Abstract
Atypical scrapie in goats has only been reported in European countries. The present study reports the identification of the first case of atypical scrapie in goats in Japan. The genotype of the animal was ALRQ/ALHQ at codons 136, 141, 154, and 171 in prion protein (PrP). Western blot analysis showed a multiplex proteinase K-resistant prion protein (PrP-res) band pattern with a band <15 kDa that was clearly distinguishable from the triplet PrP-res band pattern observed in classical scrapie cases. Histopathological and immunohistological examination showed mild vacuolation and fine granular to globular immunolabelling of disease-associated PrP in the dorsal horn of cervical spinal cord. Collectively, our results confirmed that this goat was affected by atypical scrapie.
Keywords: atypical scrapie, genotype, goat, surveillance
Prion diseases, also known as transmissible spongiform encephalopathies (TSE), are fatal neurodegenerative disorders in humans and animals [23]. The key event in the pathogenesis of these diseases is the conversion of host-encoded normal cellular prion protein (PrPC) into its pathogenic isoform (PrPSc) and its accumulation in the central nervous system [16].
Scrapie is a prion disease of small ruminants that has been documented since the 18th century [1]. To date, scrapie is recognized in two forms, classical and atypical scrapie/Nor98. Atypical scrapie/Nor98 was initially detected in Norway in 1998 [4]. Since then, atypical scrapie cases have been reported in 23 European countries [24], the U.S.A. [17], Canada [18], the Falkland Islands [8], Japan [14], Australia [6], and New Zealand [15]. Although the origin of atypical scrapie remains unclear, affected animals were older (>6 years of age) compared with classical scrapie [3], and atypical cases were detected as single cases in their flock or herd; thus, atypical scrapie is considered a sporadic prion disease or has limited transmissibility under natural conditions [11].
Atypical scrapie has been found in sheep and goats [3, 4, 6], and the genetic basis of the disease has been defined as well as classical scrapie. Animals carrying A136R154R171 and A136H154Q171 alleles are known to be relatively resistant to classical scrapie. However, these alleles are frequently found in atypical scrapie cases [13]. In addition, polymorphisms at codons 141 and 154 have been reported to be strongly linked to atypical scrapie incidences in sheep [2, 6], that is, the animals expressing A136H154Q171, A136R154R171, and A136F141R154Q171 alleles are more frequently associated with the disease [25]. On the other hand, the presence of histidine at codon 154 is a risk factor for atypical scrapie in goats [5]. Atypical scrapie in sheep has been found in almost all countries where extensive surveillance has been conducted, whereas atypical scrapie in goats has only been reported in several European counties [9, 10, 27], where its prevalence is lower than that in sheep [27].
Although 65 cases of sheep scrapie, including one atypical case that was detected in 2016, have been identified in Japan [14], no case of scrapie in goats has been published, to our knowledge. This report describes the identification of the first case of atypical goat scrapie in Japan, which was detected through the active national TSE surveillance program.
The female goat investigated was a 115-month-old mixed breed goat from Yamaguchi prefecture and had a female littermate. These goats were kept as companion animals in the same farm, not for livestock. The goat, in the present case, displayed dysstasia for a month and died on August 17, 2017, 3 days after showing astasia. Following the diagnosis of the present case, the littermate, with no symptoms, was euthanized as a suspected animal. The medulla oblongata and the cervical spinal cord (at approximately the C1 level) were collected and sent to the National Institute of Animal Health (NIAH), where TSE investigation was conducted by western blotting, which has higher sensitivity than ELISA tests (unpublished data).
Total genomic DNA was extracted from the spinal cord of the present case and its littermate. PCR amplification of a 627 base pair product of the coding region of prnp within codons 25-233 was performed by KAPATM HiFi HotStart ReadyMix (KAPA Biosystems, Woburn, MA, U.S.A.) with a primer set (forward: 5′-TCTAGCTGTCATATGAAGAAGCGACCAAAACCTGG-3′, reverse: 5′-AGCTGTGGATCCTCATCATGCCCCCCTTTGGTAATAAG-3′). The amplicons were sequenced from both ends using the same primer set by BigDye® Terminator V3.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, MA, U.S.A.) on a 3130xl Genetic analyzer (Thermo Fisher Scientific, Waltham, MA, U.S.A.). Based on comparison with a wild-type goat PrP open reading frame (ORF) (GenBank no. S82626), the amino acid sequence was predicted. The goat and its littermate had the same heterological amino acid polymorphisms as A136L141R154Q171 / A136L141H154Q171 but did not have the scrapie-resistant alleles 146S/D, 211Q, and 222K described in previous reports [7].
The accumulation of PrPSc in brain tissue was examined by western blot (WB) and immunohistochemical (IHC) analyses. For the atypical scrapie positive control, we used an obex sample from the affected sheep case (A136F141R154Q171/A136F141R154Q171) in Japan [14], and the frontal cortex samples from the affected sheep cases (UK1, A136R154R171/A136R154Q171; UK2, A136R154R171/A136H154Q171; UK3, A136R154Q171/A136R154Q171; UK4, A136H154Q171/A136H154Q171) in the United Kingdom. Following the treatment with proteinase K (PK; Roche Diagnosis Japan, Tokyo, Japan; final concentration of 40 µg/ml) for 30 min at 37°C, the samples were electrophoresed on 12% NuPAGE pre-cast gels in NuPAGE MES SDS Running buffer (Invitrogen, Carlsbad, CA, U.S.A.) or 4–20% Criterion TGX pre-cast gels in Tris-Glycine running buffer (Bio-Rad, Hercules, CA, U.S.A.) and electrotransferred onto Immobilon polyvinylidene fluoride membranes (Millipore, Darmstadt, Germany). The blotted membranes were probed with the monoclonal antibody (mAb) P4 (R-Biopharm, Darmstadt, Germany) according to a previously described method [14]. IHC for PrPSc was performed using the mAb T1 [26] or F99/97.6.1 (VMRD, Pullman, WA, U.S.A.) with pretreatment by autoclaving as previously described [21].
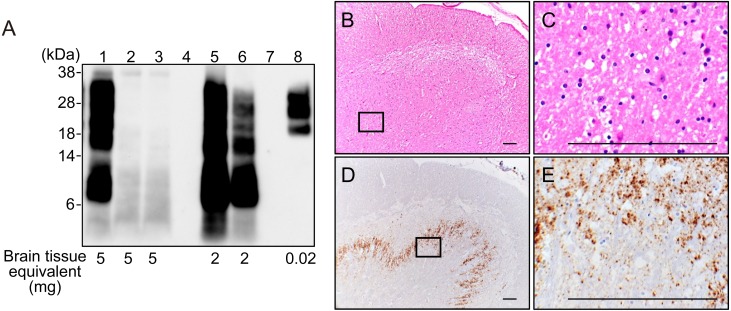
WB analysis with mAb P4 demonstrated a multiple PK-resistant PrP (PrP-res) pattern with a small PrP-res fragment below 15 kDa in the present goat case (Fig. 1A; lane 1) but not in scrapie-negative goats (Fig. 1A; lanes 2 and 3). This PrP-res pattern was similar to that in atypical scrapie cases in sheep from the U.K. (Fig. 1A; lanes 5 and 6) obtained from the Animal and Plant Health Agency (Addlestone, Surrey, U.K.) but clearly distinguishable from the triplet PrP-res pattern observed in a classical scrapie case in sheep (Fig. 1A; lane 8) archived in NIAH in Japan. The littermate of the present case was also examined by WB but tested negative for PrP-res (data not shown).
Fig. 1.
Investigation for atypical scrapie on a present goat. A: WB analysis for PrP-res in goat and sheep. Lane 1, present Japanese goat with atypical scrapie (obex); lanes 2 and 3, negative control (goat obex); lane 4, empty lane; lane 5, atypical scrapie-positive control UK2; lane 6, atypical scrapie positive control UK1; lane 7, empty lane; lane 8, classical scrapie positive control (sheep obex). The membrane was probed with monoclonal antibody (mAb) P4. Molecular markers are shown on the left (kDa). Brain tissue equivalents (mg) loaded per lane are shown on the bottom. B-E: Histopathological and immunohistochemical examination of the current goat in the cervical spinal cord. C and E is an enlarged scale of the opened boxes in B and D, respectively. Mild spongiform change with small vacuoles scattered throughout was observed in the substantia gelatinosa region of the dorsal horn on the hematoxylin and eosin stained section (B and C). Fine granular and abundant deposition of disease-associated prion proteins immunolabelled with mAb T1 were observed in the dorsal horn (D and E). Scale bar, 0.1 mm.
Since a formalin-fixed brain stem sample was unavailable, sections from the cervical spinal cord of the current case were stained with hematoxylin and eosin (HE) and examined by IHC for PrPSc deposition. Histopathological examination revealed mild spongiform change in the substantia gelatinosa region of the dorsal horn (Fig. 1B and 1C). IHC analysis showed that the fine granular to globular PrPSc immunolabelling was abundant in the dorsal horn (Fig. 1D and 1E). Less prominent globular PrPSc immunolabelling was detected throughout the spinal cord white matter. Due to the lack of other brain site samples, we could not examine the detailed PrPSc distribution in the brain.
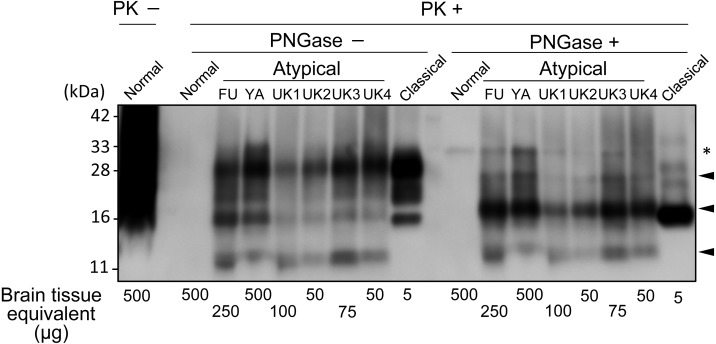
Finally, we characterized a molecular type of PrP-res using field cases of classical and atypical scrapie including the current case. Following digestion with 5 µg/ml PK, samples were treated with or without Peptide-N-Glycosidase F (PNGase; New England BioLabs Japan Inc., Japan) as described previously [12]. For clear separation of the PrP-res bands less than 33 kDa, the samples were applied on 4–20% Criterion TGX pre-cast gels in Tris-Glycine running buffer (Bio-Rad, Hercules, CA, U.S.A.) instead of a 12% polyacrylamide gel in Fig. 1. Upon deglycosylation of PrP-res, three bands were observed in all atypical cases (Fig. 2). Although the band around 11 kDa was obscure, the 25 kDa, and 17 kDa bands in this case were similar to those in other atypical scrapie cases, suggesting similarities to the same PrP-res pattern of an atypical scrapie case previously reported [2]. This PrP-res pattern was well preserved among animals possessing the different alleles of PrP. Taking into account the tissue equivalent loaded per lane, the amount of PrP-res in the atypical cases from Japan was relatively low compared to cases from the U.K. The affected goat in this case was thought to be at the terminal stage of disease. Thus, the different signal intensity might be due to the difference of tested brain regions; Japanese samples were derived from the obex while U.K. samples were obtained from the frontal cortex. Our WB data might be consistent with the previous reports that found the accumulation of PrPSc in atypical scrapie animals was observed, mainly, in the cerebral and cerebellar cortices [19, 20], with less in the brain stem. Nevertheless, our current study also demonstrated that testing obex samples with sensitive-WB with mAb P4 could diagnose atypical scrapie in goats. As this is the only reported case we must exercise caution in extrapolating our findings or in making far-reaching assumptions.
Fig. 2.
WB profiles of PrP-res with or without PNGase deglycosylation from field cases of atypical and classical scrapie. An atypical case from a Japanese sheep from Fukuoka prefecture, and the present goat case from Yamaguchi prefecture are indicated as (FU) and (YA), respectively. Atypical cases from U.K. sheep are indicated as UK1, UK2, UK3, and UK4, respectively. Arrowheads indicate PrP-res fragments from atypical cases after PNGase deglycosylation. *are non-specific signals caused by proteinase K (PK). The tissue equivalent loaded per lane is shown in the bottom of the figure. The blot was probed with mAb P4.
We believe that this is the first report of an atypical scrapie case in a goat in Japan. The age of the affected goat and an unaffected littermate in the same farm may raise the possibility that this atypical scrapie goat was a sporadic case with poor transmissibility. The animals expressing the A136H154Q171 allele, which is strongly associated with atypical scrapie [5], constitute 0.3% of goats in Japan [14]. Since this disease has demonstrated its transmissibility to other animals [22], continuous monitoring is still required for better understanding the incidence of atypical scrapie in Japan and preventing the spread of disease in advance.
Acknowledgments
We are grateful to the livestock hygiene service center of Yamaguchi prefecture for their cooperation. Expert technical assistance was provided by Naoko Tabeta, Naomi Furuya, and Junko Yamada. This work was partly supported by grants-in-aid from the Improving Food Safety and Animal Health Project of the Ministry of Agriculture, Forestry and Fisheries, Japan and by a subsidy of National Agriculture and Food Research Organization (NARO).
REFERENCES
- 1.Aguzzi A., Polymenidou M.2004. Mammalian prion biology: one century of evolving concepts. Cell 116: 313–327. doi: 10.1016/S0092-8674(03)01031-6 [DOI] [PubMed] [Google Scholar]
- 2.Arsac J. N., Andreoletti O., Bilheude J. M., Lacroux C., Benestad S. L., Baron T.2007. Similar biochemical signatures and prion protein genotypes in atypical scrapie and Nor98 cases, France and Norway. Emerg. Infect. Dis. 13: 58–65. doi: 10.3201/eid1301.060393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benestad S. L., Bratberg B.2006. Atypical scrapie-Nor98. pp. 630–634. In: Prions in Humans and Animals (Beat, Hörnlimann, Riesner, D. and Kretzschmar, H. eds.), de Gruyter, Berlin. [Google Scholar]
- 4.Benestad S. L., Sarradin P., Thu B., Schönheit J., Tranulis M. A., Bratberg B.2003. Cases of scrapie with unusual features in Norway and designation of a new type, Nor98. Vet. Rec. 153: 202–208. doi: 10.1136/vr.153.7.202 [DOI] [PubMed] [Google Scholar]
- 5.Colussi S., Vaccari G., Maurella C., Bona C., Lorenzetti R., Troiano P., Casalinuovo F., Di Sarno A., Maniaci M. G., Zuccon F., Nonno R., Casalone C., Mazza M., Ru G., Caramelli M., Agrimi U., Acutis P. L.2008. Histidine at codon 154 of the prion protein gene is a risk factor for Nor98 scrapie in goats. J. Gen. Virol. 89: 3173–3176. doi: 10.1099/vir.0.2008/004150-0 [DOI] [PubMed] [Google Scholar]
- 6.Cook R. W., Bingham J., Besier A. S., Bayley C. L., Hawes M., Shearer P. L., Yamada M., Bergfeld J., Williams D. T., Middleton D. J.2016. Atypical scrapie in Australia. Aust. Vet. J. 94: 452–455. doi: 10.1111/avj.12529 [DOI] [PubMed] [Google Scholar]
- 7.Curcio L., Sebastiani C., Di Lorenzo P., Lasagna E., Biagetti M.2016. Review: A review on classical and atypical scrapie in caprine: Prion protein gene polymorphisms and their role in the disease. Animal 10: 1585–1593. doi: 10.1017/S1751731116000653 [DOI] [PubMed] [Google Scholar]
- 8.Epstein V., Pointing S., Halfacre S.2005. Atypical scrapie in the Falkland Islands. Vet. Rec. 157: 667–668. doi: 10.1136/vr.157.21.667-c [DOI] [PubMed] [Google Scholar]
- 9.European Food Safety Authority. 2017. The European Union summary report on surveillance for the presence of transmissible spongiform encephalopathies (TSE) in 2016. EFSA J. 15: 5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fast C., Groschup M. H.2013. Classical and Atypical Scrapie in Sheep and Goats, Springer, New York. [Google Scholar]
- 11.Fediaevsky A., Tongue S. C., Nöremark M., Calavas D., Ru G., Hopp P.2008. A descriptive study of the prevalence of atypical and classical scrapie in sheep in 20 European countries. BMC Vet. Res. 4: 19. doi: 10.1186/1746-6148-4-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukuda S., Iwamaru Y., Imamura M., Masujin K., Shimizu Y., Matsuura Y., Shu Y., Kurachi M., Kasai K., Murayama Y., Onoe S., Hagiwara K., Sata T., Mohri S., Yokoyama T., Okada H.2009. Intraspecies transmission of L-type-like Bovine Spongiform Encephalopathy detected in Japan. Microbiol. Immunol. 53: 704–707. doi: 10.1111/j.1348-0421.2009.00169.x [DOI] [PubMed] [Google Scholar]
- 13.Goldmann W.2008. PrP genetics in ruminant transmissible spongiform encephalopathies. Vet. Res. 39: 30. doi: 10.1051/vetres:2008010 [DOI] [PubMed] [Google Scholar]
- 14.Imamura M., Miyazawa K., Iwamaru Y., Matsuura Y., Yokoyama T., Okada H.2017. Identification of the first case of atypical scrapie in Japan. J. Vet. Med. Sci. 78: 1915–1919. doi: 10.1292/jvms.16-0379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kittelberger R., Chaplin M. J., Simmons M. M., Ramirez-Villaescusa A., McIntyre L., MacDiarmid S. C., Hannah M. J., Jenner J., Bueno R., Bayliss D., Black H., Pigott C. J., O’Keefe J. S.2010. Atypical scrapie/Nor98 in a sheep from New Zealand. J. Vet. Diagn. Invest. 22: 863–875. doi: 10.1177/104063871002200604 [DOI] [PubMed] [Google Scholar]
- 16.Legname G., Baskakov I. V., Nguyen H. O., Riesner D., Cohen F. E., DeArmond S. J., Prusiner S. B.2004. Synthetic mammalian prions. Science 305: 673–676. doi: 10.1126/science.1100195 [DOI] [PubMed] [Google Scholar]
- 17.Loiacono C. M., Thomsen B. V., Hall S. M., Kiupel M., Sutton D., O’Rourke K., Barr B., Anthenill L., Keane D.2009. Nor98 scrapie identified in the United States. J. Vet. Diagn. Invest. 21: 454–463. doi: 10.1177/104063870902100406 [DOI] [PubMed] [Google Scholar]
- 18.Mitchell G. B., O’Rourke K. I., Harrington N. P., Soutyrine A., Simmons M. M., Dudas S., Zhuang D., Laude H., Balachandran A.2010. Identification of atypical scrapie in Canadian sheep. J. Vet. Diagn. Invest. 22: 408–411. doi: 10.1177/104063871002200310 [DOI] [PubMed] [Google Scholar]
- 19.Moore S. J., Simmons M., Chaplin M., Spiropoulos J.2008. Neuroanatomical distribution of abnormal prion protein in naturally occurring atypical scrapie cases in Great Britain. Acta Neuropathol. 116: 547–559. doi: 10.1007/s00401-008-0433-8 [DOI] [PubMed] [Google Scholar]
- 20.Nentwig A., Oevermann A., Heim D., Botteron C., Zellweger K., Drögemüller C., Zurbriggen A., Seuberlich T.2007. Diversity in neuroanatomical distribution of abnormal prion protein in atypical scrapie. PLoS Pathog. 3: e82. doi: 10.1371/journal.ppat.0030082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okada H., Iwamaru Y., Imamura M., Masujin K., Yokoyama T., Mohri S.2010. Immunohistochemical detection of disease-associated prion protein in the intestine of cattle naturally affected with bovine spongiform encephalopathy by using an alkaline-based chemical antigen retrieval method. J. Vet. Med. Sci. 72: 1423–1429. doi: 10.1292/jvms.10-0211 [DOI] [PubMed] [Google Scholar]
- 22.Okada H., Miyazawa K., Imamura M., Iwamaru Y., Masujin K., Matsuura Y., Yokoyama T.2016. Transmission of atypical scrapie to homozygous ARQ sheep. J. Vet. Med. Sci. 78: 1619–1624. doi: 10.1292/jvms.16-0259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prusiner S. B.1998. Prions. Proc. Natl. Acad. Sci. U.S.A. 95: 13363–13383. doi: 10.1073/pnas.95.23.13363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ru G.2017. Do we need to explain the occurrence of atypical scrapie? Vet. Rec. 180: 400–402. doi: 10.1136/vr.j1893 [DOI] [PubMed] [Google Scholar]
- 25.Saunders G. C., Cawthraw S., Mountjoy S. J., Hope J., Windl O.2006. PrP genotypes of atypical scrapie cases in Great Britain. J. Gen. Virol. 87: 3141–3149. doi: 10.1099/vir.0.81779-0 [DOI] [PubMed] [Google Scholar]
- 26.Shimizu Y., Kaku-Ushiki Y., Iwamaru Y., Muramoto T., Kitamoto T., Yokoyama T., Mohri S., Tagawa Y.2010. A novel anti-prion protein monoclonal antibody and its single-chain fragment variable derivative with ability to inhibit abnormal prion protein accumulation in cultured cells. Microbiol. Immunol. 54: 112–121. doi: 10.1111/j.1348-0421.2009.00190.x [DOI] [PubMed] [Google Scholar]
- 27.Vaccari G., Panagiotidis C. H., Acin C., Peletto S., Barillet F., Acutis P., Bossers A., Langeveld J., van Keulen L., Sklaviadis T., Badiola J. J., Andreéoletti O., Groschup M. H., Agrimi U., Foster J., Goldmann W.2009. State-of-the-art review of goat TSE in the European Union, with special emphasis on PRNP genetics and epidemiology. Vet. Res. 40: 48. doi: 10.1051/vetres/2009031 [DOI] [PMC free article] [PubMed] [Google Scholar]