Abstract
Emerging evidence suggests that circular RNAs (circRNAs) are linked to the development and progression of human cancers. Nevertheless, their contribution to breast cancer (BC) is still largely unknown. In the current study, we screened and identified a novel circRNA, circ-UBE2D2, which was highly expressed in BC cell lines and tissues and was closely related to aggressive clinical features and dismal prognosis. Small interfering RNA (siRNA)–mediated circ-UBE2D2 silencing notably inhibited the proliferation, migration and invasion of BC cells, whereas circ-UBE2D2 overexpression displayed opposite effects. Mechanistically, circ-UBE2D2 was able to simultaneously function as molecular sponges of miR-1236 and miR-1287 to regulate the expression of their respective target genes. Moreover, circ-UBE2D2–induced tumor-promoting effects could be effectively blocked by miR-1236 or miR-1287 in BC cells. More importantly, therapeutic delivery of cholesterol-conjugated si-circ-UBE2D2 oligonucleotides significantly delayed tumor growth in vivo. Overall, our findings indicate that circ-UBE2D2 plays an essential oncogenic role in BC, and targeting circ-UBE2D2 may be a feasible treatment for BC patients.
Introduction
As one of the most common malignancies worldwide, breast cancer (BC) is deemed to be a life-threatening disease and the leading cause of cancer-associated deaths in females (11.6%) [1]. Despite the striking breakthrough in the diagnosis and treatment of BC in the past 20 years, the 5-year survival rate of BC patients is still very dismal, mainly because of its heterogeneity and the fact that most patients are in the advanced clinical stage at the time of diagnosis [2], [3]. Therefore, an in-depth exploration of the pathogenesis of BC is strongly needed and will provide novel targets for the clinical diagnosis and treatment, thereby improving the prognosis of patients with BC.
With the completion of the Human Genome Project, scientists have surprisingly found that only about 2% of the genes in the human genome are capable of translating proteins, while the remaining 98% of genes are noncoding RNAs (ncRNAs) [4]. A large number of ncRNAs have long been considered as by-products of genomic transcription that have no definitive biological functions [5]. However, this is not the case. Recently, multiple lines of evidence showed that ncRNAs are widely involved in the regulation of various important life activities, including the occurrence and development of cancers [6]. As a member of ncRNA, circular RNA (circRNA) is mainly derived from gene exons (accounting for 80%) with a covalently closed loop structure [7]. CircRNA has different biological functions, of which the most studied is as a molecular sponge of microRNAs (miRNAs), namely, competitive endogenous RNA (ceRNA) mechanism [8]. The most prominent is CDR1as, which was identified to have more than 60 conserved binding sites of the ancient miR-7 [9], [10]. Subsequently, a wealth of studies suggests that circRNAs can act as a ceRNA to control tumorigenesis. For example, circ-BACH2 [11], circ-FNDC3B [12], circ-VANGL1 [13], circ-HIPK3 [14], circ-FAT1 [15], and circ-EIF3I [16] were proposed to participate in the development and progression of papillary thyroid carcinoma, gastric cancer, bladder cancer, colorectal cancer, osteosarcoma, and non–small cell lung cancer by sponging miR-139-5p, miR-515-5p, miR-605-3p, miR-7, miR-375, and miR-361-3p/miR-615-5p, respectively. However, the ceRNA role of circRNA in BC remains largely unclear.
In this study, we enrolled the Gene Expression Omnibus (GEO) database (GSE101124) and found a large number of dysregulated circRNAs in BC. Moreover, we identified that circ-UBE2D2 was significantly overexpressed in BC cell lines and tissues, and also characterized the clinical significance and potential molecular mechanism of circ-UBE2D2 in BC.
Material and Methods
Clinical BC Samples Collection
In the present study, we enrolled 80 patients who were diagnosed with BC and underwent surgical resection in the first affiliated hospital of Zhengzhou University. Before collecting specimens, the written informed consents were obtained from all patients. None of them received preoperative radiotherapy or chemotherapy. A total of 80 pairs of BC and adjacent normal tissues were collected, which were immediately placed into liquid nitrogen to protect RNA from degradation. The detailed clinicopathological data of the patients were presented in Table 1. All procedures in this study were approved by the ethics committee of the first affiliated hospital of Zhengzhou University.
Table 1.
The Correlation between circ-UBE2D2 Expression and Clinicopathological Features in 80 Patients with Breast Cancer
| Parameters | All Cases | circ-UBE2D2 Expression |
P Value | |
|---|---|---|---|---|
| Low | High | |||
| Age (years) | ||||
| ≤40 | 21 | 9 | 12 | .446 |
| > 40 | 59 | 31 | 28 | |
| Menopause | ||||
| Yes | 48 | 22 | 26 | .361 |
| No | 32 | 18 | 14 | |
| Tumor size | ||||
| ≤2 cm | 27 | 18 | 9 | .033 |
| >2 cm | 53 | 22 | 31 | |
| Lymph node metastasis | ||||
| No | 28 | 20 | 8 | .005 |
| Yes | 52 | 20 | 32 | |
| Clinical TNM stage | ||||
| I-II | 45 | 30 | 15 | .001 |
| III-IV | 35 | 10 | 25 | |
| Differentiation | ||||
| Well/moderate | 50 | 30 | 20 | .021 |
| Poor | 30 | 10 | 20 | |
Cell Lines and Transfection
All human BC cell lines including MCF-7, T47D, BT-474, BT-20, and MDA-MB-231 and normal breast epithelial MCF-10A cells were obtained from Chinese Academy of Sciences (Shanghai, China) and maintained in RPMI1640 or DMEM complete medium with 10% fetal bovine serum as appropriate. To knock down and overexpress circ-UBE2D2, the designed si-RNAs targeting the junction sequence of circ-UBE2D2 (Gene-Pharma, Shanghai, China) and pCD-ciR vector (Geneseed, Guangzhou, China) were obtained and transfected into BT-474 and MDA-MB-231 cells using Lipofectamine 2000 (Invitrogen, MA), respectively. Besides, miR-1236 and miR-1287 inhibitors and mimics (Gene-Pharma) were purchased to silence and ectopically express miR-1236 and miR-1287, respectively.
Quantitative Reverse Transcription PCR (qRT-PCR)
Total RNA in BC cell lines and tissues was isolated with Trizol reagent (Invitrogen), and its integrity and quantity were determined by agarose gel electrophoresis and NanoDrop1000 Spectrophotometer (NanoDrop, DE), respectively. Next, a total of 2 μg above RNA was transcribed into cDNA by using All-in-One First-Strand cDNA Synthesis Kit (GeneCopoeia, MD), followed by quantification by BlazeTaq SYBR Green qPCR mix 2.0 kit (GeneCopoeia) with 40 cycles. The expression levels of circRNAs/mRNAs and miRNAs were normalized to GAPDH and U6, respectively. All experiments were carried out in triplicate. The relative RNA expression was calculated using the 2−△△Ct method. The primers used in this study were listed in Table 2.
Table 2.
The Primer Sequences for qRT-PCR Analysis
| Gene | Direction | Sequence |
|---|---|---|
| Circ-UBE2D2 | Forward | AATGGCAGCATTTGTCTTGA |
| Reverse | GCCCCTGTGAGTAAGCTACG | |
| miR-1236 | Forward | CAGTGAGTGACAGGGGAA |
| Reverse | CCAGTTTTTTTTTTTTTTTCCCCAT | |
| miR-1287 | Forward | GTGCTGGATCAGTGGTTC |
| Reverse | GTCCAGTTTTTTTTTTTTTTTGACTC | |
| HOXB7 | Forward | CGTCCCTGCCTACAAATCAT |
| Reverse | GCAAACGCACAAGAAGTTTG | |
| MTA2 | Forward | TTTCACGCCATGGATACCTT |
| Reverse | AGGGCCTCCTCAAATAGCAT | |
| ZEB1 | Forward | CACCAGATGCATTTTCACAA |
| Reverse | GTGTAACTGCACAGGGAGCA | |
| KLF8 | Forward | GGTGTCCACGTCAACATCTG |
| Reverse | CATGGGCAGAGACTGCACTA | |
| AFP | Forward | GCCTTTCTGGAAGAACTTTGC |
| Reverse | CTGGAGTGGGCTTTTTGTGT | |
| p21 | Forward | GACACCACTGGAGGGTGACT |
| Reverse | CAGGTCCACATGGTCTTCCT | |
| PIK3CB | Forward | ACCCAATGTTCAACCTCCTT |
| Reverse | CCCTGGGTCACAACTTCTTG | |
| ANGPT1 | Forward | GAGAACCTTCAAGGCTTGGTT |
| Reverse | GACTGTGTCCATCAGCTCCA | |
| GAGE1 | Forward | GAACCAGCAACTCAACGTCA |
| Reverse | TTCACCTCCTCTGGATTTGG | |
| CD105 | Forward | CATCCGTCTCCGAGTTCCT |
| Reverse | ACAGCAGGCTCACACAGTTG | |
| ATF6α | Forward | GGGAGTCACACAGCTCCCTA |
| Reverse | TGAACCGACATTTCTCATGG | |
| EGFR | Forward | TTACGCTCCCTCAAGGAGAT |
| Reverse | TGCAGCTGTTTTCACCTCTG | |
| GAPDH | Forward | ACCCAGAAGACTGTGGATGG |
| Reverse | TTCAGCTCAGGGATGACCTT | |
| U6 | Forward | CTCGCTTCGGCAGCACA |
| Reverse | AACGCTTCACGAATTTGCGT |
Cell Proliferation Assay
Cell viability was assessed by cell counting kit-8 (CCK-8) assay. Concretely, the transfected BT-474 and MDA-MB-231 cells were plated into 96-well plates and conventionally cultured for 24, 48, and 72 hours. Then, each well was treated with 10 μl of CCK-8 reagent (Dojindo, Kumamoto, Japan) for 3 hours at 37°C. The absorbance at 450 nm was recorded by a microplate spectrophotometer.
Cell Migration and Invasion Assays
The Transwell chambers (Corning, NY) coated without and with Matrigel (Becton Dickinson, CA) were employed to test the migratory and invasive capacities of BC cells, respectively. Briefly, 100 μl of treated BT-474 and MDA-MB-231 cells was seeded into the chambers upon 24-well plates at a density of 2.5 × 105 cells per well, along with 600 μl of the complete medium underneath the chambers. After 18 hours, the cells that did not migrate and invade into the lower surface of chambers were wiped off by a cotton swab, while the migrated and invaded cells on the lower surface were fixed with methanol for 5 minutes and stained with Giemsa for 10 minutes, and finally the number of stained cells in four random fields under a light microscope was counted.
Cell Apoptosis Assay
The evaluation of cell apoptosis was conducted by Annexin V-PE/7-AAD kit (Becton Dickinson) based on manufacturer’s manual. In brief, the transfected BT-474 and MDA-MB-231 cells were digested with trypsinase without EDTA and washed with PBS three times. Next, 500 μl of binding buffer was added to resuspend cells, followed by dark incubation with 5 μl of Annexin V-PE and 7-AAD solution on ice for 10 minutes. Lastly, the number of apoptotic cells was analyzed by flow cytometry (Becton Dickinson).
In Vivo Tumorigenicity Assay and Immunohistochemistry (IHC)
A total of 15 5-week-old male BALB/c nude mice were purchased from Chinese Academy of Sciences (Shanghai) and randomly divided into 3 groups with 5 mice in each group. Then, the nude mice were subcutaneously injected with 1 × 107 MDA-MB-231 cells and routinely cultured until the tumors were visible (about 7 days), followed by intratumorally injection with cholesterol-conjugated oligonucleotides (si-NC, si-circ-UBE2D2#1, and si-circ-UBE2D2#2) 3 times a week for 2 weeks. Thirty-five days later, the subcutaneous tumors of each mice were obtained and weighed, followed by IHC staining for Ki-67 (#ab209897, Abcam). The protocol of IHC staining was described previously [17].
Dual-Luciferase Reporter Assay
To determine the probability of the binding between circ-UBE2D2 and miR-1236/miR-1287, the full length of circ-UBE2D2 with wild-type (Luc-circ-UBE2D2-WT) or mutant (Luc-circ-UBE2D2-Mut) predicted miR-1236/miR-1287 binding sequence was synthesized and subcloned into pEZX-MT06 vector (GeneCopoeia) to perform luciferase reporter assay. BT-474 and MDA-MB-231 cells with 60%-70% confluency were treated with above luciferase reporter vector and miR-1236/miR-1287 or control mimics using Lipofectamine 2000 (Invitrogen) for 48 hours. The luciferase activity of each group was tested by Luc-Pair Duo-Luciferase Assay Kit 2.0 (GeneCopoeia) with GloMax 20/20 luminescence detector (Promega, WI).
Statistical Analysis
The statistical data in our study were all analyzed by IBM SPSS 24.0 software and were displayed as mean ± standard deviation (SD) representing at least three effective independent repeated experiments. Differences between groups were tested by Student’s t or one-way analysis as appropriate. Kaplan-Meier plot was utilized to assess the survival time of BC patients with circ-UBE2D2 low or high expression. *P < .05, **P < .01, ***P < .001.
Results
Circ-UBE2D2 in clinical samples
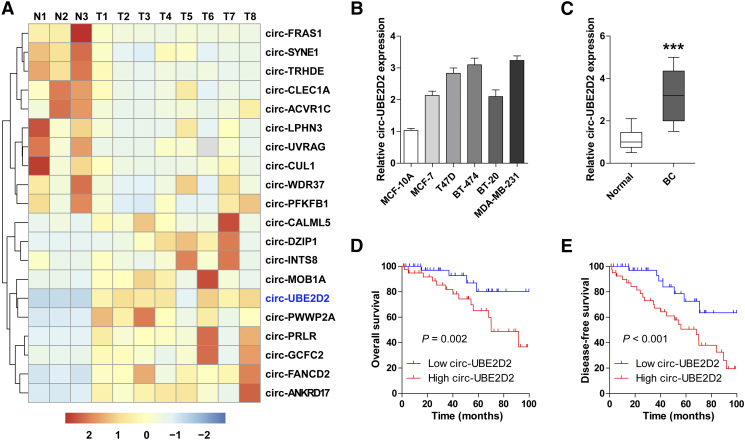
To identify the aberrant circRNAs in BC, we enrolled and analyzed the GEO database (GSE101124) containing eight BC and three normal tissues. As shown in Figure 1A, circ-UBE2D2 displayed the largest differential expression change between BC and normal tissues. We then collected BC tissues and cell lines to confirm this finding; the qRT-PCR results showed that circ-UBE2D2 was remarkably overexpressed in BC cell lines as well as tissues when compared with the corresponding normal controls (Figure 1, B and C). Next, BC patients were divided into two groups with high and low circ-UBE2D2 expression according to the median expression value of circ-UBE2D2. We found that BC patients with high circ-UBE2D2 expression exhibited larger tumor size, lymph node metastasis, advanced clinical TNM stage, and poor differentiation than those with low circ-UBE2D2 expression (Table 1). Moreover, shorter overall and disease-free survival times were observed in the high–circ-UBE2D2 expression group as compared with the low–circ-UBE2D2 expression group, as demonstrated by Kaplan-Meier plot (Fig. 1, D and E). Taken together, these results suggest that circ-UBE2D2 is highly expressed in BC and linked to poor prognosis.
Fig. 1.
Circ-UBE2D2 is frequently increased in BC cell lines and tissues. (A) The hierarchical clustering map showing the top 10 upregulated and downregulated circRNAs in 8 BC and 3 normal tissues (GSE101124). (B, C) qRT-PCR analysis of the expression of circ-UBE2D2 in BC cell lines (B) and tissues (C). (D, E) The overall (D) and disease-free (E) survival curves of BC patients with low and high circ-UBE2D2 expression. ***P < .001.
Knockdown of circ-UBE2D2 Suppresses BC Cell Proliferation, Migration, and Invasion In Vitro as well as Tumor Growth In Vivo
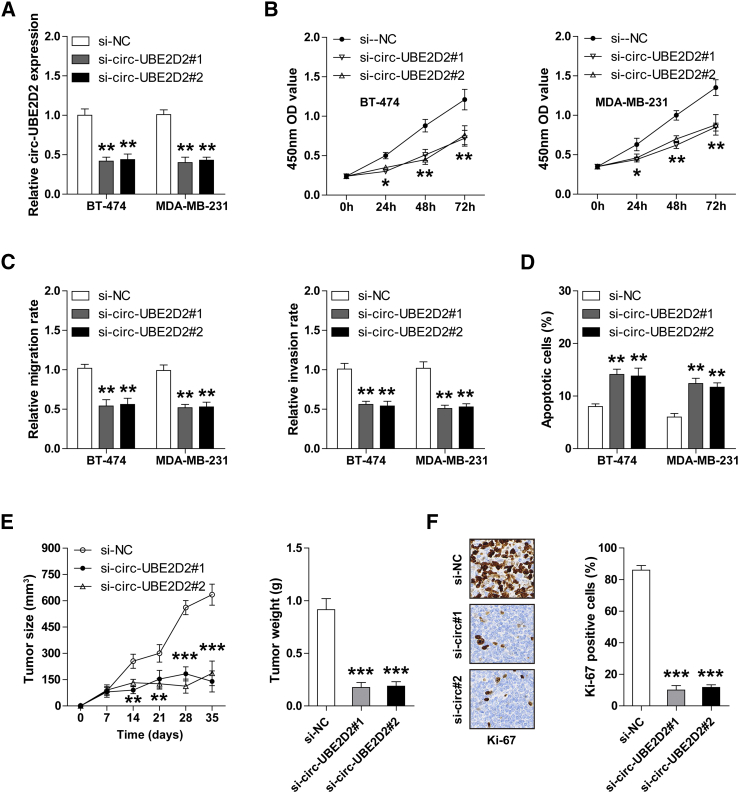
We designed two siRNAs targeting circ-UBE2D2 (si-circ-UBE2D2#1 and si-circ-UBE2D2#2), and the qRT-PCR results revealed that both of them could effectively reduce the expression level of circ-UBE2D2 in BT-474 and MDA-MB-231 cells (Figure 2A). Then, we assessed the proliferative, migratory, and invasive capability of BC cells after circ-UBE2D2 silencing. The CCK-8 results showed that circ-UBE2D2-knockdown BT-474 and MDA-MB-231 cells displayed slower proliferation rate compared to control cells (Figure 2B). Similarly, depletion of circ-UBE2D2 attenuated the ability of BT-474 and MDA-MB-231 cells to migrate and invade (Figure 2C). In addition, the number of apoptotic cells increased after circ-UBE2D2 knockdown (Figure 2D).
Fig. 2.
Depletion of circ-UBE2D2 significantly inhibits the aggressive phenotype of BC cells in vitro and delays tumor growth in vivo. (A) qRT-PCR analysis of circ-UBE2D2 expression in BT-474 and MDA-MB-231 cells transfected with si-NC, si-circ-UBE2D2#1, and si-circ-UBE2D2#2. (B-D) CCK-8 (B), Transwell migration/invasion (C), and flow cytometric apoptotic (D) assays in BT-474 and MDA-MB-231 cells after circ-UBE2D2 knockdown. (E) The volume and weight of subcutaneous tumors in nude mice in the indicated groups (si-NC, si-circ-UBE2D2#1, and si-circ-UBE2D2#2). (F) IHC staining with Ki-67 antibody in subcutaneous tumors in nude mice in the indicated groups (si-NC, si-circ-UBE2D2#1, and si-circ-UBE2D2#2). NC = negative control. *P < .05, **P < .01, ***P < .001.
To explore whether circ-UBE2D2 also functioned in vivo, we established the xenograft tumor model and intratumorally injected cholesterol-conjugated si-circ-UBE2D2 and control oligonucleotides. As expected, smaller tumor volume and weight were observed in the two circ-UBE2D2-knockdown groups than those in the control group (Figure 2E). Besides, this result was also confirmed by IHC staining for Ki-67 proliferation index (Figure 2F).
Overall, these in vitro and in vivo findings indicate that circ-UBE2D2 is an oncogene in BC and targeting it may effectively retard BC aggressive progression.
Circ-UBE2D2 Functions as a Sponge of miR-1236 and miR-1287
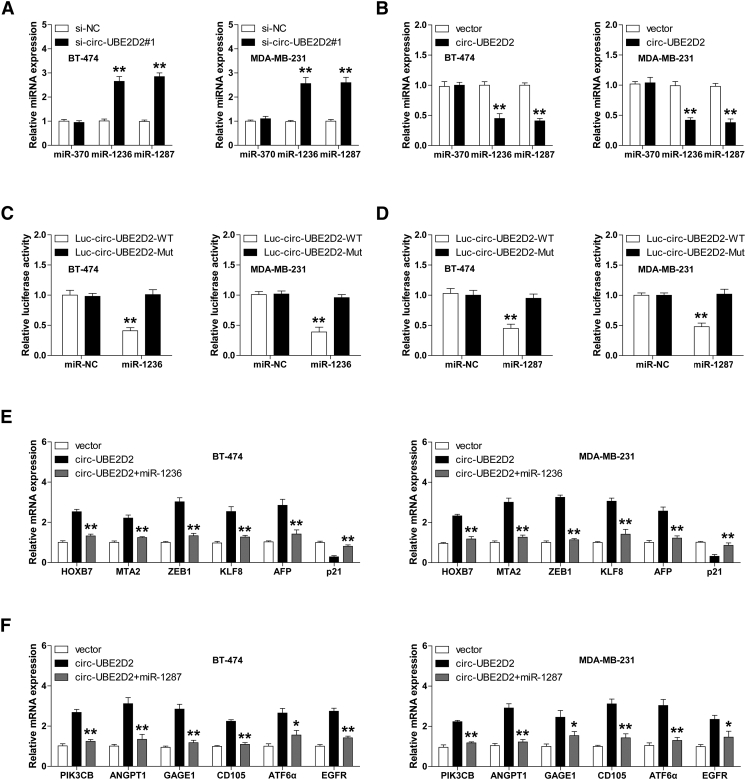
Numerous studies have shown that circRNA is able to sponge miRNAs to inhibit its activity. To identify the circ-UBE2D2-binding miRNAs, we analyzed the online CIRCinteractome database (https://circinteractome.nia.nih.gov) and found that there are two predicted binding sequences of miR-370, miR-1236 and miR-1287 on circ-UBE2D2, respectively. We then manipulated circ-UBE2D2 expression to test whether it could alter the expression levels of these three miRNAs in BT-474 and MDA-MB-231 cells. The qRT-PCR results showed that circ-UBE2D2 silencing and overexpression, respectively, increased and decreased the expression of miR-1236 and miR-1287, but not miR-370 (Figure 3, A and B). Furthermore, the luciferase reporter assay was carried out to evaluate the interaction between circ-UBE2D2 and miR-1236/miR-1287. As shown in Figure 3, C and D, overexpression of miR-1236 or miR-1287 significantly inhibited the luciferase activity of wild-type circ-UBE2D2 vector but did not affect that of mutant one in BT-474 and MDA-MB-231 cells. In addition, we also examined the expression levels of downstream target genes of miR-1236 or miR-1287. The qRT-PCR and Western blot results displayed that ectopic expression of circ-UBE2D2 notably upregulated the expression of HOXB7, MTA2, ZEB1, KLF8, and AFP, whereas it downregulated p21 expression; these genes were all targeted by miR-1236 according to previous studies, and miR-1236 overexpression could evidently block the above effect caused by circ-UBE2D2 (Figure 3E, Supplementary Figure 1). Likewise, the miR-1287 target genes’ expression levels including PIK3CB, ANGPT1, GAGE1, CD105, ATF6α, and EGFR were all uniformly increased after circ-UBE2D2 overexpression, and this increase was significantly abolished by miR-1287 overexpression in BT-474 and MDA-MB-231 cells (Figure 3F, Supplementary Figure 1). Collectively, these results demonstrate that circ-UBE2D2 harbors miR-1236/miR-1287 binding capacity in BC cells.
Fig. 3.
Circ-UBE2D2 functions as a sponge of miR-1236 and miR-1287. (A, B) qRT-PCR analysis of the expression of miR-370, miR-1236, and miR-1287 in circ-UBE2D2-knockdown (A)/overexpressing (B) BT-474 and MDA-MB-231 cells. (C, D) The luciferase activity of BT-474 and MDA-MB-231 cells cotransfected with wild-type or mutant circ-UBE2D2 luciferase vector and control or miR-1236 (C)/miR-1287 (D) mimics. (E, F) qRT-PCR analysis of the expression of reported miR-1236 (E)/miR-1287 (F) target genes in circ-UBE2D2–overexpressing BT-474 and MDA-MB-231 cells transfected with or without miR-1236/miR-1287. NC = negative control. *P < .05, **P < .01.
Manipulating the Expression of miR-1236 or miR-1287 Effectively Reverses circ-UBE2D2–Induced Effects
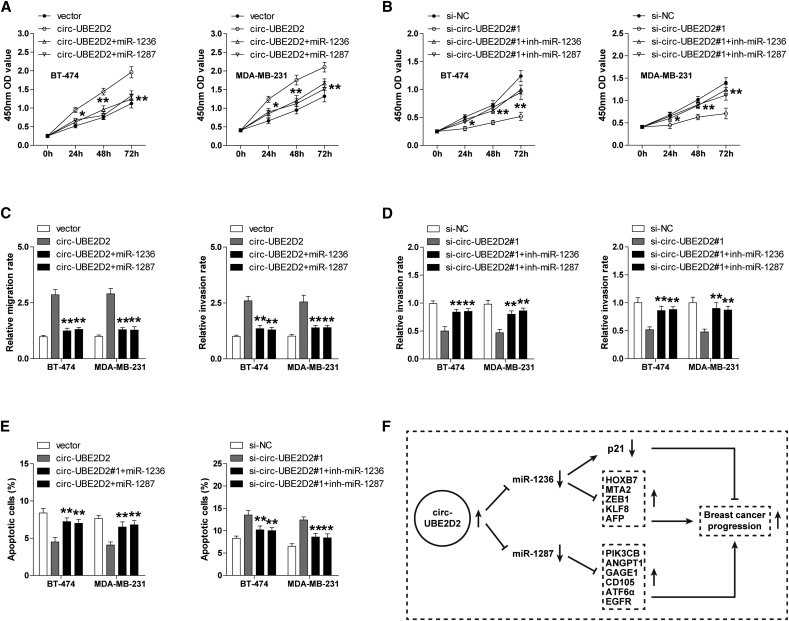
Next, functional experiments were conducted to assess whether miR-1236 and miR-1287 mediated the tumor-promoting effect of circ-UBE2D2 in BC. The results showed that enforced expression of circ-UBE2D2 markedly promoted the proliferation, migration, and invasion of BT-474 and MDA-MB-231 cells and inhibited cell apoptosis (Figure 4, A, C, and E); however, these effects were counteracted by miR-1236 or miR-1287 overexpression (Figure 4, A, C, and E). Similarly, the tumor-inhibiting effect caused by circ-UBE2D2 silencing could be significantly abrogated by miR-1236 or miR-1287 knockdown (Figure 4, B, D, and E). Altogether, these data suggest that circ-UBE2D2 exerts the carcinogenic role, at least partially, through miR-1236 and miR-1287 in BC.
Fig. 4.
miR-1236 or miR-1287 reverses the effects induced by circ-UBE2D2 in BC cells. (A, B) CCK-8 assay in circ-UBE2D2–overexpressing (A)/knockdown (B) BT-474 and MDA-MB-231 cells transfected with miR-1236/miR-1287 mimics (A) or inhibitors (B). (C, D) Transwell migration/invasion assay in circ-UBE2D2–overexpressing (C)/knockdown (D) BT-474 and MDA-MB-231 cells transfected with miR-1236/miR-1287 mimics (C) or inhibitors (D). (E) Cell apoptosis assay with flow cytometry in circ-UBE2D2–overexpressing/knockdown BT-474 and MDA-MB-231 cells transfected with miR-1236/miR-1287 mimics or inhibitors. (F) The schematic diagram of circ-UBE2D2 as a molecular sponge of miR-1236 and miR-1287 to accelerate the progression of BC.
Discussion
CircRNA, as a special class of noncoding RNA, is emerging as a key regulator in cancer progression. In the current study, by analyzing the GEO database (GSE101124), we found a novel circRNA circ-UBE2D2 that was significantly upregulated in eight BC tissues as compared with three normal tissues, which was further validated by qRT-PCR in the collected 80 pairs of BC and paracancerous tissues. Moreover, the upregulation of circ-UBE2D2 was closely associated with advanced clinicopathological features and poor prognosis. The functional experiments showed that manipulation of circ-UBE2D2 expression affected the biological phenotypes of BC cells, including proliferation, apoptosis, migration and invasion. Further mechanism studies indicated that circ-UBE2D2 exerted the tumor-promoting effect mainly through acting as a ceRNA via competitively binding with miR-1236 and miR-1287 (Figure 4F). Therefore, our findings highlight the biological relevance of circRNA in BC, and the crucial oncogenic role of circ-UBE2D2 suggests its potential as a molecular targeted therapy for patients with BC.
Recent studies show that circRNAs are abundant in eukaryotes and dysregulated in many human diseases, including cancer [18]. And some dysregulated circRNAs have been reported to engage BC tumorigenesis by functioning as tumor suppressors or oncogenes. For example, circ-TADA2As [19], circ-ASS1 [20], circ-LIFR [21], and circ-ITCH [22] were proposed to be dramatically decreased in BC tissues and cell lines and restrained the proliferation and metastasis of BC by acting as tumor suppressors. On the contrary, circ-EPSTI1 [23], circ-AGFG1 [24], circ-BARD1 [25], circ-UBAP2 [26], and circ-ANKS1B [27] were shown to be highly expressed in BC and contributed to BC aggressive progression by serving as oncogenes. These above studies reveal that circRNAs are the pivotal regulators of BC. Herein, by data mining of GSE101124 database, we also found a large number of aberrantly expressed circRNAs in BC and further characterized the important role of circ-UBE2D2 in promoting BC progression. Whether other dysregulated circRNAs in our study, such as circ-FRAS1, circ-ANKRD17, and circ-DZIP1, are also involved in the carcinogenic process of BC and whether there is some cross talk between these reported dysregulated BC-related circRNAs are worth further investigation.
Accumulating evidence demonstrates that circRNAs harbor different biological functions, including interaction with proteins, regulation of host gene expression, adsorption of miRNAs, and translation of proteins, etc. [28]. Among them, the most studied is the ceRNA mechanism, that is, circRNAs act as a sponge of miRNAs to attenuate the inhibitory effect of miRNAs on their target genes [29]. Likewise, in this study, we identified that miR-1236 and miR-1287 were the direct targets of circ-UBE2D2, and circ-UBE2D2 could alter the expression of miR-1236/miR-1287 downstream target genes by abundantly sponging miR-1236 and miR-1287 in BC. miR-1236 and miR-1287 were both proved as the tumor suppressors by targeting the different genes in various cancers. Concretely, miR-1236 could directly interact with the 3′-untranslated region of oncogenes such as HOXB7, MTA2, ZEB1, KLF8, and AFP to inhibit their expression [30], [31], [32]. In addition, miR-1236 could also bind to the promoter of the tumor suppressor p21 to elevate its expression [33]. And the oncogenes including PIK3CB, ANGPT1, GAGE1, CD105, ATF6α, and EGFR were identified as the targets of miR-1287 [34], [35], [36], [37]. Consistently, we found that circ-UBE2D2 respectively upregulated and downregulated the expression of above-mentioned oncogenes and tumor suppressors by reducing the activity of miR-1236 and miR-1287, and the tumor-promoting effects caused by circ-UBE2D2 were abolished by miR-1236 and miR-1287, implying that miR-1236 and miR-1287 are the key mediators of circ-UBE2D2 in BC. Further research is needed to explore the role of this circ-UBE2D2/miR-1236/ miR-1287 axis in other malignancies.
It is noteworthy that siRNA-based therapeutic approaches have achieved tremendous success in the treatment of cancer patients [38]. Here, we found that RNA delivery of cholesterol-conjugated si-circ-UBE2D2 oligonucleotides could significantly suppress tumor growth in vivo, suggesting that patients with BC may also benefit from treatments that target circ-UBE2D2. Future studies are required to overcome the challenges such as off-target effect, specificity, and uptake rate before it being applied to the clinical treatment [38].
In summary, our data convincingly indicate that circ-UBE2D2 functions as an oncogene by sponging and inhibiting the tumor suppressor miR-1236 and miR-1287 in BC, meanwhile providing a promising prognostic biomarker and druggable target for BC patients.
Disclosure Statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by grant from National Natural Science Foundation of China (no. 81272371), the Major Project of Science and Technology in Henan Province (no. 161100311400), and the Outstanding Talents in Science and Technology Innovation in Henan Province (184200510013).
Footnotes
Circ-UBE2D2 in Breast Cancer.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ponde NF, Zardavas D, Piccart M. Progress in adjuvant systemic therapy for breast cancer. Nat Rev Clin Oncol. 2019;16:27–44. doi: 10.1038/s41571-018-0089-9. [DOI] [PubMed] [Google Scholar]
- 3.McKenna MT, Weis JA, Brock A, Quaranta V, Yankeelov TE. Precision medicine with imprecise therapy: computational modeling for chemotherapy in breast cancer. Transl Oncol. 2018;11:732–742. doi: 10.1016/j.tranon.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeke JD, Church G, Hessel A, Kelley NJ, Arkin A, Cai Y, Carlson R, Chakravarti A, Cornish VW, Holt L. Genome engineering. The genome project-write. Science. 2016;353:126–127. doi: 10.1126/science.aaf6850. [DOI] [PubMed] [Google Scholar]
- 5.Kaikkonen MU, Adelman K. Emerging roles of non-coding RNA transcription. Trends Biochem Sci. 2018;43:654–667. doi: 10.1016/j.tibs.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Carey KT, Wickramasinghe VO. Regulatory potential of the RNA processing machinery: implications for human disease. Trends Genet. 2018;34:279–290. doi: 10.1016/j.tig.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 7.Wilusz JE. A 360 degrees view of circular RNAs: from biogenesis to functions. Wiley Interdiscip Rev RNA. 2018;9 doi: 10.1002/wrna.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 10.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 11.Cai X, Zhao Z, Dong J, Lv Q, Yun B, Liu J, Shen Y, Kang J, Li J. Circular RNA circBACH2 plays a role in papillary thyroid carcinoma by sponging miR-139-5p and regulating LMO4 expression. Cell Death Dis. 2019;10:184. doi: 10.1038/s41419-019-1439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Wang X, Huang H, Wang Y, Zhang F, Wang S. Hsa_circ_0067997 promotes the progression of gastric cancer by inhibition of miR-515-5p and activation of X chromosome-linked inhibitor of apoptosis (XIAP) Artif Cells Nanomed Biotechnol. 2019;47:308–318. doi: 10.1080/21691401.2018.1553787. [DOI] [PubMed] [Google Scholar]
- 13.Zeng Z, Zhou W, Duan L, Zhang J, Lu X, Jin L, Yu Y. Circular RNA circ-VANGL1 as a competing endogenous RNA contributes to bladder cancer progression by regulating miR-605-3p/VANGL1 pathway. J Cell Physiol. 2019;234:3887–3896. doi: 10.1002/jcp.27162. [DOI] [PubMed] [Google Scholar]
- 14.Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, Sun H, Pan Y, He B, Wang S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Liu G, Huang K, Jie Z, Wu Y, Chen J, Chen Z, Fang X, Shen S. CircFAT1 sponges miR-375 to promote the expression of Yes-associated protein 1 in osteosarcoma cells. Mol Cancer. 2018;17:170. doi: 10.1186/s12943-018-0917-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Nan A, Zhang N, Jia Y, Li X, Ling Y, Dai J, Zhang S, Yang Q, Yi Y. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non–small cell lung cancer. Mol Cancer. 2019;18:13. doi: 10.1186/s12943-019-0943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Razi SS, Kazemi T, Lotfipour F, Mohammad HA, Shanehbandi D, Hallaj-Nezhadi S, Baradaran B. Gene therapy with IL-12 induced enhanced anti-tumor activity in fibrosarcoma mouse model. Artif Cells Nanomed Biotechnol. 2016;44:1988–1993. doi: 10.3109/21691401.2015.1129618. [DOI] [PubMed] [Google Scholar]
- 18.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu JZ, Shao CC, Wang XJ, Zhao X, Chen JQ, Ouyang YX, Feng J, Zhang F, Huang WH, Ying Q. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10:175. doi: 10.1038/s41419-019-1382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou JC, Xu Z, Zhong SL, Zhang HD, Jiang LH, Chen X, Zhu LP, Li J, Zhou SY, Yang SJ. Circular RNA circASS1 is downregulated in breast cancer cells MDA-MB-231 and suppressed invasion and migration. Epigenomics-Uk. 2019;11:199–213. doi: 10.2217/epi-2017-0167. [DOI] [PubMed] [Google Scholar]
- 21.Yan L, Zheng M, Wang H. Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR-492. Cancer Manag Res. 2019;11:1033–1041. doi: 10.2147/CMAR.S186857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang ST, Liu LB, Li XM, Wang YF, Xie PJ, Li Q, Wang R, Wei Q, Kang YH, Meng R, et al. Circ-ITCH regulates triple-negative breast cancer progression through the Wnt/beta-catenin pathway. Neoplasma 2018. [DOI] [PubMed]
- 23.Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D, Yang L, Wang J, Tang H, Xie X. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics. 2018;8:4003–4015. doi: 10.7150/thno.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang R, Xing L, Zheng X, Sun Y, Wang X, Chen J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer. 2019;18:4. doi: 10.1186/s12943-018-0933-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Zhao J, Zou H, Han C, Ma J, Zhao J, Tang J. Circlular RNA BARD1 (Hsa_circ_0001098) overexpression in breast cancer cells with TCDD treatment could promote cell apoptosis via miR-3942/BARD1 axis. Cell Cycle. 2018;17:2731–2744. doi: 10.1080/15384101.2018.1556058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S, Li Q, Wang Y, Li X, Wang R, Kang Y, Xue X, Meng R, Wei Q, Feng X. Upregulation of circ-UBAP2 predicts poor prognosis and promotes triple-negative breast cancer progression through the miR-661/MTA1 pathway. Biochem Biophys Res Commun. 2018;505:996–1002. doi: 10.1016/j.bbrc.2018.10.026. [DOI] [PubMed] [Google Scholar]
- 27.Zeng K, He B, Yang BB, Xu T, Chen X, Xu M, Liu X, Sun H, Pan Y, Wang S. The pro-metastasis effect of circANKS1B in breast cancer. Mol Cancer. 2018;17:160. doi: 10.1186/s12943-018-0914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu S, Liu Z, Yang X, Zhou J, Yu H, Zhang R, Li H. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018;414:301–309. doi: 10.1016/j.canlet.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 30.Duan X, Liu D, Wang Y, Chen Z. Circular RNA hsa_circ_0074362 promotes glioma cell proliferation, migration, and invasion by attenuating the inhibition of miR-1236-3p on HOXB7 expression. DNA Cell Biol. 2018;37:917–924. doi: 10.1089/dna.2018.4311. [DOI] [PubMed] [Google Scholar]
- 31.An JX, Ma MH, Zhang CD, Shao S, Zhou NM, Dai DQ. miR-1236-3p inhibits invasion and metastasis in gastric cancer by targeting MTA2. Cancer Cell Int. 2018;18:66. doi: 10.1186/s12935-018-0560-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bian T, Jiang D, Liu J, Yuan X, Feng J, Li Q, Zhang Q, Li X, Liu Y, Zhang J. miR-1236-3p suppresses the migration and invasion by targeting KLF8 in lung adenocarcinoma A549 cells. Biochem Biophys Res Commun. 2017;492:461–467. doi: 10.1016/j.bbrc.2017.08.074. [DOI] [PubMed] [Google Scholar]
- 33.cancer Thoma C Kidney. RNA activation in RCC: p21 and miR-1236 are a promising pair. Nat Rev Urol. 2015;12:598. doi: 10.1038/nrurol.2015.259. [DOI] [PubMed] [Google Scholar]
- 34.Schwarzenbacher D, Klec C, Pasculli B, Cerk S, Rinner B, Karbiener M, Ivan C, Barbano R, Ling H, Wulf-Goldenberg A. MiR-1287-5p inhibits triple negative breast cancer growth by interaction with phosphoinositide 3-kinase CB, thereby sensitizing cells for PI3Kinase inhibitors. Breast Cancer Res. 2019;21:20. doi: 10.1186/s13058-019-1104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Hu J, Li L, Cai S, Zhang H, Zhu X, Guan G, Dong X. Upregulated circular RNA circ_0016760 indicates unfavorable prognosis in NSCLC and promotes cell progression through miR-1287/GAGE1 axis. Biochem Biophys Res Commun. 2018;503:2089–2094. doi: 10.1016/j.bbrc.2018.07.164. [DOI] [PubMed] [Google Scholar]
- 36.Ishiy F, Fanganiello RD, Kobayashi GS, Kague E, Kuriki PS, Passos-Bueno MR. CD105 is regulated by hsa-miR-1287 and its expression is inversely correlated with osteopotential in SHED. Bone. 2018;106:112–120. doi: 10.1016/j.bone.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 37.Wolter M, Werner T, Malzkorn B, Reifenberger G. Role of microRNAs located on chromosome arm 10q in malignant gliomas. Brain Pathol. 2016;26:344–358. doi: 10.1111/bpa.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh A, Trivedi P, Jain NK. Advances in siRNA delivery in cancer therapy. Artif Cells Nanomed Biotechnol. 2018;46:274–283. doi: 10.1080/21691401.2017.1307210. [DOI] [PubMed] [Google Scholar]