncol Rep. 2012;27:356–362. doi: 10.3892/or.2011.1552. [DOI] [PubMed] [Google Scholar]
- 58.Botelho MC, et al. The role of estrogens and estrogen receptor signaling pathways in cancer and infertility: the case of schistosomes. Trends Parasitol. 2015;31:246–250. doi: 10.1016/j.pt.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 59.Botelho MC, et al. Tumour-like phenotypes in urothelial cells after exposure to antigens from eggs of Schistosoma haematobium: an oestrogen-DNA adducts mediated pathway? Int J Parasitol. 2013;43:17–26. doi: 10.1016/j.ijpara.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 60.Mann VH, Morales ME, Rinaldi G, Brindley PJ. Culture for genetic manipulation of developmental stages of Schistosoma mansoni. Parasitology. 2010;137:451–462. doi: 10.1017/S0031182009991211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grubman SA, et al. Regulation of intracellular pH by immortalized human intrahepatic biliary epithelial cell lines. Am J Physiol. 1994;266:G1060–1070. doi: 10.1152/ajpgi.1994.266.6.G1060. [DOI] [PubMed] [Google Scholar]
- 62.Masters JR, et al. Tissue culture model of transitional cell carcinoma: characterization of twenty-two human urothelial cell lines. Cancer Res. 1986;46:3630–3636. [PubMed] [Google Scholar]
- 63.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang S, Liu Z, Wang L, Zhang X. NF-kappaB signaling pathway, inflammation and colorectal cancer. Cell Mol Immunol. 2009;6:327–334. doi: 10.1038/cmi.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deltcheva E, Nimmo R. RUNX transcription factors at the interface of stem cells and cancer. Biochem J. 2017;474:1755–1768. doi: 10.1042/BCJ20160632. [DOI] [PubMed] [Google Scholar]
- 68.Rennoll S, Yochum G. Regulation of MYC gene expression by aberrant Wnt/beta-catenin signaling in colorectal cancer. World J Biol Chem. 2015;6:290–300. doi: 10.4331/wjbc.v6.i4.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wahlstrom T, Henriksson MA. Impact of MYC in regulation of tumor cell metabolism. Biochim Biophys Acta. 2015;1849:563–569. doi: 10.1016/j.bbagrm.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 70.Vogt PK. Fortuitous convergences: the beginnings of JUN. Nat Rev Cancer. 2002;2:465–469. doi: 10.1038/nrc818. [DOI] [PubMed] [Google Scholar]
- 71.Colomer Carlota, Marruecos Laura, Vert Anna, Bigas Anna, Espinosa Lluis. NF-κB Members Left Home: NF-κB-Independent Roles in Cancer. Biomedicines. 2017;5(4):26. doi: 10.3390/biomedicines5020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Serasanambati M, Chilakapati S. Function of Nuclear Factor kappa B (NF-kB) in human diseases - a review. South Indian Journal of Biological Sciences. 2016;2:368–387. doi: 10.22205/sijbs/2016/v2/i4/103443. [DOI] [Google Scholar]
- 73.Liu W, et al. The molecular effect of metastasis suppressors on Src signaling and tumorigenesis: new therapeutic targets. Oncotarget. 2015;6:35522–35541. doi: 10.18632/oncotarget.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Talamonti MS, Roh MS, Curley SA, Gallick GE. Increase in activity and level of pp60c-src in progressive stages of human colorectal cancer. J Clin Invest. 1993;91:53–60. doi: 10.1172/JCI116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu S., Xu-Monette Z. Y., Tzankov A., Green T., Wu L., Balasubramanyam A., Liu W.-m., Visco C., Li Y., Miranda R. N., Montes-Moreno S., Dybkaer K., Chiu A., Orazi A., Zu Y., Bhagat G., Richards K. L., Hsi E. D., Choi W. W. L., Zhao X., van Krieken J. H., Huang Q., Huh J., Ai W., Ponzoni M., Ferreri A. J. M., Zhou F., Slack G. W., Gascoyne R. D., Tu M., Variakojis D., Chen W., Go R. S., Piris M. A., Moller M. B., Medeiros L. J., Young K. H. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;121(20):4021–4031. doi: 10.1182/blood-2012-10-460063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim S, et al. MYC and BCL2 overexpression is associated with a higher class of Memorial Sloan-Kettering Cancer Center prognostic model and poor clinical outcome in primary diffuse large B-cell lymphoma of the central nervous system. BMC Cancer. 2016;16:363. doi: 10.1186/s12885-016-2397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hwang Ki-Tae, Kim Kwangsoo, Chang Ji Hyun, Oh Sohee, Kim Young A, Lee Jong Yoon, Jung Se Hee, Choi In Sil. BCL2 Regulation according to Molecular Subtype of Breast Cancer by Analysis of The Cancer Genome Atlas Database. Cancer Research and Treatment. 2018;50(3):658–669. doi: 10.4143/crt.2017.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bodenstine TM, et al. Maspin: molecular mechanisms and therapeutic implications. Cancer Metastasis Rev. 2012;31:529–551. doi: 10.1007/s10555-012-9361-0. [DOI] [PubMed] [Google Scholar]
- 79.Abraham RT. The ATM-related kinase, hSMG-1, bridges genome and RNA surveillance pathways. DNA Repair (Amst) 2004;3:919–925. doi: 10.1016/j.dnarep.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 80.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 81.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 82.Mendoza M, Mandani G, Momand J. The MDM2 gene family. Biomol Concepts. 2014;5:9–19. doi: 10.1515/bmc-2013-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu SB, Shen ZF, Guo YJ, Cao LX, Xu Y. PML silencing inhibits cell proliferation and induces DNA damage in cultured ovarian cancer cells. Biomed Rep. 2017;7:29–35. doi: 10.3892/br.2017.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 85.Bernardi R, Papa A, Pandolfi PP. Regulation of apoptosis by PML and the PML-NBs. Oncogene. 2008;27:6299–6312. doi: 10.1038/onc.2008.305. [DOI] [PubMed] [Google Scholar]
- 86.LeBlanc H, et al. Tumor-cell resistance to death receptor–induced apoptosis through mutational inactivation of the proapoptotic Bcl-2 homolog Bax. Nat Med. 2002;8:274–281. doi: 10.1038/nm0302-274. [DOI] [PubMed] [Google Scholar]
- 87.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/S0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 88.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Engelmann D, Putzer BM. The dark side of E2F1: in transit beyond apoptosis. Cancer Res. 2012;72:571–575. doi: 10.1158/0008-5472.CAN-11-2575. [DOI] [PubMed] [Google Scholar]
- 90.Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14:518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.