Abstract
The oyster microbiome is thought to contribute to the pathogenesis of mass mortality disease in Pacific oysters, associated with OsHV-1. As filter-feeders, oysters host a microbiota that can be influenced by the estuarine environment. This may alter susceptibility to OsHV-1 infections, causing variable mortality. This study aimed at: (1) differences in the microbiome of Pacific oysters with a common origin but grown in geographically distinct estuaries; (2) evaluating changes occurring in the microbiota, especially in Vibrio, and (3) differential responses of the oyster microbiome, in response to an OsHV-1 infection. Pacific oysters sourced from a single hatchery but raised separately in Patonga Creek, Shoalhaven River and Clyde River of NSW, Australia, were used and challenged with OsHV-1. The initial microbiome composition was different in the three batches and changed further, post-injection (p < 0.05). The Patonga oysters with the highest mortality also had higher OsHV-1 and Vibrio quantities compared to the other two batches (p < 0.05). The higher initial bacterial diversity in Patonga oysters decreased in moribund oysters which was not observed in the other two batches (p < 0.05). The microbiome of survivors of OsHV-1 infection and negative control oysters of two batches, did not show any changes with the relevant pre-challenged microbiome. A strong correlation was observed between the OsHV-1 and Vibrio quantities in OsHV-1 infected oysters (r = 0.6; p < 0.001). In conclusion, the Pacific oyster microbiome differed in different batches despite a common hatchery origin. Different microbiomes responded differently with a differential outcome of OsHV-1 challenge. The higher Vibrio load in oysters with higher OsHV-1 content and higher mortality, suggests a role in Vibrio in the pathogenesis of this mortality disease. This study provided insights of the potential of different estuarine environments to shape the Pacific oyster microbiome and how different microbiomes are associated with different outcomes of OsHV-1 infection.
Keywords: Bioinformatics, Microbiology, Molecular biology, Virology
1. Introduction
Pacific oysters (Crassostrea gigas) make the greatest contribution to global oyster production with 625,925 tonnes per annum worth US$ 1.3 billion out of a total oyster production of 5.2 million tonnes worth US$ 4.2 billion (FAO, 2014). In Australia, edible oyster farming contributes approximately 38% to the total marine mollusc production, with an estimated 11.3 thousand tonnes produced in 2015–2016 (Mobsby and Koudah, 2017). However, outbreaks of severe and widespread oyster mortality have greatly impacted Pacific oyster farming in Australia (Jenkins et al., 2013; Paul-Pont et al., 2014; de Kantzow et al., 2017), New Zealand (Keeling et al., 2014) and in Europe (Segarra et al., 2010; Garcia et al., 2011; Peeler et al., 2012; Roque et al., 2012; Clegg et al., 2014).
Outbreaks of summer mortalities in Pacific oysters were first reported in Japan in 1950 (Takeuchi et al., 1960) and since then this syndrome affecting adult oysters was reported in many parts of the world, including Italy (Takeuchi et al., 1960), USA (Glude, 1974) and France (Maurer et al., 1986). In France, mass mortality events have also been reported in C. gigas spats since 1993 (Renault et al., 1994a), which were later identified to be associated with Ostreid herpesvirus-1 (OsHV-1) (Davison et al., 2005; Garcia et al., 2011). The reference genotype of OsHV-1 (Le Deuff and Renault, 1999; Davison et al., 2005) and related genotypes have been identified as the prominent pathogens which caused C. gigas mortalities in France from 1991 to 2008 (Martenot et al., 2011; Renault et al., 2012). However, at the end of spring 2008, widespread mortalities were reported in France which killed billions of young oysters and a genomic variant of OsHV-1, called μVar, was identified from these outbreaks (Segarra et al., 2010; Renault et al., 2012). Since then, severe disease of Pacific oysters associated with the variant and closely related microvariant genotypes were associated with disease events observed in different parts of the world, including Australia (Jenkins et al., 2013; Paul-Pont et al., 2015; Whittington et al., 2015), New Zealand (Renault et al., 2012), Ireland (Peeler et al., 2012; Clegg et al., 2014), Spain (Roque et al., 2012) and Scandinavia (Mortensen et al., 2016).
Despite the different genotypes of OsHV-1 and their causal relationship with oyster mortality, Pacific oyster mortality events are most often considered a result of complex interactions between the physiological status of the oysters, the environment and multiple pathogens (Samain et al., 2007; Alfaro et al., 2018; Pernet et al., 2018; de Lorgeril et al., 2018). Considering the age of oysters, spat and juvenile oysters are generally more susceptible to OsHV-1 infection and related mortality events (Renault et al., 1994b; Schikorski et al., 2011), while processes such as spawning which causes physiological stress also predispose oysters to mortality (Samain et al., 2007; Garcia et al., 2011). Furthermore, research has revealed a role of oyster genotype in susceptibility to the viral infections and mortality events (Dégremont, 2011; Azéma et al., 2017b; de Lorgeril et al., 2018). Meanwhile, environmental factors such as elevated seawater temperature (Petton et al., 2013; Renault et al., 2014), alterations in salinity (Soletchnik et al., 2007; Fuhrmann et al., 2016) and pH of water (Fuhrmann et al., 2019) have also been reported to affect the outcome of OsHV-1 infections.
Reinforcing the complex interactions between oysters, the environment and pathogen factors, it is notable that microvariant genotypes of OsHV-1 can be present in Pacific oysters without associated mortality (Dundon et al., 2011; Shimahara et al., 2012; Hick et al., 2016). Not all host and environmental factors that contribute to the pathogenesis related to infectious processes and subsequent oyster mortality are clearly understood. The role of bacteria in summer mortality of oysters was questioned after demonstration of high loads of Vibrio spp. in the haemolymph of moribund oysters (Lipp et al., 1976). Several subsequent studies report isolation/detection of pathogenic Vibrio spp. from Pacific oysters during mass mortality outbreaks such as Vibrio splendidus (Lacoste et al., 2001; Le Roux et al., 2002; Pernet et al., 2012), Vibrio aestuarianus (Garnier et al., 2008) and Vibrio harveyi (Saulnier et al., 2010). Recently, Pacific oysters exposed to a mass mortality outbreak and subsequently treated with antibiotics demonstrated delayed and reduced mortality compared to oysters that were not treated with antibiotics (Petton et al., 2015b). The suggestion of a role for bacteria present in the oyster microbiome or environment in the pathogenesis was supported by a rapid increase in the cultivable Vibrio count preceding OsHV-1 replication in the same outbreak (Petton et al., 2015b). Recently, de Lorgeril et al. (2018) demonstrated that infections caused by the variant μVar result in immunosuppression which shifts the oyster microbiome to allow opportunistic infections from bacteria such as Vibrio.
The Pacific oyster microbiome is dominated by Proteobacteria (Hernández-Zárate and Olmos-Soto, 2006; Fernandez-Piquer et al., 2012; Trabal Fernández et al., 2014). The results of recent 16S rRNA gene studies suggest that Pseudomonas and Vibrio are not abundant members of the oyster microbiome (Trabal Fernández et al., 2014; Lokmer and Wegner, 2015; King et al., 2018). Instead, members of the phylum Bacteroidetes, phylum Firmicutes (Trabal Fernández et al., 2014) and Arcobacter spp. (Fernandez-Piquer et al., 2012) have been shown to be abundant in the oyster microbiome. The microbiome composition can be influenced by oyster genetics (Wegner et al., 2013) and can change under various conditions that cause stress in oysters, including temperature (Lokmer and Wegner, 2015), translocation (Lokmer et al., 2016a), infection (Green and Barnes, 2010) and antibiotic stress (Lokmer et al., 2016b). The immediate environment can influence the microbiome composition of oysters, owing in part to their filter-feeding (Le Roux et al., 2016; Lokmer et al., 2016a). During times of stress, many of the environmental bacteria that are taken in may opportunistically colonize the oyster (Green and Barnes, 2010).
It is interesting to investigate how the microbiome composition of Pacific oysters with a common genetic background is affected by the environment they live in, and whether alterations in the microbiome can contribute to a differential response to the infection caused by OsHV-1. The aim of the current study was to evaluate changes in the Pacific oyster microbiome with changes in their environment and during progression of an experimental OsHV-1 infection. Specifically, studies were conducted to: (1) assess differences in the bacterial communities in Pacific oysters with a common origin but grown in three geographically distinct estuaries; (2) to evaluate changes occurring in the microbiota, with special reference to Vibrio populations and in response to OsHV-1 infection; and (3) to evaluate the responses of different batches of oysters with distinctly different life history, to an experimental OsHV-1 infection, conducted in a controlled laboratory environment.
2. Materials & methods
2.1. Oysters and their husbandry
Healthy triploid Pacific oysters (n = 348; 30–80 mm shell length) sourced from a commercial hatchery (Shellfish Culture Ltd., Tasmania) were used in this study. Different batches were grown under commercial farming conditions for 7 months in Patonga Creek, Hawkesbury River (juveniles, 10 months old, n = 116), or for 11–12 months in Shoalhaven River (adults, 14 months old, Goodnight Oysters, Greenwell Point, n = 116) or the Clyde River (adults, 16 months old, Signature Oysters, n = 116). These are 3 geographically separated estuaries in NSW, Australia which are impacted by a variety of human activities in addition to oyster farming. None of these oysters had been previously exposed to OsHV-1 based on surveillance for freedom from OsHV-1 in these estuaries and all oysters were certified negative for OsHV-1 by the competent government authority at the time of interstate transport from the hatchery to NSW. Further, the oysters were confirmed negative by testing a sample (n = 12) of each batch using a real-time quantitative PCR (qPCR) assay. The oysters were transported to a physical containment level 2 aquatic facility at the University of Sydney and acclimated for 3 days before experimental infection.
The oysters from each batch were randomly allocated to eight replicate 15 L tanks (n = 14 oysters per tank), making 24 tanks in total. A static water system was maintained with aerated, artificial seawater (ASW; Red Sea® salt, 30–31 ppt salinity) at 22 °C. Six of the tanks for each batch were designated for challenge with OsHV-1 while 2 tanks were designated for oysters challenged with the negative control.
Temperature data-loggers (Thermocron®) recorded the temperature every 30 min in selected tanks. The pH of water was maintained at 8.2 (range: 8.0–8.2) and ammonia, nitrite and nitrate levels were maintained <0.25 ppm. These water quality parameters were monitored daily, and adjustments were performed as required, by water exchange. Oysters were fed with a maintenance ration of commercial algae diet (Shellfish Diet, 1800; Reed Mariculture) daily in the morning. Mortality was recorded with twice daily monitoring for 7 days. Dead oysters were removed and stored at −80 °C until molecular studies were conducted.
2.2. Source of infective OsHV-1
A cryopreserved, filtered oyster tissue homogenate with confirmed OsHV-1 infection was used as the inoculum. The tissue homogenate was stored as multiple aliquots, at -80 °C with 10% v/w fetal bovine serum (Gibco) and 10% v/v glycerol. The inoculum was prepared from diseased oysters from a previous OsHV-1 related mortality event and had been used in previous experimental infections (Paul-Pont et al., 2015). The negative control was prepared as a cryopreserved, filtered tissue homogenate prepared from apparently healthy Pacific oysters from a disease-free population (Evans et al., 2015). The cryopreserved inoculums were thawed at 4 °C and diluted by 1:100 in distilled water (Ultrapure™) to provide the dose described below.
2.3. Challenge with OsHV-1
Oysters were immersed in a solution of MgCl2 (50 g/l) for 4–6h until relaxation of the adductor muscles caused the valve to open. A 100 μl aliquot of the diluted OsHV-1 inoculum (6.5 × 105 OsHV-1 DNA copies μl−1) was injected into the adductor muscle, using a 1 ml syringe and a 25-gauge needle. The control oysters were injected with a diluted, filtered oyster tissue homogenate that was negative for OsHV-1. After injection, oysters were transferred to the same designated tanks without feed for 8 hours. Physical separation and procedural care were taken to prevent cross contamination between tanks.
2.4. Sampling
Twelve oysters (n = 4 from each batch) were sampled on Day 0 prior to injection. Thereafter, 2 live oysters per tank were sampled on Days 1, 2, 3, 5 and 7. Samples were maintained on ice until processing. Dead oysters were sampled if identified by non-responsive gaping when exposed to external stimuli including air exposure.
2.5. OsHV-1 DNA quantification
Oysters were carefully shucked and equal portions of gill and mantle tissue (0.08–0.12 g total) were excised and placed in a 1.5 ml tube containing 1 ml of distilled water (Ultrapure™) and 0.4 g of 0.1 mm zirconia-silica beads (Biospec Products, Daintree Scientific). The tubes were kept at -80 °C until homogenization by bead-beating with a TissueLyser II (Qiagen) at a frequency of 30 Hz for 2 × 2 min cycles with 180° rotation of the tubes. The tubes were then centrifuged at 900 g for 10 min and 50 μl of the supernatant was used for nucleic acid extraction.
Nucleic acids were purified from 50 μl of clarified gill and mantle homogenates using the MagMax™-96 Viral RNA Isolation Kit (Thermo Fisher Scientific) with a BioSprint-96 ™ magnetic particle processor (Qiagen) using the AM1836 deep-well standard program (Thermo Fisher Scientific). Purified nucleic acids were eluted in 75 μl elution buffer and were stored at -20 °C until OsHV-1 quantification.
The number of copies of the B-region of the OsHV-1 genome was determined relative to a plasmid DNA standard according to a method described by Paul-Pont et al. (2013), adapted from a qPCR assay using hydrolysis probe chemistry (Martenot et al., 2010). Samples were tested in duplicate 25 μL reactions prepared with Path-ID qPCR master mix (Life Technologies), the primers OsHV1BF and OsHV1B4 and the fluorescent probe, OsHV1ProbeB (Table 1) and tested using a Mx3000P Multiplex Quantitative PCR System (Stratagene).
Table 1.
Primers and probes used in this study.
| Code of primer/probe | Sequence (5’→3′) | Reference |
|---|---|---|
| OsHV1BF | 5′- GTCGCATCTTTGGATTTAACAA-3′ | Martenot et al. (2010) |
| OsHV1B4 | 5′-ACTGGGATCCGACTGACA AC-3′ | Martenot et al. (2010) |
| OsHV1ProbeB | 5′-6FAM-TGCCCCTGTCATCTTGAGGTATAGACAATC-TAMRA-3′ | Martenot et al. (2010) |
| Vib1 | 5′-GGCGTAAAGCGCATGCAGGT-3′ | Vezzulli et al. (2012) |
| Vib2 | 5′-GAAATT CTACCCCCCTCTACAG-3′ | Vezzulli et al. (2012) |
| Nadak_rRNAF | 5′-TCCTACGGGAGGCAGCAGT-3′ | Nadkarni et al. (2002) |
| Nadak_rRNAR | 5′-GGACTACCAGGGTATCTAATCCTGTT-3′ | Nadkarni et al. (2002) |
| Nadak_rRNA probe | 5′-6FAM-CGTATTACCGCGGCTGCTGGCAC-BHQ1-3′ | Nadkarni et al. (2002) |
| 27F | 5′-AGAGTTTGATCMTGGCTCAG-3′ | Lane (1991) |
| 519R | 5′-GWATTACCGCGGCKGCTG-3′ | Lane (1991) |
| 1492R | 5′-TACCTTGTTACGACTT-3′ | Weisburg et al. (1991) |
2.6. Bacterial studies
Each sample was prepared from soft tissues remaining after the gill and mantle biopsy and after removal of the digestive gland. The tissues were transferred into a pre-weighed stomacher bag (Interscience, France), weighed and disrupted by stomaching with 4× (w/v) sterile ASW, in a bag mixer (MiniMix, Interscience, France) for 1 min, at maximum (9) speed. On Day 0, individual oysters were homogenized and thereafter, tissues from 4 oysters from each treatment and the same batch were pooled and homogenized. A coarsely clarified homogenate (5 ml) was obtained by collecting the material filtered through the inner mesh (porosity <250 μm) of the stomaching bag (BagPage®, Interscience, France). These homogenates were used directly for bacterial culture and stored at -80 °C for molecular studies of bacterial content.
Preparation of homogenates for bacterial molecular studies was by bead beating as previously described, with 150 μl of homogenates, 390 μl lysis/binding solution (MagMax™ - 96 Viral RNA Isolation Kit) in a 2 ml microcentrifuge tube containing 0.4 g of silica-zirconia beads. Homogenates were centrifuged at 900 g for 10 min and 180 μl of the supernatant was used for the nucleic acid purification with the MagMax™-96 Viral Isolation Kit. Nucleic acids were stored at -20 °C until bacterial DNA quantification.
2.6.1. Isolation, identification and quantification of cultivable vibrio and total bacteria
Ten microliters each from each fresh tissue homogenate was spread on a marine salt agar-blood (MSA-B) plate and a thiosulphate-citrate-bile salts-sucrose (TCBS) agar (Oxoid, UK) plate separately, using sterile disposable spreaders (Copan, Brescia, Italy). MSA-B was made using 20 g of BD® tryptone soy agar, 7.5 g NaCl, 500 ml of deionized, ultrapure water and 15 ml of sterile, anticoagulated, sheep blood for each 500 ml of MSA-B (Buller, 2014). The inoculated culture plates were incubated at 23 °C for 24h (MSA-B) or 48hr (TCBS agar) in a refrigerated incubator (LMS Ltd, UK).
Bacterial colonies on MSA-B plates and TCBS plates were counted and the number of colony forming units (CFU)/g of oyster tissue were calculated (total cultivable bacterial count, TCBC; total cultivable Vibrio count, TCVC). Colony morphology on TCBS plates was recorded based on visual assessment of size and colour. Single colonies of dominant morphotypes on TCBS agar were directly inoculated into nutrient broth with 2% NaCl and incubated at 23 °C overnight. Broth cultures (0.85 ml) were mixed with glycerol (0.15 ml) and stored at -80 °C. Species identification of select cryopreserved Vibrio cultures was later performed at the Animal Health Laboratories, Department of Agriculture, Western Australia, using biochemical methods.
2.6.2. Quantification of total vibrio spp. DNA by qPCR
The qPCR assay described by Vezzulli et al. (2012) was adapted for quantification of Vibrio spp. The primers Vib1 and Vib2 (Table 1) amplify a target between position 567 and 680 (based on the Escherichia coli numbering system) of the 16S ribosomal RNA gene which is specific for the genus Vibrio [Vezzulli et al. (2012); Thompson et al. (2004)].
Samples were tested in duplicate 20 μl reactions prepared with Fast SYBR® Green Master Mix (Applied Biosystems), using a 7500 Fast RT-PCR system (Applied Biosystems, Foster City, CA). Each reaction contained 10 μl of Fast SYBR® Green Master Mix (2x), 0.2 μM of each primer, 5 μl of neat template DNA and sterile, nuclease free water. The thermocycling programme was: initial denaturation at 95 °C for 20 s; 40 cycles of denaturation at 95 °C for 3 s and annealing at 58 °C for 30 s; melt curves of the final PCR products were generated and automatically analysed from 60 °C to 95 °C at 1 °C intervals.
Quantitative standards and positive control samples were prepared from genomic DNA of Vibrio alginolyticus isolated from oysters used in this study. The Vibrio gene copy numbers in the standards were estimated by determining the CFU/ml in V. alginolyticus cultures at stationary phase. A spread plate method enumerated colonies by spreading serial dilutions of the V. alginolyticus broth culture on TCBS agar plates (Buck and Cleverdon, 1960; Herbert, 1990). Each of 6 ten-fold dilutions was spread on three replicate plates. The total Vibrio count per PCR sample was calculated by dividing the total Vibrio 16S rDNA copy number by the presumptive number of 16S rDNA copies per Vibrio genome [n = 9 (Klappenbach et al., 2001; Thompson et al., 2004; Vezzulli et al., 2012),]. The total Vibrio count per oyster tissue homogenate was expressed as number of Vibrio genome equivalents per mg of oyster tissue.
Quantification of Vibrio spp. DNA in samples was determined by standard curve quantitation method, using 7500 software v2.3 (Applied Biosystems) for a 10-fold serial dilution containing between 4.65 × 101 and 4.65 × 106 copies of Vibrio rRNA gene. A PCR run was considered valid when there was no amplification of no template controls; amplification of both replicates of the positive control with a cycle threshold (Ct) within the range of the standard curve; and standard curve with r2 > 0.99 and efficiency between 90 and 110%. Samples exhibiting an exponential increase in SYBR fluorescence signal in both replicates with a Ct value > 15 and <35 and a dissociation curve (Tm ranging from 77.7 to 79.1 °C) that conformed to that of the positive control, were considered for quantification of total Vibrio DNA.
2.6.3. Quantification of total bacterial DNA by qPCR
The qPCR assay based on TaqMan® chemistry described by Nadkarni et al. (2002) was used to quantify total bacteria. The primers Nadak_rRNAF and Nadak_rRNAR and probe, Nadak_rRNA (Table 1) target position 331 to 797 of 16S rRNA gene (Escherichia coli numbering system) which is highly conserved across domain Bacteria.
Purified nucleic acids were tested in duplicate 25 μl PCR reactions prepared in TaqMan™ Fast Universal PCR Master Mix (2×) (Thermofisher Scientific), using Fast 7500 Real-time PCR system (Applied Biosystems). Each reaction contained 12.5 μl of 2× Master Mix, 0.9 μM of each primer and 0.1 μM probe, 5 μl of neat template DNA and sterile, nuclease free water. The thermocycling program was: initial denaturation of 95 °C for 10 min; 40 cycles of denaturation at 95 °C for 15 s, and annealing at 60 °C for 1 min.
The standard was prepared from nucleic acids purified from a non-pathogenic Escherichia coli strain 2038 culture that was quantified by spread plate method. Gene copy numbers were estimated from CFU/ml in E. coli culture. PCR runs were analyzed by the standard curve quantitation method (Applied Biosytems) based on amplification of a 10-fold serial dilution containing between 1.84 × 101 and 1.84 × 108 copies of E. coli rRNA gene. A PCR run was considered valid when there was no amplification of negative controls; amplification of both replicates of the positive control with a cycle threshold (Ct) within the range of the standard curve; and standard curve with r2 > 0.99 and efficiency between 90 and 110%. Samples exhibiting an exponential increase in the fluorescence signal in both replicates with a Ct value > 15 and <35, were considered for quantification of the total bacteria DNA. Total bacteria quantity of samples was expressed as the number of bacteria genome equivalents per mg oyster tissue. This was calculated by dividing the total 16S rDNA gene copies by the presumptive number of 16S rDNA copies per Proteobacteria genome [n = 3.5, Kormas (2011); Vezzulli et al. (2012)], based on the species of bacteria of the phyla that dominate the oyster microbiota (Prieur et al., 1990; Wegner et al., 2013; Lokmer et al., 2016a).
2.7. Microbiome analysis by high throughput 16S rRNA gene sequencing
The microbial community composition was identified by high-throughput sequencing (MiSeq, Illumina) of the hypervariable region V1–V3 of the 16S rRNA gene. Twenty-six nucleic acid extracts were selected for analysis, to represent 5 different OsHV-1 infection and disease states for each of the 3 batches of oysters (Table 2).
Table 2.
Cohorts of oysters used for microbiome analysis (On average, n = 8 oysters were evaluated per batch/cohort).
| Cohort | Description |
|---|---|
| A | Pacific oysters before OsHV-1 challenge |
| B | Apparently healthy oysters soon after OsHV-1 challenge |
| C | Challenged oysters at or near death |
| D | Surviving oysters at the completion of a 7-day OsHV-1 challenge period |
| E | Unchallenged control oysters at the end of a 7-day trial period |
Initially, the V1–V8 hypervariable region of the 16S rRNA gene was amplified using the primers 27F and 1492R [Table 1 (Weisburg et al., 1991);]. Reactions were prepared with the Expand High Fidelity PCR System (Roche®, Germany) and used a BioRad T100™ thermal cycler (Applied Biosystems). The PCR amplification was carried out in two stages to overcome inhibition and minimize the impact of PCR bias on diversity profiling. The first stage of amplification was carried out with 8 replicates per nucleic acid sample in a 50 μl reaction containing 200 μM of each dNTP, 0.3 μM of each primer, 5 μl of template DNA, 0.75 μl of Expand High Fidelity enzyme mix, 5 μl of Expand High Fidelity buffer (10x, 15mM MgCl2) and sterile, nuclease free water. The PCR program was: initial denaturation of 95 °C for 5 min; 30 cycles of denaturation at 94 °C for 15 s, annealing at 50 °C for 30 s and elongation at 72 °C for 1 min; final elongation of 72 °C for 7 min. Replicate PCR products were pooled, mixed by vortexing and 5 μl from the amplicon pool was used as template in stage 2 amplification using the same PCR conditions. Products were visualised by gel electrophoresis using 2% agarose gels containing RedSafe (Intron, Biotechnology).
Appropriate size PCR products were purified using the QIAamp DNA minikit (Qiagen) and submitted to the Australian Genome Research Facility (AGRF). PCR amplicons generated using primers 27F (Lane, 1991) and 519R (Lane et al. (1985); Table 1), using AmpliTaq Gold 360 Master mix (Life Technologies, Australia). Indexing the amplicons was performed with TaKaRa Taq DNA Polymerase (Clontech) and the resulting amplicons were measured by fluorometry (Invitrogen™ Picogreen) and normalised to provide an equimolar pool of amplicons. These were quantified by qPCR and sequenced with Illumina MiSeq 300bp paired end chemistry.
2.7.1. Bioinformatic analyses
Paired-ends reads were assembled by aligning the forward and reverse reads using PEAR v0.9.5 (Zhang et al., 2014), primer sequences were trimmed and USEARCH ver. 8.0.1623 (Edgar, 2010; Edgar et al., 2011) was used for quality filtering and to remove full-length duplicate sequences and singletons. Chimeric sequences were detected against the reference database (RDP Gold database) and discarded. Sequences were clustered into operational taxonomic units (OTUs) at 97% similarity and taxonomy was assigned using the RDP classifier (Liu and Wong, 2013) as implemented in QIIME, against the Greengenes database ver. 13_8 (Aug 2013) (DeSantis et al., 2006). An OTU table was generated by calculating the absolute abundance of each identified OTU for each sample, based on the number of sequenced reads. OTUs with an abundance of less than 10 reads per sample were discarded.
2.8. Data analyses
Survival analyses were conducted using the Kaplan-Meier estimate (Kaplan and Meier, 1958), notwithstanding an experiment design focused on longitudinal sampling. Oysters collected during random sampling and surviving oysters at the end of the experiment were censored at the respective observation time. Survival time was measured as days from the onset of the injection when oysters were injected with the OsHV-1 inoculum. Each batch of oysters was compared using a mixed Cox regression model (Cox, 1972), taking into account the random effect of tank, using RStudio ver. 3.4.4 (R Core Team, 2018). The proportionality of hazards (PH) was checked based on Schoenfeld residuals (Schoenfeld, 1982).
The OsHV-1 DNA quantity, total bacteria, total Vibrio, TCBC and TCVC in oysters were compared between batches, between live and dead oysters of the same batch, between days after OsHV-1 challenge in a batch and with relevant control groups. Data were log10 transformed for normal distribution. Kruskal-Wallis test was used for OsHV-1 data analysis and the analyses of bacterial data in oysters before OsHV-1 challenge. The bacterial data (total bacteria, total Vibrio, TCBC and TCVC) in oysters during the viral challenge period were done using separate generalized linear models (GzLM, SPSS Statistics ver. 22; IBM SPSS Cooperation, Somers, NY, USA). The main effects of batch including the geographic origin of oysters (Patonga Creek, Shoalhaven River, Clyde River), treatment (OsHV-1 injected or negative control), outcome of infection (live or dead) and the days post-injection (Day 1, 2, 3, 5 and 7) were tested together with possible interactions. Post-hoc pairwise mean comparisons were made using the least significant difference method. The results were presented as geometric means and their corresponding 95% confidence intervals of OsHV-1 genome equivalents, bacterial genome equivalents or Vibrio genome equivalents/mg, CFU values/g (for TCBC and TCVC), respectively. Finally, the correlation between OsHV-1 load and Vibrio gene copies in oyster tissues was tested using Spearman's correlation coefficient (SPSS Statistics ver. 22.0). Significance was set at p < 0.05 for all statistical analyses.
The relative abundance of bacterial families in each sample was graphically represented using 100% stacked 2-D column graphs (Excel, Microsoft). Diversity analyses were performed with Paleontological Statistics (PAST) software ver. 3.16 (Hammer et al., 2001). Alpha diversity was assessed by means of Shannon's diversity index and Simpson's evenness index. Dissimilarity of bacterial community structure between samples from different batches and different cohorts (beta diversity) was visualized by non-metric multidimensional scaling (n-MDS) based on the two-dimensional Bray-Curtis dissimilarity index (BC). One-way permutational multivariate analysis of variance (PERMANOVA) was used for statistical analysis of beta diversity. Significance was set at p < 0.05 for all statistical analyses.
3. Results
3.1. Mortality of oysters
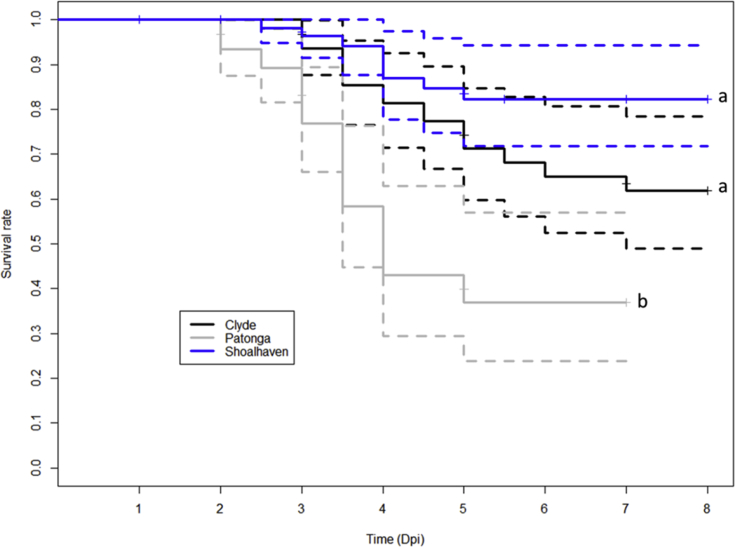
The onset of mortality occurred 2 days post-OsHV-1 injection and the cumulative mortality was different for the three batches of oysters (Fig. 1, Table 3, p < 0.05). The highest cumulative mortality was seen in the batch from Patonga Creek (63.1%) compared to oysters from Clyde River (38.1%) and Shoalhaven River (17.7%). No mortality was observed in the negative control oysters.
Fig. 1.
Kaplan-Meier survival curves for each batch (Clyde River, Patonga Creek, Shoalhaven River) of oysters challenged with OsHV-1 (solid line) and 95% confidence intervals (dashed line). Each batch comprised n = 73–86 oysters at the onset of the viral challenge and n = 12 live oysters were sampled from each batch on days 1, 2, 3, 5, and 7. Different letters (a and b) indicate significance (p < 0.05) identified by a mixed Cox regression model.
Table 3.
Odds of mortality in oysters injected with OsHV-1 depending on the batch of oysters. (Odd's ratio, standard error (SE), the 95% confidence interval of the risk ratio and p-value).
| Contrasts | Odd's ratio | SE | 95% Confidence Interval (CI) | p-value |
|---|---|---|---|---|
| Patonga vs Clyde | 3.00 | 0.32 | 1.61–5.54 | <0.001 |
| Shoalhaven vs Clyde | 0.48 | 0.42 | 0.15–1.53 | 0.21 |
| Shoalhaven vs Patonga | 0.16 | 0.41 | 0.06–0.46 | <0.001 |
Patonga, Patonga Creek; Shoalhaven, Shoalhaven River; Clyde, Clyde River.
3.2. Quantification of OsHV-1 DNA by qPCR
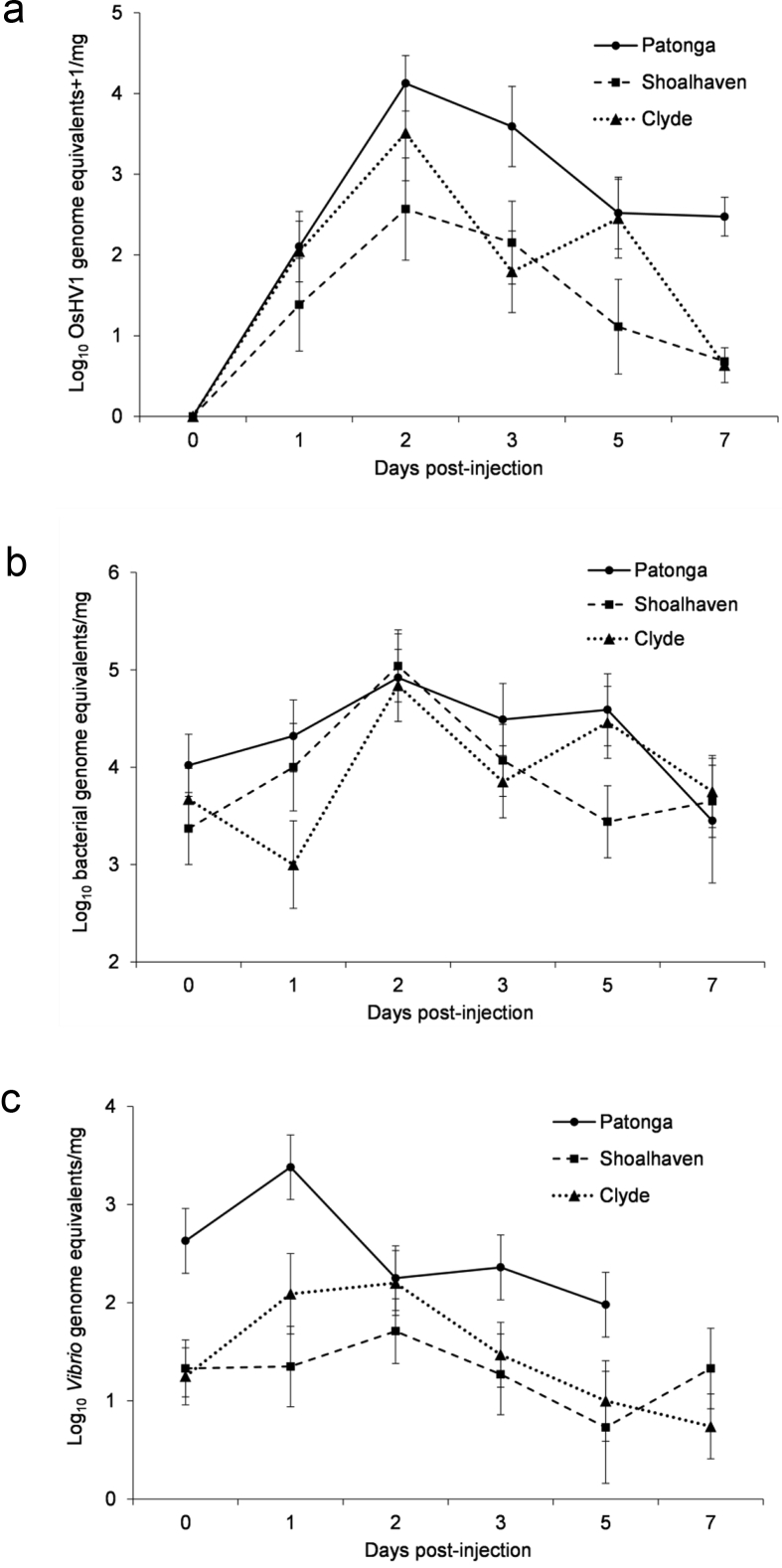
The live oysters from Patonga Creek that were sampled during the infection trial had higher OsHV-1 DNA concentrations compared to their counterparts from the other two locations (Table 4; p < 0.05). This batch also had the highest mortality and a higher OsHV-1 DNA concentration at the time of death compared to dead/moribund oysters from the Clyde River (Table 4; p < 0.05). In all three batches, longitudinal random sampling of live oysters indicated that the quantity of OsHV-1 DNA increased on Day 2 compared to Day 1 post-injection (Fig. 2a), which coincided with the onset of mortality. All dead oysters were positive for OsHV-1 DNA and OsHV-1 DNA was not detected in oysters injected with the negative control inoculum. The quantity of OsHV-1 DNA in moribund/dead oysters was higher than in live, apparently healthy oysters sampled at random on Days 1, 2, 3, 5, and 7 post-injection (Table 4; p < 0.001).
Table 4.
Mean OsHV-1 concentrations in oyster tissues (geometric means and their corresponding 95% confidence intervals) during the infection.
| Status of oysters | Mean OsHV-1 concentration (OsHV-1 genome equivalents/mg) | 95% Confidence Interval (CI) |
|---|---|---|
| Live oysters: | 1.61 × 102 | 8.24 × 101–3.14 × 102 |
| Patonga Creek | 1.22 × 103∗ | 2.95 × 102–5.04 × 103 |
| Shoalhaven River | 4.05 × 101 | 1.61 × 101–1.02 × 102 |
| Clyde River | 1.31 × 102 | 4.45 × 101–3.88 × 102 |
| Dead oysters: | 1.36 × 105 | 8.48 × 104–2.20 × 105 |
| Patonga Creek | 2.25 × 105∗ | 1.20 × 105–4.22 × 105 |
| Shoalhaven River | 5.55 × 104 | 8.15 × 103–3.77 × 105 |
| Clyde River | 9.69 × 104 | 4.75 × 104–1.98 × 105 |
OsHV-1 concentration in live Patonga oysters was higher than the other two batches. It was also higher in dead Patonga oysters compared to dead oysters from Clyde River.
Fig. 2.
Temporal distribution of (a) OsHV-1 viral load (log10 OsHV-1 genome equivalents +1/mg), (b) Total bacterial load (bacterial genome equivalents/mg) and (c) Total Vibrio load (Vibrio genome equivalents/mg tissue) in live oysters that were sampled longitudinally on Days 0, 1, 2, 3, 5 and 7 post-OsHV-1 injection. All three batches showed a significant/nearly significant increase in OsHV-1 concentration on Day 2 compared to Day 1 (Patonga, p < 0.05; Shoalhaven, p = 0.053; Clyde, p = 0.052). The Patonga oysters also showed an increase in total Vibrio load on Day 1 compared to Day 2 post-viral injection (p < 0.05). All data are represented as log10 means ±SE.
3.3. Bacterial quantity
There was no difference in any bacteriological parameter (total bacteria load, total Vibrio load, TCBC and TCVC) between the three batches of oysters, at the beginning of the infection trial (p > 0.05 for each parameter). Considering temporal dynamics during the infection, the total Vibrio load and TCVC in live oysters from Patonga were higher on Day 1 compared to Day 2 post-viral injection (Fig. 2c; p < 0.05). There was no such trend in the other two batches (p > 0.05). Overall, the highest Vibrio quantity was seen in oysters from Patonga during the viral infection (Table 6; p < 0.05). The batches from Shoalhaven and Clyde Rivers demonstrated increases in the total bacterial load in live oysters on Day 2 as opposed to Day 1 post-injection (Fig. 2b; p < 0.05). The total bacterial concentration and the total Vibrio concentration were higher in the tissues of dead or moribund oysters compared to apparently healthy oysters sampled on Days 1, 2, 3, 5, and 7 post-injection (Table 6; p < 0.05). Interestingly, no change in total bacteria and total Vibrio was observed in the negative control oysters from all 3 batches, during the trial (Table 5; p > 0.05).
Table 6.
Total bacterial load and total Vibrio load (geometric means† and their corresponding 95% confidence intervals, CI) in oysters during the viral infection.
| Status of oysters | Mean bacterial loada (95% CI) | Mean Vibrio loadb (95% CI) |
|---|---|---|
| Live oysters: | 1.08 × 104 (7.89 × 103–1.47 × 104) | 4.24 × 101 (3.14 × 101–5.73× 101) |
| Patonga Creek | 2.72 × 104 (1.75 × 104–4.20 × 104) | 2.88 × 102 (1.91 × 102–4.36 × 102)* |
| Shoalhaven River | 1.48 × 104 (9.17 × 103–2.38 × 104) | 1.89 × 101 (1.16 × 101–3.07 × 101) |
| Clyde River | 1.86 × 104 (1.20 × 104–2.89 × 104) | 1.15 × 102 (7.74 × 101–1.72 × 102) |
| Dead oysters: | 9.24 × 104 (5.66 × 104–1.51 × 105) | 4.19 × 102 (2.69 × 102–6.54 × 102) |
*The mean Vibrio load in Patonga oysters were higher than in the other two batches (p < 0.05).
Bacterial genome equivalents/mg.
Vibrio genome equivalents/mg.
Geometric means were derived from back-transformed model means.
Table 5.
Results of Generalised Linear Model (GzLM) analysis of total bacterial load and total Vibrio load with the effects of geographic origin of oysters (batch), OsHV-1 challenge (treatment), days post-OsHV-1 injection (day) and outcome of infection (status) accounted for in the model.
| Total bacterial load |
Total Vibrio load |
|||||
|---|---|---|---|---|---|---|
| df | X2 | p-value | df | X2 | p-value | |
| Intercept | 1 | 2609.97 | <0.001 | 1 | 644.97 | <0.001 |
| Batch | 2 | 1.05 | 0.593 | 2 | 52.69 | <0.001 |
| Treatment | 1 | 1.85 | 0.174 | 1 | 2.40 | 0.121 |
| Day | 6 | 28.61 | <0.001 | 6 | 19.39 | 0.004 |
| Status | 1 | 30.78 | <0.001 | 1 | 36.41 | <0.001 |
| Batch × Group × Day × Status | 28 | 108.62 | <0.001 | 24 | 118.97 | <0.001 |
Note: df, degree of freedom; X2, Wald Chi Square.
Although changes were noted with the Vibrio load in oysters challenged with OsHV-1, the average relative percentage of Vibrio spp. from the total bacterial population, remained low with 0.26% in pre-challenged oysters and reaching a maximum of 5.25% on Day 1 post-injection. Overall, a strong correlation was observed between the OsHV-1 DNA load and the total Vibrio load in OsHV-1 infected oyster tissues (r = 0.6; p < 0.001). Representative Vibrio isolates from the OsHV-1 infected oysters were identified as Vibrio alginolyticus (3/6) and Vibrio mediterranei (2/6), by biochemical methods.
3.4. Bacterial community structure in different batches of oysters
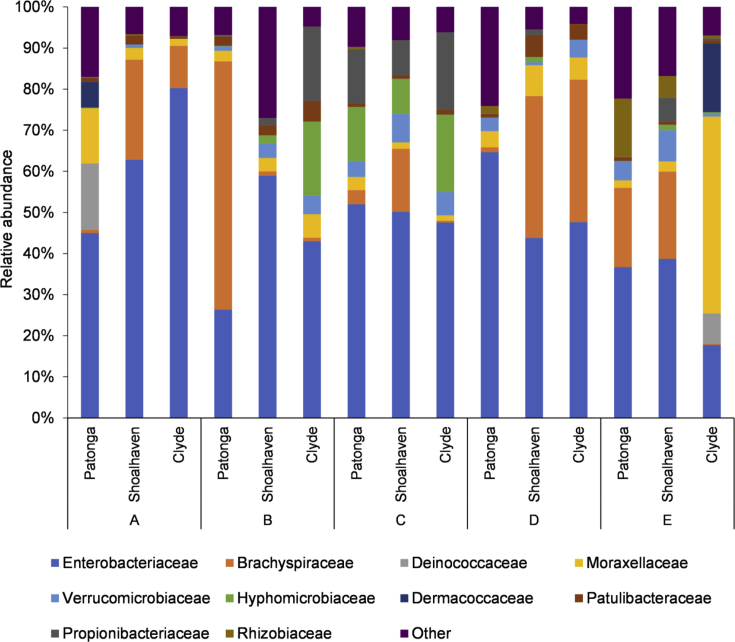
Both Shannon's diversity index (H) and Simpson's evenness index (S) were different between the oysters grown in the three different estuaries, before acclimation to the laboratory conditions and OsHV-1 challenge (p < 0.05). Both H and S were highest in oysters from Patonga Creek (H = 1.77, S= 0.74) compared to those from the Clyde River (H = 0.77, S = 0.33) and Shoalhaven River (H = 1.14, S = 0.54). While the relative abundance of family Brachyspiraceae was quite low (0.8%) in the batch from Patonga Creek, a comparatively higher relative abundance was seen in the other two batches (Shoalhaven, 24.3%; Clyde 10.3%; Fig. 3). In contrast, the relative abundance of family Moraxellaceae was higher in oysters from Patonga Creek (13.5%) compared to the other two batches (Shoalhaven, 2.9%; Clyde 1.7%). Further, the relative abundance of family Deinococcaceae was observed to be 16.1 % in the oysters from Patonga Creek while it was negligible in the other two batches (Fig. 3).
Fig. 3.
Relative abundance of bacterial families in oysters farmed in different estuaries: before OsHV-1 challenge (A); apparently healthy oysters 24h after OsHV-1 challenge (B); challenged oysters at or near death (C); surviving oysters at the completion of a 7-day OsHV-1 challenge period (D); unchallenged control oysters at the end of the trial period (E). Patonga, oysters from Patonga Creek; Shoalhaven, oysters from Shoalhaven River; Clyde, oysters from Clyde River. The bacterial families that constituted >5% of an individual sample and were found in at least two samples are presented, and the rest are indicated as ‘others’.
3.5. Changes in bacterial community during OsHV-1 infection
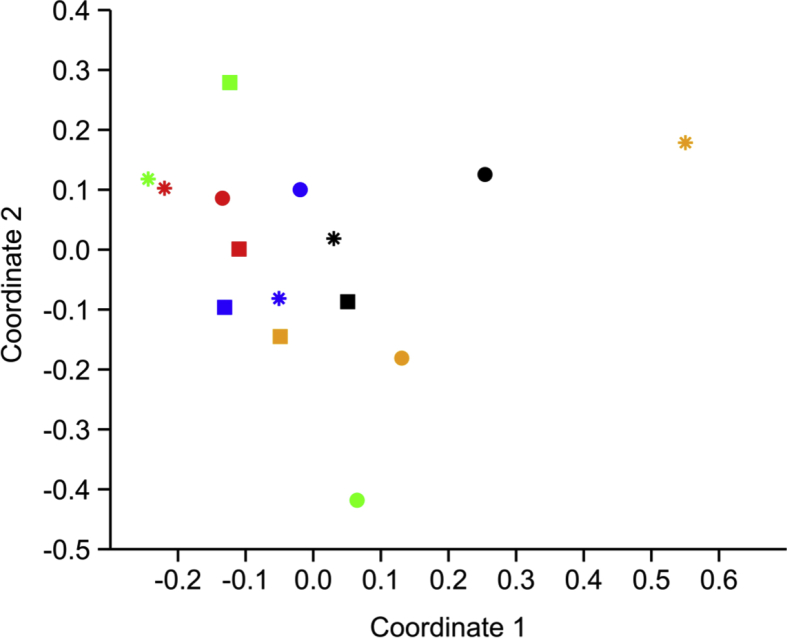
The relative abundance of bacterial families changed in all three batches of oysters, 24 h post-OsHV-1 injection. However, the direction of changes was different between batches. For example, the relative percentage of family Brachyspiraceae increased up to 60.4 % in Patonga oysters while it decreased to 3.2% and 2.4% in oysters from Shoalhaven and Clyde Rivers, respectively (see section 3.4 for initial values; Fig. 3). Moreover, the family Hyphomicrobiaceae which was either absent or present in negligible amounts in the pre-challenge oysters was seen to increase in oysters from Shoalhaven River (2.1%) and Clyde River (18.1%). This was not observed in Patonga oysters. During the OsHV-1 infection, the dead/moribund oysters from Patonga Creek showed a decrease in bacterial diversity (H = 1.66, S = 0.68) compared to pre-challenge (H = 1.77, S = 0.74; p < 0.05). However, the dead or moribund oysters from the other two locations, with an originally lower diversity, did not demonstrate such a decrease (p > 0.05). Furthermore, the bacterial community structure in Patonga and Shoalhaven oysters that survived the OsHV-1 challenge at the end of the 7-day trial, were similar to their counterparts in the pre-challenged cohort (Fig. 4, p > 0.05). Moreover, the bacterial community structure in Patonga and Clyde oysters that were challenged with the negative control, were not different from the pre-challenge counterparts, at the end of 7 days in the laboratory.
Fig. 4.
Non-metric multidimensional scaling based on two-dimensional Bray-Curtis dissimilarity. Colours indicate different cohorts: black (A), before OsHV-1 challenge; green (B), 24h after viral challenge; red (C), dead/moribund oysters; blue (D), survived oysters; yellow (E), control oysters at the end of the experiment. Shapes indicate different locations of origin of the oysters: circle, Patonga Creek; square, Shoalhaven River; star, Clyde River.
4. Discussion
The present study discusses how the bacterial community composition of Pacific oysters with a common hatchery origin can be influenced by the geographic site at which they are grown. It also discusses how these influences are related to the changes in the Pacific oyster microbiome and OsHV-1 load during an experimental OsHV-1 infection conducted in a controlled environment. Further, the study the provides insights as to how these differences in microbiome composition affect the outcome of the OsHV-1 infection, in addition to the impact of age factor.
Our results showed that the bacterial community composition of the Pacific oyster microbiome was different between the oysters grown in the three geographically distinct estuaries despite their common hatchery origin. The filter-feeding pattern of oysters leading to the uptake of environmental microbes with their feed can be attributed to these differences in microbiomes of oysters with a common genetic origin. The study involved three batches of oysters from two broad age categories (juvenile oysters (n = 1) and adult (n = 2). The adult batches were of approximately equal ages (14 and 16 months). In addition to the difference of age between the juveniles and adults, we also observed a difference between the microbiome composition of the two batches of adult oysters, indicating an influence of farming environment in shaping the microbiome composition. Further studies are required to focus on controlling other factors such as age to further evaluate the influence of environment on the microbiome. A metagenomic study in Crassostrea virginica revealed differences in microbiomes in oysters reared in different sites (Ossai et al., 2017) while conventional bacterial studies conducted using C. virginica have also shown that the bacterial composition in oysters varies with their environment (Prieur et al., 1990). However, it is also important to note that the microbiome composition is also influenced by host-related factors as well as by interactions within the microbiome, in the long run (Lokmer et al., 2016b).
In the present study, we observed changes in the oyster microbiome soon after the viral challenge. These changes may primarily reflect the initial response of the microbiome to the viral infection. Having said that, it is interesting to note that the microbiome of Patonga oysters behaved differently to the other two batches along with a higher OsHV-1 DNA concentration in tissues and higher mortality. The difference in the mortality pattern was consistent with previous studies (Renault et al., 1995; Whittington et al., 2015b; Hick et al., 2018) whereby juvenile oysters (<1 year) had higher mortality than adult oysters (>1 year). However, it was interesting to note that there was no difference in mortality between the two batches of adults despite a higher concentration of OsHV-1 in Clyde oysters which can be corresponded to differences noted in the initial microbiome composition. Moreover, the oysters from Patonga Creek demonstrated the highest initial bacterial diversity and showed a decrease in bacterial diversity during the moribund stage. This was not seen with the other two batches, indicating a differential response of the microbiome. In a study conducted by Wegner et al. (2013), Pacific oysters with initially high microbial diversity had shown a decrease in diversity following disturbance due to heat stress, whereas oysters with originally lower diversity did not show such a decrease after exposure to heat. Low bacterial diversity has repeatedly been found with impaired health in oysters (Garnier et al., 2007; Green and Barnes, 2010). Thus, the decrease in bacterial diversity which precedes mortality can be considered a good indicator in declining health in oysters (Wegner et al., 2013; Lokmer and Wegner, 2015). Furthermore, studies involving pathogenic Vibrio infections in Pacific oysters have shown bacterial community structure disruption in dead/moribund oysters, characterized by very low diversity (Lokmer and Wegner, 2015).
The changes in the microbiome that were observed in oysters challenged with the virus, were not seen in the control oysters from two batches, in our study. The bacterial community composition of those control oysters was not different from their pre-challenged counterparts, at the end of 7 days in the laboratory. As the control oysters were also subjected to muscle relaxation and was given an injection, the disturbance to the microbiome prior to the viral challenge, appear to be transient. In contrast, the changes induced by OsHV-1 infection seem to remain and can be considered as an indication of a disease state. Oyster families susceptible to OsHV-1 showed disruption of the bacterial community structure when cohabitated with OsHV-1-injected oysters (de Lorgeril et al., 2018).
Despite the inter-batch differences observed in bacterial community composition after 24 h of the viral challenge, the total bacterial load remained same. The persistent bacterial community composition at higher taxonomic levels regardless of the changes in lower taxonomic levels (Lokmer et al., 2016a) can be attributed to this static bacterial load. In the present study, the Patonga oysters displayed the highest total Vibrio load with a peak on Day 1 post-injection. This peak may be associated with the higher and early (on Day 2 post-injection) mortality in Patonga oysters which was not seen in oysters from the other two batches. While the Shoalhaven oysters reported few mortalities, no mortalities were observed in Clyde oysters as early as Day 2 post-injection. Although we could not test the pathogenicity of the Vibrio isolates of our study, it is possible that this increase in Vibrio, to have a role in the final cumulative mortality. Stress-related shifts in the oyster microbiome resulting in increase in the potentially pathogenic bacteria are thought to contribute to oyster mortality (Lokmer and Wegner, 2015).
Although differences in temporal Vibrio dynamics and the total Vibrio load were noted between batches, we observed a low average relative abundance of Vibrio spp. in all batches, throughout the course of the viral infection. This was consistent with previous studies reporting oyster mortality events in the field (King et al., 2018) as well as experimental conditions (Lokmer and Wegner, 2015). Trabal et al. (2012) also reports a very low abundance of Vibrio in gut tissues obtained from healthy Pacific oysters. Contrary to the conventional knowledge that identifies Vibrio spp. as one of the dominating bacterial genera in oysters (Colwell and Liston, 1960; Prieur et al., 1990), a large proportion of the oyster microbiome cannot be cultivated by standard procedures (Romero and Espejo, 2001; Fernandez-Piquer et al., 2012). Previous literature on Eastern oysters (C. virginica) and Sydney rock oysters (Saccostrea glomerata) also report low relative abundances of Vibrio spp. (Green and Barnes, 2010; Ossai et al., 2017). Nevertheless, Vibrio spp. can have an impact on the host health, despite its low relative abundance (Thurber et al., 2009).
We also observed a wider range of OsHV-1 DNA concentrations (102 - 106 viral copies/mg oyster tissue) in oysters that died during this infection trial. Previous field and lab studies have demonstrated viral concentrations exceeding 106-107 copies mg/tissue to be associated with moribund/dead oysters infected with OsHV-1 (Pepin et al., 2008; Oden et al., 2011; Paul-Pont et al., 2014). As highlighted by Paul-Pont et al. (2015), variations in the OsHV-1 DNA concentration in moribund oyster tissues can be expected due to various reasons such as genetic background, age, physiology and life history. Despite the highest OsHV-1 DNA concentration in the youngest batch (10 months), the lowest OsHV-1 concentration was not reported from the oldest batch (16 months) but a younger batch (14 months), in this study. This indicated a potential role of factors other than age in determining the physiological response of oysters to OsHV-1. Apart from the role played by age in OsHV-1 infections (Paul-Pont et al., 2013b; Hick et al., 2018), the differences in the microbiome composition among the batches may have differentially influenced the viral multiplication in these organisms. Recent studies on OsHV-1 disease have highlighted the role of size and other factors such as life history and genetics, on oyster mortality (Petton et al., 2015a; Azéma et al., 2017a, 2017b). Interactions between the age and size of oysters in determining the susceptibility of Pacific oysters to OsHV-1-related mortality (Hick et al., 2018) suggest complex interactions of a range of factors other than age, in causing mortality.
There was a strong correlation between the OsHV-1 DNA load and Vibrio load in infected oysters, in the present study. Conventional culture methods used in Vibrio quantification have not seen such a correlation between the Vibrio concentration and the OsHV-1 DNA load in oysters exposed to a field mortality outbreak (Petton et al., 2015b). Lemire et al. (2015) have shown that the non-virulent strains of Vibrio spp. inhabiting healthy oysters get progressively replaced by phylogenetically coherent, virulent strains of Vibrio during the onset of a natural infection. These non-virulent strains are also expected to facilitate the disease caused by the virulent strains. Thus, the difference in correlation of Vibrio gene copy number and cultivable Vibrio load, to OsHV-1 DNA load and can be attributed to the ability of the PCR assay in detecting non-cultivable Vibrio strains (Cai et al., 2006), or the non-virulent strains of Vibrio that were dead and were replaced by pathogenic Vibrio strains.
The apparently healthy oysters used in our study were injected with a bacteria-free, partially purified tissue homogenate carrying OsHV-1 and were maintained in artificial seawater throughout the experimental period. Therefore, all Vibrio strains quantified and isolated in this study should originate from the oyster microbiome. Vibrio alginolyticus that was isolated in our study have been previously reported in mass mortality events of Pacific oysters (Go et al., 2017). As mentioned above, we could not test the pathogenicity of the Vibrio isolates of our study nor could we identify the same species with 16S rRNA metabarcoding as the genetic resolution did not permit identification of Vibrio at species level. Testing the pathogenicity of Vibrio isolates may have provided valuable clues to identify the pathogenic role of Vibrio spp. present in the Pacific oyster microbiome, during OsHV-1 infections. In an experiment where oysters exposed to a field mortality outbreak and subsequently treated with chloramphenicol, Petton et al. (2015b) demonstrated that a high load of OsHV-1 DNA (≥108 viral copies/mg tissue) is not sufficient for a full expression of the disease by demonstrating a delayed and reduced oyster mortality in the absence of bacteria. In this regard, the indication that the pathogenic species or strains of Vibrio in healthy oysters become pathogenic when there is a primary viral infection, needs to be studied further. Moreover, it is important to investigate whether the mass mortality events reported among Pacific oysters are caused by several different infectious diseases or by a single polymicrobial disease, by targeting Vibrio spp. alone.
5. Conclusion
The composition of the bacterial community in Pacific oysters grown in different geographic sites was different despite their common hatchery origin. In addition to the differences related to age, different outcomes of OsHV-1 challenge were seen between same-aged batches, with differences in the initial microbiome. The initial bacterial community composition did not change in some batches of oysters that survived OsHV-1 challenge and in oysters challenged with the negative control. The higher Vibrio load in oysters with higher OsHV-1 DNA content and higher mortality suggests a role of Vibrio in mortality associated with OsHV-1 infections. Further studies are needed to evaluate the direct and indirect influences of Vibrio and other members of the resident microbial community of oysters during OsHV-1 infection.
Declarations
Author contribution statement
Erandi Pathirana, Marine Fuhrmann: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Richard Whittington, Paul Hick: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This study was funded by the Australian Government through the Fisheries Research & Development Corporation (FRDC) and the University of Sydney, Australia.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
Broken Bay Oysters, NSW, Signature Oysters, NSW and Goodnight Oysters, Greenwell Point, NSW are thanked for providing oysters for this study. Dr. Olivia Evans, Alison Tweedie, Slavicka Patten and Craig Kristo of the University of Sydney, Camden, are thanked for technical assistance. Sydney Informatics Hub of the University of Sydney is acknowledged for providing high performance computing facilities for bioinformatic analysis.
References
- Alfaro A.C., Nguyen T.V., Merien F. The complex interactions of Ostreid herpesvirus 1, Vibrio bacteria, environment and host factors in mass mortality outbreaks of Crassostrea gigas. Rev. Aquac. 2018;1:21. [Google Scholar]
- Azéma P., Maurouard E., Lamy J.-B., Dégremont L. The use of size and growing height to improve Crassostrea gigas farming and breeding techniques against OsHV-1. Aquaculture. 2017;471:121–129. [Google Scholar]
- Azéma P., Lamy J.-B., Boudry P., Renault T., Travers M.-A., Dégremont L. Genetic parameters of resistance to Vibrio aestuarianus, and OsHV-1 infections in the Pacific oyster, Crassostrea gigas, at three different life stages. Genet. Sel. Evol. 2017;49:23. doi: 10.1186/s12711-017-0297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck J.D., Cleverdon R.C. The spread plate as a method for the enumeration of marine bacteria. Limnol. Oceanogr. 1960;5:78–80. [Google Scholar]
- Buller N.B. Cabi; Oxfordshire, UK: 2014. Bacteria and Fungi from Fish and Other Aquatic Animals: a Practical Identification Manual. [Google Scholar]
- Cai T., Jiang L., Yang C., Huang K. Application of real-time PCR for quantitative detection of Vibrio parahaemolyticus from seafood in eastern China. FEMS Immunol. Med. Microbiol. 2006;46:180–186. doi: 10.1111/j.1574-695X.2005.00016.x. [DOI] [PubMed] [Google Scholar]
- Clegg T.A., Morrissey T., Geoghegan F., Martin S.W., Lyons K., Ashe S., More S.J. Risk factors associated with increased mortality of farmed Pacific oysters in Ireland during 2011. Prev. Vet. Med. 2014;113:257–267. doi: 10.1016/j.prevetmed.2013.10.023. [DOI] [PubMed] [Google Scholar]
- Colwell R., Liston J. Microbiology of shellfish. Bacteriological study of the natural flora of Pacific oysters (Crassostrea gigas) Appl. Microbiol. 1960;8:104–109. doi: 10.1128/am.8.2.104-109.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D.R. Regression models and life tables. J. R. Stat. Ser. Soc. B Stat. Methodol. 1972;20:187–220. [Google Scholar]
- Davison A.J., Trus B.L., Cheng N., Steven A.C., Watson M.S., Cunningham C., Le Deuff R.-M., Renault T. A novel class of herpesvirus with bivalve hosts. J. Gen. Virol. 2005;86:41–53. doi: 10.1099/vir.0.80382-0. [DOI] [PubMed] [Google Scholar]
- de Kantzow M.C., Hick P.M., Dhand N.K., Whittington R.J. Risk factors for mortality during the first occurrence of pacific oyster mortality syndrome due to Ostreid herpesvirus–1 in Tasmania, 2016. Aquaculture. 2017;468:328–336. [Google Scholar]
- de Lorgeril J., Lucasson A., Petton B., Toulza E., Montagnani C., Clerissi C., Vidal-Dupiol J., Chaparro C., Galinier R., Escoubas J.-M. Immune-suppression by OsHV-1 viral infection causes fatal bacteraemia in Pacific oysters. Nat. Commun. 2018;9:4215. doi: 10.1038/s41467-018-06659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dégremont L. Evidence of herpesvirus (OsHV-1) resistance in juvenile Crassostrea gigas selected for high resistance to the summer mortality phenomenon. Aquaculture. 2011;317:94–98. [Google Scholar]
- DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundon W.G., Arzul I., Omnes E., Robert M., Magnabosco C., Zambon M., Gennari L., Toffan A., Terregino C., Capua I., Arcangeli G. Detection of Type 1 Ostreid Herpes variant (OsHV-1 μvar) with no associated mortality in French-origin Pacific cupped oyster Crassostrea gigas farmed in Italy. Aquaculture. 2011;314:49–52. [Google Scholar]
- Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans O., Hick P., Dhand N., Whittington R.J. Transmission of Ostreid herpesvirus-1 in Crassostrea gigas by cohabitation: effects of food and number of infected donor oysters. Aquac Environ Interact. 2015;7:281–295. [Google Scholar]
- FAO . Food and Agriculture Organization of the United Nations; Rome: 2014. FAO Yearbook - Fishery & Aquaculture Statistics 2012. 2014. [Google Scholar]
- Fernandez-Piquer J., Bowman J.P., Ross T., Tamplin M.L. Molecular analysis of the bacterial communities in the live Pacific oyster (Crassostrea gigas) and the influence of postharvest temperature on its structure. J. Appl. Microbiol. 2012;112:1134–1143. doi: 10.1111/j.1365-2672.2012.05287.x. [DOI] [PubMed] [Google Scholar]
- Fuhrmann M., Richard G., Quéré C., Petton B., Pernet F. Low pH reduced survival of the oyster Crassostrea gigas exposed to the Ostreid herpesvirus 1 by altering the metabolic response of the host. Aquaculture. 2019;503:167–174. [Google Scholar]
- Fuhrmann M., Petton B., Quillien V., Faury N., Morga B., Pernet F. Salinity influences disease-induced mortality of the oyster Crassostrea gigas and infectivity of the Ostreid herpesvirus 1 (OsHV-1) Aquacult Env Interac. 2016;8:543–552. [Google Scholar]
- Garcia C., Thébault A., Dégremont L., Arzul I., Miossec L., Robert M., Chollet B., François C., Joly J.-P., Ferrand S. Ostreid herpesvirus 1 detection and relationship with Crassostrea gigas spat mortality in France between 1998 and 2006. Vet. Res. 2011;42:73. doi: 10.1186/1297-9716-42-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier M., Labreuche Y., Nicolas J.L. Molecular and phenotypic characterization of Vibrio aestuarianus subsp. francensis subsp. nov., a pathogen of the oyster Crassostrea gigas. Syst. Appl. Microbiol. 2008;31:358–365. doi: 10.1016/j.syapm.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Garnier M., Labreuche Y., Garcia C., Robert M., Nicolas J.L. Evidence for the involvement of pathogenic bacteria in summer mortalities of the Pacific oyster Crassostrea gigas. Microb. Ecol. 2007;53:187–196. doi: 10.1007/s00248-006-9061-9. [DOI] [PubMed] [Google Scholar]
- Glude J. Proceedings of the Third US–Japan Meeting on Aquaculture at Tokyo, Japan. 1974. A summary report of Pacific coast oyster mortality investigations 1965–1972. [Google Scholar]
- Go J., Deutscher A.T., Spiers Z.B., Dahle K., Kirkland P.D., Jenkins C. Mass mortalities of unknown aetiology in pacific oysters Crassostrea gigas in port stephens, New south wales, Australia. Dis. Aquat. Org. 2017;125:227–242. doi: 10.3354/dao03146. [DOI] [PubMed] [Google Scholar]
- Green T., Barnes A. Bacterial diversity of the digestive gland of Sydney rock oysters, Saccostrea glomerata infected with the paramyxean parasite, Marteilia sydneyi. J. Appl. Microbiol. 2010;109:613–622. doi: 10.1111/j.1365-2672.2010.04687.x. [DOI] [PubMed] [Google Scholar]
- Hammer Ø., Harper D., Ryan P. PAST-palaeontological statistics, ver. 1.89. Palaeontol electron. 2001;4:1–9. [Google Scholar]
- Herbert R. Methods for enumerating microorganisms and determining biomass in natural environments. In: Grigorova R., Norris J., editors. Methods in Microbiology. Elsevier Science Publishing Co. Inc.; San Diego, United States: 1990. pp. 1–39. [Google Scholar]
- Hernández-Zárate G., Olmos-Soto J. Identification of bacterial diversity in the oyster Crassostrea gigas by fluorescent in situ hybridization and polymerase chain reaction. J. Appl. Microbiol. 2006;100:664–672. doi: 10.1111/j.1365-2672.2005.02800.x. [DOI] [PubMed] [Google Scholar]
- Hick P., Evans O., Looi R., English C., Whittington R.J. Stability of Ostreid herpesvirus-1 (OsHV-1) and assessment of disinfection of seawater and oyster tissues using a bioassay. Aquaculture. 2016;450:412–421. [Google Scholar]
- Hick P.M., Evans O., Rubio A., Dhand N.K., Whittington R.J. Both age and size influence susceptibility of Pacific oysters (Crassostrea gigas) to disease caused by Ostreid herpesvirus -1 (OsHV-1) in replicated field and laboratory experiments. Aquaculture. 2018;489:110–120. [Google Scholar]
- Jenkins C., Hick P., Gabor M., Spiers Z., Fell S.A., Gu X., Read A., Go J., Dove M., O'Connor W., Kirkland P.D., Frances J. Identification and characterisation of an ostreid herpesvirus-1 microvariant (OsHV-1 μ-var) in Crassostrea gigas (Pacific oysters) in Australia. Dis. Aquat. Org. 2013;105:109–126. doi: 10.3354/dao02623. [DOI] [PubMed] [Google Scholar]
- Kaplan E.L., Meier P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958;53:457–481. [Google Scholar]
- Keeling S., Brosnahan C., Williams R., Gias E., Hannah M., Bueno R., McDonald W., Johnston C. New Zealand juvenile oyster mortality associated with Ostreid herpesvirus-1 an opportunistic longitudinal study. Dis. Aquat. Org. 2014;109:231–239. doi: 10.3354/dao02735. [DOI] [PubMed] [Google Scholar]
- King W.L., Jenkins C., Go J., Siboni N., Seymour J.R., Labbate M. Characterisation of the pacific oyster microbiome during a summer mortality event. Microb. Ecol. 2018;1:11. doi: 10.1007/s00248-018-1226-9. [DOI] [PubMed] [Google Scholar]
- Klappenbach J.A., Saxman P.R., Cole J.R., Schmidt T.M. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 2001;29:181–184. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormas K.A. Interpreting diversity of Proteobacteria based on 16S rRNA gene copy number. In: Sezenna M., editor. Proteobacteria: Phylogeny, Metabolic Diversity and Ecological Effects. Nova Publishers; Hauppauge, NY: 2011. pp. 73–89. [Google Scholar]
- Lacoste A., Jalabert F., Malham S., Cueff A., Gelebart F., Cordevant C., Lange M., Poulet S. A Vibrio splendidus strain is associated with summer mortality of juvenile oysters Crassostrea gigas in the Bay of Morlaix (North Brittany, France) Dis. Aquat. Org. 2001;46:139–145. doi: 10.3354/dao046139. [DOI] [PubMed] [Google Scholar]
- Lane D. 16S/23S rRNA sequencing. In: Stackebrandt E., Goodfellow M., editors. Nucleic Acid Techniques in Bacterial Systematics. John Wiley and Sons; Chichester, UK: 1991. pp. 115–175. [Google Scholar]
- Lane D.J., Pace B., Olsen G.J., Stahl D.A., Sogin M.L., Pace N.R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U.S.A. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Deuff R.-M., Renault T. Purification and partial genome characterization of a herpes-like virus infecting the Japanese oyster, Crassostrea gigas. J. Gen. Virol. 1999;80:1317–1322. doi: 10.1099/0022-1317-80-5-1317. [DOI] [PubMed] [Google Scholar]
- Le Roux F., Wegner K.M., Polz M.F. Oysters and vibrios as a model for disease dynamics in wild animals. Trends Microbiol. 2016;24:568–580. doi: 10.1016/j.tim.2016.03.006. [DOI] [PubMed] [Google Scholar]
- Le Roux F., Gay M., Lambert C., Waechter M., Poubalanne S., Chollet B., Nicolas J.-L., Berthe F. Comparative analysis of Vibrio splendidus-related strains isolated during Crassostrea gigas mortality events. Aquat. Living Resour. 2002;15:251–258. [Google Scholar]
- Lemire A., Goudenege D., Versigny T., Petton B., Calteau A., Labreuche Y., Le Roux F. Populations, not clones, are the unit of Vibrio pathogenesis in naturally infected oysters. ISME J. 2015;9:1523. doi: 10.1038/ismej.2014.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipp P., Brown B., Liston J., Chew K. Recent findings on the summer diseases of Pacific oysters. Proc. Natl. Shellfish. Assoc. 1976:9–10. [Google Scholar]
- Liu K.-L., Wong T.-T. Naïve Bayesian classifiers with multinomial models for rRNA taxonomic assignment. IEEE ACM Trans. Comput. Biol. Bioinform. 2013;10 doi: 10.1109/TCBB.2013.114. 1-1. [DOI] [PubMed] [Google Scholar]
- Lokmer A., Wegner K.M. Hemolymph microbiome of Pacific oysters in response to temperature, temperature stress and infection. ISME J. 2015;9:670–682. doi: 10.1038/ismej.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokmer A., Kuenzel S., Baines J.F., Wegner K.M. The role of tissue-specific microbiota in initial establishment success of Pacific oysters. Environ. Microbiol. 2016;18:970–987. doi: 10.1111/1462-2920.13163. [DOI] [PubMed] [Google Scholar]
- Lokmer A., Goedknegt M.A., Thieltges D.W., Fiorentino D., Kuenzel S., Baines J.F., Wegner K.M. Spatial and temporal dynamics of pacific oyster hemolymph microbiota across multiple scales. Front. Microbiol. 2016;7:1367. doi: 10.3389/fmicb.2016.01367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martenot C., Oden E., Travaille E., Malas J.P., Houssin M. Comparison of two real-time PCR methods for detection of Ostreid herpesvirus 1 in the Pacific oyster Crassostrea gigas. J. Virol. Methods. 2010;170:86–89. doi: 10.1016/j.jviromet.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Martenot C., Oden E., Travaillé E., Malas J.-P., Houssin M. Detection of different variants of Ostreid herpesvirus 1 in the Pacific oyster, Crassostrea gigas between 2008 and 2010. Virus Res. 2011;160:25–31. doi: 10.1016/j.virusres.2011.04.012. [DOI] [PubMed] [Google Scholar]
- Maurer D., Comps M., His E. Characteristics of the summer mortality of the Crassostrea gigas oyster in the Arcachon basin. Haliotis. 1986;15:309–317. [Google Scholar]
- Mobsby, D., Koudah, A., 2017. Australian Fisheries and Aquaculture Statistics 2016. In: Fisheries Research and Development Corporation Project 2017-095. ABARES, Department of Agriculture and Water Resources, Canberra, Australia.
- Mortensen S., Strand Å., Bodvin T., Alfjorden A., Skår C.K., Jelmert A., Aspán A., Sælemyr L., Naustvoll L.-J., Albretsen J. Summer mortalities and detection of Ostreid herpesvirus microvariant in Pacific oyster Crassostrea gigas in Sweden and Norway. Dis. Aquat. Org. 2016;117:171–176. doi: 10.3354/dao02944. [DOI] [PubMed] [Google Scholar]
- Nadkarni M.A., Martin F.E., Jacques N.A., Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- Oden E., Martenot C., Berthaux M., Travaillé E., Malas J.P., Houssin M. Quantification of Ostreid herpesvirus-1 (OsHV-1) in Crassostrea gigas by real-time PCR: determination of a viral load threshold to prevent summer mortalities. Aquaculture. 2011;317:27–31. [Google Scholar]
- Ossai S., Ramachandran P., Ottesen A., Reed E., DePaola A., Parveen S. Microbiomes of American oysters (Crassostrea virginica) harvested from two sites in the chesapeake Bay. Genome Announc. 2017;5 doi: 10.1128/genomeA.00729-17. e00729-00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul-Pont I., Dhand N.K., Whittington R.J. Influence of husbandry practices on OsHV-1 associated mortality of Pacific oysters Crassostrea gigas. Aquaculture. 2013;412–413:202–214. [Google Scholar]
- Paul-Pont I., Evans O., Dhand N.K., Whittington R.J. Experimental infections of Pacific oyster Crassostrea gigas using the Australian Ostreid herpesvirus-1 (OsHV-1) μVar strain. Dis. Aquat. Org. 2015;113:137–147. doi: 10.3354/dao02826. [DOI] [PubMed] [Google Scholar]
- Paul-Pont I., Evans O., Dhand N.K., Rubio A., Coad P., Whittington R. Descriptive epidemiology of mass mortality due to Ostreid herpesvirus-1 (OsHV-1) in commercially farmed Pacific oysters (Crassostrea gigas) in the Hawkesbury River estuary, Australia. Aquaculture. 2014;422:146–159. [Google Scholar]
- Peeler E.J., Reese R.A., Cheslett D.L., Geoghegan F., Power A., Thrush M.A. Investigation of mortality in Pacific oysters associated with Ostreid herpesvirus-1 μVar in the Republic of Ireland in 2009. Prev. Vet. Med. 2012;105:136–143. doi: 10.1016/j.prevetmed.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Pepin J.-F., Riou A., Renault T. Rapid and sensitive detection of Ostreid herpesvirus 1 in oyster samples by real-time PCR. J. Virol. Methods. 2008;149:269–276. doi: 10.1016/j.jviromet.2008.01.022. [DOI] [PubMed] [Google Scholar]
- Pernet F., Barret J., Le Gall P., Corporeau C., Dégremont L., Lagarde F., Pépin J.-F., Keck N. Mass mortalities of Pacific oysters Crassostrea gigas reflect infectious diseases and vary with farming practices in the Mediterranean Thau lagoon, France. Aquac Environ Interact. 2012;2:215–237. [Google Scholar]
- Pernet F., Fuhrmann M., Petton B., Mazurié J., Bouget J.-F., Fleury E., Daigle G., Gernez P. Determination of risk factors for herpesvirus outbreak in oysters using a broad-scale spatial epidemiology framework. Sci. Rep. 2018;8:10869. doi: 10.1038/s41598-018-29238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petton B., Pernet F., Robert R., Boudry P. Temperature influence on pathogen transmission and subsequent mortalities in juvenile Pacific oysters Crassostrea gigas. Aquac Environ Interact. 2013;3:257–273. [Google Scholar]
- Petton B., Boudry P., Alunno-Bruscia M., Pernet F. Factors influencing disease-induced mortality of Pacific oysters Crassostrea gigas. Aquac Environ Interact. 2015;6:205–222. [Google Scholar]
- Petton B., Bruto M., James A., Labreuche Y., Alunno-Bruscia M., Le Roux F. Crassostrea gigas mortality in France: the usual suspect, a herpes virus, may not be the killer in this polymicrobial opportunistic disease. Front. Microbiol. 2015;6:686. doi: 10.3389/fmicb.2015.00686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieur D., Nicolas J., Plusquellec A., Vigneulle M. Interactions between bivalve mollusks and bacteria in the marine-environment. Oceanogr. Mar. Biol. 1990;28:277–352. [Google Scholar]
- Renault T., Cochennec N., Le Deuff R.-M., Chollet B. Herpes-like virus infecting Japanese oyster (Crassostrea gigas) spat. Bull. Eur. Assoc. Fish Pathol. 1994;14:64–66. [Google Scholar]
- Renault T., Le Deuff R.-M., Cochennec N., Maffart P. Herpesviruses associated with mortalities among Pacific oyster, Crassostrea gigas, in France-Comparative study. Revue de Médecine Vétérinaire. 1994;145:735–742. [Google Scholar]
- Renault T., Le Deuff R., Cochennec N., Chollet B., Maffart P. Herpes-like viruses associated with high mortality levels in larvae and spat of Pacific oysters, Crassostrea gigas: a comparative study, the thermal effects on virus detection in hatchery-reared larvae, reproduction of the disease in axenic larvae. Vet. Res. 1995;26:539–543. [PubMed] [Google Scholar]
- Renault T., Bouquet A.L., Maurice J.-T., Lupo C., Blachier P. Ostreid herpesvirus 1 infection among Pacific oyster (Crassostrea gigas) spat: relevance of water temperature to virus replication and circulation prior to the onset of mortality. Appl. Environ. Microbiol. 2014;80:5419–5426. doi: 10.1128/AEM.00484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault T., Moreau P., Faury N., Pepin J.-F., Segarra A., Webb S. Analysis of clinical Ostreid herpesvirus 1 (Malacoherpesviridae) specimens by sequencing amplified fragments from three virus genome areas. J. Virol. 2012;86:5942–5947. doi: 10.1128/JVI.06534-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero J., Espejo R.T. The prevalence of noncultivable bacteria in oysters (Tiostrea chilenensis Philippi, 1845) J. Shellfish Res. 2001;20:1235–1240. [Google Scholar]
- Roque A., Carrasco N., Andree K., Lacuesta B., Elandaloussi L., Gairin I., Rodgers C., Furones M. First report of OsHV-1 microvar in Pacific oyster (Crassostrea gigas) cultured in Spain. Aquaculture. 2012;324:303–306. [Google Scholar]
- Samain J.-F., Degremont L., Soletchnik P., Haure J., Bédier E., Ropert M., Moal J., Huvet A., Bacca H., Van Wormhoudt A. Genetically based resistance to summer mortality in the Pacific oyster (Crassostrea gigas) and its relationship with physiological, immunological characteristics and infection processes. Aquaculture. 2007;268:227–243. [Google Scholar]
- Saulnier D., De Decker S., Haffner P., Cobret L., Robert M., Garcia C. A large-scale epidemiological study to identify bacteria pathogenic to Pacific oyster Crassostrea gigas and correlation between virulence and metalloprotease-like activity. Microb. Ecol. 2010;59:787–798. doi: 10.1007/s00248-009-9620-y. [DOI] [PubMed] [Google Scholar]
- Schikorski D., Renault T., Saulnier D., Faury N., Moreau P., Pépin J.-F. Experimental infection of Pacific oyster Crassostrea gigas spat by Ostreid herpesvirus 1: demonstration of oyster spat susceptibility. Vet. Res. 2011;42:27. doi: 10.1186/1297-9716-42-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld D. Partial residuals for the proportional hazards regression model. Biometrika. 1982;69:239–241. [Google Scholar]
- Segarra A., Pepin J.F., Arzul I., Morga B., Faury N., Renault T. Detection and description of a particular Ostreid herpesvirus 1 genotype associated with massive mortality outbreaks of Pacific oysters, Crassostrea gigas, in France in 2008. Virus Res. 2010;153:92–99. doi: 10.1016/j.virusres.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Shimahara Y., Kurita J., Kiryu I., Nishioka T., Yuasa K., Kawana M., Kamaishi T., Oseko N. Surveillance of type 1 Ostreid herpesvirus (OsHV-1) variants in Japan. Fish Pathol. 2012;47:129–136. [Google Scholar]
- Soletchnik P., Ropert M., Mazurié J., Fleury P.G., Le Coz F. Relationships between oyster mortality patterns and environmental data from monitoring databases along the coasts of France. Aquaculture. 2007;271:384–400. [Google Scholar]
- Takeuchi T., Takemoto Y., Matsubara T. Hiroshima-ken Suisan Shikenjo Hokoku (Report of Hiroshima Prefectural Fisheries Experimental Station); 1960. Haematological Study of Bacteria Affected Oysters; pp. 1–7. [Google Scholar]
- Thompson J.R., Randa M.A., Marcelino L.A., Tomita-Mitchell A., Lim E., Polz M.F. Diversity and dynamics of a north atlantic coastal Vibrio community. Appl. Environ. Microbiol. 2004;70:4103–4110. doi: 10.1128/AEM.70.7.4103-4110.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber R.V., Willner-Hall D., Rodriguez-Mueller B., Desnues C., Edwards R.A., Angly F., Dinsdale E., Kelly L., Rohwer F. Metagenomic analysis of stressed coral holobionts. Appl. Environ. Microbiol. 2009;11:2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- Trabal Fernández N., Mazón-Suástegui J.M., Vázquez-Juárez R., Ascencio-Valle F., Romero J. Changes in the composition and diversity of the bacterial microbiota associated with oysters (Crassostrea corteziensis, Crassostrea gigas and Crassostrea sikamea) during commercial production. FEMS Microbiol. Ecol. 2014;88:69–83. doi: 10.1111/1574-6941.12270. [DOI] [PubMed] [Google Scholar]
- Trabal N., Mazon-Suastegui J.M., Vazquez-Juarez R., Asencio-Valle F., Morales-Bojorquez E., Romero J. Molecular analysis of bacterial microbiota associated with oysters (Crassostrea gigas and and Crassostrea corteziensis) in different growth phases at two cultivation sites. Microb. Ecol. 2012;64:555–569. doi: 10.1007/s00248-012-0039-5. [DOI] [PubMed] [Google Scholar]
- Vezzulli L., Brettar I., Pezzati E., Reid P.C., Colwell R.R., Hofle M.G., Pruzzo C. Long-term effects of ocean warming on the prokaryotic community: evidence from the vibrios. ISME J. 2012;6:21–30. doi: 10.1038/ismej.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner K.M., Volkenborn N., Peter H., Eiler A. Disturbance induced decoupling between host genetics and composition of the associated microbiome. BMC Microbiol. 2013;13:252. doi: 10.1186/1471-2180-13-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington R.J., Dhand N.K., Evans O., Paul-Pont I. Further observations on the influence of husbandry practices on OsHV-1 μVar mortality in Pacific oysters Crassostrea gigas: age, cultivation structures and growing height. Aquaculture. 2015;438:82–97. [Google Scholar]
- Zhang J., Kobert K., Flouri T., Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]