Abstract
A systematic validation of reference genes is a pre-requisite for the proper normalization of gene transcripts. In the present study, the annotated sequences from black pepper (Piper nigrum L.) leaf transcriptome were used as reference genes namely actin (PnACT), glyceraldehyde phosphate dehydrogenase (PnGAPDH), β-tubulin (PnTUB), ubiquitin conjugating enzyme (PnUBCE), 18srRNA and elongation factor-1-α (PnElF) to identify the stable reference gene. We focused the selection of stable reference gene on important biotic stress (Phytophthora) with different algorithms (geNorm, NormFinder and BestKeeper) along with Reffinder which resulted in identification of PnGAPDH and PnUBCE as stable genes. Norm qPCR (R package) was also used to estimate the stability of the selected genes. We elucidated the expression patterns of a target gene PnBGLU which codes for 1,3 beta glucanase with most stable as well as least stable reference genes by which the importance of selecting the stable gene for gene expression studies in this system was emphasized. The mean expression levels of PnBGLU was significantly overestimated and misinterpreted when least stable reference gene was used as normalizer. The selected reference genes on further analysis of the expression dynamics of PnBGLU among resistant and susceptible genotypes showed PnGAPDH as the suitable reference gene for P. nigrum–P. capsici pathosystem.
Electronic supplementary material
The online version of this article (10.1007/s12298-019-00653-9) contains supplementary material, which is available to authorized users.
Keywords: Biotic stress, Gene expression, Housekeeping genes, Validation
Introduction
Gene expression analysis is the basis for the functional genomics of any gene as it unveils the mechanisms involved in the response of plants to environmental stress. The reference genes used as internal control for any real time PCR based expression analysis are mostly cellular maintenance genes which are involved in basic cellular processes and believed to be not affected by the environmental stress. There is evidence that transcript levels of certain housekeeping genes may also vary considerably under variable conditions (Vandesompele et al. 2002). Statistical algorithms such as geNorm, NormFinder, BestKeeper and Reffinder (Vandesompele et al. 2002; Andersen et al. 2004; Pfaffl et al. 2004) have been developed for reference gene transcript normalization in the recent past, but unfortunately this robust approach is under-utilized in many crop plants. Instead, some putative genes with housekeeping activity are taken as reference genes without proper validation. Accurate measurement of gene expression by real time PCR relies on the selection of valid reference genes for transcript normalization which should be unaffected by experimental conditions (Kozera and Rapacz 2013). Upon pathogen attack, plants essentially activate selective defense responses according to the type of pathogen. Phytophthora capsici belongs to the group hemibiotrophs and its pathosystem is vigorously studied in model plants viz., Nicotiana & Arabidopsis. The defense mechanism although flows through some specific signaling pathways, the degree and pattern of involvement of genes related in the pathway might differ between different plant species and it is essential to study individual pathosystem of interest.
Black pepper popularly known as King of spices, native to India, is an export oriented important spice crop grown in tropical countries. The foot rot disease caused by oomycete P. capsici contributes to major crop loss as it infects the vine both in the nursery and in field (Anandaraj and Sharma 1990). The elucidation of interaction of P. nigrum–P. capsici pathosystem is still in its infancy with the analysis of 1,3-beta-glucanase Jebakumar et al. (2001), phenyl ammonia lyase (PAL) Nazeem et al. (2008) between resistant and susceptible varieties. The omics efforts have led to the generation of the black pepper root transcriptome Gordo et al. (2012), leaf transcriptome upon inoculation with P. capsici (Johnson et al. 2012; Hao et al. 2016), fruit transcriptome (Hu et al. 2015) and leaf transcriptome with the major focus on simple sequence repeats (SSR) bearing miRNA (Joy et al. 2013). The leaf transcriptome of IISR Shakthi, a moderately resistant variety to this pathogen (Johnson et al. 2012) is an excellent resource for understanding the biological functions of the defense related genes. The pathogenesis related genes from plants would become candidate genes towards controlling the pathogen infection in future. To achieve this, gene expression analysis is of paramount importance. This needs suitable reference genes that can serve as an internal control for studying the dynamics of defense related genes upon pathogen infection. In this crop, there is no detailed study on reference genes for this stress. In light of this, the present study was undertaken by using the annotated locus viz., GAPDH, actin, ubiquitin conjugating enzyme, elongation factor 1-α, beta-tubulin and 18srRNA from black pepper transcriptome (http://220.227.138.212/TRDB; NCBI ID: PRJNA314826) and validating it towards its use as reference genes for the expression studies for this particular stress condition. In addition to the selection of stable reference gene, the expression profile of 1,3-beta-glucanase, was also analyzed by the selected reference gene from resistant and susceptible varieties of black pepper for the pathogen. We have taken the glucanase gene as our target gene in our study since the apparent role of 1,3 beta glucanase is found to be mainly against fungi/oomycete, it is induced only upon infection (Egea et al. 1999; Wakelin and Leung 2009) and also the protein have been found in seeds or leaves (Sou and Leung 2001; Saikia et al. 2005).
Materials and methods
Plants and in planta inoculation
The resistant (IISR Shakthi) and susceptible (Subhakara) varieties of black pepper against foot rot caused by P. capsici were raised (3–4 leaf stage) in sterile potting mixture (1:1:1) (Soil:Sand:FYM) maintained in the greenhouse. A virulent isolate of P. capsici (05–06) obtained from National repository of Phytophthora, Indian Institute of Spices Research, Kozhikode was used to challenge inoculate the plants. The experiment was conducted in three replications for each hpi (hrs post inoculation). Inoculum plugs of 3 mm size from 72 h old mycelial culture of P. capsici grown on carrot agar media were placed on the lower surface of the leaves and moist cotton strip was placed over the inoculum plug and was secured using cello tape. Plants with moist cotton on the lower surface of the leaves served as control.
RNA extraction, cDNA synthesis
Leaf samples (4th leaf from each plant) were collected from IISR Shakthi and Subhakara at different time points (0.5, 1, 2 4, 8, 16, 24, 48 and 72 hpi) from 3 biological replicates. The necrotic regions (spots) if any were removed, the leaves were flash frozen in liquid nitrogen and kept in − 80 °C. Total RNA was extracted from both IISR Shakthi and Subhakara from uninoculated as well as challenge inoculated leaves using Trizol reagent (Invitrogen), treated with DNase I (Fermentas) to avoid genomic DNA contamination. One microgram of each RNA samples was reverse transcribed to cDNA using oligodT (18) primers (Fermentas). The purity and concentration of the total RNA was analyzed by denaturing gel electrophoresis and spectrophotometer.
Primer design
Five genes (loci) viz., PnACT, PnGAPDH, PnTUB, PnE1F, PnUBCE, and PnBGLU from P. nigrum–P. capsici transcriptome database were retrieved. The P. nigrum 18srRNA sequence was retrieved from NCBI database (AH001736). The primers were designed with parameters: 150–200 bp maximum length, optimal Tm at 60 °C, GC% between 45 and 65% using primer 3plus software (Table 1).
Table 1.
Primers sequence of six internal control genes and gene of interest
| Annotated gene name | Gene function | Piper TrDB locus | Arabidopsis orthologs | Primer sequence (5′–3′) | Amplicon length (bp) | Amplification efficiency | R2 value |
|---|---|---|---|---|---|---|---|
| Actin (PnACT) | Cytoskeletal structural protein | 24661 | AT5G09810.1 | F: CTGGTGTCATGGTTGGAATG | 157 | 118 | 0.97 |
| R: GGTGCGACAACCTTGATTTT | |||||||
| GAPDH (PnGAPDH) | Oxidoreductase in glycolysis and gluconeogenesis | 35657 | AT1G79530.1 | F: ATGAAGGATTGGCGAGGTGG | 121 | 118 | 0.99 |
| R: AGGCCATTCCAGTGAGCTTC | |||||||
| β-tubulin (PnTUB) | GTP binding, GTPase activity, structural molecule activity | 32265 | AT2G29550.1 | F: ACTGTTGAGAGCCACGTGAG | 136 | 107 | 0.98 |
| R: GTAACCTGCTGTCTCCGCTT | |||||||
| 18srRNA (Pn18s) | Encodes small subunit of ribosomal RNA | M82469.1 | AT3G41768.1 | F: GAAAGACGAACAACTGCGAAAG | 118 | 117 | 0.99 |
| R: TGGTCGGCATCGTTTATGG | |||||||
| e1F (PnE1F) | Enzymatic delivery of aminoacyl t-RNA to ribosome | 221 | AT1G07940.2 | F: AAAGGTGACGACCATTCCAG | 232 | 95 | 0.99 |
| R: TCCCATCTCAGGTTTTGAGG | |||||||
| Ubiquitin congugating enzyme (PnUBCE) | Protein ligase activity, acid-aminoacid activity, ATP binding | 10974 | AT1G75440.1 | F:TTTGAGTTTCATCTGCATCAGC | 135 | 124 | 0.98 |
| R: TGAAACCACCACCTAGTCTCCT | |||||||
| β-1,3 Glucanase (PnBGLU) | Pathogenesis related protein | 3839 | F: GGGCAACGAACAGATTCCTA | 122 | 113 | 0.99 | |
| R: TGTGAACTGCAGTGGAGACC |
Two step: quantitative real-time PCR analysis
The qRT-PCR expression analysis was performed using SYBR green in 72 well plate Rotor Gene Q real time PCR system (Qiagen, Germany). Standard curves were generated for all the reference genes using a fivefold serial dilution of cDNA pool and PCR efficiency was calculated (Table 1, Supplementary Figure 1). The reaction conditions were: 94 °C for 5 min, 35 cycles of 94 °C for 30 s and 60 °C for 30 s. Expressions of all the six reference genes were monitored at 0.5, 1, 2, 4, 8, 12, 16, 24, 48 and 72 hpi respectively. For each reaction, 100 ng cDNA, 1 µM of each primer and 10 µl of SYBR green master mix (Qiagen) were added to make final volume of 20 µl per sample. Three replicates for the qRT-PCR were performed for biological replicates and the mean value was considered. A melt curve analysis was performed for the six reference gene primers.
Verification of amplified products by sequencing
The qRT-PCR products were separated on a 2% agarose gel and purified using QIA quick gel extraction kit (Germany) and sequenced in order to verify their sequence. A local BLAST search was done with the Piper transcriptome database to match the sequences to the annotated transcripts of the selected genes in the transcriptome along with NCBI BLAST searches. The Arabidopsis Initiative Resource (TAIR) BLAST 2.2.8 was done to find the Arabidopsis orthologs locus for the reference gene sequences.
Data acquisition
GenEX software version 6.0.1.612 was used to analyze the data. Reffinder was used to get an aggregate ranking of candidate reference genes used in the study. The relative stability of these genes was also determined in NormqPCR (R package) (Perkins et al. 2012).
Validation of candidate reference genes
The best (PnGAPDH), least stable gene (PnACT) were used to quantify the gene expression of PnBGLU at 0.5, 1, 2, 4, 8, 12, 16, 24, 48 and 72 hpi respectively from both the genotypes. Standard curves were generated using a fivefold serial dilution of the cDNA pool and PCR efficiency was calculated. The sequence of PnBGLU obtained from Piper TrDB was used to validate the normalizer gene by qRT-PCR with the verification of amplicon sequence with local search with Piper TrDB and BLAST searches with NCBI database. The reaction conditions were: 94 °C for 5 min, 35 cycles of 94 °C for 30 s and 60 °C for 30 s.
Utility of selected reference genes among genotypes
A relative expression level of PnBGLU was quantified in IISR Shakthi and Subhakara using a combination of reference genes PnGAPDH and PnUBCE at different time points of inoculation with P. capsici was done. The comparison of relative expression of PnBGLU was calculated using Pfaffl model (Pfaffl 2001). REST2009 software (Corbett Research Pvt. Ltd), which statistically tests qPCR data for pairwise differences between groups, was used.
Results
Verification of amplicons, primer specificity and PCR efficiency analyses
Melt curve analyses were performed following qRT-PCR to confirm specific amplification of all the samples which gave a single peak for each gene. The electrophoretic separation of amplicons produced a single fragment of the expected sizes (118–232 bp), with no primer dimer products. The sequenced amplicons had the same sequence as those in the Piper transcriptome database and local BLAST analysis of the obtained sequence proved their identity and the genes were named as (Piper nigrum genes) PnGAPDH, PnUBCE, PnTUB, Pn18S, PnACT, PnE1F. The Arabidopsis orthologs locus was found out by TAIR BLAST 2.2.8 analysis of the reference gene sequences. Standard curves were generated using a fivefold serial dilution of the cDNA pool with a linear regression coefficient of 0.986–0.999 and the PCR efficiencies ranged from 95 to 124%, confirming the suitability of the primer pairs in the qRT-PCR-based quantification. The identified Piper genes were taken as candidate reference genes for the transcript profiling. The cq values are the basic result of any qPCR. The qRT-PCR expression profiles of reference genes are presented as quantification cycle (Cq) values (Supplementary Figure 2). With the quantitation cycle values (cq), we attempted the stability analysis with the following algorithms.
Gene expression stability analyses
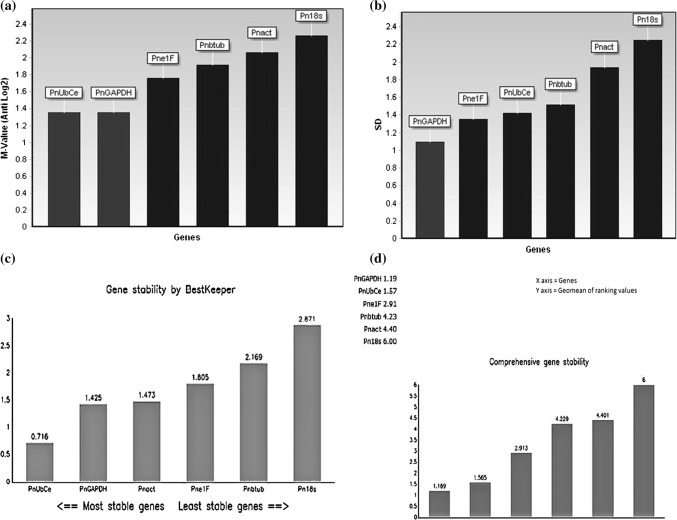
The expression stability was analysed by geNorm, NormFinder, BestKeeper. Reffinder gave a comprehensive ranking of different algorithms used (Fig. 1a–d). NormqPCR software from R package also was used.
Fig. 1.
Expression stability of 6 reference genes by a geNorm, b NormFinder, c BestKeeper, d Reffinder
geNorm analysis
The raw Ct values were transformed into quantities for relative comparison in geNorm. geNorm recommends the practice of using multiple internal control genes instead of single normalizer based on the low M value which indicates more stable gene expression. Stepwise exclusion of least stable genes allowed ranking of reference genes and below default geNorm threshold of 1.5 is considered to be having stable expression (Table 2). Across the samples, geNorm algorithm yielded PnGAPDH and PnUBCE as most stably expressed while, Pn18S as least stable.
Table 2.
Statistical analysis of the expression data of six candidate reference genes
| Gene name | BestKeeper coefficient of correlation (r) | NormFinder stability value | geNorm expression stability (M) |
|---|---|---|---|
| PnGAPDH | 0.808 | 1.090 | 1.348 |
| PnUbCE | 0.688 | 1.417 | 1.348 |
| PnACT | 1.940 | 2.071 | |
| PnTUB | 1.512 | 1.922 | |
| Pn18s | 2.247 | 2.273 | |
| PnE1F | 1.355 | 1.770 |
NormFinder analysis
NormFinder algorithm based on variance estimation approach, was used to reanalyze the stability of expression among reference genes which gives the reliable indicator of stability as the standard deviation when groups are not considered. NormFinder calculates accumulated standard deviation. Although the rankings generated by NormFinder slightly varied from geNorm, GAPDH still remained as best gene.
BestKeeper analysis
The BestKeeper program is based on standard deviation (SD), coefficient of correlation (r), coefficient of variance (CV) of Cq values. Genes with SD > 1 were considered as inconsistent. The analysis of the data using variations [SD (± Ct) and CV (%Ct)] for all the candidate genes showed that PnGAPDH and PnUBCE with low SD values (1.42 and 0.72) while PnE1F, PnTUB, PnACT and Pn18S with the high SD values (1.81, 2.17, 1.47 and 2.87) respectively. The analysis of the inter reference gene relation was done between the shortlisted genes. BestKeeper analysis ranked PnUbCE and PnGAPDH as most stable genes.
Overall ranking order and selection of optimal reference gene
Reffinder integrates four major computational programs viz., geNorm, NormFinder, BestKeeper and comparative delta Ct method and generates a comprehensive ranking by assigning weight to each candidate reference genes and calculating geometric mean of their weights (Fig. 1d; Table 2).
NormqPCR
The relative stability of these genes was determined in NormqPCR (R package) using the select HKs function, which can take either “geNorm” or “NormFinder” as an argument. In geNorm, the pairwise comparisons of each gene with every other gene were performed to determine their relative stability in gene expression. Lower M values represent genes with more stable expression across specimens being compared. In geNorm, the average pairwise gene expression variation of each potential housekeeping gene is compared to all other evaluated reference genes. The genes that demonstrate the least variance in comparison with all other genes are ranked as the most stable genes and are therefore likely to be the best reference genes. We have found two best housekeeping genes i.e., PnGAPDH and PnUBCE. After finding the best housekeeping genes, we have subtracted the mean of the Cq reference values for the gene from the other genes in each sample, in order to normalise them and allow a direct comparison of gene expression between different sample types (by calculating the 2−∆∆Cq value). To perform relative quantification between different sample types, the 2−∆∆Cq values should be calculated by subtracting the ∆Cq value for a given gene for the case sample from the control sample. The 2−∆∆Cq values were calculated using the deltaDeltaCq function in NormqPCR. This will return a list of all target genes with their corresponding values.
Reference gene validation of PnBGLU expression
To validate the suitability of reference genes identified in the study, we assessed the expression profile of 1,3-beta-glucanase locus (Locus_3839) from IISR Shakthi and Subhakara infected with P. capsici (05–06). Melt curve analyses, qRT PCR amplification, sequencing of amplified product (113 bp) as done for reference genes. The sequence of amplicons was matched with Piper transcriptome database along with the BLAST analysis of obtained sequence proving their identity to locus_3839 and was named as PnBGLU. Standard curves were generated using a fivefold serial dilution of cDNA pool with a linear regression coefficient of 0.99699 and estimated PCR efficiency of 113%. The expression pattern varied with reference genes with varying stability. Mean expression level of PnBGLU at 72 hpi with stable PnUBCE & PnGAPDH reference genes were 7.152 & 8.5 fold respectively. Whereas with unstable PnACT as reference gene, the expression level of PnBGLU was 571.788 (Fig. 2a–c).
Fig. 2.
Comparative expression of PnBGLU transcript using reference genes aPnUBCE, bPnACT and cPnGAPDH
Discussion
The histone H3 was identified as fruit tissue specific reference gene in lysine metabolism related gene expression in Piper nigrum L. cv. ‘Reyin No. 1’ (Hu et al. 2015). The selection was made based on the analysis on stability of histone H3, ubiquitin-7, cyclophilin, polyubiquitin-1, polyubiquitin-2, glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH), and actin for reference gene using geNORM analysis. The ubiquitin gene was used to analyze the polyphenol pathway genes in two different species of black pepper viz., resistant (Piper flaviflorum) and susceptible (Piper nigrum cv. Reyin-1) (Hao et al. 2016) while the 18sRNA was used in analysis of gene expression in Phytophthora susceptible variety Panniyur 1 (Mahadevan et al. 2016). The present study attempted a detailed analysis to identify the reference gene suitable for gene expression analysis in Piper nigrum species that are resistant and susceptible to P. capsici infection using different statistical algorithms viz., geNorm, NormFinder, BestKeeper, Reffinder and NormqPCR (R package).
We selected the annotated loci of well-known reference gene sequences from the transcriptome database, validated with sequencing reactions and named with prefix ‘Pn’ to denote as Piper nigrum genes. qRT-PCR procedures were optimized for all the selected reference genes.
In order to achieve best results, the common rule of Best3 can be applied using at least three reference genes (Bustin et al. 2009), three different validation programs (Jacob et al. 2013) and three biological replicates for each genotype (Pawłowicz et al. 2012). In this direction, we have used six reference genes, three different algorithms and three technical replicates of pooled biological replicates. We have employed three different statistical algorithms (geNorm, NormFinder and BestKeeper) to analyze the expression stability of the selected reference genes and online tool Reffinder to obtain comprehensive ranking. Based on the rankings from each algorithm, Reffinder assigned an appropriate weight to individual genes and according to the aggregate ranking of the whole data set, PnGAPDH and PnUBCE were the stable genes which are the same result from NormqPCR.
When the suitability of selected genes [most stable genes (PnUBCE, PnGAPDH) and least stable gene (PnACT)] was checked for the expression of an important defense related protein gene 1,3-beta-glucanase from resistant (IISR Shakthi), susceptible (Subhakara) variety, the most stable genes revealed that maximum transcript abundance of PnBGLU in IISR Shakthi occurs at 72 hpi. In Subhakara, down regulation of PnBGLU was observed from 0.5 hpi till 72 hpi. But normalization with least stable genes depicted a picture that PnBGLU expression in case of Subhakara as up regulation from 0.5 hpi onwards and at 72 hpi, it showed 90 fold increase in mean expression level. According to the available reports, this pattern of PnBGLU expression in susceptible may not be an appreciable expression of that gene since the PnBGLU expression is only inducible and also in late hours of infection (Egea et al. 1999; Jebakumar et al. 2001). The expression of PnBGLU, showed steady up regulation pattern in resistant genotype with PnGAPDH, while it was not appreciable with PnUBCE though it is taken as stable gene from the results of algorithms. The present study suggests that the qPCR results on transcript abundance were affected by stability of reference genes and hence the selection of suitable reference gene is a must in any gene expression studies.
This result implicates that the accumulation of 1,3-beta-glucanase to a greater extent with early expression in IISR Shakthi attributes for its resistance to P. capsici. As the transcripts were analyzed from the non-necrotic area from the varieties, our finding also tells the specific induction of this enzyme is confined to the resistant variety. Induced expression of glucanase by P. capsici in black pepper evidenced (Nazeem et al. 2008) by western blot analysis was studied in resistant (Kalluvally) and the susceptible (Panniyur-1) varieties. In both the varieties, the expression of glucanase was observed only after infection by P. capsici. The glucanase expression was absent in the healthy or uninfected plants.
Jebakumar et al. (2001) reported the presence of this enzyme from 48 hpi during infection by P. capsici in resistant variety IISR Shakthi but the susceptible varieties (Subhakara and Panniyur 1) failed to get the reaction. Our result is in accordance with this study with the expression of glucanase only in resistant variety. According to Zhang et al. (2013), the expression of CaBGLU was higher in the leaves than in the roots in capsicum infected with P. capsici. Initially the expression was down regulated and after 12 hpi, it increased with maximum expression at 72 hpi. The expression level was higher in resistant, moderately resistant varieties compared to susceptible. We also found the maximum expression at 72 hpi in moderately resistant variety compared to susceptible.
In conclusion, we have shown the possibility to identify excellent reference genes from a non-model crop Piper nigrum. The PnGAPDH as most stable reference gene showed in this study would enable the gene expression studies in black pepper and other plant—P. capsici pathosystem. The differential expression of 1,3-beta glucanase elucidated in two test genotypes using the validated reference gene provides the underlying mechanism of defense against the oomycete pathogen in black pepper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Agarose gel showing the amplification of reference genes: (1) PnACT (2) PnGAPDH (3) PnTUB (4) Pn18S, (5) PnUBCE (6) PnE1F (PNG 127 kb)
Box plot graphs of Cq values for each reference genes tested at different time points after inoculation with P. capsici (TIFF 239 kb)
Acknowledgements
The authors are thankful to Mrs. Rosana Babu for primer designing, Mr. Jayarajan for statistical analysis. The authors extend their gratitude to Indian Council of Agriculture Research (ICAR) for financial support through Outreach programme on Phytophthora, Fusarium and Ralstonia diseases of horticultural and field crops (PhytoFuRa).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Anandaraj M, Sharma YR. A simple baiting technique to detect and isolate Phytophthora capsici (P. palmivora MF4) from soil. Mycol Res. 1990;94:1003–1004. doi: 10.1016/S0953-7562(09)81321-X. [DOI] [Google Scholar]
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Kubista M, Mueller R, Nolan T, Pfaffl M, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Egea C, Dickinson MJ, Candela M, Candela ME. β-1,3-glucanase isoenzymes and genes in resistant and susceptible pepper (Capsicum annuum) cultivars infected with Phytophthora capsici. Physiol Plant. 1999;107:312–318. doi: 10.1034/j.1399-3054.1999.100308.x. [DOI] [Google Scholar]
- Gordo SMC, Pinheiro DG, Moreira ECO, Rodrigues SM, Poltronieri MC, DeLemos OF, Da Silva IT, Ramos RTJ, Silva A, Schneider H, Silva WA, Sampaio I, Darnet S. High throughput sequencing of black pepper root transcriptome. BMC Plant Biol. 2012;12:168. doi: 10.1186/1471-2229-12-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao C, Xia Z, Fan R, Tan L, Hu L, Wu B, Wu H. De novo transcriptome sequencing of black pepper (Piper nigrum L.) and an analysis of genes involved in phenylpropanoid metabolism in response to Phytophthora capsici. BMC Genom. 2016;17:822. doi: 10.1186/s12864-016-3155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Hao C, Fan R, Wu B, Tan L, Wu H. De novo assembly and characterization of fruit transcriptome in black pepper (Piper nigrum) PLoS ONE. 2015;10(6):e0129822. doi: 10.1371/journal.pone.0129822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Guertler R, Naim S, Nixdorf S, Fedier A, Hacker NF, Heinzelmann-Schwarz V. Careful selection of reference genes is required for reliable performance of RT-qPCR in human normal and cancer cell lines. PLoS ONE. 2013;8:e5918. doi: 10.1371/annotation/0a53f994-bfe2-45db-9dbb-97fdfea023c5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jebakumar SR, Anandaraj M, Sarma YR. Induction of PR proteins and defense related enzymes in black pepper due to inoculation with Phytophthora capsici. Ind Phytopathol. 2001;54:23–28. [Google Scholar]
- Johnson KG, Vijeshkumar IP, Anandaraj M. Transcriptomics approaches for gene discovery in plants—a case study in Piper. Agrotechnology. 2012;1:2. [Google Scholar]
- Joy N, Asha S, Mallika V, Soniya EV. De novo transcriptome sequencing reveals a considerable bias of simple sequence repeats towards the downstream of ‘Pre-miRNAs’ of black pepper. PLoS ONE. 2013;8:e56694. doi: 10.1371/journal.pone.0056694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozera B, Rapacz M. Reference genes in real-time PCR. J Appl Genet. 2013;54:391–406. doi: 10.1007/s13353-013-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan C, Krishnan A, Saraswathy GG, Surendran A, Jaleel A, Sakuntala M. Transcriptome-assisted label-free quantitative proteomics analysis reveals novel insights into Piper nigrum—Phytophthora capsici Phytopathosystem. Front Plant Sci. 2016;7:785. doi: 10.3389/fpls.2016.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazeem PA, Achuthan CR, Babu TD, Parabu GV, Girija D, Keshavachandran R, Samiyappan R. Expression of pathogenesis related proteins in black pepper (Piper nigrum L.) in relation to Phytophthora foot rot disease. J Trop Agric. 2008;46:45–51. [Google Scholar]
- Pawłowicz I, Kosmala A, Rapacz M. Expression pattern of the psbO gene and its involvement in acclimation of the photosynthetic apparatus during abiotic stresses in Festuca arundinacea and F. pratensis. Acta Physiol Plant. 2012;34:1915–1924. doi: 10.1007/s11738-012-0992-0. [DOI] [Google Scholar]
- Perkins JR, McMahon SB, Bennett DLH, Orengo C, Kohl M. ReadqPCR and NormqPCR: R packages for the reading, quality checking and normalisation of RT-qPCR quantification cycle (Cq) data. BMC Genom. 2012;13:296. doi: 10.1186/1471-2164-13-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Saikia R, Singh BP, Kumar R, Arora DK. Detection of pathogenesis-related proteins–chitinase and beta-1, β-glucanase in induced chickpea. Curr Sci. 2005;89:659–663. [Google Scholar]
- Sou YY, Leung DWM. Elevation of extracellular β-1,3-glucanase and chitinase activities in rose in response to treatment with acibenzolar-S-methyl and infection by D. rosae. J Plant Physiol. 2001;158:971–976. doi: 10.1078/0176-1617-00300. [DOI] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Bio. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakelin AM, Leung DWM. β-1,3-glucanase activity in the stigma of healthy petunia flowers. Biol Plant. 2009;53:69–74. doi: 10.1007/s10535-009-0010-6. [DOI] [Google Scholar]
- Zhang YL, Li DW, Gong ZH, Wang JE, Yin YX, Ji JJ. Genetic determinants of the defense response of resistant and susceptible pepper (Capsicum annuum) cultivats infected with Phytophthora capsici (Oomycetes; Pythiaceae) Genet Mol Res. 2013;12:3605–3621. doi: 10.4238/2013.September.13.5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Agarose gel showing the amplification of reference genes: (1) PnACT (2) PnGAPDH (3) PnTUB (4) Pn18S, (5) PnUBCE (6) PnE1F (PNG 127 kb)
Box plot graphs of Cq values for each reference genes tested at different time points after inoculation with P. capsici (TIFF 239 kb)