Abstract
Background
An anatomical covariance analysis (ACA) enables to elucidate inter-regional connections on a group basis, but little is known about the connections among white matter structures or among gray and white matter structures. Effect of including multiple magnetic resonance imaging (MRI) modalities into ACA framework in detecting white-to-white or gray-to-white connections is yet to be investigated.
New method
Proposed extended anatomical covariance analysis (eACA), analyzes correlations among gray and white matter structures (multi-structural) in various types of imaging modalities (T1-weighted images, T2 maps obtained from dual-echo sequences, and diffusion tensor images (DTI)). To demonstrate the capability to detect a disruption of the correlation network affected by pathology, we applied the eACA to two groups of cognitively-normal elderly individuals, one with (PiB+) and one without (PiB-) amyloid deposition in their brains.
Results
The volume of each anatomical structure was symmetric and functionally related structures formed a cluster. The pseudo-T2 value was highly homogeneous across the entire cortex in the PiB- group, while a number of physiological correlations were altered in the PiB + group. The DTI demonstrated unique correlation network among structures within the same phylogenetic portions of the brain that were altered in the PiB + group.
Comparison with Existing Method
The proposed eACA expands the concept of existing ACA to the connections among the white matter structures. The extension to other image modalities expands the way in which connectivity may be detected.
Conclusion
The eACA has potential to evaluate alterations of the anatomical network related to pathological processes.
Keywords: Neuroscience, Magnetic resonance imaging, Correlation network, Positron emission tomography, Amyloid, Anatomical covariance, T2 relaxation, Diffusion tensor imaging
1. Introduction
The brain is an organ that consists of basic functional elements, wiring that connects the units to form purposeful and efficient functional modules, and systems to support and maintain the working environment for cells in the brain. The gray matter forms the functional units and the white matter fibers, as wires, connect these units. Advancements in neuroimaging technologies and neurophysiology allow the in vivo observation of the connections among gray matter structures, which has evolved into an emerging concept of the connectome. Among various methods to infer connections among brain regions, anatomical covariance analysis (ACA) (Alexander-Bloch et al., 2013; Evans, 2013; Lerch et al., 2006) has been emerged to infer inter-regional connections on a group basis. The ACA is based on an assumption that anatomical structures that form a functional unit are tightly connected with axons and synapses, and therefore, their biological properties are similar. For example, if region A and B are connected, a person with high gray matter density in the region A also has the region B with high gray matter density (and vice versa). Such relationships, termed correlation networks, might be genetically defined, or could be acquired later in life through experience, training, or degeneration caused by various diseases. One of the well-described disease-related anatomical correlations is diaschisis, which was originally defined as a dysfunction of areas that are connected to a damaged brain area, but can also result in atrophy of the areas connected to the damaged area (e.g., crossed cerebellar atrophy (Jimenez-Caballero, 2012; Menon, 2012; Olsen et al., 1963)). For the past decade, efforts have been made to systematically and comprehensively investigate the correlations of cortical thickness, volume, density, or myelination (Ma and Zhang, 2017) among entire cortical areas, which was summarized in (Evans, 2013). The pattern of anatomical correlation is similar to that observed in resting-state fMRI analysis, but varies depending on the stage of brain development (Raznahan et al., 2011) and aging (Brickman et al., 2007). Disease-related anatomical covariance has been elucidated in various neurological diseases with brain atrophy, including Alzheimer's disease (He et al., 2008), frontotemporal dementia (Hafkemeijer et al., 2016), semantic dementia (Seeley et al., 2009), progressive non-fluent aphasia (Seeley et al., 2009), corticobasal syndrome (Seeley et al., 2009), multiple sclerosis (He et al., 2009), and temporal lobe epilepsy (Bernhardt et al., 2008), as well as disorders with less morphological abnormalities, such as schizophrenia (Wible et al., 1995, 2001) or fibromyalgia (Kim et al., 2015).
To systemically and comprehensively apply the ACA strategy to capture the anatomical features of the brain, several attempts have been made to analyze correlation network among white matter as well as gray matter structures. Such methods utilized the white matter parcellation based on a projection of cortical areas (Mitelman et al., 2005) or white matter regions-of-interest derived from voxel-based group comparison (Hulshoff Pol et al., 2004). However, existing approaches cover limited amount of preselected white matter areas or ignore boundary of the white matter structures that should be defined by orientation of fiber bundles. Therefore, structure-by-structure correlation network of the white matter area still need to be elucidated.
In this study, we extended the ACA method in two directions: multi-structural and multi-modality. Our approach, the extended anatomical covariance analysis (eACA) method, analyzed correlations among whole brain areas, including gray and white matter structures (multi-structural) in various types of imaging modalities (multi-modality). To parcellate both gray and white matter structures, we applied the multi-atlas label fusion method (Wu et al., 2016). Following this multi-dimensional extension in the image domain, the biological rationale for the “connectome” seen in the eACA needs to be re-defined. Connectome originally implies the connection among or within gray matter structures where neurons exist. The spatial extension to the white matter structures expands the concept to the connections between the white matter structures and between the gray and the white matter structures, in addition to the conventional gray-to-gray connections. The extension to multiple image modalities expands the way in which connectivity may be detected, which was originally based only on the regional property of the cortical gray morphology measured from T1-weighted images. We included diffusion MRI-based measures, fractional anisotropy (FA), and the sum of the three eigenvalues (trace) as markers of white matter integrity or the rotationally invariant magnitude of water diffusion within brain tissue, both of which are known to be affected by brain development (Dubois et al., 2008; Faria et al., 2010; Huppi and Dubois, 2006; Schmithorst et al., 2002, 2005), aging (Sexton et al., 2014; Xie et al., 2016) or diseases (for reviews (Dong et al., 2004; Lerner et al., 2014; Rollins, 2007; Sundgren et al., 2004). An estimate of T2 relaxation time (pseudo-T2 or pT2, as referred to in (Nakamura et al., 2014)), was included as a measure by which to evaluate the status of myelination and macromolecules (Du et al., 2012; Miot-Noirault et al., 1997; Mosher and Dardzinski, 2004).
We hypothesized that the re-defined connectome in the eACA could be applied to capture the alterations in the brain correlation network, especially related to brain pathologies seen in both gray and white matter structures. To test this hypothesis, we investigated whether the eACA could detect correlation networks related to amyloid deposition in cognitively normal individuals. Increasing evidence indicates that amyloid deposition is seen in 20–40% of cognitively normal individuals over 65 years of age (Aizenstein et al., 2008; Bouwman et al., 2009; Mintun et al., 2006; Peskind et al., 2006; Shaw et al., 2009). Amyloid pathology is known to alter both gray (Chang et al., 2015; Ten Kate et al., 2018; Tijms et al., 2016) and white matter structures (Kantarci et al., 2014; Molinuevo et al., 2014) through the neuronal toxicity (Bloom, 2014; Iadanza et al., 2018) and vascular changes (Dallaire-Theroux et al., 2017; Lee et al., 2018). Recent studies suggest that whether subtle anatomical alterations related to amyloid deposition are MRI-detectable or not depends on the demographics of the target population and methods applied to analyze MRIs. The previous findings include atrophy located in the cingulate cortex (Storandt et al., 2009), the temporal neocortex (Chetelat et al., 2012), the hippocampal areas (Hsu et al., 2015), the inferior frontal cortex (Oh et al., 2014), and the basal forebrain (Grothe et al., 2014), or no atrophy (Bourgeat et al., 2010). Based on our hypothesis that the eACA could provide unique anatomical features that are difficult to capture through conventional regional comparison, we investigated the correlation network of the amyloid-positive and amyloid-negative normal cognitive groups to determine whether the eACA could detect differences between the two groups.
2. Methods
2.1. Participants
All participants enrolled in this study were selected from the BIOCARD study (Biomarkers of Cognitive Decline Among Normal Individuals (http://www.biocard-se.org) (Albert et al., 2014), which aims to identify biomarkers associated with the progression from normal cognitive status to cognitive impairment or dementia, with a particular focus on Alzheimer's disease. These analyses are based on a subset of participants who have been scanned with 3T neuroimaging protocols since 2015, including high-resolution anatomical MRI and amyloid positron emission tomography (PET). In these analyses, 95 participants were scanned with multi-modal MR imaging (T1-and T2-weighted images, and diffusion tensor images), and the Pittsburgh compound, B (PiB-PET)). Eight subjects were excluded because of a voxel size coding error or multi-modal mis-coregistration that caused miscalculation of the distribution volume ratio (DVR) map of PiB-PET scanning. The remaining 87 subjects were selected for the subsequent processing. Table 1 summarizes the demographic information of the participants involved in this study. The research protocols of the BIOCARD study were approved by local institutional review boards. All subjects signed written, informed consent for their participation.
Table 1.
Characteristics of subjects.
| Characteristric | PiB- (n = 63) | PiB+ (n = 24) | Statistic | p value |
|---|---|---|---|---|
| Age (y) | 66.5 ± 10.7 | 70.4 ± 5.4 | 2.23† | 0.029* |
| Sex (female) | 42 (67%) | 15 (63%) | 0.013§ | 0.910 |
| MMSE1 | 29.5 ± 0.7 | 29.2 ± 2.1 | 0.713† | 0.483 |
| Total CVLT2 | 65.3 ± 9.2 | 61.4 ± 14.2 | 1.26† | 0.218 |
| cDVR3 | 0.95 ± 0.04 | 1.23 ± 0.16 | 8.373† | <0.001*** |
* p < 0.05 and *** p < 0.001.
MMSE = Mini-Mental State Examination.
CVLT = California Verbal Learning Test.
cDVR = mean cortical distribution volume ratio.
t statistic in Welch's two-sample t-test.
χ2 in chi-squared test.
2.2. Scan protocol
MRI scans were obtained on a Philips Achieva 3.0T scanner. T1-weighted structural images were obtained from the three-dimensional (3D) magnetization-prepared rapid gradient-echo (MPRAGE) sequence (repetition time [TR] = 6.8 ms, echo time [TE] = 3.1 ms, inversion time [TI] = 843 ms, field of view [FOV] = 256 × 256, thickness/gap = 1.2/1.2 mm, flip angle = 8, sagittal plane). Proton density and T2-weighted images were obtained using a double-echo fast spin echo sequence (TR = 4.2 s, TE1/TE2 = 28/80 ms, FOV = 212 × 212, thickness/gap = 2.2/2.2 mm, flip angle = 90, axial plane), from which T2 maps were generated. To obtain diffusion tensor images, b0 and diffusion weighted images were acquired from a spin echo sequence (TR = 7.5 s, TE = 75 ms, FOV = 260 × 260, thickness/gap = 2.2/2.2 mm, flip angle = 90, b-value = 700, number of gradients = 33, axial plane).
Dynamic 11C-PiB PET studies were performed on a GE scanner in 3D mode. To minimize motion during scanning, participants were fitted with a thermoplastic head-mask. After an IV bolus injection of a mean (SD) 14.7 (0.8) mCi of 11C-PiB, PET data were scanned immediately and acquired according to the protocol: 4×0.25; 8×0.5; 9×1, 2×3; and 10 × 5 min (70 min total, 33 frames). Dynamic images were reconstructed using filtered back-projection with a ramp filter (image size = 128 × 128, pixel size = 2 × 2 mm, slice thickness = 4.25 mm), yielding a spatial resolution of approximately 4.5 mm full width at half maximum (FWHM). Overall quality control review was performed by one of the co-authors (K.O.).
2.3. Image processing
The pipeline of the proposed eACA was summarized in Fig. 1. Details of each step will be described explicitly below.
Fig. 1.
Schematic flowchart of the proposed extended anatomical covariance analysis. A: parcel-based metrics for each image modality were extracted by image preprocessing, coregistration and brain parcellation, PiB+/PiB- grouping was achieved based on cortical DVR images, B: parcel-by-parcel correlation matrices of PiB+ and PiB- group were generated for each image modality, C: hierarchical clustering was performed on each correlation matrix, D: significant between-group difference for each parcel-pair correlation was evaluated by permutation test. The x-axis is the absolute difference of inter-parcel correlation coefficients between groups.
2.3.1. T1-weighted images
To remove potential confounders of low-frequency-intensity non-uniformity present in the MR images, N4ITK bias correction was applied on the 3D T1-weighted images (Mintun et al., 2006; Tustison et al., 2010).
2.3.2. pT2 maps
The T2 maps (Duncan et al., 1996) were calculated from proton-weighted images (first echo) and T2-weighted images (second echo) obtained from the double-echo fast spin echo sequence. Pixel-by-pixel T2 maps were obtained from the two-echo T2 images by
Where S1 and S2 are the first echo and second echo image intensities, with echo times TE1 and TE2, respectively. Rigid transformation implemented in SPM12 (cost function = Normalized Mutual Information; reslicing with trilinear interpolation; no wrapping) was applied to transform the T2-weighted images (S2) to the corresponding T1-weighted images; then, the transformation matrix was applied to co-register the T2 maps to the corresponding T1-weighted images. We used the term “pT2,” since the dual-echo T2 estimation is regarded as semi-quantitative compared to that based on multi-echo sequences with 16 ≤ echos.
2.3.3. DTI
The tensor calculation function implemented in MRICloud (www.MRICloud.org) (Mori et al., 2016) was used to calculate a tensor field from the raw b0 and diffusion weighted images. The tensor-calculation pipeline adopted a state-of-the-art corrections scheme for simultaneous corrections of motion and distortion by eddy current using a cost function based on tensor-fitting residuals and gradient-specific distortion-modeling (Li et al., 2013). The pipeline also applied voxel-by-voxel outlier rejections for occasional intensity loss due to local elastic motions. The MRICloud generated scalar maps, such as the fractional anisotropy (FA) and the trace maps, from the tensor field. An experienced neurologist performed the second quality check, based on visual inspection of the color-coded orientation maps calculated from the tensor field, to ensure the data had no noticeable motion artifacts. The DTIs were linearly co-registered to the corresponding T1-weighted images using rigid transformation, in two steps: 1) co-registration of the b0 image to the corresponding T2-weighted image; and 2) co-registration of the result of 1) to the corresponding T1-weighted image, by applying the transformation matrix obtained from T2-to T1-weighted image co-registration (2.3.2). This concatenated approach was adopted based on the fact that b0 images share very similar contrast with T2-weighted images.
2.3.4. PET
The time frames of each dynamic PiB-PET scan were aligned to the average of the first 2 min to correct for motion (Jenkinson et al., 2002). For registration purposes, we obtained static images by averaging the first 20 min of each dynamic PET scan. The 3D T1-weighted images were rigidly registered onto their corresponding 20-minute average. We generated a study-specific template using T1-weighted images in a diffeomorphic registration framework (Avants et al., 2010b), and then segmented this template using FreeSurfer (version 5.1, http://surfer.nmr.mgh.harvard.edu) (Desikan et al., 2006). The appropriate MRI template was registered onto each subject's T1-weighted image using diffeomorphic registration (Avants et al., 2008). The FreeSurfer segmentation in template space was transformed accordingly onto the PET scan. Consecutive transformations were concatenated so that FreeSurfer segmentations were interpolated only once. Distribution volume ratio (DVR) images were calculated in the native space of each PET image using the simplified reference tissue model with the cerebellar gray matter as reference tissue (Zhou et al., 2007). The brain parcellation map generated by the MRICloud (see 2.5 for more details) on the T1-weighted MRI space was transformed on the DVR image by applying the transformation matrix obtained from the rigid transformation of the T1-weighted image to the 20-minute average PET image. The cDVR was calculated as the average of the DVR values of the cingulate, frontal, parietal (including precuneus), lateral temporal, and the lateral occipital cortical regions, excluding the sensorimotor strip.
2.4. Definition of amyloid-positive and amyloid-negative based on DVR
To separate the 87 participants into individuals with minimal amyloid deposition (PiB-) and individuals with elevated levels of amyloid deposition (PiB-), a mean cDVR threshold of 1.02 was used, estimated by the best intersection of the probability density functions of the PiB+ and PiB- groups based on a Gaussian mixture model (Bilgel et al., 2016).
2.5. MRI parcellation and matrix generation
The eACA was performed on an atlas-based matrix, which was created by an anatomical parcellation map (a set of labels that follow anatomical ontology) on each T1-weighted image space. The parcellation map was generated by a multi-atlas label-fusion method, in which multiple brain MRIs with the parcellation map are transformed to a target image, and the transformed parcels are fused (label fusion) to create the anatomical parcellation map specific to the target image. The “T1-multi-atlas segmentation” function implemented in the MRICloud was applied to parcellate each of the T1-weighted images into a parcellation map that contained 285 ontological parcels, including both gray and white matter structures (Wu et al., 2016), which was suitable for this study targeting both gray and white matter structures. The function offers a fully automated multi-atlas image parcellation algorithm, which combines the image transformation algorithm, Large Deformation Diffeomorphic Metric Mapping (LDDMM) (Christensen et al., 1997; Granander and Miller, 1996; Miller et al., 1997; Oishi et al., 2009), and the atlas-fusion algorithm based on the joint-label fusion method (Wang et al., 2013), along with a local search algorithm (Coupe et al., 2011). The multi-atlas label-fusion algorithm relies on a collection of pre-parcellated T1-weighted brain MRIs as teaching files (atlas library version 9B, which includes 26 atlases that contain MRIs from cognitively-normal individuals and individuals with cognitive impairments or dementia; 50–90 years old (Wu et al., 2016)) to achieve high parcellation accuracy against a wide range of atrophic changes seen in patients with memory impairments or dementia (Liang et al., 2015; Zu et al., 2017). The parcels are listed in the Appendix Table 1. The parcellation map generated on the T1-weighted image was applied to a co-registered pT2 map and DTI to perform parcel-by-parcel quantification of the volume, pT2, FA, and trace. The atlas-based image parcellation and quantification method was chosen for two reasons: information integration and dimension reduction. Introducing a common ontological framework among the T1-weighted image, the pT2 map, and DTI enables the integration of multimodality information, and can effectively reduce the dimension of the data space while minimizing the loss of meaningful information, which is essential to avoid the curse of dimensionality (Miller et al., 2013; Mori et al., 2013).
The normalized volume of each parcel was calculated by dividing the volume of each parcel by that of the whole brain. Parcels that were located outside the brain parenchyma, such as the cerebrospinal fluid space, skull, bone marrow, scalp, and the choroid plexus, or that were too small for accurate identification, such as the mammillary body, fimbria, optic tracts, substructures in the brainstem and the subcallosal and rostral white matter around the anterior cingulate cortex (ACC), were excluded from the eACA. The parcellation map was further modified by removing non-brain structures, such as the ventricles and sulci, and applied to other image modalities that were co-registered to the T1-weighted images, to quantify the pT2, FA, and trace values of each parcel (see Fig. 1A). To eliminate the potential effects of partial volume (e.g., contamination of signals from non-brain structures) or that of slight misregistration due to geometric distortion, we excluded pixels with a pT2 value of more than 200 ms, a trace value of more than 0.0045 mm2s-1 (to exclude voxels that potentially include signal from the cerebrospinal fluid), and a trace value of less than 0.0015 mm2s-1 (to exclude voxels that potentially include an artifact related to spatial distortion due to adjacent air cavity) from the quantification. The above-mentioned procedures resulted in a volume matrix (87 individuals × 172 parcels), a pT2 matrix (87 individuals × 142 parcels), a FA matrix (87 individuals × 142 parcels), and a trace matrix (87 individuals × 142 parcels). Overall quality control review was performed by one of the authors (K.O.).
2.6. Correction for age and sex
Age and sex are known to affect the local volume of the brain, DTI-derived measures, and functional connectivity (Barnes et al., 2010). An adjustment for age and sex is thus necessary for the eACA. We adopted the method proposed by (Dukart et al., 2011) to adjust for the confounding effects. Essentially, confounding variables (age and sex) were modeled as predictors to determine all parcels on each MR modality (normalized volume, pT2, FA, and trace) based on the PiB- group. The following general linear model for a single image modality was solved for the parameters and ,
In the above model, and are age and sex vectors for PiB- subjects. is a raw image feature vector for each parcel, is the image features without effects of age and sex. To obtain image features for the PiB + group, the estimated parameters and were applied to general linear model in terms of .
Given that the PiB- group was significantly younger than the PiB + group (Table 1), we also rebuilt the above linear model by adjusting confounding variables based on both PiB+ and PiB- groups.
2.7. Group-wise anatomical correlation matrix
The anatomical correlation was defined as statistical associations in volume or image-intensity between brain structures defined by the parcellation map (see Section 4). Pearson correlation coefficients across subjects within one group (PiB- or PiB+) were calculated to form the anatomical correlation matrix (N×N, where N represents the number of parcels) for each modality, such as the normalized volume matrices (172×172) and other intensity matrices (pT2, FA, trace, 142×142), as shown in Fig. 1B. Since the obtained interregional correlation matrices were symmetric, only the lower triangle part of each matrix, including N×(N-1)/2 connections served our interests.
2.8. The agglomerative hierarchical cluster analysis (clustergram)
To explicitly describe the pattern of each interregional correlation matrix, we further applied agglomerative hierarchical cluster analysis for each correlation matrix. Clustering was generated by grouping parcels with similar correlation vectors (1×N) according to minimum linkage based on correlation distance between every pair, as described in (Liu et al., 2012). The hierarchical tree was cut by a prefix threshold (60% of maximum linkage across all parcels) for all modalities. The threshold was defined based on the volume matrix, in an attempt to decrease the threshold of maximum linkage to increase number of clusters, while setting a constraint of a minimum number of parcels (two parcels) included in each cluster.
2.9. The eACA and statistical comparisons between PiB+ and PiB-
To investigate the effects of brain amyloid deposition on the anatomical correlation, we compared the anatomical correlation matrices between the PiB- and PiB + groups. Based on previous research that suggested strong correlations among functionally connected anatomical areas, as observed in healthy cohorts (Chen et al., 2008; Lerch et al., 2006), and among areas vulnerable to specific diseases, as observed in cohorts of diseases (Bernhardt et al., 2008; Hafkemeijer et al., 2016; He et al., 2008, 2009; Seeley et al., 2009), we hypothesized that the anatomical correlations in normal subjects would be disrupted by amyloid deposition. To extract this pairwise correlation from all parcel-pairs (N×(N-1)/2), a permutation test was applied to assess the significance of each inter-parcel correlation coefficient between PiB- and PiB+ (Bassett et al., 2008). Permutation was repeated for 5000 times to construct sampling distribution of all correlation coefficients under the null hypothesis that observed correlation differences were not determined by true group factor. A p < 0.05 was considered significant difference after the false discovery rate (FDR) correction (Benjamini and Hochberg, 1995). The results were represented in a Manhattan plot for each image modality, where parcel pairs were depicted along the horizontal axis, and the negative logarithm of the p-value for each parcel pair was depicted on the vertical axis. Under a limited number of permutations (5,000 in our study), the estimated p value could be zero when the effect size is large. On such occasions, we used p = 0.01 instead for the visualization.
3. Results
3.1. The eACA
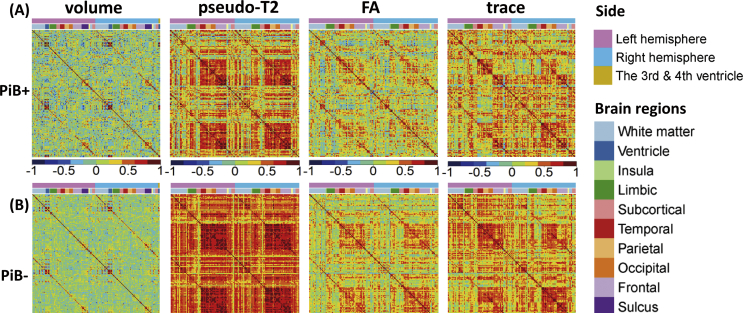
The correlation matrices derived from multiple image modalities are demonstrated in Fig. 2, in which the correlation coefficients of each combination of parcels were color-coded from blue (-1), to green (0), to red (1). The findings for each modality are summarized in Sections 3.3–3.6, below.
Fig. 2.
Correlation matrices for various image modalities. A: PiB + group, B: PiB- group. For each correlation matrix, the structural labels along each axis for volume, pseudo-T2, FA, and trace are presented in the color bar above the matrix.
3.2. The hierarchical cluster analysis
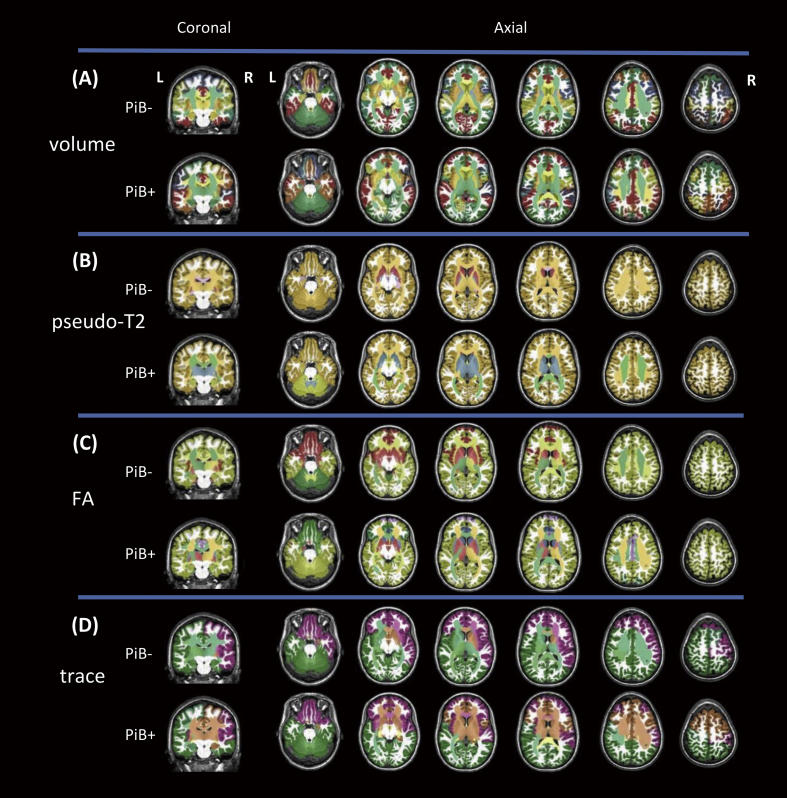
The hierarchical clustering results for each correlation matrix were mapped to a brain template, as shown in Fig. 3. Parcels with a similar set of covaried parcels were clustered and represented by colors that were arbitrarily assigned. The details of cluster components across every image modality are listed in Appendix Table 2 – 9. The findings for each modality are summarized in Sections 3.3–3.6, below.
Fig. 3.
The agglomerative hierarchical cluster analysis for each correlation matrix. A: volume, B: pseudo-T2, C: FA, D: trace. Clustering was generated by grouping parcels with similar correlation vectors according to the minimum linkage between each pair. The clusters are represented by colors that were arbitrarily assigned.
3.3. Normalized volume
The hierarchical clustering identified 23 clusters for the PiB- group, which are visualized in (Fig. 3A – PiB-), and which had a left-right symmetric appearance. The limbic structures (hippocampus, amygdala, cingulum, fornix, stria terminalis, parahippocampal gyrus, and entorhinal cortex) formed a single cluster. In contrast, 18 clusters were identified for the PiB + group, which are visualized in Fig. 3A – PiB+, and which showed a more asymmetric appearance than the PiB- group, especially in the orbitofrontal, parietal, and occipital areas.
No significant difference in the correlation coefficients between the PiB- and the PiB + groups was identified among 14706 (172×171/2) parcel-pairs of normalized volume (Fig. 4A).
Fig. 4.
Manhattan plot of anatomical correlation with various image modalities. A: volume, B: trace, C: FA, D: pseudo-T2, E: Scatterplot of correlation between the right entorhinal cortex and the right cerebellum with p < 0.001 (PiB- > PiB+). Each dot in the Manhattan plots represents significance for different correlation coefficients between a pair of anatomical structures. The blue horizontal line represents p = 0.05 (FDR-corrected).
3.4. pT2 maps
The hierarchical clustering identified seven clusters for the PiB- group, which are visualized in Fig. 3B – PiB-, and show that all the cortical and deep white matter parcels were clustered to form a single component (Fig. 3B – PiB-). In contrast, five clusters were identified for the PiB + group, which are visualized in Fig. 3B– PiB+, and which indicated two major cortical components, the cerebrum and the cerebellum (Fig. 3B– PiB+). The pT2 values of the parcels of the entire cerebrum were highly correlated.
Among 10011 (142×141/2) parcel-pairs of pT2 values, the correlation coefficients of 45 parcel-pairs were significantly higher in the PiB- group than in the PiB + group (Fig. 4E). Most of the significant parcel-pairs were related to the cerebellum or the hippocampus. Among them, one of the most significant correlations was identified the right entorhinal cortex and the right cerebellum (r for PiB + group = -0.242, r for PiB- group = 0.66, corrected p < 0.001), as shown in Fig. 4F. The complete list of parcel-pairs with significantly different correlations between groups is demonstrated in Table 2.
Table 2.
Pairs of structures with a significant difference in the correlation coefficient between the PiB+ and the PiB- groups. NA: not applicable (non of the pairs demonstrated sifnificant difference).
| Image modality | Pair of structures |
Correlation coefficient for PiB+ | Correlation coefficient for PiB- | Corrected p value | |
|---|---|---|---|---|---|
| Name of parcel1 | Name of parcel1 | ||||
| Volume | NA | NA | NA | NA | NA |
| pT2 | ST_L | PLIC_L | -0.312 | 0.483 | 0.044 |
| Hippo_L | IO_R | -0.043 | 0.680 | 0.044 | |
| Hippo_L | EC_L | -0.269 | 0.651 | 0.044 | |
| Hippo_L | IFO_L | -0.165 | 0.671 | <0.001 | |
| Hippo_L | MF_L | 0.003 | 0.665 | <0.001 | |
| Hippo_L | IF_opercularis_L | -0.003 | 0.648 | 0.044 | |
| Hippo_L | IF_triangularis_L | -0.185 | 0.701 | <0.001 | |
| Hippo_L | IF_triangularis_R | -0.047 | 0.630 | <0.001 | |
| Caudate_tail_L | EC_L | -0.581 | 0.450 | <0.001 | |
| MO_L | SCR_L | -0.315 | 0.478 | <0.001 | |
| Cerebellum_L | CGC_L | -0.314 | 0.614 | <0.001 | |
| Cerebellum_L | EC_L | -0.312 | 0.703 | 0.044 | |
| Cerebellum_L | ACR_L | -0.430 | 0.609 | <0.001 | |
| Cerebellum_L | SCR_L | -0.404 | 0.562 | <0.001 | |
| Cerebellum_L | SF_L | -0.201 | 0.707 | 0.044 | |
| Cerebellum_L | SF_PFC_L | -0.282 | 0.693 | <0.001 | |
| Cerebellum_L | CGC_R | -0.341 | 0.605 | 0.044 | |
| Cerebellum_L | EC_R | -0.383 | 0.694 | <0.001 | |
| Cerebellum_L | IFO_R | -0.324 | 0.688 | <0.001 | |
| Cerebellum_L | ACR_R | -0.447 | 0.596 | 0.044 | |
| Cerebellum_L | PoC_R | -0.024 | 0.755 | <0.001 | |
| Cerebellum_L | SF_PFC_R | -0.286 | 0.670 | <0.001 | |
| PVWa_R | A_L | -0.353 | 0.455 | <0.001 | |
| PVWa_R | MO_L | -0.282 | 0.457 | <0.001 | |
| PVWa_R | A_R | -0.211 | 0.507 | 0.044 | |
| PVWa_R | IO_R | -0.356 | 0.430 | 0.044 | |
| SCR_R | CerebellumWM_L | -0.127 | 0.703 | 0.044 | |
| Caudate_tail_R | IO_R | -0.143 | 0.630 | 0.044 | |
| Caudate_tail_R | EC_R | -0.340 | 0.465 | 0.044 | |
| Caudate_tail_R | CGC _L | -0.344 | 0.421 | 0.044 | |
| Caudate_tail_R | EC_L | -0.512 | 0.462 | <0.001 | |
| Fu_R | PLIC_L | -0.427 | 0.449 | <0.001 | |
| PCC_R | PLIC_L | -0.358 | 0.432 | <0.001 | |
| Cerebellum_R | EC_L | -0.301 | 0.678 | 0.044 | |
| Cerebellum_R | ACR_L | -0.342 | 0.599 | <0.001 | |
| Cerebellum_R | SCR_L | -0.437 | 0.552 | <0.001 | |
| Cerebellum_R | CGC_L | -0.179 | 0.584 | 0.044 | |
| Cerebellum_R | IF_triangularis_R | -0.038 | 0.631 | 0.044 | |
| Cerebellum_R | SF_L | -0.110 | 0.687 | <0.001 | |
| Cerebellum_R | SF_pole_R | -0.052 | 0.609 | 0.044 | |
| Cerebellum_R | SF_PFC_L | -0.109 | 0.682 | <0.001 | |
| Cerebellum_R | EC_R | -0.242 | 0.661 | <0.001 | |
| Cerebellum_R | ACR_R | -0.305 | 0.581 | <0.001 | |
| Cerebellum_R | PrC_R | -0.008 | 0.737 | 0.044 | |
| Cerebellum_R | SF_PFC_R | -0.155 | 0.648 | <0.001 | |
| FA | MO_L | RLIC_L | -0.455 | 0.407 | <0.001 |
| CGC_R | IT_R | -0.584 | 0.531 | <0.001 | |
| MFO_L | Subgenual_ACC_L | -0.391 | 0.556 | <0.001 | |
| SCC_R | MF_R | -0.079 | 0.576 | <0.001 | |
| Trace | NA | NA | NA | NA | NA |
Abbreviation of brain structures is explained in Appendix Table 1.
3.5. DTI
3.5.1. FA
The hierarchical clustering identified seven clusters for the PiB- group, which are visualized in Fig. 3C – PiB-. The FA values of the parcels within the neocortex (a cluster marked by yellow), within the paleocortex (piriform cortex, periamygdalar area, and olfactory areas; a cluster marked by red), and within the diencephalon-rhombencephalon complex (cerebellum-thalamus; a cluster marked by green), were all highly correlated. In contrast, eight clusters were identified for the PiB + group. The paleocortex cluster seen in the PiB- was fragmented and a new cluster was seen in the deep white matter area. Moreover, most of the cerebrum and cerebellar cortices were highly correlated.
Among 10011 (142×141/2) parcel-pairs of FA values, four parcel-pairs of FA between the right cingulum and the right interior temporal gyrus (r for PiB + group = -0.584, r for PiB- group = 0.531, corrected p < 0.001), the left retrolenticular part of internal capsule and the left middle occipital gyrus (r for PiB + group = -0.455, r for PiB- group = 0.407, corrected p < 0.001), the right splenium of corpus callosum and the right middle frontal gyrus (r for PiB + group = -0.079, r for PiB- group = 0.576, corrected p < 0.001), and the left subgenual anterior cingulate cortex and the left middle fronto-orbital gyrus (r for PiB + group = -0.391, r for PiB- group = 0.556, corrected p < 0.001) were identified as correlation coefficients that were significantly lower in the PiB + group than in the PiB- group, as shown in Fig. 4C and Table 2.
3.5.2. Trace
The hierarchical clustering identified nine clusters for the PiB- group, as visualized in Fig. 3D – PiB-. Among them, two major clusters were observed in the cortex; one related to the frontopolar-orbitofrontal and the right frontotemporal areas (a cluster marked by purple in Fig. 3D), and another related to the parieto-occipital and left frontotemporal areas (a cluster marked by green in Fig. 3D). In contrast, eight clusters were identified for the PiB + group, as visualized in Fig. 3D – PiB+. An additional cluster in the cortical area, the dorsal frontal cluster, which was not seen in the PiB- group, was seen in the PiB + group (marked by brown in Fig. 3D – PiB+).
No significant difference in the correlation coefficients between the PiB- and the PiB + groups was identified among 10011 (142×141/2) parcel-pairs of trace values (Fig. 4B).
3.6. Confounding adjustment based on the entire cohort
Even after replacing the covariates from PiB- with those from the entire cohort in the eACA confounding adjustment, the significant pairs of structures remained the same for the volume and DTI (shown in Appendix Table 10) analysis. The only exception was seen in the pT2. Thirty of 46 significant pairs of structures for pT2 were unaffected by adjustment changes. As described in Section 3.4, most of the significant parcel-pairs were still related to the cerebellum or the hippocampus.
4. Discussion
The eACA method described here provides the ability to visualize a “normal correlation network” and the disruption related to amyloid deposition in a comprehensive way, via the correlation matrix and the clustergram. Anatomical correlation is assumed to be modulated by developmental coordination, synaptic maturation, and other neuropathologies (Alexander-Bloch et al., 2013; Hof and Morrison, 2004; Montembeault et al., 2016). Previous studies about anatomical covariance related to neurological diseases have primarily targeted on cortical thickness (Avants et al., 2010a), surface area (Palaniyappan et al., 2011), and gray (Seeley et al., 2009) and white matter volumes (Hulshoff Pol et al., 2004). Our study is the first, to our knowledge, to systematically explore the whole-brain anatomical covariance associated with amyloid pathology in a cognitively normal population. The primary finding was evidence for a disruption of the physiological intrinsic brain network apparent on pT2 values and FA in the PiB + group.
4.1. Cross-modality comparisons in the eACA
Overall, the eACA results demonstrated that each modality had a unique pattern of physiological correlations. The number of clusters and the patterns observed in this study were based on 60% of the maximum linkage in the hierarchical tree (Liu et al., 2012), although the method to define an optimal modality-specific threshold has not been fully investigated. Further studies are needed to investigate the optimal threshold to capture the landscape of anatomical correlations, the molecular and cellular underpinnings of the macroscopic pattern observed in our study, and the mechanisms of covariate patterns specific to each modality.
4.2. Normalized volume
The left-right symmetric appearance of the physiological correlation would suggest strong connections between bilateral corresponding anatomical areas via commissural fibers (Mechelli et al., 2005). The result of hierarchical clustering indicated that the clusters obtained from the PiB- group represented functional units formed by regionally connected areas, such as a cluster that involves the Papez circuit (the entorhinal cortex, parahippocampal gyrus, hippocampus, fornix, cingulum, thalamus, hypothalamus, and the stria terminalis) and the amygdala, which are crucial in memory modulation (Hamann et al., 1999; Markowitsch, 1997). Although such symmetric and functionally meaningful clusters seen in the PiB- were disrupted in the clustergram obtained from the PiB+, the pairwise correlation analysis demonstrated that the physiological correlation seen in the PiB- group was preserved in the PiB + group.
4.3. pT2 maps
T2 or pT2 values in the brain are known to be affected by various micro-environmental factors of the brain tissue, especially the existence of myelin and macromolecules (Du et al., 2012; Miot-Noirault et al., 1997) (Mosher and Dardzinski, 2004). In this study, pT2 values of entire brain parcels were well correlated in both the PiB- and the PiB + groups, except for a dissociation between the cerebrum and the cerebellum observed in the PiB + group.
The pairwise correlation analysis demonstrated that the physiological correlation seen in the PiB- group was widely altered in the PiB + group. Among the 71 parcel-pairs with alteration, 47 pairs were related to the cerebellum, indicating stable pT2 values for the cerebellum independent of the pT2 values of the cerebral structures. This finding was congruent with the results of clustering analysis, suggesting that, similar to the evaluation of amyloid-PET (Catafau et al., 2016), the pT2 value of the cerebellum could be a reference for the evaluation of amyloid deposition in the cerebrum. Namely, for the PiB- group, pT2 value among the regions connected to the cerebellum was homogeneous (little intra-individual variation) but the inter-individual variation of the pT2 value was substantial. For the PiB + group, this cerebellum-related correlation network was disrupted. Seven pairs were related to the hippocampus, indicating consistently low pT2 values of the hippocampus, independent of the pT2 values of other structures. The scatterplot (Fig. 4F) clearly demonstrates the strength of the eACA approach, in which the two groups cannot be differentiated based on the pT2 of each structure, but a difference in the regression lines can distinguish the groups. Interestingly, for the cerebellum- and hippocampus-related correlation networks, the PiB + group demonstrated the negative correlation between pair of regions, while PiB- group always demonstrated strong positive correlation. The implication is not straightforward, but this tendency seen in the PiB + group: pT2 value of the cerebellum or hippocampus tends to be lower for the individuals with higher pT2 value in the connected structures, might indicate physiological or histopathological alterations that result in lower pT2 value in these regions. Indeed, both cerebellum and hippocampus are known as structures with altered BOLD response related to amyloid pathology (Kennedy et al., 2018; Quiroz et al., 2010).
4.4. DTI
4.4.1. FA
The left-right symmetric appearance of the physiological correlation was remarkable in FA, which suggested strong connections between bilateral corresponding anatomical areas via commissural fibers.
The pairwise correlation analysis demonstrated that the physiological correlation between the cingulum bundle and the inferior temporal lobe, and between the superior longitudinal fasciculus and the superior parietal lobule, were altered in the PiB + group. A relationship between amyloid burden and altered DTI measures in the selective white matter bundles, particularly the cingulum has been repeatedly reported in amnestic MCI (Douaud et al., 2013; Mito et al., 2018; Svard et al., 2017) and even in cognitively-normal elderly (Gold et al., 2014; Molinuevo et al., 2014; Racine et al., 2014; Rieckmann et al., 2016). The dissociation between the altered white matter bundles and preserved cortical structure might cause the altered physiological correlation.
4.4.2. Trace
There was a striking physiological asymmetry seen in the trace clusters. The left language areas were clustered with the bilateral prefrontal area and the bilateral parieto-occipital areas, while the right side of the corresponding areas were clustered with basal and polar aspects of the bilateral frontal lobe, the pattern of which follows the macroscopic anatomical asymmetries seen in the normal brain (Lancaster et al., 2003; Toga and Thompson, 2003). Although the right dominant cluster was separated into two sub-clusters in the PiB + group, the pairwise correlation analysis demonstrated no differences between the PiB- and PiB + groups, indicating the robustness of the trace value to amyloid deposition.
4.5. Limitations
Several limitations should be noted. First, the ground-truth of the physiological intrinsic brain network seen in this study is unknown. This lack of ground-truth is a common problem among connectome- or connectivity-related studies. Validations based on histology-MRI comparative studies are highly anticipated. Second, differences in the eACA between the PiB- and the PiB + groups should be interpreted as features that are related to amyloid deposition in the brain, but which may not be a definitive indication of future Alzheimer's disease, since it is not yet known which of these individuals might progress to mild cognitive impairment or dementia. There is a potential that the results of eACA might vary, depending on other physiopathological factors, such as genotype (Chen et al., 2011; Panizzon et al., 2012) and Tau deposition (see 4.5). Further classification of the study population might be needed in a future study. Third, interpretation of the hierarchical clustering followed by visualization of difference between groups was descriptive; while the permutation test applied to the correlation matrices was intended to identify group differences, the clustering approach was intended to capture spatial distribution of the correlation network. Forth, the generalizability of the proposed eACA approach to images obtained on different scanners or scan protocols needs to be evaluated. In particular, it is known that scalar values derived from DTI are affected by scan parameters. External validation of the results should be performed in the future. Fifth, DTI-to-T1-weighted image co-registration was affected by geometric distortion. Although we applied thresholds (see Section 2.5) to ameliorate the effect of cerebrospinal fluid contamination or signal dropout due to susceptibility artifacts, the effects of distortion and misregistration might have remained in the results. Sixth, each modality has its own noise patterns and variations in measurements related to both biological and methodological effects, as seen in Fig. 4. However, there is no general consensus about the appropriate ways to set modality-specific thresholds to detect pathological changes. As an initial attempt, we applied the same statistical threshold for all modalities, but the biologically optimal method for the modality-specific thresholding has yet to be investigated. Seventh, the small sample size of our study might have resulted in low sensitivity to detect mild anatomical features related to amyloid deposition. Ultimately, to investigate the long-term dynamics of the altered physiological correlation, longitudinal studies are necessary.
5. Conclusion
In conclusion, we adopted an eACA approach to describe the physiological correlation identified in a comparison of cognitively normal individuals with and without evidence of significant amyloid deposition. The pattern of physiological correlation was modality-specific, and congruent with previous research based on conventional ACA. The eACA approach could detect altered physiological correlation in T2 and FA in cognitively normal elderly with amyloid deposition. The eACA has the potential to evaluate alterations of anatomical status related to pathological processes.
Declarations
Author contribution statement
Chenfei Ye, Kenichi Oishi: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Timothy Brown: Analyzed and interpreted the data.
Murat Bilgel, Johnny Hsu: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Marilyn Albert, Ting Ma, Michael I. Miller, Susumu Mori: Contributed reagents, materials, analysis tools or data.
Brian Caffo: Conceived and designed the experiments; Analyzed and interpreted the data.
Funding statement
This work was made possible by the Dana Foundation Clinical Neuroscience Research Program, grant P41EB015909 from the National Institutes of Health, the Pilot Project Discovery Program from the Johns Hopkins Individualized Health Initiative (inHealth), the Fakhri Rad BriteStar award from the Department of Radiology Johns Hopkins University School of Medicine, a grant from the Basic Research Foundation of Shenzhen Science and Technology Program (JCYJ20150403161923510), the Basic Research Foundation Key Project Track of Shenzhen Science and Technology Program (JCYJ20170413110656460), and was supported, in part, by the Intramural Research Program of the National Institute on Aging.
Competing interest statement
The authors declare the following conflict of interests: Dr Miller and Dr Mori are the co-founders, and Dr Oishi is a consultant, for Anatomy Works. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
Additional information
No additional information is available for this paper.
Acknowledgements
We would like to acknowledge the contributions of the Geriatric Psychiatry Branch (GPB) of the intramural program of the NIMH who initiated the study (PI: Dr. Trey Sunderland). We are particularly indebted to Dr. Karen Putnam, who has provided ongoing documentation of the GPB study procedures and the data files received from NIMH. We also thank Ms. Mary McAllister for help with manuscript editing. Data collection was supported by Biomarkers of Cognitive Decline Among Normal Individuals: the BIOCARD Study (BIOCARD) (U19-AG033655) funded by the National Institute on Aging (NIA). The BIOCARD Study consists of seven cores with the following members: (1) the Administrative Core (Marilyn Albert and Barbara Rodzon); (2) the Clinical Core (Marilyn Albert, Rebecca Gottesman, Ned Sacktor, Scott Turner, Leonie Farrington, Maura Grega, Daniel D'Agostino, Gay Rudow, Scott Rudow); (3) the Imaging Core (Michael Miller, Susumu Mori, Tilak Ratnanather, Timothy Brown, Anthony Kolasny, Kenichi Oishi, William Schneider, Laurent Younes); (4) the Biospecimen Core (Richard O'Brien, Abhay Moghekar, Jacqueline Darrow); (5) the Informatics Core (Roberta Scherer, David Shade, Ann Ervin, Jennifer Jones, Matt Toepfner, Hamadou Coulibaly, April Broadnax, Lisa Lassiter); the (6) Biostatistics Core (Mei-Cheng Wang, Daisy Zhu, Jiangxia Wang); and (7) the Neuropathology Core (Juan Troncoso, Olga Pletnikova, Gay Rudow, Karen Fisher).We are grateful to the members of the BIOCARD Scientific Advisory Board who provided continued oversight and guidance regarding the conduct of the study, including: Drs. John Csernansky, David Holtzman, David Knopman, Walter Kukull, and Kevin Grimm, as well as Drs. John Hsiao and Laurie Ryan, who provided oversight on behalf of the NIA, respectively. We would also like to thank the members of the BIOCARD Resource Allocation Committee who provided ongoing guidance regarding the use of the biospecimens collected as part of the study, including: Drs. Constantine Lyketsos, Carlos Pardo, Gerard Schellenberg, Leslie Shaw, Madhav Thambisetty, and John Trojanowski.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Aizenstein H.J., Nebes R.D., Saxton J.A., Price J.C., Mathis C.A., Tsopelas N.D., Ziolko S.K., James J.A., Snitz B.E., Houck P.R., Bi W., Cohen A.D., Lopresti B.J., DeKosky S.T., Halligan E.M., Klunk W.E. Frequent amyloid deposition without significant cognitive impairment among the elderly. Arch. Neurol. 2008;65:1509–1517. doi: 10.1001/archneur.65.11.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M., Soldan A., Gottesman R., McKhann G., Sacktor N., Farrington L., Grega M., Turner R., Lu Y., Li S., Wang M.C., Selnes O. Cognitive changes preceding clinical symptom onset of mild cognitive impairment and relationship to ApoE genotype. Curr. Alzheimer Res. 2014;11:773–784. doi: 10.2174/156720501108140910121920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch A., Giedd J.N., Bullmore E. Imaging structural co-variance between human brain regions. Nat. Rev. Neurosci. 2013;14:322–336. doi: 10.1038/nrn3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants B.B., Cook P.A., Ungar L., Gee J.C., Grossman M. Dementia induces correlated reductions in white matter integrity and cortical thickness: a multivariate neuroimaging study with sparse canonical correlation analysis. Neuroimage. 2010;50:1004–1016. doi: 10.1016/j.neuroimage.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants B.B., Epstein C.L., Grossman M., Gee J.C. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants B.B., Yushkevich P., Pluta J., Minkoff D., Korczykowski M., Detre J., Gee J.C. The optimal template effect in hippocampus studies of diseased populations. Neuroimage. 2010;49:2457–2466. doi: 10.1016/j.neuroimage.2009.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes J., Ridgway G.R., Bartlett J., Henley S.M., Lehmann M., Hobbs N., Clarkson M.J., MacManus D.G., Ourselin S., Fox N.C. Head size, age and gender adjustment in MRI studies: a necessary nuisance? Neuroimage. 2010;53:1244–1255. doi: 10.1016/j.neuroimage.2010.06.025. [DOI] [PubMed] [Google Scholar]
- Bassett D.S., Bullmore E., Verchinski B.A., Mattay V.S., Weinberger D.R., Meyer-Lindenberg A. Hierarchical organization of human cortical networks in health and schizophrenia. J. Neurosci. 2008;28:9239–9248. doi: 10.1523/JNEUROSCI.1929-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- Bernhardt B.C., Worsley K.J., Besson P., Concha L., Lerch J.P., Evans A.C., Bernasconi N. Mapping limbic network organization in temporal lobe epilepsy using morphometric correlations: insights on the relation between mesiotemporal connectivity and cortical atrophy. Neuroimage. 2008;42:515–524. doi: 10.1016/j.neuroimage.2008.04.261. [DOI] [PubMed] [Google Scholar]
- Bilgel M., An Y., Zhou Y., Wong D.F., Prince J.L., Ferrucci L., Resnick S.M. Individual estimates of age at detectable amyloid onset for risk factor assessment. Alzheimers Dement. 2016;12:373–379. doi: 10.1016/j.jalz.2015.08.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom G.S. Amyloid-beta and tau: the trigger and bullet in Alzheimer disease pathogenesis. JAMA Neurol. 2014;71:505–508. doi: 10.1001/jamaneurol.2013.5847. [DOI] [PubMed] [Google Scholar]
- Bourgeat P., Chetelat G., Villemagne V.L., Fripp J., Raniga P., Pike K., Acosta O., Szoeke C., Ourselin S., Ames D., Ellis K.A., Martins R.N., Masters C.L., Rowe C.C., Salvado O., Group A.R. Beta-amyloid burden in the temporal neocortex is related to hippocampal atrophy in elderly subjects without dementia. Neurology. 2010;74:121–127. doi: 10.1212/WNL.0b013e3181c918b5. [DOI] [PubMed] [Google Scholar]
- Bouwman F.H., Schoonenboom N.S., Verwey N.A., van Elk E.J., Kok A., Blankenstein M.A., Scheltens P., van der Flier W.M. CSF biomarker levels in early and late onset Alzheimer's disease. Neurobiol. Aging. 2009;30:1895–1901. doi: 10.1016/j.neurobiolaging.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Brickman A.M., Habeck C., Zarahn E., Flynn J., Stern Y. Structural MRI covariance patterns associated with normal aging and neuropsychological functioning. Neurobiol. Aging. 2007;28:284–295. doi: 10.1016/j.neurobiolaging.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Catafau A.M., Bullich S., Seibyl J.P., Barthel H., Ghetti B., Leverenz J., Ironside J.W., Schulz-Schaeffer W.J., Hoffmann A., Sabri O. Cerebellar amyloid-beta plaques: how frequent are they, and do they influence 18F-florbetaben SUV ratios? J. Nucl. Med. 2016;57:1740–1745. doi: 10.2967/jnumed.115.171652. [DOI] [PubMed] [Google Scholar]
- Chang Y.T., Huang C.W., Chang Y.H., Chen N.C., Lin K.J., Yan T.C., Chang W.N., Chen S.F., Lui C.C., Lin P.H., Chang C.C. Amyloid burden in the hippocampus and default mode network: relationships with gray matter volume and cognitive performance in mild stage Alzheimer disease. Medicine (Baltim.) 2015;94 doi: 10.1097/MD.0000000000000763. e763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.H., Panizzon M.S., Eyler L.T., Jernigan T.L., Thompson W., Fennema-Notestine C., Jak A.J., Neale M.C., Franz C.E., Hamza S., Lyons M.J., Grant M.D., Fischl B., Seidman L.J., Tsuang M.T., Kremen W.S., Dale A.M. Genetic influences on cortical regionalization in the human brain. Neuron. 2011;72:537–544. doi: 10.1016/j.neuron.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J., He Y., Rosa-Neto P., Germann J., Evans A.C. Revealing modular architecture of human brain structural networks by using cortical thickness from MRI. Cerebr. Cortex. 2008;18:2374–2381. doi: 10.1093/cercor/bhn003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetelat G., Villemagne V.L., Villain N., Jones G., Ellis K.A., Ames D., Martins R.N., Masters C.L., Rowe C.C., Group A.R. Accelerated cortical atrophy in cognitively normal elderly with high beta-amyloid deposition. Neurology. 2012;78:477–484. doi: 10.1212/WNL.0b013e318246d67a. [DOI] [PubMed] [Google Scholar]
- Christensen G.E., Joshi S.C., Miller M.I. Volumetric transformation of brain anatomy. IEEE Trans. Med. Imaging. 1997;16:864–877. doi: 10.1109/42.650882. [DOI] [PubMed] [Google Scholar]
- Coupe P., Manjon J.V., Fonov V., Pruessner J., Robles M., Collins D.L. Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation. Neuroimage. 2011;54:940–954. doi: 10.1016/j.neuroimage.2010.09.018. [DOI] [PubMed] [Google Scholar]
- Dallaire-Theroux C., Callahan B.L., Potvin O., Saikali S., Duchesne S. Radiological-pathological correlation in Alzheimer's disease: systematic review of antemortem magnetic resonance imaging findings. J. Alzheimer's Dis. 2017;57:575–601. doi: 10.3233/JAD-161028. [DOI] [PubMed] [Google Scholar]
- Desikan R.S., Segonne F., Fischl B., Quinn B.T., Dickerson B.C., Blacker D., Buckner R.L., Dale A.M., Maguire R.P., Hyman B.T., Albert M.S., Killiany R.J. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Dong Q., Welsh R.C., Chenevert T.L., Carlos R.C., Maly-Sundgren P., Gomez-Hassan D.M., Mukherji S.K. Clinical applications of diffusion tensor imaging. J. Magn. Reson. Imaging. 2004;19:6–18. doi: 10.1002/jmri.10424. [DOI] [PubMed] [Google Scholar]
- Douaud G., Menke R.A., Gass A., Monsch A.U., Rao A., Whitcher B., Zamboni G., Matthews P.M., Sollberger M., Smith S. Brain microstructure reveals early abnormalities more than two years prior to clinical progression from mild cognitive impairment to Alzheimer's disease. J. Neurosci. 2013;33:2147–2155. doi: 10.1523/JNEUROSCI.4437-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F., Cooper A., Cohen B.M., Renshaw P.F., Ongur D. Water and metabolite transverse T2 relaxation time abnormalities in the white matter in schizophrenia. Schizophr. Res. 2012;137:241–245. doi: 10.1016/j.schres.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois J., Dehaene-Lambertz G., Perrin M., Mangin J.F., Cointepas Y., Duchesnay E., Le Bihan D., Hertz-Pannier L. Asynchrony of the early maturation of white matter bundles in healthy infants: quantitative landmarks revealed noninvasively by diffusion tensor imaging. Hum. Brain Mapp. 2008;29:14–27. doi: 10.1002/hbm.20363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukart J., Schroeter M.L., Mueller K., Alzheimer's Disease Neuroimaging I. Age correction in dementia–matching to a healthy brain. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022193. e22193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan J.S., Bartlett P., Barker G.J. Technique for measuring hippocampal T2 relaxation time. AJNR Am. J. Neuroradiol. 1996;17:1805–1810. [PMC free article] [PubMed] [Google Scholar]
- Evans A.C. Networks of anatomical covariance. Neuroimage. 2013;80:489–504. doi: 10.1016/j.neuroimage.2013.05.054. [DOI] [PubMed] [Google Scholar]
- Faria A.V., Zhang J., Oishi K., Li X., Jiang H., Akhter K., Hermoye L., Lee S.K., Hoon A., Stashinko E., Miller M.I., van Zijl P.C., Mori S. Atlas-based analysis of neurodevelopment from infancy to adulthood using diffusion tensor imaging and applications for automated abnormality detection. Neuroimage. 2010;52:415–428. doi: 10.1016/j.neuroimage.2010.04.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold B.T., Zhu Z., Brown C.A., Andersen A.H., LaDu M.J., Tai L., Jicha G.A., Kryscio R.J., Estus S., Nelson P.T., Scheff S.W., Abner E., Schmitt F.A., Van Eldik L.J., Smith C.D. White matter integrity is associated with cerebrospinal fluid markers of Alzheimer's disease in normal adults. Neurobiol. Aging. 2014;35:2263–2271. doi: 10.1016/j.neurobiolaging.2014.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granander U., Miller M.I. Computational anatomy: an emerging discipline. Stat. Comput. Graph. Newsl. 1996;7:3–8. [Google Scholar]
- Grothe M.J., Ewers M., Krause B., Heinsen H., Teipel S.J., Alzheimer's Disease Neuroimaging I. Basal forebrain atrophy and cortical amyloid deposition in nondemented elderly subjects. Alzheimers Dement. 2014;10:S344–353. doi: 10.1016/j.jalz.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafkemeijer A., Moller C., Dopper E.G., Jiskoot L.C., van den Berg-Huysmans A.A., van Swieten J.C., van der Flier W.M., Vrenken H., Pijnenburg Y.A., Barkhof F., Scheltens P., van der Grond J., Rombouts S.A. Differences in structural covariance brain networks between behavioral variant frontotemporal dementia and Alzheimer's disease. Hum. Brain Mapp. 2016;37:978–988. doi: 10.1002/hbm.23081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann S.B., Ely T.D., Grafton S.T., Kilts C.D. Amygdala activity related to enhanced memory for pleasant and aversive stimuli. Nat. Neurosci. 1999;2:289–293. doi: 10.1038/6404. [DOI] [PubMed] [Google Scholar]
- He Y., Chen Z., Evans A. Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer's disease. J. Neurosci. 2008;28:4756–4766. doi: 10.1523/JNEUROSCI.0141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Dagher A., Chen Z., Charil A., Zijdenbos A., Worsley K., Evans A. Impaired small-world efficiency in structural cortical networks in multiple sclerosis associated with white matter lesion load. Brain. 2009;132:3366–3379. doi: 10.1093/brain/awp089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof P.R., Morrison J.H. The aging brain: morphomolecular senescence of cortical circuits. Trends Neurosci. 2004;27:607–613. doi: 10.1016/j.tins.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Hsu P.J., Shou H., Benzinger T., Marcus D., Durbin T., Morris J.C., Sheline Y.I. Amyloid burden in cognitively normal elderly is associated with preferential hippocampal subfield volume loss. J. Alzheimer's Dis. 2015;45:27–33. doi: 10.3233/JAD-141743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulshoff Pol H.E., Schnack H.G., Mandl R.C., Cahn W., Collins D.L., Evans A.C., Kahn R.S. Focal white matter density changes in schizophrenia: reduced inter-hemispheric connectivity. Neuroimage. 2004;21:27–35. doi: 10.1016/j.neuroimage.2003.09.026. [DOI] [PubMed] [Google Scholar]
- Huppi P.S., Dubois J. Diffusion tensor imaging of brain development. Semin. Fetal Neonatal Med. 2006;11:489–497. doi: 10.1016/j.siny.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Iadanza M.G., Jackson M.P., Hewitt E.W., Ranson N.A., Radford S.E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 2018;19(12):755–773. doi: 10.1038/s41580-018-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M., Bannister P., Brady M., Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Jimenez-Caballero P.E. Crossed cerebellar atrophy: Update. J. Neurosci. Rural Pract. 2012;3:235–236. doi: 10.4103/0976-3147.102588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarci K., Schwarz C.G., Reid R.I., Przybelski S.A., Lesnick T.G., Zuk S.M., Senjem M.L., Gunter J.L., Lowe V., Machulda M.M., Knopman D.S., Petersen R.C., Jack C.R., Jr. White matter integrity determined with diffusion tensor imaging in older adults without dementia: influence of amyloid load and neurodegeneration. JAMA Neurol. 2014;71:1547–1554. doi: 10.1001/jamaneurol.2014.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy K.M., Foster C.M., Rodrigue K.M. Increasing beta-amyloid deposition in cognitively healthy aging predicts nonlinear change in BOLD modulation to difficulty. Neuroimage. 2018;183:142–149. doi: 10.1016/j.neuroimage.2018.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Kim J., Loggia M.L., Cahalan C., Garcia R.G., Vangel M.G., Wasan A.D., Edwards R.R., Napadow V. Fibromyalgia is characterized by altered frontal and cerebellar structural covariance brain networks. Neuroimage Clin. 2015;7:667–677. doi: 10.1016/j.nicl.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster J.L., Kochunov P.V., Thompson P.M., Toga A.W., Fox P.T. Asymmetry of the brain surface from deformation field analysis. Hum. Brain Mapp. 2003;19:79–89. doi: 10.1002/hbm.10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Zimmerman M.E., Narkhede A., Nasrabady S.E., Tosto G., Meier I.B., Benzinger T.L.S., Marcus D.S., Fagan A.M., Fox N.C., Cairns N.J., Holtzman D.M., Buckles V., Ghetti B., McDade E., Martins R.N., Saykin A.J., Masters C.L., Ringman J.M., Frster S., Schofield P.R., Sperling R.A., Johnson K.A., Chhatwal J.P., Salloway S., Correia S., Jack C.R., Jr., Weiner M., Bateman R.J., Morris J.C., Mayeux R., Brickman A.M., Dominantly Inherited Alzheimer N. White matter hyperintensities and the mediating role of cerebral amyloid angiopathy in dominantly-inherited Alzheimer's disease. PLoS One. 2018;13 doi: 10.1371/journal.pone.0195838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch J.P., Worsley K., Shaw W.P., Greenstein D.K., Lenroot R.K., Giedd J., Evans A.C. Mapping anatomical correlations across cerebral cortex (MACACC) using cortical thickness from MRI. Neuroimage. 2006;31:993–1003. doi: 10.1016/j.neuroimage.2006.01.042. [DOI] [PubMed] [Google Scholar]
- Lerner A., Mogensen M.A., Kim P.E., Shiroishi M.S., Hwang D.H., Law M. Clinical applications of diffusion tensor imaging. World Neurosurg. 2014;82:96–109. doi: 10.1016/j.wneu.2013.07.083. [DOI] [PubMed] [Google Scholar]
- Li Y., Shea S.M., Lorenz C.H., Jiang H., Chou M.C., Mori S. Image corruption detection in diffusion tensor imaging for post-processing and real-time monitoring. PLoS One. 2013;8 doi: 10.1371/journal.pone.0049764. e49764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z., He X., Ceritoglu C., Tang X., Li Y., Kutten K.S., Oishi K., Miller M.I., Mori S., Faria A.V. Evaluation of cross-protocol stability of a fully automated brain multi-atlas parcellation tool. PLoS One. 2015;10 doi: 10.1371/journal.pone.0133533. e0133533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhu X.H., Qiu P., Chen W. A correlation-matrix-based hierarchical clustering method for functional connectivity analysis. J. Neurosci. Methods. 2012;211:94–102. doi: 10.1016/j.jneumeth.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Zhang N. Cross-population myelination covariance of human cerebral cortex. Hum. Brain Mapp. 2017;38:4730–4743. doi: 10.1002/hbm.23698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitsch H.J. Varieties of memory: systems, structures, mechanisms of disturbance. Neurol. Psychiatry Brain Res. 1997;5:37–56. [Google Scholar]
- Mechelli A., Friston K.J., Frackowiak R.S., Price C.J. Structural covariance in the human cortex. J. Neurosci. 2005;25:8303–8310. doi: 10.1523/JNEUROSCI.0357-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon B. Crossed cerebellar atrophy. J. Neurosci. Rural Pract. 2012;3:422–423. doi: 10.4103/0976-3147.102650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M., Banerjee A., Christensen G., Joshi S., Khaneja N., Grenander U., Matejic L. Statistical methods in computational anatomy. Stat. Methods Med. Res. 1997;6:267–299. doi: 10.1177/096228029700600305. [DOI] [PubMed] [Google Scholar]
- Miller M.I., Faria A.V., Oishi K., Mori S. High-throughput neuro-imaging informatics. Front. Neuroinf. 2013;7:31. doi: 10.3389/fninf.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintun M.A., Larossa G.N., Sheline Y.I., Dence C.S., Lee S.Y., Mach R.H., Klunk W.E., Mathis C.A., DeKosky S.T., Morris J.C. [11C]PIB in a nondemented population: potential antecedent marker of Alzheimer disease. Neurology. 2006;67:446–452. doi: 10.1212/01.wnl.0000228230.26044.a4. [DOI] [PubMed] [Google Scholar]
- Miot-Noirault E., Barantin L., Akoka S., Le Pape A. T2 relaxation time as a marker of brain myelination: experimental MR study in two neonatal animal models. J. Neurosci. Methods. 1997;72:5–14. doi: 10.1016/s0165-0270(96)00148-3. [DOI] [PubMed] [Google Scholar]
- Mitelman S.A., Buchsbaum M.S., Brickman A.M., Shihabuddin L. Cortical intercorrelations of frontal area volumes in schizophrenia. Neuroimage. 2005;27:753–770. doi: 10.1016/j.neuroimage.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Mito R., Raffelt D., Dhollander T., Vaughan D.N., Tournier J.D., Salvado O., Brodtmann A., Rowe C.C., Villemagne V.L., Connelly A. Fibre-specific white matter reductions in Alzheimer's disease and mild cognitive impairment. Brain. 2018 Mar 1;141(3):888–902. doi: 10.1093/brain/awx355. [DOI] [PubMed] [Google Scholar]
- Molinuevo J.L., Ripolles P., Simo M., Llado A., Olives J., Balasa M., Antonell A., Rodriguez-Fornells A., Rami L. White matter changes in preclinical Alzheimer's disease: a magnetic resonance imaging-diffusion tensor imaging study on cognitively normal older people with positive amyloid beta protein 42 levels. Neurobiol. Aging. 2014;35:2671–2680. doi: 10.1016/j.neurobiolaging.2014.05.027. [DOI] [PubMed] [Google Scholar]
- Montembeault M., Rouleau I., Provost J.S., Brambati S.M., Alzheimer's Disease Neuroimaging I. Altered gray matter structural covariance networks in early stages of Alzheimer's disease. Cerebr. Cortex. 2016;26:2650–2662. doi: 10.1093/cercor/bhv105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S., Oishi K., Faria A.V., Miller M.I. Atlas-based neuroinformatics via MRI: harnessing information from past clinical cases and quantitative image analysis for patient care. Annu. Rev. Biomed. Eng. 2013;15:71–92. doi: 10.1146/annurev-bioeng-071812-152335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S., Wu D., Ceritoglu C., Li Y., Kolasny A., Vaillant M.A., Faria A.V., Oishi K., Miller M.I. MRICloud: delivering high-throughput MRI neuroinformatics as cloud-based software as a service. Comput. Sci. Eng. 2016;18:21–35. [Google Scholar]
- Mosher T.J., Dardzinski B.J. Cartilage MRI T2 relaxation time mapping: overview and applications. Semin. Musculoskelet. Radiol. 2004;8:355–368. doi: 10.1055/s-2004-861764. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Brown R.A., Araujo D., Narayanan S., Arnold D.L. Correlation between brain volume change and T2 relaxation time induced by dehydration and rehydration: implications for monitoring atrophy in clinical studies. Neuroimage Clin. 2014;6:166–170. doi: 10.1016/j.nicl.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., Habeck C., Madison C., Jagust W. Covarying alterations in Abeta deposition, glucose metabolism, and gray matter volume in cognitively normal elderly. Hum. Brain Mapp. 2014;35:297–308. doi: 10.1002/hbm.22173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K., Faria A., Jiang H., Li X., Akhter K., Zhang J., Hsu J.T., Miller M.I., van Zijl P.C., Albert M., Lyketsos C.G., Woods R., Toga A.W., Pike G.B., Rosa-Neto P., Evans A., Mazziotta J., Mori S. Atlas-based whole brain white matter analysis using large deformation diffeomorphic metric mapping: application to normal elderly and Alzheimer's disease participants. Neuroimage. 2009;46:486–499. doi: 10.1016/j.neuroimage.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen S., Hansen E., Plum C.M. Leuko-encephalopathy with crossed cerebro-cerebellar hemiatrophy. Acta Neurol. Scand. 1963;39:343–355. doi: 10.1111/j.1600-0404.1963.tb05335.x. [DOI] [PubMed] [Google Scholar]
- Palaniyappan L., Mallikarjun P., Joseph V., White T.P., Liddle P.F. Regional contraction of brain surface area involves three large-scale networks in schizophrenia. Schizophr. Res. 2011;129:163–168. doi: 10.1016/j.schres.2011.03.020. [DOI] [PubMed] [Google Scholar]
- Panizzon M.S., Fennema-Notestine C., Kubarych T.S., Chen C.H., Eyler L.T., Fischl B., Franz C.E., Grant M.D., Hamza S., Jak A., Jernigan T.L., Lyons M.J., Neale M.C., Prom-Wormley E.C., Seidman L., Tsuang M.T., Wu H., Xian H., Dale A.M., Kremen W.S. Genetic and environmental influences of white and gray matter signal contrast: a new phenotype for imaging genetics? Neuroimage. 2012;60:1686–1695. doi: 10.1016/j.neuroimage.2012.01.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peskind E.R., Li G., Shofer J., Quinn J.F., Kaye J.A., Clark C.M., Farlow M.R., DeCarli C., Raskind M.A., Schellenberg G.D., Lee V.M., Galasko D.R. Age and apolipoprotein E*4 allele effects on cerebrospinal fluid beta-amyloid 42 in adults with normal cognition. Arch. Neurol. 2006;63:936–939. doi: 10.1001/archneur.63.7.936. [DOI] [PubMed] [Google Scholar]
- Quiroz Y.T., Budson A.E., Celone K., Ruiz A., Newmark R., Castrillon G., Lopera F., Stern C.E. Hippocampal hyperactivation in presymptomatic familial Alzheimer's disease. Ann. Neurol. 2010;68:865–875. doi: 10.1002/ana.22105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine A.M., Adluru N., Alexander A.L., Christian B.T., Okonkwo O.C., Oh J., Cleary C.A., Birdsill A., Hillmer A.T., Murali D., Barnhart T.E., Gallagher C.L., Carlsson C.M., Rowley H.A., Dowling N.M., Asthana S., Sager M.A., Bendlin B.B., Johnson S.C. Associations between white matter microstructure and amyloid burden in preclinical Alzheimer's disease: a multimodal imaging investigation. Neuroimage Clin. 2014;4:604–614. doi: 10.1016/j.nicl.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A., Lerch J.P., Lee N., Greenstein D., Wallace G.L., Stockman M., Clasen L., Shaw P.W., Giedd J.N. Patterns of coordinated anatomical change in human cortical development: a longitudinal neuroimaging study of maturational coupling. Neuron. 2011;72:873–884. doi: 10.1016/j.neuron.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckmann A., Van Dijk K.R., Sperling R.A., Johnson K.A., Buckner R.L., Hedden T. Accelerated decline in white matter integrity in clinically normal individuals at risk for Alzheimer's disease. Neurobiol. Aging. 2016;42:177–188. doi: 10.1016/j.neurobiolaging.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins N.K. Clinical applications of diffusion tensor imaging and tractography in children. Pediatr. Radiol. 2007;37:769–780. doi: 10.1007/s00247-007-0524-z. [DOI] [PubMed] [Google Scholar]
- Schmithorst V.J., Wilke M., Dardzinski B.J., Holland S.K. Correlation of white matter diffusivity and anisotropy with age during childhood and adolescence: a cross-sectional diffusion-tensor MR imaging study. Radiology. 2002;222:212–218. doi: 10.1148/radiol.2221010626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmithorst V.J., Wilke M., Dardzinski B.J., Holland S.K. Cognitive functions correlate with white matter architecture in a normal pediatric population: a diffusion tensor MRI study. Hum. Brain Mapp. 2005;26:139–147. doi: 10.1002/hbm.20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley W.W., Crawford R.K., Zhou J., Miller B.L., Greicius M.D. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62:42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton C.E., Walhovd K.B., Storsve A.B., Tamnes C.K., Westlye L.T., Johansen-Berg H., Fjell A.M. Accelerated changes in white matter microstructure during aging: a longitudinal diffusion tensor imaging study. J. Neurosci. 2014;34:15425–15436. doi: 10.1523/JNEUROSCI.0203-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L.M., Vanderstichele H., Knapik-Czajka M., Clark C.M., Aisen P.S., Petersen R.C., Blennow K., Soares H., Simon A., Lewczuk P., Dean R., Siemers E., Potter W., Lee V.M., Trojanowski J.Q., Alzheimer's Disease Neuroimaging I. Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann. Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storandt M., Mintun M.A., Head D., Morris J.C. Cognitive decline and brain volume loss as signatures of cerebral amyloid-beta peptide deposition identified with Pittsburgh compound B: cognitive decline associated with Abeta deposition. Arch. Neurol. 2009;66:1476–1481. doi: 10.1001/archneurol.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundgren P.C., Dong Q., Gomez-Hassan D., Mukherji S.K., Maly P., Welsh R. Diffusion tensor imaging of the brain: review of clinical applications. Neuroradiology. 2004;46:339–350. doi: 10.1007/s00234-003-1114-x. [DOI] [PubMed] [Google Scholar]
- Svard D., Nilsson M., Lampinen B., Latt J., Sundgren P.C., Stomrud E., Minthon L., Hansson O., van Westen D. The effect of white matter hyperintensities on statistical analysis of diffusion tensor imaging in cognitively healthy elderly and prodromal Alzheimer's disease. PLoS One. 2017;12 doi: 10.1371/journal.pone.0185239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Kate M., Visser P.J., Bakardjian H., Barkhof F., Sikkes S.A.M., van der Flier W.M., Scheltens P., Hampel H., Habert M.O., Dubois B., Tijms B.M. Gray matter network disruptions and regional amyloid beta in cognitively normal adults. Front. Aging Neurosci. 2018;10:67. doi: 10.3389/fnagi.2018.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijms B.M., Kate M.T., Wink A.M., Visser P.J., Ecay M., Clerigue M., Estanga A., Garcia Sebastian M., Izagirre A., Villanua J., Martinez Lage P., van der Flier W.M., Scheltens P., Sanz Arigita E., Barkhof F. Gray matter network disruptions and amyloid beta in cognitively normal adults. Neurobiol. Aging. 2016;37:154–160. doi: 10.1016/j.neurobiolaging.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Toga A.W., Thompson P.M. Mapping brain asymmetry. Nat. Rev. Neurosci. 2003;4:37–48. doi: 10.1038/nrn1009. [DOI] [PubMed] [Google Scholar]
- Tustison N.J., Avants B.B., Cook P.A., Zheng Y., Egan A., Yushkevich P.A., Gee J.C. N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging. 2010;29:1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Suh J.W., Das S.R., Pluta J.B., Craige C., Yushkevich P.A. Multi-atlas segmentation with joint label fusion. IEEE Trans. Pattern Anal. Mach. Intell. 2013;35:611–623. doi: 10.1109/TPAMI.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wible C.G., Anderson J., Shenton M.E., Kricun A., Hirayasu Y., Tanaka S., Levitt J.J., O'Donnell B.F., Kikinis R., Jolesz F.A., McCarley R.W. Prefrontal cortex, negative symptoms, and schizophrenia: an MRI study. Psychiatry Res. 2001;108:65–78. doi: 10.1016/s0925-4927(01)00109-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wible C.G., Shenton M.E., Hokama H., Kikinis R., Jolesz F.A., Metcalf D., McCarley R.W. Prefrontal cortex and schizophrenia. A quantitative magnetic resonance imaging study. Arch. Gen. Psychiatr. 1995;52:279–288. doi: 10.1001/archpsyc.1995.03950160029007. [DOI] [PubMed] [Google Scholar]
- Wu D., Ma T., Ceritoglu C., Li Y., Chotiyanonta J., Hou Z., Hsu J., Xu X., Brown T., Miller M.I., Mori S. Resource atlases for multi-atlas brain segmentations with multiple ontology levels based on T1-weighted MRI. Neuroimage. 2016;125:120–130. doi: 10.1016/j.neuroimage.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S., Zhang Z., Chang F., Wang Y., Zhou Z., Guo H. Subcortical white matter changes with normal aging detected by multi-shot high resolution diffusion tensor imaging. PLoS One. 2016;11 doi: 10.1371/journal.pone.0157533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Resnick S.M., Ye W., Fan H., Holt D.P., Klunk W.E., Mathis C.A., Dannals R., Wong D.F. Using a reference tissue model with spatial constraint to quantify [11C]Pittsburgh compound B PET for early diagnosis of Alzheimer's disease. Neuroimage. 2007;36:298–312. doi: 10.1016/j.neuroimage.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu C., Wang Z., Zhang D., Liang P., Shi Y., Shen D., Wu G. Robust multi-atlas label propagation by deep sparse representation. Pattern Recognit. 2017;63:511–517. doi: 10.1016/j.patcog.2016.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.