Abstract
Background
Results of previous studies on the association of the CpG island methylator phenotype (CIMP) with colorectal cancer (CRC) prognosis were inconsistent and mostly based on different CIMP definitions. The current study aimed to comprehensively investigate the associations between DNA methylation on genes previously used to define CIMP status with CRC survival.
Results
Patients with CRC followed up for a median of 5.2 years were divided into a study cohort (n = 568) and a validation cohort (n = 308). DNA methylation was measured in tumor tissue using the Illumina Infinium HumanMethylation450 BeadChip and restricted to 43 genes used to define CIMP status in previous studies. Cox proportional hazard regression models were used to estimate adjusted hazard ratios (HR) and 95% confidence intervals (CI) of survival after CRC, including adjustment for tumor stage, microsatellite instability, and BRAF mutation status. In the study cohort, ten CpG sites were identified to be associated with CRC survival. Seven of these ten CpG sites were also associated with CRC survival in the validation cohort and were used to construct a prognostic score. CRC patients with a prognostic score of the lowest methylation level showed poorer disease-specific survival compared with patients with the highest methylation level in both the study cohort and the validation cohort (HR = 3.11 and 95% CI = 1.97–4.91, and HR = 3.06 and 95% CI = 1.71–5.45, respectively).
Conclusions
A CpG panel consisting of seven CpG sites was found to be strongly associated with CRC survival, independent from important clinical factors and mutations associated with CIMP. Further studies are required to confirm these findings.
Electronic supplementary material
The online version of this article (10.1186/s13148-019-0703-4) contains supplementary material, which is available to authorized users.
Keywords: Colorectal cancer, Survival, DNA methylation, CpG site, Prognosis
Background
Colorectal cancer (CRC) is one of the most common cancers worldwide with a 5-year survival rate of more than 60% [1]. Survival after CRC is largely dependent on the disease stage at diagnosis, but there is accumulating evidence that somatic mutations and epigenetic changes play a significant role in the progression of CRC and already guide clinical decision-making [2, 3]. As an important regulation approach in epigenetics, aberrant DNA methylation was extensively observed in various cancers including CRC. High-level methylation of CpG islands in the promoter region (CpG island methylator phenotype (CIMP)) can induce the silencing of the tumor-suppressor genes, which can stimulate tumor onset and progression [4–6]. Colorectal tumors showing such methylation were consistently associated with patient and tumor characteristics, such as older age, proximal location, and microsatellite instability (MSI-H) [7]. However, studies on the association of CIMP status and survival after CRC were inconsistent [8, 9]. In addition to methodological issues in some of the studies, studies mostly used different marker sets to define CIMP, which likely contributed to the inconsistency of results [10, 11].
In previous studies, genes used to define CIMP status were either selected because they were cancer suppressor genes silenced by methylation in the promoter region or because they were differentially methylated in tumor tissue and normal tissue [5, 7, 12]. Although high methylation of CpG islands in the promoter region is presumed to silence these genes, other aberrant methylations on these genes may also influence their activity. Meanwhile, high-throughput assays are available to comprehensively assess methylation of CpGs on CIMP-related genes [13, 14]. We therefore aimed to investigate the association between methylation of CpG sites on genes previously used to define CIMP status with CRC survival.
Results
Patient cohort
Of 702 patients in the study cohort and 366 patients in the validation cohort, 134 patients and 58 patients, respectively, were excluded because of missing information on important covariates (chemotherapy, MSI status, or BRAF mutation status). Finally, 568 patients in the study cohort and 308 patients in the validation cohort were included in the analyses. The median follow-up time was 5.2 years in both cohorts. The characteristics of the patients included in the study cohort and validation cohort were very similar. Slight differences were observed in the distribution of the education level, alcohol consumption, lymph node count, chemotherapy frequency, and KRAS mutation (Additional file 1: Table S1).
Selection of CIMP-related genes
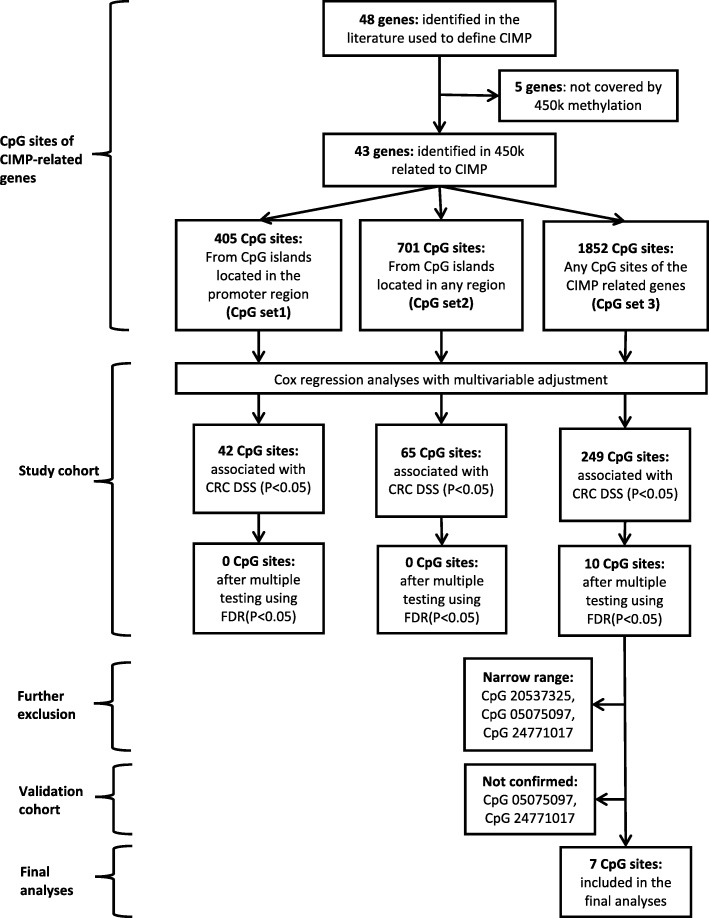
The literature search yielded 48 genes that were used to define CIMP in previous studies [15, 16]. Five of these 48 genes were not covered by the 450k methylation array but were also rarely used for the definition of CIMP (Additional file 1: Table S2). Within the 43 CIMP genes investigated in this study, 405 CpGs were located in CpG islands of the promoter region (CpG set 1), 701 CpGs were located in CpG islands of any region (CpG set 2), and 1852 CpGs were located anywhere on the genes (CpG set 3) (Fig. 1).
Fig. 1.
Flow chart of CIMP-related gene selection and identification of prognostic CpG sites associated with disease-specific survival. Cox regression analyses adjusted for age, sex, tumor location, tumor stage, chemotherapy, BRAF mutation, and microsatellite instability
Identification of prognostic CpGs
In adjusted analyses, 42 CpGs from CpG set 1 and 65 CpGs from CpG set 2 were found to be associated with disease-specific survival (DSS) (p value<0.05). However, after correction for multiple testing, none of these CpG sites was associated with DSS anymore. By further extension to CpGs anywhere on the genes (CpG set 3), 10 of the 249 associated CpG sites showed significant associations with DSS after correction for multiple testing.
Three of the 10 CpGs had β values with a range from the 10th to the 90th percentile of less than 0.1 which was considered too narrow to define distinct groups (cg05075097, cg24771017, and cg20537325). Two of the three CpGs were also not associated with DSS in the validation cohort (cg05075097 and cg24771017). Therefore, seven CpGs (cg16935707, cg05481217, cg08044454, cg01552551, cg24311416, cg02425108, and cg15659052) confirmed in the validation cohort were included in the final analyses (Fig. 1, Additional file 1: Table S3).
Associations of identified CpG sites with survival
All seven CpGs showed similar associations with DSS in the study cohort and the validation cohort (Fig. 2, Additional file 1: Figure S1). Compared to tertile group 3, tertile group 1 (lowest methylation) showed significantly poorer survival for all the seven CpGs in the study cohort. Very similar results were observed in the validation cohort, except for cg02425108, where the association was somewhat weaker (Table 1).
Fig. 2.
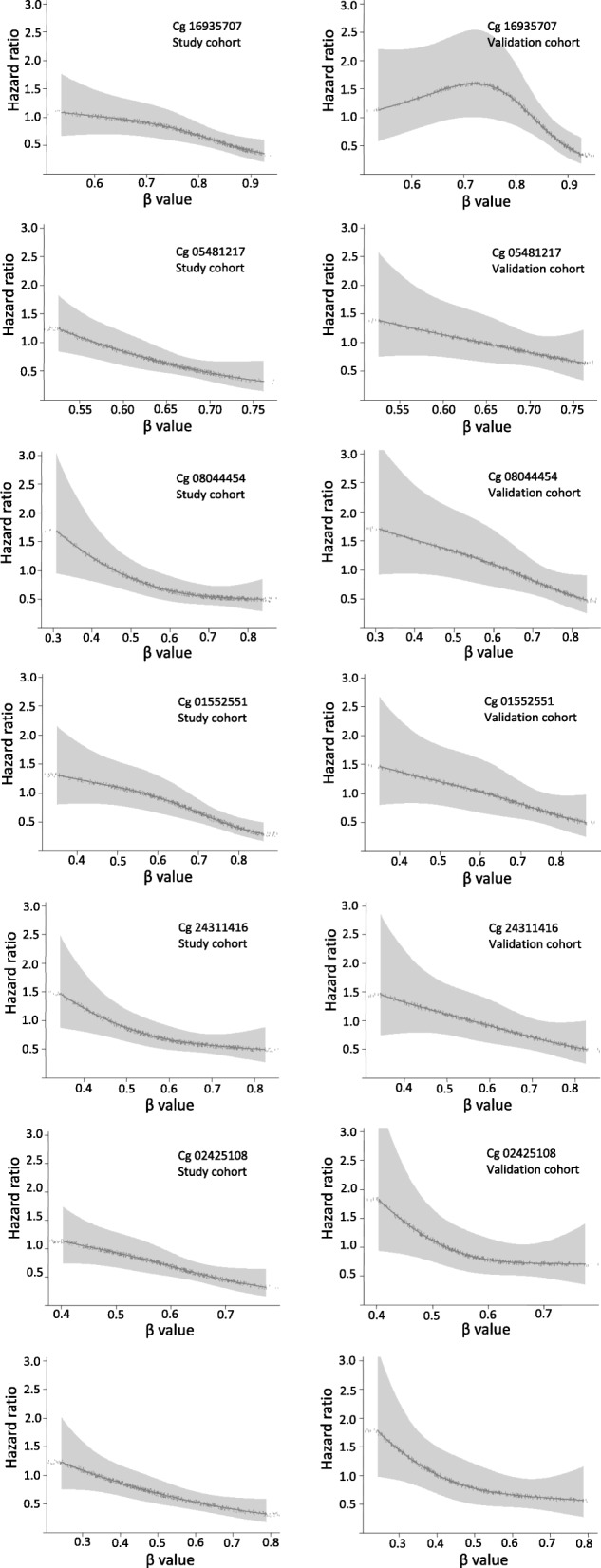
Dose-response association of methylation levels and disease-specific survival in the study cohort and the validation cohort. Restricted cubic splines analyses with adjustment for age, sex, tumor location, tumor stage, chemotherapy, BRAF, mutation and microsatellite instability
Table 1.
Association of methylation at the seven identified CpG sites with disease-specific survival in the study cohort and the validation cohort
| CpG sites | Methylation level | Study cohort | Validation cohort | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | Deaths (%) | HR (95% CI)a | p value of trend | N | Deaths (%) | HR (95% CI)a | p value of trend | ||
| cg16935707 | β valueb | 568 | 1.43 (1.22–1.67) | < 0.0001 | 308 | 1.33 (1.09–1.61) | 0.0046 | ||
| Tertile 3 | 193 | 45 (23) | Ref. | 125 | 27 (22) | Ref. | |||
| Tertile 2 (0.84342) | 182 | 38 (21) | 1.53 (0.98–2.39) | 99 | 25 (25) | 1.95 (1.08–3.52) | |||
| Tertile 1 (0.72736) | 193 | 53 (27) | 2.22 (1.46–3.36) | 0.0002 | 84 | 30 (36) | 2.12 (1.20–3.74) | 0.0095 | |
| cg05481217 | β valueb | 568 | 1.43 (1.22–1.67) | < 0.0001 | 308 | 1.30 (1.06–1.60) | 0.0115 | ||
| Tertile 3 | 187 | 36 (19) | Ref. | 173 | 42 (24) | Ref. | |||
| Tertile 2 (0.66890) | 183 | 36 (20) | 1.29 (0.79–2.08) | 78 | 17 (22) | 1.27 (0.71–2.27) | |||
| Tertile 1 (0.61265) | 198 | 64 (32) | 2.23 (1.46–3.42) | 0.0002 | 57 | 23 (40) | 2.02 (1.18–3.45) | 0.0125 | |
| cg08044454 | β valueb | 568 | 1.44 (1.22–1.69) | < 0.0001 | 308 | 1.39 (1.14–1.70) | 0.0014 | ||
| Tertile 3 | 195 | 40 (21) | Ref. | 110 | 20 (18) | Ref. | |||
| Tertile 2 (0.71507) | 195 | 41 (21) | 1.25 (0.80–1.97) | 98 | 28 (29) | 1.87 (1.03–3.42) | |||
| Tertile 1 (0.57145) | 178 | 55 (31) | 2.23 (1.44–3.46) | 0.0003 | 100 | 34 (34) | 2.48 (1.39–4.44) | 0.0021 | |
| cg01552551 | β valueb | 568 | 1.43 (1.22–1.68) | < 0.0001 | 308 | 1.28 (1.04–1.58) | 0.0215 | ||
| Tertile 3 | 189 | 28 (15) | Ref. | 104 | 25 (24) | Ref. | |||
| Tertile 2 (0.72226) | 189 | 49 (26) | 1.94 (1.21–3.10) | 95 | 22 (23) | 1.38 (0.76–2.51) | |||
| Tertile 1 (0.59834) | 190 | 59 (31) | 2.50 (1.58–3.95) | < 0.0001 | 109 | 35 (32) | 2.17 (1.24–3.79) | 0.0065 | |
| cg24311416 | β valueb | 568 | 1.43 (1.21–1.71) | < 0.0001 | 308 | 1.32 (1.04–1.66) | 0.0209 | ||
| Tertile 3 | 190 | 40 (21) | Ref. | 118 | 27 (23) | Ref. | |||
| Tertile 2 (0.65694) | 185 | 38 (21) | 1.03 (0.64–1.64) | 90 | 23 (26) | 1.06 (0.59–1.89) | |||
| Tertile 1 (0.53430) | 193 | 58 (30) | 2.20 (1.45–3.35) | 0.0002 | 100 | 32 (32) | 2.00 (1.17–3.43) | 0.0135 | |
| cg02425108 | β valueb | 568 | 1.40 (1.19–1.65) | < 0.0001 | 308 | 1.32 (1.04–1.66) | 0.0200 | ||
| Tertile 3 | 209 | 35 (17) | Ref. | 104 | 27 (26) | Ref. | |||
| Tertile 2 (0.62299) | 175 | 38 (22) | 1.78 (1.12–2.85) | 107 | 28 (26) | 1.32 (0.75–2.31) | |||
| Tertile 1 (0.52523) | 184 | 63 (34) | 2.44 (1.59–3.74) | < 0.0001 | 97 | 27 (28) | 1.68 (0.95–2.96) | 0.0727 | |
| cg15659052 | β valueb | 568 | 1.38 (1.17–1.63) | 0.0001 | 308 | 1.37 (1.09–1.71) | 0.0062 | ||
| Tertile 3 | 183 | 30 (16) | Ref. | 113 | 25 (22) | Ref. | |||
| Tertile 2 (0.53788) | 193 | 38 (20) | 2.12 (1.28–3.51) | 92 | 17 (18) | 1.55 (0.80–2.99) | |||
| Tertile 1 (0.40182) | 192 | 68 (35) | 2.34 (1.50–3.67) | 0.0002 | 103 | 40 (39) | 1.99 (1.16–3.34) | 0.0118 | |
Abbreviations: HR hazard ratio, CI confidence interval
aCox regression analyses adjusted for age, sex, tumor stage, tumor location, chemotherapy, MSI status, and BRAF mutation status
bHR calculated for per unit decrease of the Z score of the β values
Associations of the prognostic methylation score with survival
The coefficient values for the calculation of the coefficient score were − 1.24524 for cg16935707, − 1.32717 for cg05481217, − 1.23953 for cg08044454, − 1.15191 for cg01552551, − 0.62350 for cg24311416, − 0.53487 for cg02425108, and − 0.17845 for cg15659052. Using this score based on methylation at seven CpG sites, low methylation was strongly associated with poorer DSS compared with high methylation in the study cohort (HR = 3.16; 95% CI = 2.08–4.80) and in the validation cohort (HR = 2.75; 95% CI = 1.56–4.85) (Table 2). Patients with intermediate methylation scores also showed poorer DSS compared to patients with high methylation scores; however, the trend was not statistically significant.
Table 2.
Association of the coefficient score with disease-specific survival in the study cohort and the validation cohort
| Patient groups | Subgroups | Tertile groups (cutoff points) | Study cohort | Validation cohort | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | Deaths (%) | HR (95% CI)a | p value of trend | N | Deaths (%) | HR (95% CI)a | p value of trend | |||
| All patients | Tertile 3 | 195 | 36 (18) | Ref. | 124 | 25 (20) | Ref. | |||
| Tertile 2 (− 4.32247) | 188 | 31 (16) | 1.31 (0.80–2.15) | 89 | 22 (25) | 1.66 (0.91–3.05) | ||||
| Tertile 1 (− 3.92950) | 185 | 69 (37) | 3.16 (2.08–4.80) | < 0.0001 | 95 | 35 (37) | 2.75 (1.56–4.85) | 0.0004 | ||
| Microsatellite instability | MSS | Tertile 3 | 175 | 35 (20) | Ref. | 114 | 25 (22) | Ref. | ||
| Tertile 2 (− 4.32247) | 161 | 29 (18) | 1.31 (0.79–2.17) | 79 | 21 (27) | 1.63 (0.88–3.01) | ||||
| Tertile 1 (− 3.92950) | 167 | 66 (40) | 3.11 (2.04–4.76) | < 0.0001 | 89 | 33 (37) | 2.65 (1.49–4.72) | 0.0009 | ||
| MSI-H | Tertile 3 | 20 | 1 (5) | Ref. | 10 | 0 (0) | Ref. | |||
| Tertile 2 (− 4.32247) | 27 | 2 (7) | NC | 10 | 1 (10) | NC | ||||
| Tertile 1 (− 3.92950) | 18 | 3 (17) | NC | NC | 6 | 2 (33) | NC | NC | ||
| BRAF mutation | Negative | Tertile 3 | 185 | 34 (18) | Ref. | 115 | 22 (19) | Ref. | ||
| Tertile 2 (− 4.32247) | 171 | 28 (16) | 1.23 (0.74–2.07) | 81 | 19 (23) | 1.53 (0.81–2.91) | ||||
| Tertile 1 (− 3.92950) | 166 | 61 (37) | 3.24 (2.09–5.02) | < 0.0001 | 93 | 34 (37) | 2.72 (1.53–4.85) | 0.0007 | ||
| Positive | Tertile 3 | 10 | 2 (20) | Ref. | 9 | 3 (33) | Ref. | |||
| Tertile 2 (− 4.32247) | 17 | 3 (18) | NC | 8 | 3 (38) | NC | ||||
| Tertile 1 (− 3.92950) | 19 | 8 (42) | NC | NC | 2 | 1 (50) | NC | NC | ||
| Tumor location | Proximal colon | Tertile 3 | 59 | 5 (8) | Ref. | 40 | 10 (25) | Ref. | ||
| Tertile 2 (− 4.32247) | 80 | 11 (14) | 2.60 (0.87–7.84) | 27 | 6 (22) | 0.99 (0.31–3.13) | ||||
| Tertile 1 (− 3.92950) | 71 | 25 (35) | 6.92 (2.46–19.4) | < 0.0001 | 41 | 14 (34) | 2.05 (0.79–5.34) | 0.1176 | ||
| Distal colon | Tertile 3 | 71 | 19 (27) | Ref. | 41 | 5 (12) | Ref. | |||
| Tertile 2 (− 4.32247) | 49 | 5 (10) | 0.73 (0.25–2.14) | 26 | 8 (31) | 1.76 (0.51–6.09) | ||||
| Tertile 1 (− 3.92950) | 56 | 20 (36) | 2.03 (1.05–3.94) | 0.0477 | 21 | 8 (38) | 6.00 (1.61–22.4) | 0.0098 | ||
| Rectum proximal | Tertile 3 | 65 | 12 (18) | Ref. | 43 | 10 (23) | Ref. | |||
| Tertile 2 (− 4.32247) | 59 | 15 (25) | 1.36 (0.62–3.01) | 36 | 8 (22) | 3.48 (0.97–12.4) | ||||
| Tertile 1 (− 3.92950) | 58 | 24 (41) | 3.59 (1.64–7.87) | 0.0012 | 33 | 13 (39) | 4.02 (1.38–11.7) | 0.0129 | ||
Abbreviations: CI confidence interval, HR hazard ratio, NC not calculated (< 5 patients)
aCox regression adjusted for age, sex, tumor stage, tumor location, chemotherapy, MSI status, and BRAF mutation status
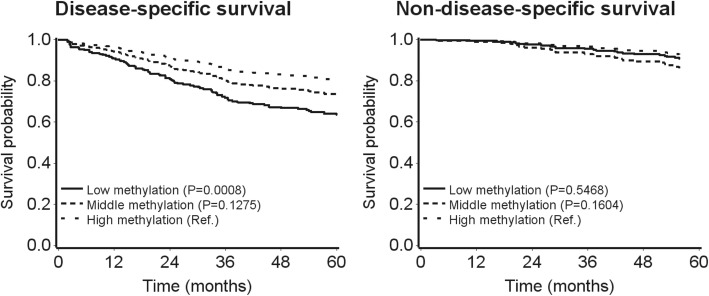
Unadjusted Kaplan–Meier curves of the association of the prognostic score with DSS and non-DSS showed poorer survival in the lowest tertile group only (survival rates after 36 months were 68% and 87% for patients in the lowest and highest tertile of the prognostic score, respectively; Additional file 1: Figure S2). Using direct adjusted survival curves, the middle and lowest tertile score were increasingly strongly associated with poorer DSS, whereas no association was found for CRC patients who died of other causes (Fig. 3). In addition, by adding our CpG panel to the model constructed by the clinical and molecular factors, the AIC decreased from 1461 to 1433 in the study cohort and from 823 to 812 in the validation cohort. As most of the CRC patients died of CRC, the association of the methylation panel with overall survival was similar with the association for DSS (data not shown).
Fig. 3.
Direct survival curves of the coefficient score with disease-specific survival and non-disease-specific survival in the validation cohort (adjusted for age, sex, tumor location, tumor stage, chemotherapy, BRAF mutation, and microsatellite instability)
In subgroup analyses stratified by tumor location, MSI status, and BRAF mutation status, similar results were observed for the association between the prognostic scores and DSS (Table 2). The prognostic score was not related to clinical and tumor characteristics of the CRC patients such as age, sex, education level, family history of CRC, lifetime regular active smoking status, tumor location, cancer stage, lymph node count, chemotherapy, KRAS mutation, BRAF mutation, and MSI status in both study and validation cohort. Only for chemotherapy heterogeneity was observed in the study cohort due to dissimilar distribution in the tertile 3 group. However, this was not observed in the validation cohort (Additional file 1: Table S4).
Discussion
In this population-based patient cohort study, we identified seven CpG sites on 43 genes previously used to define CIMP status in the published literature that were independently associated with DSS. At each of the seven CpG sites, lower methylation levels were strongly associated with poorer survival after CRC. The associations observed in the study cohort and the validation cohort were very similar, suggesting that results might be reproducible in other cohorts, too. By incorporating the seven CpGs into a prognostic CpG score we constructed a potential prediction panel for CRC prognosis.
As one of the most widely investigated features of CRC, CIMP was inconsistently associated with poorer outcome in previous studies and the CIMP definitions used were largely different [8, 17]. Mostly, studies focused on CpG sites in the promoter region, where hypermethylation is the aberrant type in cancer cells. In the present study, all seven identified CpGs were selected from CIMP-related genes which we assumed was a more effective approach compared to an epigenome-wide analysis. Recently, a genome-wide methylation study reported a prognostic classifier for non-metastatic CRC consisting of 20 CpGs which also showed poorer survival with lower levels of methylation at the associated CpG sites [18]. The identified CpGs in our study did not overlap, and our score showed a slightly stronger prognostic effect. Among the seven prognostic CpGs identified, cg16935707, cg05481217, and cg24311416 are located in the gene bodies, cg08044454 and cg15659052 are located in the 5′UTR region, cg02425108 is located in the 3′UTR region, and only cg01552551 is located in the S-shore of a CpG island which belongs to the promoter region. At all seven CpG sites, lower methylation instead of higher methylation was associated with poorer survival. Although the molecular mechanism of the association between hypomethylation of CpGs and poorer CRC survival is not known, methylation in the gene body, where hypomethylation is the aberrant type, was found to contribute to cancer-causing somatic and germline mutations in previous studies [19, 20]. In a study on primary glioblastoma, Nagarajan et al. found recurring gene body promoter hypomethylation events which may alter the transcriptional landscape of glioblastoma through the activation of a limited number of normally silenced promoters within gene bodies, in at least one case leading to expression of an oncogenic protein [21].
The seven identified CpGs are located on three different genes: ELMO1 (cg05481217, cg08044454, cg01552552, and cg15659052), CADM1 (cg16935707 and cg24311416), and ADAMTS1 (cg02425108). ELMO1 (engulfment and cell motility 1) together with Dock180 as a complex plays an important role in promoting cancer cell invasion which has been shown for gliomas, breast cancer, and ovarian cancer [22–24]. Methylation markers on ELMO1 were reported to be associated with cancers such as oropharyngeal squamous cell carcinoma and glioblastoma. One other smaller study which adjusted for mucinous type and location only investigated methylation in ELMO1 along with other epigenetic markers in a panel and found worse prognosis in KRAS mutated cancers [25–27]. Cell adhesion molecule 1 (CADM1/TSLC1) is a putative tumor suppressor involved in cell adhesion, proliferation, and apoptosis [28, 29]. The epigenetic inactivation of CADM1/TSLC1 was found as a frequent alteration in the development of CRC and correlates with late stages of the disease [30]. ADAMTS1 (a disintegrin and metalloproteinase with thrombospondin type 1 motifs) is an extracellular matrix metalloproteinase with protease activity and antiangiogenic activity [31]. ADAMTS1 was identified as an epigenetically deregulated gene in CRC, and it was suggested that ADAMTS1 could play an important role in tumor growth and metastasis [32]. All three genes involved are tumor suppressor genes that seem to have effects on tumor growth and metastasis. The alteration of methylation of these genes may play an important role in tumor progression and in the prognosis of CRC patients.
To our knowledge, this is the first study that comprehensively investigates the association between methylation of CpG sites on CIMP-related genes and CRC prognosis in a large patient cohort which is not restricted to specific gene regions. Besides DNA methylation, other molecular pathological features such as MSI, BRAF mutation, and single-nucleotide polymorphisms (SNPs) were frequently investigated for their prognostic value in CRC. For example, MSI-H was found to be a marker of better prognosis among CRC patients in a meta-analysis (OR = 0.58; 95% CI 0.47–0.72), and patients with BRAF mutation were found to have significantly worse progression-free survival compared with BRAF wild-type patients (HR = 1.33; 95% CI 1.12–1.57) [33, 34]. However, as the prognostic values of MSI and BRAF mutation seem to be affected by each other, it was suggested that these markers should be considered jointly [35]. A genome-wide analysis of SNPs illustrated that several SNPs were statistically associated with poorer survival of CRC, with the strongest associations noted for rs209489 (HR = 1.8, p = 7.6 × 10−10). However, this result has not been validated in an independent cohort yet [36]. Compared with these biomarkers, all the seven prognostic CpGs were selected through multivariable Cox regression with adjustment considering tumor stage, MSI status, and BRAF mutation status as confounders. As validation in an independent cohort yielded very similar results, the panel might be a tool for outcome prediction but further validation in large studies is needed. Besides outcome prediction, further studies will be needed to investigate if and to what extent the CpG score is also useful for the prediction of therapy response.
Both the study cohort and validation cohort were derived from a large population-based study including 876 unselected patients with complete information on clinical and molecular characteristics. Despite the adequate size of the cohort population, the statistical power was still limited in many of the subgroup analyses. Also, the prognostic CpGs identified were selected from CIMP-related genes. It is likely that CpGs of other genes could also be associated with CRC survival, and incorporating these additional sites may further improve the prognostic use of our panel. Accordingly, more large studies on the association between methylation of CpG sites and CRC outcomes are needed without restrictions to specific gene regions. Also, although the CpG sites were confirmed and similarly associated in the validation cohort, external validation in another study is still needed to establish the prognostic score.
Conclusion
In summary, our study found that DNA methylations at seven CpG sites on CIMP-related genes were strongly associated with CRC prognosis independent of cancer stage, MSI status, BRAF mutation status, and other important factors that affect the outcome of CRC. The prognostic score based on methylation levels at these seven CpG sites strongly predicted CRC survival in the study cohort and the validation cohort. If confirmed in other cohorts, this CpG panel could be a novel tool for CRC outcome prediction and may have relevance for clinical decision-making. Still, other large studies are needed to confirm our findings and to elaborate its potential utility.
Methods
Study population
Patients with CRC were recruited into the DACHS (Darmkrebs: Chancen der Verhütung durch Screening) study, a large ongoing population-based cohort study on CRC with long-term follow-up [37]. Patients histologically confirmed CRC who were at least 30 years old and physically and mentally able to participate in a personal interview were enrolled in 22 hospitals in the Rhine–Neckar–Odenwald region of southwestern Germany. Sociodemographic and lifestyle information was collected by trained interviewers using a standardized questionnaire during face-to-face interviews. Additionally, discharge letters, pathology reports, and endoscopy reports were collected at baseline. After 3 years, information on the patients’ therapy was requested from the patients’ physicians. After 5 years, survivors were contacted to complete a standardized questionnaire. In addition, at both follow-ups, disease recurrence was assessed. Also, data on vital status were obtained from the population registries and causes of death were verified by death certificates from the health authorities. More details on the study design, data collection, and follow-up have been reported previously [38, 39].
In the present study, only patients recruited between 2003 and 2007 with available information on DNA methylation were included. The study was approved by the ethics committees of the Medical Faculty of the University of Heidelberg and of the Medical Chambers of Baden–Wuerttemberg and Rhineland–Palatinate.
Tumor sample analyses
DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) tumor samples of the patients under microscopic control of unstained slides and was prepared using the DNeasy tissue kit (Qiagen, Hilden, Germany) [40]. Methylation of tissue DNA was analyzed using the Illumina Human Methylation 450 BeadChip (Illumina, San Diego, CA, USA) following the manufacturer’s instructions. Tumors of patients examined in the pathology institutes in Heilbronn, Ludwigshafen, Mannheim, and Speyer (study cohort) were analyzed more than 1 year apart from tumors of patients examined in the pathology institute in Heidelberg (validation cohort). Methylation signals at CpG sites were converted into β values (methylated signal/(unmethylated signal + methylated signal)). β values ranged from 0 to 1: 0 represents totally unmethylated and 1 represents totally methylated. In data pre-processing, the following probes were excluded: probes targeting the X and Y chromosomes, probes containing a single-nucleotide polymorphism (dbSNP132 Common) within five base pairs, probes not mapping uniquely to the human reference genome (hg19) allowing for one mismatch, and probes that have failed in more than 10% of the samples based on the detection p value (detection p value > 0.01). Data was normalized by pre-processing in the R package “minfi” [41].
MSI status was determined using a mononucleotide marker panel (BAT25, BAT26, and CAT25) [42]. KRAS mutation was determined by single-stranded conformational polymorphism technique using the same DNA sample [43]. The expression of BRAF V600E was determined by immunohistochemical analyses in sections of tissue microarray blocks and evaluated by two pathologists independently.
Statistical analysis
Genes used to define CIMP in previous studies were identified by a literature review [15, 16]. We focused on CIMP genes because CIMP is an established marker in CRC to differentiate between CRC pathways. We assumed that methylation on these genes could be more relevant than on other genes when investigating associations with survival. Also, by focusing on CIMP genes, the number of association tests is lower which increases the chance to find associations even after adjustment for multiple testing.
In a split-sample approach, we used a larger study cohort to investigate associations of CpG sites on CIMP-related genes with DSS and a smaller validation cohort to validate our findings. Hazard ratios (HR) and 95% confidence intervals (95% CI) were calculated by multivariable Cox proportional hazard regression with adjustment for age, sex, tumor stage, tumor location, chemotherapy, MSI status, and BRAF mutation status. In addition, a correction for late entry defined as the potentially delayed time between the date of diagnosis and the date of enrolment was included in the adjusted model, as well as a time-dependent variable of age and chemotherapy. We used three different sets of CpGs on the CIMP genes to test for associations: CpG set 1 consisted of CpGs located in the CpG islands of promoter regions, CpG set 2 included any CpG in CpG islands, and CpG set 3 included any CpG in any region on the genes (Fig. 1). In the study cohort, CpGs were deemed to be associated with CRC prognosis only if they passed the correction for multiple testing by Benjamini–Hochberg (false discovery rates (FDR) < 0.05) and if the β value range was not too narrow (≥ 0.1) to ensure reproducibility.
The CpGs associated with DSS in the study cohort were then analyzed in the validation cohort using the same adjusted Cox model. The association between tertiles of β values and DSS analyzed in the study cohort was analyzed in the validation cohort using the tertile cutoffs of the study cohort. Dose-response curves of methylation levels at the identified CpGs were plotted to illustrate the association with DSS by restricted cubic spline regression adjusting for the confounders mentioned above. The CpGs confirmed in the validation cohort were used to investigate individual associations and to construct a prognostic score using the coefficient score. A heatmap illustrating the methylation level of the selected CpGs and their association with CRC survival was generated using the R package “pheatmap”.
The coefficient score was calculated by summing the product of the β value of each validated CpG with its corresponding coefficient value in the Cox regression. Tertiles of the sum were used to build three groups with tertile 3 representing patients with the highest methylation at the selected CpGs sites, which was used as the reference (among the identified CpGs, lower methylation was associated with higher mortality). The coefficient value and tertile cutoff point identified in the study cohort were used for the analyses in the validation cohort. Direct adjusted survival curves (adjusting for the same covariates) and Kaplan–Meier curves were plotted to illustrate the association of the prognostic score with DSS and non-DSS (deaths due to other causes) over time. The associations of clinical and molecular features with the coefficient score in the study and the validation cohort were analyzed by chi-square test. The Akaike information criterion (AIC) was computed using R, software version 3.5.0. All other statistical analyses were performed with SAS, software version 9.4 (SAS Institute Inc., Cary, NC, USA).
Additional file
Table S1. Characteristics of patients in the study cohort and the validation cohort. Table S2. Genes used to define CIMP status in previous studies that were available on the 450k methylation array. Table S3. CpG sites identified in the study cohort that were included or excluded in the final analyses. Table S4. Association of the coefficient score with characteristics of colorectal cancer patients in the study cohort and the validation cohort. Figure S1. Heatmaps showing the methylation level of the seven CpG sites across all the patients in the study cohort (A) and the validation cohort (B), respectively. Figure S2. Unadjusted Kaplan–Meier curves of the association of the coefficient score with disease-specific survival and non-disease-specific survival in the validation cohort. (DOCX 2549 kb)
Acknowledgements
The authors thank the study participants and the interviewers who collected the data. The authors also thank the following hospitals and cooperating institutions that recruited patients for this study: Chirurgische Universitätsklinik Heidelberg, Klinik am Gesundbrunnen Heilbronn, St. Vincentiuskrankenhaus Speyer, St. Josefskrankenhaus Heidelberg, Chirurgische Universitätsklinik Mannheim, Diakonissenkrankenhaus Speyer, Krankenhaus Salem Heidelberg, Kreiskrankenhaus Schwetzingen, St. Marienkrankenhaus Ludwigshafen, Klinikum Ludwigshafen, Stadtklinik Frankenthal, Diakoniekrankenhaus Mannheim, Kreiskrankenhaus Sinsheim, Klinikum am Plattenwald Bad Friedrichshall, Kreiskrankenhaus Weinheim, Kreiskrankenhaus Eberbach, Kreiskrankenhaus Buchen, Kreiskrankenhaus Mosbach, Enddarmzentrum Mannheim, Kreiskrankenhaus Brackenheim, and Cancer Registry of Rhineland-Palatinate, Mainz. We are also very grateful for the support of the pathologies in the provision of tumor samples: Institut für Pathologie, Universitätsklinik Heidelberg; Institut für Pathologie, Klinikum Heilbronn; Institut für Angewandte Pathologie, Speyer; Pathologisches Institut, Universitätsklinikum Mannheim; Institut für Pathologie, Klinikum Ludwigshafen; Institut für Pathologie, Klinikum Stuttgart; and Institut für Pathologie, Klinikum Ludwigsburg.
We thank Ute Handte-Daub, Ansgar Brandhorst, Marina Gernold, and Bettina Walter for their excellent technical assistance.
Abbreviations
- AIC
Akaike information criterion
- CI
Confidence interval
- CIMP
CpG island methylator phenotype
- CpGs
CpG sites
- CRC
Colorectal cancer
- DSS
Disease-specific survival
- HR
Hazard ratio
- MSI
Microsatellite instability
- MSI-H
High-level microsatellite instability
Authors’ contributions
MJ, YZ, JC, MH, and HBr made substantial contributions to the conception and design of the study. MJ and MH contributed to the development of methodology. MH, HBr, JC, LJ, VW, MM, KT, WR, MB, EH, MK, AU, BB, and HBl contributed to the acquisition of data. MJ, DE, and MH contributed to the analysis and interpretation of data. MJ and MH contributed to the draft of the article. All authors contributed to the critical revision of the article. All authors approved the final version of the manuscript.
Funding
This work was supported by the German Research Council (BR 1704/6-1, BR 1704/6-3, BR 1704/6-4, CH 117/1-1, HO 5117/2-1, HE 5998/2-1, KL 2354/3-1, RO 2270/8-1, and BR 1704/17-1); the Interdisciplinary Research Program of the National Center for Tumor Diseases (NCT), Germany; the DKFZ-Heidelberg Center for Personalized Oncology (DKFZ-HIPO, H052); and the German Federal Ministry of Education and Research (01KH0404, 01ER0814, 01ER0815, 01ER1505A, and 01ER1505B).
The funders of the study had no role in study design, collection, analysis or interpretation of data, or writing of the report.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study was approved by the ethics committees of the Medical Faculty of the University of Heidelberg (310/2001) and of the State Medical Boards of Baden–Wuerttemberg (M-198-02) and Rhineland–Palatinate (837.419.02 (3637)).
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RG, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017. [DOI] [PubMed]
- 2.Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Cote JF, Tomasic G, Penna C, Ducreux M, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66(8):3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 3.Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouche O, Landi B, Louvet C, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol Off J Am Soc Clin Oncol. 2008;26(3):374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 4.Kim YS, Deng G. Epigenetic changes (aberrant DNA methylation) in colorectal neoplasia. Gut and liver. 2007;1(1):1–11. doi: 10.5009/gnl.2007.1.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96(15):8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 7.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38(7):787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 8.Bae JM, Kim JH, Cho NY, Kim TY, Kang GH. Prognostic implication of the CpG island methylator phenotype in colorectal cancers depends on tumour location. Br J Cancer. 2013;109(4):1004–1012. doi: 10.1038/bjc.2013.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58(1):90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleven AH, Derks S, Draht MX, Smits KM, Melotte V, Van Neste L, Tournier B, Jooste V, Chapusot C, Weijenberg MP, et al. CHFR promoter methylation indicates poor prognosis in stage II microsatellite stable colorectal cancer. Clin Cancer Res. 2014;20(12):3261–3271. doi: 10.1158/1078-0432.CCR-12-3734. [DOI] [PubMed] [Google Scholar]
- 11.Hokazono K, Ueki T, Nagayoshi K, Nishioka Y, Hatae T, Koga Y, Hirahashi M, Oda Y, Tanaka M. A CpG island methylator phenotype of colorectal cancer that is contiguous with conventional adenomas, but not serrated polyps. Oncol Let. 2014;8(5):1937–1944. doi: 10.3892/ol.2014.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9(3):305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2014;6(6):692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 14.Naumov VA, Generozov EV, Zaharjevskaya NB, Matushkina DS, Larin AK, Chernyshov SV, Alekseev MV, Shelygin YA, Govorun VM. Genome-scale analysis of DNA methylation in colorectal cancer using Infinium HumanMethylation450 BeadChips. Epigenetics. 2013;8(9):921–934. doi: 10.4161/epi.25577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia M, Gao X, Zhang Y, Hoffmeister M, Brenner H. Different definitions of CpG island methylator phenotype and outcomes of colorectal cancer: a systematic review. Clin Epigenetics. 2016;8:25. doi: 10.1186/s13148-016-0191-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashktorab H, Rahi H, Wansley D, Varma S, Shokrani B, Lee E, Daremipouran M, Laiyemo A, Goel A, Carethers JM, et al. Toward a comprehensive and systematic methylome signature in colorectal cancers. Epigenetics. 2013;8(8):807–815. doi: 10.4161/epi.25497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juo YY, Johnston FM, Zhang DY, Juo HH, Wang H, Pappou EP, Yu T, Easwaran H, Baylin S, van Engeland M, et al. Prognostic value of CpG island methylator phenotype among colorectal cancer patients: a systematic review and meta-analysis. Ann Oncol. 2014;25(12):2314–2327. doi: 10.1093/annonc/mdu149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gundert M, Edelmann D, Benner A, Jansen L, Jia M, Walter V, Knebel P, Herpel E, Chang-Claude J, Hoffmeister M, et al. Genome-wide DNA methylation analysis reveals a prognostic classifier for non-metastatic colorectal cancer (ProMCol classifier) Gut. 2019;68(1):101–110. doi: 10.1136/gutjnl-2017-314711. [DOI] [PubMed] [Google Scholar]
- 19.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 20.Witte T, Plass C, Gerhauser C. Pan-cancer patterns of DNA methylation. Genome Med. 2014;6(8):66. doi: 10.1186/s13073-014-0066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagarajan RP, Zhang B, Bell RJ, Johnson BE, Olshen AB, Sundaram V, Li D, Graham AE, Diaz A, Fouse SD, et al. Recurrent epimutations activate gene body promoters in primary glioblastoma. Genome Res. 2014;24(5):761–774. doi: 10.1101/gr.164707.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Yang L, Fu H, Yan J, Wang Y, Guo H, Hao X, Xu X, Jin T, Zhang N. Association between Galphai2 and ELMO1/Dock180 connects chemokine signalling with Rac activation and metastasis. Nat Commun. 2013;4:1706. doi: 10.1038/ncomms2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jarzynka MJ, Hu B, Hui KM, Bar-Joseph I, Gu W, Hirose T, Haney LB, Ravichandran KS, Nishikawa R, Cheng SY. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Res. 2007;67(15):7203–7211. doi: 10.1158/0008-5472.CAN-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh H, Negishi M. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature. 2003;424(6947):461–464. doi: 10.1038/nature01817. [DOI] [PubMed] [Google Scholar]
- 25.Yagi K, Akagi K, Hayashi H, Nagae G, Tsuji S, Isagawa T, Midorikawa Y, Nishimura Y, Sakamoto H, Seto Y, et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 2010;16(1):21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- 26.Michaelsen Signe, Aslan Derya, Urup Thomas, Poulsen Hans, Grønbæk Kirsten, Broholm Helle, Kristensen Lasse. DNA Methylation Levels of the ELMO Gene Promoter CpG Islands in Human Glioblastomas. International Journal of Molecular Sciences. 2018;19(3):679. doi: 10.3390/ijms19030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren S, Gaykalova D, Wang J, Guo T, Danilova L, Favorov A, Fertig E, Bishop J, Khan Z, Flam E, et al. Discovery and development of differentially methylated regions in human papillomavirus-related oropharyngeal squamous cell carcinoma. Int J Cancer. 2018;143(10):2425–2436. doi: 10.1002/ijc.31778. [DOI] [PubMed] [Google Scholar]
- 28.van Baars R, van der Marel J, Snijders PJ, Rodriquez-Manfredi A, ter Harmsel B, van den Munckhof HA, Ordi J, del Pino M, van de Sandt MM, Wentzensen N, et al. CADM1 and MAL methylation status in cervical scrapes is representative of the most severe underlying lesion in women with multiple cervical biopsies. Int J Cancer. 2016;138(2):463–471. doi: 10.1002/ijc.29706. [DOI] [PubMed] [Google Scholar]
- 29.Fukami T, Fukuhara H, Kuramochi M, Maruyama T, Isogai K, Sakamoto M, Takamoto S, Murakami Y. Promoter methylation of the TSLC1 gene in advanced lung tumors and various cancer cell lines. Int J Cancer. 2003;107(1):53–59. doi: 10.1002/ijc.11348. [DOI] [PubMed] [Google Scholar]
- 30.Chen K, Wang G, Peng L, Liu S, Fu X, Zhou Y, Yu H, Li A, Li J, Zhang S, et al. CADM1/TSLC1 inactivation by promoter hypermethylation is a frequent event in colorectal carcinogenesis and correlates with late stages of the disease. Int J Cancer. 2011;128(2):266–273. doi: 10.1002/ijc.25356. [DOI] [PubMed] [Google Scholar]
- 31.Choi JE, Kim DS, Kim EJ, Chae MH, Cha SI, Kim CH, Jheon S, Jung TH, Park JY. Aberrant methylation of ADAMTS1 in non-small cell lung cancer. Cancer Genet Cytogenet. 2008;187(2):80–84. doi: 10.1016/j.cancergencyto.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Lind GE, Kleivi K, Meling GI, Teixeira MR, Thiis-Evensen E, Rognum TO, Lothe RA. ADAMTS1, CRABP1, and NR3C1 identified as epigenetically deregulated genes in colorectal tumorigenesis. Cell Oncol. 2006;28(5-6):259–272. doi: 10.1155/2006/949506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guastadisegni C, Colafranceschi M, Ottini L, Dogliotti E. Microsatellite instability as a marker of prognosis and response to therapy: a meta-analysis of colorectal cancer survival data. Eur J Cancer. 2010;46(15):2788–2798. doi: 10.1016/j.ejca.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 34.Venderbosch S, Nagtegaal ID, Maughan TS, Smith CG, Cheadle JP, Fisher D, Kaplan R, Quirke P, Seymour MT, Richman SD, et al. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: a pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS studies. Clin Cancer Res. 2014;20(20):5322–5330. doi: 10.1158/1078-0432.CCR-14-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seppala TT, Bohm JP, Friman M, Lahtinen L, Vayrynen VM, Liipo TK, Ristimaki AP, Kairaluoma MV, Kellokumpu IH, Kuopio TH, et al. Combination of microsatellite instability and BRAF mutation status for subtyping colorectal cancer. Br J Cancer. 2015;112(12):1966–1975. doi: 10.1038/bjc.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phipps AI, Passarelli MN, Chan AT, Harrison TA, Jeon J, Hutter CM, Berndt SI, Brenner H, Caan BJ, Campbell PT, et al. Common genetic variation and survival after colorectal cancer diagnosis: a genome-wide analysis. Carcinogenesis. 2016;37(1):87–95. doi: 10.1093/carcin/bgv161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia M, Jansen L, Walter V, Tagscherer K, Roth W, Herpel E, Kloor M, Blaker H, Chang-Claude J, Brenner H, et al. No association of CpG island methylator phenotype and colorectal cancer survival: population-based study. Br J Cancer. 2016;115(11):1359–1366. doi: 10.1038/bjc.2016.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmeister M, Jansen L, Rudolph A, Toth C, Kloor M, Roth W, Blaker H, Chang-Claude J, Brenner H. Statin use and survival after colorectal cancer: the importance of comprehensive confounder adjustment. J Natl Cancer Inst. 2015;107(6):djv045. doi: 10.1093/jnci/djv045. [DOI] [PubMed] [Google Scholar]
- 39.Brenner H, Chang-Claude J, Jansen L, Knebel P, Stock C, Hoffmeister M. Reduced risk of colorectal cancer up to 10 years after screening, surveillance, or diagnostic colonoscopy. Gastroenterology. 2014;146(3):709–717. doi: 10.1053/j.gastro.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 40.Warth A, Kloor M, Schirmacher P, Blaker H. Genetics and epigenetics of small bowel adenocarcinoma: the interactions of CIN, MSI, and CIMP. Mod Pathol. 2011;24(4):564–570. doi: 10.1038/modpathol.2010.223. [DOI] [PubMed] [Google Scholar]
- 41.Pidsley R, CC YW, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Findeisen P, Kloor M, Merx S, Sutter C, Woerner SM, Dostmann N, Benner A, Dondog B, Pawlita M, Dippold W, et al. T25 repeat in the 3’ untranslated region of the CASP2 gene: a sensitive and specific marker for microsatellite instability in colorectal cancer. Cancer Res. 2005;65(18):8072–8078. doi: 10.1158/0008-5472.CAN-04-4146. [DOI] [PubMed] [Google Scholar]
- 43.Blaker H, Helmchen B, Bonisch A, Aulmann S, Penzel R, Otto HF, Rieker RJ. Mutational activation of the RAS-RAF-MAPK and the Wnt pathway in small intestinal adenocarcinomas. Scand J Gastroenterol. 2004;39(8):748–753. doi: 10.1080/00365520410005847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Characteristics of patients in the study cohort and the validation cohort. Table S2. Genes used to define CIMP status in previous studies that were available on the 450k methylation array. Table S3. CpG sites identified in the study cohort that were included or excluded in the final analyses. Table S4. Association of the coefficient score with characteristics of colorectal cancer patients in the study cohort and the validation cohort. Figure S1. Heatmaps showing the methylation level of the seven CpG sites across all the patients in the study cohort (A) and the validation cohort (B), respectively. Figure S2. Unadjusted Kaplan–Meier curves of the association of the coefficient score with disease-specific survival and non-disease-specific survival in the validation cohort. (DOCX 2549 kb)
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.