BACE1 is the rate-limiting enzyme in amyloid-β generation. Zhu et al. report that selective inhibition of matrix metallopeptidase 13 (MMP13) decreases BACE1 levels and amyloid-β load, and improves cognitive performance in a mouse model of Alzheimer’s disease. MMP13 inhibition may have therapeutic potential via translational regulation of BACE1.
Keywords: Alzheimer’s disease, BACE1, CL82198, eIF4B, MMP13
Abstract
MMP13 (matrix metallopeptidase 13) plays a key role in bone metabolism and cancer development, but has no known functions in Alzheimer’s disease. In this study, we used high-throughput small molecule screening in SH-SY5Y cells that stably expressed a luciferase reporter gene driven by the BACE1 (β-site amyloid precursor protein cleaving enzyme 1) promoter, which included a portion of the 5′ untranslated region (5′UTR). We identified that CL82198, a selective inhibitor of MMP13, decreased BACE1 protein levels in cultured neuronal cells. This effect was dependent on PI3K (phosphatidylinositide 3-kinase) signalling, and was unrelated to BACE1 gene transcription and protein degradation. Further, we found that eukaryotic translation initiation factor 4B (eIF4B) played a key role, as the mutation of eIF4B at serine 422 (S422R) or deletion of the BACE1 5′UTR attenuated MMP13-mediated BACE1 regulation. In APPswe/PS1E9 mice, an animal model of Alzheimer’s disease, hippocampal Mmp13 knockdown or intraperitoneal CL82198 administration reduced BACE1 protein levels and the related amyloid-β precursor protein processing, amyloid-β load and eIF4B phosphorylation, whereas spatial and associative learning and memory performances were improved. Collectively, MMP13 inhibition/CL82198 treatment exhibited therapeutic potential for Alzheimer’s disease, via the translational regulation of BACE1.
Introduction
Alzheimer’s disease is a common degenerative disease in the elderly. The aetiology of Alzheimer’s disease is considered as multifactorial and includes oxidative stress, neuroinflammation and synaptic failure (Querfurth and LaFerla, 2010). Histopathological lesions include senile plaques, which are composed mainly of amyloid-β protein, and neurofibrillary tangles due to abnormal phosphorylated tau protein levels (Roland and Helmut, 2009). As amyloid-β remains one of the diagnostic biomarkers that differentiates Alzheimer’s disease from other dementias (Jack et al., 2018), targeting amyloid-β or BACE1 (beta-site amyloid precursor protein cleaving enzyme 1) has become an important therapeutic approach (Graham et al., 2017).
BACE1 is the rate-limiting enzyme for amyloid-β generation (Vassar et al., 2009). Although BACE1 deficiency rescues cognitive impairment in animal studies (Ohno et al., 2004; Hussain et al., 2007; Hu et al., 2018), selective BACE1 inhibitors have failed in clinical trials to date (Hawkes, 2017; Zhu et al., 2018). Evidence has suggested that BACE1 has more than 40 membrane protein substrates and plays an important role in synaptic plasticity and cognitive function (Filser et al., 2015; Zhu et al., 2018); therefore, complete BACE1 inhibition might cause harmful side effects (Menting and Claassen, 2014). Due to the multifactorial aetiology of Alzheimer’s disease, inhibiting BACE1 may still not be enough to halt disease progression (Scheltens et al., 2016). It remains unclear whether BACE1 inhibitors would be ultimately beneficial for patients with Alzheimer’s disease (Graham et al., 2017; Coimbra et al., 2018).
Current ongoing clinical trials are focused on inflammation, neuroprotection and neurotransmitters, among others. Disease-modifying therapy has emerged as an alternative strategy (Cummings et al., 2017). It is worth noting that BACE1 deregulation is closely associated with disease mechanisms (Chami and Checler, 2012). For example, in reactive astrocytes triggered by neuroinflammation, Bace1 transcription is enhanced (Frost and Li, 2017). Oxidative stress markers that contribute to cell death are positively correlated with BACE1 activity (Tamagno et al., 2002; Chami and Checler, 2012). Calcium disturbances also activate Bace1 transcription (Kusakawa et al., 2000; Wen et al., 2008). Interestingly, in Alzheimer’s disease brains, BACE1 protein levels, but not mRNA levels, are significantly increased (Holsinger et al., 2002; Preece et al., 2003; Yang et al., 2003), highlighting the importance of post-transcriptional mechanisms in the pathophysiology of Alzheimer’s disease (Sun et al., 2012). In this respect, identifying the potential mechanisms that regulate BACE1 at the translational level might provide an alternative strategy for understanding and treating Alzheimer’s disease.
MMP13 (matrix metallopeptidase 13, or collagenase 3) is located primarily in the extracellular matrix (ECM) (Huntley, 2012). It has been reported that MMP13 plays a key role in bone metabolism and cartilage homeostasis, as well as in tumour invasion and metastasis (Leeman et al., 2002; Li and Johnson, 2011). The substrates of MMP13 include type I–IV collagen and perlecan (Leeman et al., 2002), which are major components of the basement membrane that play a key role in neurological diseases (Wlodarczyk et al., 2011; Yurchenco, 2011). Early changes in the ECM have been identified in Alzheimer’s disease (Lepelletier et al., 2017), but direct evidence of the involvement of MMP13 is lacking. Descriptive studies have shown that MMP13 is increased in ischaemic brains (Cuadrado et al., 2009) and in microglia treated with amyloid-β (Ito et al., 2007).
The current study offers new insights into the translational regulation of BACE1 through MMP13 signalling and provides evidence that reducing BACE1 protein levels instead of completely inhibiting BACE1 represents an alternative strategy for Alzheimer’s disease treatment.
Materials and methods
Reagents and cell culture
MMP13 inhibitors WAY170523 (Tocris Bioscience), CL82198 (Cayman) and SC205756 (Santa Cruz Biotechnology), FGF/PDGF/VEGF RTK inhibitor 341610 (Merk Millipore), PI3 kinase inhibitors LY294002 (Sigma Aldrich), eIF4E/eIF4G interaction inhibitor 4EGI1 (Selleck), protease inhibitor MG132 (Sigma), the transcriptional inhibitor actinomycin D (ActD, Sigma) and protein biosynthesis inhibitor cycloheximide (CHX, Sigma) were dissolved in dimethyl sulphoxide (DMSO) and lysosomal inhibitor chloroquine (CQ, Sigma) was dissolved in sterilized double-distilled H2O. The ultimate concentration of DMSO was not more than 1:2000. All chemicals were used in the concentrations and treatment time indicated in the figure legends.
Cell lines of HEK293 (human embryonic kidney), SH-SY5Y (human neuroblastoma) and HT22 (mouse hippocampal neuron) were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Gibco) supplemented with 10% foetal bovine serum (FBS, Gibco) and 100 U/ml penicillin and 100 µg/ml streptomycin. HEK293 cells stably expressing amyloid-β precursor protein (HEK-APP) were cultivated in complete DMEM with 200 µg/ml G418. After treatment with CL82198, the conditioned media was collected and cleared by centrifugation at 12 000g for 20 min at 4°C to remove cellular debris.
Primary cortical neurons were prepared from Sprague-Dawley rat embryos at Day 17–18 and treated with 0.25% trypsin for 15 min as described previously (Zha et al., 2017). In brief, digested tissues were dissociated by trituration and plated on a 6-well plate at a density of 1.0 × 106 cells/well, and the cultures were maintained in DMEM for 24 h. Subsequently, the medium was replaced with fresh Neurobasal™ medium (Invitrogen) supplemented with 2% B27 (Invitrogen). Half of the medium was removed and replenished with the fresh Neurobasal™ medium every 2–3 days. Neurons were treated with 0.1, 0.5, 1, 2, 5 and 10 μM SC205756 for 48 h. All procedures were carried out in accordance with the Chongqing Medical University guidelines for the care and use of laboratory animals. All cells were incubated at 37°C and 5% CO2.
Gene constructs and RNA interference
Human pCMV-eIF4B (eukaryotic translation initiation factor 4B) plasmid was purchased from Origene. S422R mutation was made using Mut Express MultiS Fast Mutagenesis Kit (Vazyme). Primer sequences were: F: 5′-GGACAGGAAGGGAGTCATCACAAACTGGGACCTCCACCAC-3′, and R: 5′-GTGATGACTCCCTTCCTGTCCTCGACCGTTCCCGTTCCTGA-3′. Human pcDNA4-BACE1 plasmid containing the entire human BACE1 coding region but did not contain 5′UTR (−5′UTR) was acquired from Dr W.H. Zhou (Chongqing Medical University). pcDNA4-BACE1/+5′UTR containing the 5′UTR of BACE1 (NM_012104) was amplified and cloned into pcDNA4 vector using the following primers: F: 5′-CCGCTCGAGTCCCCAGCCCGCCCGGGAGCT-3′, R: 5′-CCGGAATTCGGTGGGCCCCGGCCTTCGGG-3′. The restriction sites were XhoI and EcoRI (Lammich et al., 2004). All constructs were confirmed by DNA sequencing (Zhu et al., 2012). pCMV6-MMP13 was purchased from Origene. Transient transfections were performed using Lipofectamine™ 2000 (Invitrogen). Lentiviral particles bearing Mmp13 shRNA were purchased from GeneChem. The following Mmp13 shRNA sequences were used: Mmp13 shRNA-1: 5′-GAGCACTACTTGAAATCAT-3′ (Meierjohann et al., 2010) and Mmp13 shRNA-2: 5′-GUGACCUUAUGUUUAUCUU-3′ (NM_008607).
The siRNA oligonucleotides for human BACE1 were synthesized by Shanghai GenePharma Co. Ltd, and the non-targeting control siRNA (NC) was used as the negative control. The following siRNA oligonucleotides were used: siBACE1-1 (5′-UGGACUGCAAGGAGUACAAtt-3′), siBACE1-6 (5′-GACUGUGGCUACAACAUUCtt-3′) (Singer et al., 2005) and NC (5′-UUCUCCGAACGUGUCACGUtt-3′). HEK293 cells were transfected with 60 nM siRNAs for 6 h using Lipofectamine™ 2000 (Invitrogen) according to the manufacturer’s protocol. Forty-eight hours after transfection, inhibition of target genes was confirmed by western blotting.
Luciferase activity assay
HEK293 cells were seeded onto 96-well plates for 24 h before transfection. Cells were then transiently transfected with 200 ng luciferase reporter plasmid bearing 758 bp (−691 to +67) nucleotides of human BACE1 promoter (pGL4.21-BACE1) or empty plasmid pGL4.10 (a negative control, driven by SV40, Promega) using Lipofectamine™ 2000 according to the manufacturer’s instructions. Twenty-four hours after transfection, cells were treated with 5 μM CL82198 or 10 nM WAY170523 for 48 h, and DMSO was used as a negative control. The luciferase activities were measured using a luciferase assay kit (Promega) according to the manufacturer’s instructions and the relative fold changes of luciferase activity were normalized to pGL4.10 as internal standard.
Human brain tissues
Cortical brain samples (grey matter) were obtained from Sydney Brain Bank (Neuroscience Research Australia) and NIH NeuroBiobank (USA) (Supplementary Table 1). The study was approved by the Ethics Committee for Human Research at Chongqing Medical University. All procedures were conducted in accordance with the Declaration of Helsinki.
Animal treatment and protein preparation
All protocols were approved by the Commission of Chongqing Medical University for ethics of experiments on animals and were conducted in accordance with international standards. APPswe/PS1E9 transgenic mice (APP/PS1, B6C3) were obtained from Jackson Laboratory imported by Nanjing University (#004462). Controls were generated from littermates that did not have Alzheimer’s disease phenotype. Six-month-old mice were intraperitoneally injected with CL82198 (10 mg/kg) or saline every other day for 2 months (Wang et al., 2013). For the Mmp13 shRNA experiment, mice at 7 months of age were anaesthetized with 10% chloral hydrate (50 mg/kg, i.p.) before being mounted on a stereotaxic apparatus, and the cannulas (30-gauge stainless steel tubing) were implanted into the hippocampus (anterior-posterior: −2.0 mm, medial-lateral: ±1.5, dorsal-ventral: −2.0 mm). After 7 days recovery, 2 μl lentivirus particles (1.5 × 1012 viral genomes/ml) were injected into the hippocampus bilaterally at a rate of 0.1 μl/min (Hsiao et al., 2014).
Mice were euthanized with isofluorane anaesthesia and then transcardially perfused with ice-cold phosphate-buffered saline (PBS). Brains were rapidly isolated and sonicated in RIPA buffer (Cell Signaling Technology). Soluble extracellular proteins were collected from homogenized lysates after centrifugation at 16 000g for 1 h at 4°C. The supernatants were used for analyses of secreted APP forms (sAPPα/β). Pellet was used for insoluble material extraction using 70% formic acid (Sigma) as described previously (Zhu et al., 2011; Obregon et al., 2012). For total protein extraction, cells or brain tissues were homogenized in ice-cold lysis buffer [50 mM Tris-HCl, pH 8.0, 140 mM NaCl; 1.5 mM MgCl2; 0.5 % NP-40 with complete protease inhibitor cocktail (Roche)]. Samples were sonicated on ice and centrifuged at 16 000g for 10 min. Protein concentration was measured in the supernatant by BCA Protein Assay (Dingguo).
Western blotting assay
Following the sample preparation as described above, protein samples were separated on 8% SDS-PAGE or 16.5% Tris-tricine gels [for C-terminal fragment (CTF)], and were transferred onto 0.45-μm or 0.22-μm (for CTF) PVDF membranes (Millipore). The monoclonal or polyclonal antibodies used were BACE1 (Abcam, 1:1000); APP-full length and β-CTF, α-CTF (A8717, Sigma, 1:3000); sAPPα (6E10, Covance, 1:1000); sAPPβ (Covance, 1:500); ADAM10 (Abcam, 1:1000); phospho-eIF4B (Ser422) (Cell Signaling Technology, 1:1000); eIF4B (Proteintech, 1:2000); MMP13 (Santa Cruz, 1:500); PSEN1 (Proteintech, 1:500); neprilysin (Proteintech, 1000); IDE (Proteintech, 1:1000); phospho-Akt (Ser473) (Proteintech, 1:500); Akt (Proteintech, 1:1000); PI3K (phosphatidylinositide 3-kinase; Proteintech, 1:500); GAPDH (Proteintech, 1:10 000); β-actin (Proteintech, 1:2000). The secondary antibodies were goat anti-rabbit or anti-mouse horseradish peroxidase-labelled antibodies (Proteintech, 1:5000). The membranes were visualized using an ECL reagent (Thermo) and a Fusion FX5 image analysis system (Vilber Lourmat). Relative protein intensities were calculated using Quantity One® software (Bio-Rad).
Quantitative real-time polymerase chain reaction
Total RNA from cells or tissues was extracted using TRIzol® reagent (Invitrogen). First-strand cDNA was generated using 1 μg of total RNA with reverse transcriptase kit (TaKaRa). The mRNA expression of human BACE1 or mouse Mmp13 was detected by quantitative real-time PCR (qPCR) with SYBR® Premix Ex Taq™ II (TaKaRa) on an Eppendorf detection system (Fan et al., 2016). The reaction mixture (20 μl total) consisted of 10 μl 2× SYBR® mix, 8 μl nuclease-free water, 0.5 μl of primers (human BACE1, F: 5′-TCTGTCGGAGGGAGCATGAT-3′, R: 5′-GCAAACGAAGGTTGGTGGT-3′; or mouse Mmp13, F: 5′-TACCATCCTG CGACTCTTGC-3′, R: 5′-TTCACCCACATCAGGCACTC-3′; or human/mouse ACTB: F: 5′-CACGATGGAGGGGCCGGACTCATC-3′, R: 5′-TAAAGACCTCTATGCCAACACAG T-3′) and 1 μl diluted cDNA. The reactions were performed as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 34 s. After the reactions were completed, the temperature was increased from 60°C to 95°C to obtain the melting curve. Actb (β-actin) was used as an internal control.
ELISA
Soluble and insoluble amyloid-β40 and amyloid-β42 or PI3K activity were measured by sandwich ELISA using commercially available kits (Cusabio).
Morris water maze
The Morris water maze test included four platform trials per day for five consecutive days, and a probe trial on the sixth day. In the platform trials, each mouse was allowed 60 s to find the platform. If the mouse could not locate the platform, we guided it to the platform and allowed it to rest there for 15 s. The escape latency was measured. In probe trials, the platform was removed, and each mouse was given 60 s to locate where the platform was originally placed. Performance was video-recorded and analysed by image analysing software (ANY-maze, Stoelting). Data were analysed by two-way ANOVA, followed by Tukey’s post hoc test.
Contextual and cued fear conditioning
Mice were tested in a 3-day paradigm as described previously (Cheng et al., 2014). On the first day, the mice were placed in the training chamber to acclimate for 120 s, then they were presented with a 30 s pure tone (2800 Hz, 85 dB, 30 s) that co-terminated with 2 s foot shock (0.45 mA). This procedure was repeated three times with an intertrial interval of 120 s. On the second day, the mice were returned to the training chamber for 10 min without exposure to either the tone or foot shock, to measure contextual freezing. On the last day, the mice were placed in the training chamber for 120 s, then presented with a 30 s pure tone (2800 Hz) without the 2 s foot shock. Behaviour was recorded by video camera and freezing data were measured using FreezeScan software.
Immunohistochemistry
Hemibrains were drop-fixed in 4% paraformaldehyde. Frozen parasagittal sections (30 mm) from cryopreserved brains were cut using a sliding microtome. Sections were stained with anti-amyloid-β antibody (6E10) or ionized calcium-binding adaptor molecule 1 (Iba1, Proteintech) antibody, developed by DAB immunohistochemistry (Zhongshan Golden Bridge), counterstained with haematoxylin. The total number of 6E10- or Iba1-positive plaques in each section was quantified by ImageJ software and the average number of plaques per section was calculated (8–11 sections/mouse). The total number of plaques in the sections from each mouse were then visually counted at low magnification with a microscope and plaque numbers were calculated (Luo et al., 2016).
Statistical analysis
All data are presented as the mean ± SEM from three independent experiments. The statistical comparisons between the two groups were tested using Student’s t-test. The comparisons among groups were tested using one-way or two-way analysis of variance (ANOVA).
Data availability
The data that support the findings of present study are available from the corresponding author on reasonable request.
Results
CL82198, a specific MMP13 inhibitor, reduces BACE1 protein levels
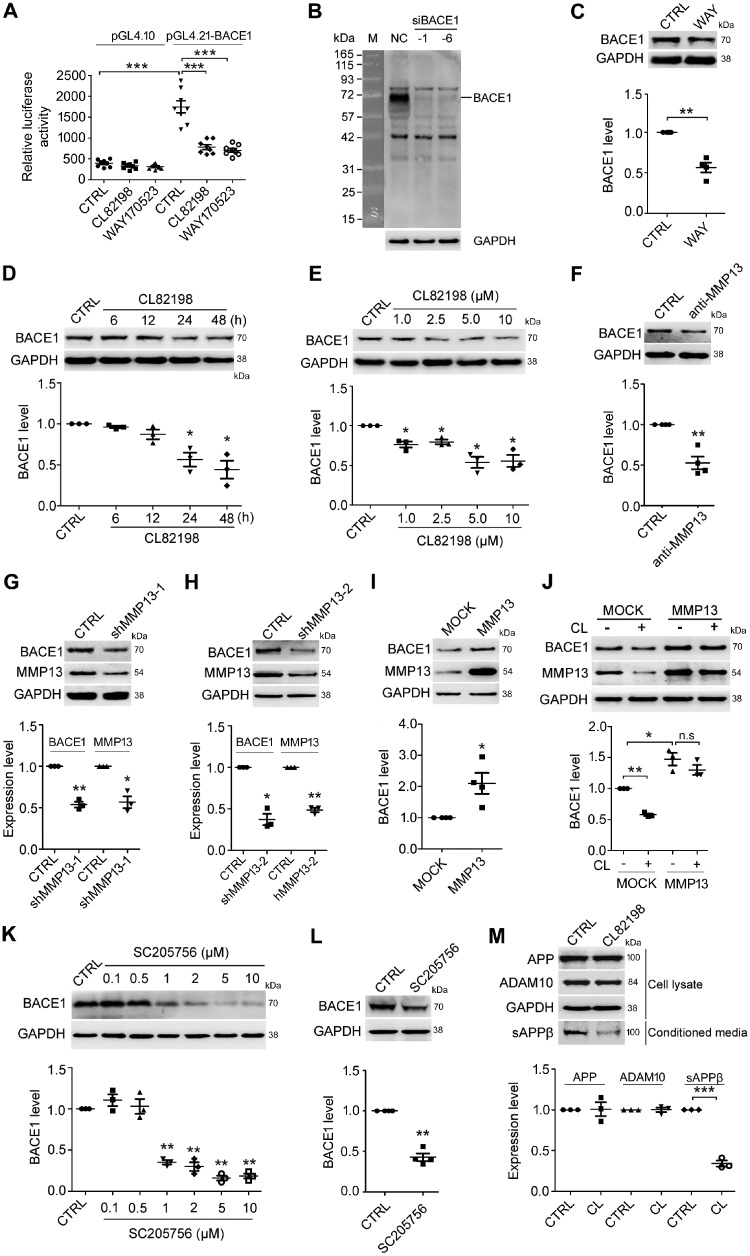
As shown in Fig. 1A, HEK293 cells transiently transfected with the BACE1 promoter showed a dramatic increase in luciferase activity compared with those transfected with the plasmid vector pGL4.10. Two MMP13 inhibitors, WAY170523 and CL82198, significantly decreased BACE1 promoter-driven luciferase activity without having an effect on cells transfected with pGL4.10. Western blotting analysis showed that BACE1 protein levels were dramatically decreased when BACE1 was knocked down with siRNA, indicating that the BACE1 antibody was specific for BACE1 at ∼70kD (Fig. 1B). As expected, WAY170523 significantly decreased BACE1 protein levels in HEK293 cells (Fig. 1C). Because WAY170523 may inhibit MMP1 and MMP9 (Li and Johnson, 2011) in addition to MMP13, we determined whether CL82198, which does not have an effect on MMP1 and MMP9 (Chen et al., 2000; Li and Johnson, 2011; Wang et al., 2013), could also reduce BACE1 levels. As shown in Fig. 1D and E, CL82198 reduced BACE1 protein levels starting at 24 h of treatment at concentrations ranging from 1 μM to 10 μM in SH-SY5Y (human neuroblastoma) cells. We then tested whether the direct manipulation of MMP13 could affect BACE1 protein expression. As shown in Fig. 1F, an MMP13-neutralizing antibody decreased BACE1 protein levels in SH-SY5Y cells. In addition, Mmp13 knockdown with two different shRNAs in HT22 (mouse hippocampal) cells and MMP13 overexpression in HEK293 cells led to a significant decrease and increase of BACE1 protein levels, respectively (Fig. 1G–I). To validate that MMP13 indeed mediates the effects of CL82198 on BACE1 further, we next carried out a rescue experiment. As shown in Fig. 1J, the effect of CL82198 on BACE1 protein expression could be diminished by overexpressing MMP13 in HEK293 cells. It has been reported that another reagent, SC205756 [pyrimidine-4,6-dicarboxylic acid, bis-(4-fluoro-3-methyl-benzylamide)], also exhibits MMP13 inhibitory effects (Nishimura et al., 2012; Duncan et al., 2016). Thus, we further assessed the effects of SC205756 on BACE1. Indeed, in primary cultured neurons (Fig. 1K) and HEK293 cells (Fig. 1L), SC205756 significantly reduced BACE1 protein expression. As BACE1 is a key enzyme in APP processing, we next examined whether CL82198 may be functional in APP proteolysis. In HEK-APP cells incubated with CL82198 for 48 h, soluble APPβ (sAPPβ) levels were significantly reduced, while APP and ADAM10 levels were not altered (Fig. 1M). These results indicated that MMP13 effectively regulated BACE1 protein expression in multiple cell lines and primary neurons and that the effect of CL82198 was mediated by MMP13.
Figure 1.
CL82198 inhibits BACE1 protein expression through MMP13. (A) HEK293 cells were transfected with the luciferase reporter plasmids pGL4.21-BACE1 and pGL4.10 (negative control) in the absence or presence of 5 μM CL82198 or 10 nM WAY170523 for 48 h. Luciferase assays were performed with a GloMax 96 microplate luminometer. Firefly luciferase activity was normalized to that of the plasmid pGL4.10. (B) BACE1 was knocked down with BACE1 siRNA (siBACE-1 and -6) or control siRNA (NC) in HEK293 cells for 48 h. A dramatic decrease in the amount of BACE1 protein is shown at ∼70 kD. M = protein marker. (C) HEK293 cells were treated with WAY170523 (WAY, 10 nM) for 48 h, while control cells were treated with DMSO (CTRL). (D) SH-SY5Y cells were treated with 5 μM CL82198 for 6, 12, 24 and 48 h. (E) SH-SY5Y cells were treated with 1, 2.5, 5 and 10 μM of CL82198 for 48 h. (F) SH-SY5Y cells were treated with an MMP13 neutralizing antibody (anti-MMP13, 1:500) or control rabbit IgG antibody (CTRL) for 48 h. (G–I) Mmp13 was knocked down with shRNA-1 (shMMP13-1, G) or shRNA-2 (shMMP13-2, H) in HT22 cells for 72 h or was overexpressed with an MMP13 vector in HEK293 cells for 48 h (I). CTRL = control shRNA; MOCK = control vector. (J) HEK293 cells were transfected with either the control vector (MOCK) or MMP13 vector for 48 h in the absence or presence of 5 μM CL82198 (CL). Cell lysates were subjected to western blotting analysis. Representative western blots for BACE1 are shown on the top, and quantifications are shown below. (K) Primary cultured cortical neurons were treated with 0.1, 0.5, 1, 2, 5 and 10 μM SC205756 for 48 h. (L) HEK293 cells were treated with 1 μM SC205756 for 48 h. (M) HEK-APP cells were treated with 5 µM CL82198 (CL) for 48 h. Cell lysates were prepared and subjected to western blotting analysis for APP and ADAM10. sAPPβ was analysed in conditioned media using an sAPPβ antibody. All values were normalized to CTRL or MOCK (1.0) within each experiment. The error bars are the SEM. n.s. = no significant difference; *P < 0.05, **P < 0.01, ***P < 0.001 (ANOVA, n = 3 or 4).
PI3K signaling is involved in MMP13-mediated regulation of BACE1
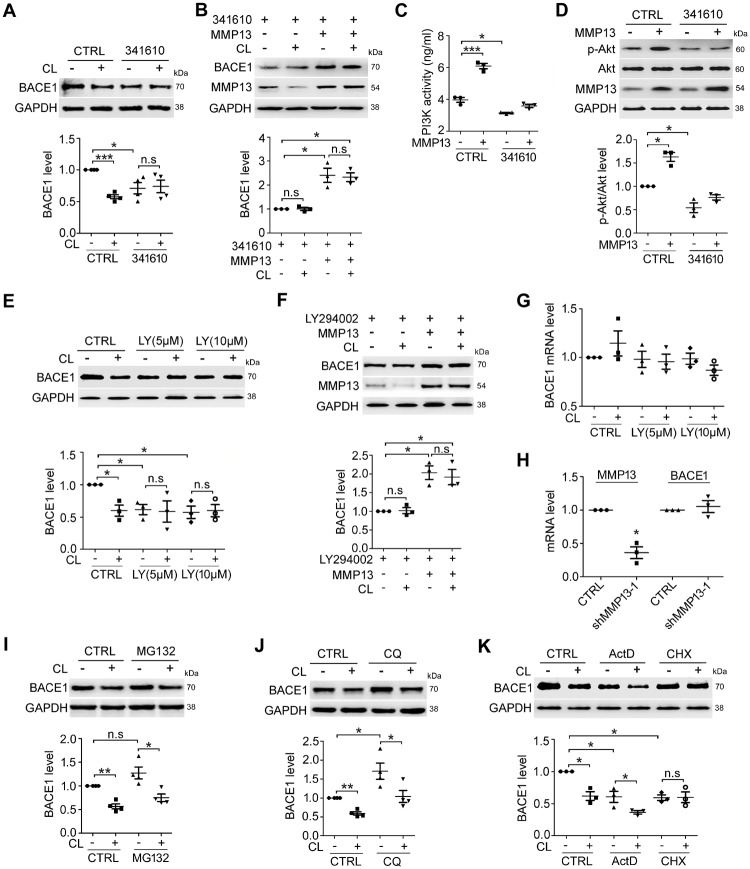
MMP13 is known to promote receptor tyrosine kinase (RTK)-driven migration in melanocytes and melanoma cells (Meierjohann et al., 2010). Studies have also demonstrated that RTK-PI3K signalling regulates a variety of cellular functions, including differentiation, survival and motility (Nakada et al., 2013; Hevner, 2015; Regad, 2015). To determine whether RTK-PI3K is involved in the MMP13-mediated regulation of BACE1 protein expression, we first assessed the effect of 341610, an FGF/PDGF/VEGF RTK inhibitor (Renhowe et al., 2009), in SH-SY5Y cells. As shown in Fig. 2A, 341610 treatment alone significantly reduced the basal protein levels of BACE1. In the presence of 341610, BACE1 inhibition by CL82198 was attenuated. Rescue experiments further showed that in SH-SY5Y cells treated with 341610, BACE1 protein levels were significantly increased by MMP13 overexpression, which also attenuated the effect of CL82198 (Fig. 2B). MMP13 seemed to directly activate PI3K signalling because PI3K activity (Fig. 2C) and phospho-Akt (p-Akt) levels (Fig. 2D) were significantly higher in MMP13-overexpressing cells than in vehicle control cells. Treatment with 341610 reduced PI3K activity and p-Akt under control conditions and attenuated the effect of MMP13 on PI3K/p-Akt (Fig. 2C and D). Consistently, treatment with the PI3K inhibitor LY294002 alone significantly reduced BACE1 protein levels and diminished the effect of CL82198 on BACE1 (Fig. 2E). In the presence of LY294002, MMP13 overexpression reversed the downregulation of BACE1, and the effect of CL82198 on BACE1 was attenuated (Fig. 2F). These results indicated that RTK-PI3K signalling was involved in MMP13-mediated BACE1 protein regulation.
Figure 2.
MMP13-mediated BACE1 regulation involves PI3K signalling and is unrelated to BACE1 transcription and protein degradation. (A) BACE1 protein levels in SH-SY5Y cells treated with 5 μM CL82198 (CL) for 48 h in the absence (CTRL) or presence of the RTK inhibitor 341610 (4 μM). (B) SH-SY5Y cells were transfected with either control or MMP13 vector for 48 h in the absence or presence of 5 μM CL82198 (CL) and 4 μM 341610. (C) SH-SY5Y cells were transfected with either control or MMP13 vector for 48 h in the absence or presence of 4 μM 341610, and PI3K activity was measured by ELISA. (D) p-Akt protein in SH-SY5Y cells transfected with either control or MMP13 vector for 48 h in the absence or presence of 4 μM 341610. (E) BACE1 protein levels in SH-SY5Y cells treated with 5 μM CL82198 (CL) for 48 h in the absence (CTRL) or presence of the PI3K inhibitor LY294002 (LY, 5 and 10 μM). (F) SH-SY5Y cells were transfected with either control or MMP13 vector for 48 h in the absence or presence of 5 μM CL82198 (CL) and LY294002 (LY, 5 and 10 μM). (G) Relative BACE1 mRNA levels in SH-SY5Y cells treated with 5 μM CL82198 (CL) for 48 h in the absence (CTRL) or presence of LY294002 (LY, 5 and 10 μM). (H) Relative Bace1 mRNA levels in HT22 cells treated with vehicle (CTRL) or Mmp13 shRNA-1 for 72 h. (I and J) BACE1 protein levels in SH-SY5Y cells treated with 5 μM CL82198 (CL) for 48 h in the absence (CTRL) or presence of 1 μM MG132 (G) or 100 μM CQ (H). (K) BACE1 protein levels in SH-SY5Y cells treated with 5 μM CL82198 (CL) for 48 h in the absence (CTRL) or presence of the transcriptional inhibitor actinomycin D (ActD, 0.1 μM) or the protein synthesis inhibitor cycloheximide (CHX, 5 μM). All values were normalized to CTRL (1.0) within each experiment. The error bars are the SEM. n.s. = no significant difference; *P < 0.05, **P < 0.01, ***P < 0.001 (ANOVA, n = 3 or 4).
Reduced BACE1 protein levels may result from gene transcription inhibition, protein degradation enhancement or protein synthesis reduction. Thus, we determined whether BACE1 was controlled at the transcriptional level. As shown in Fig. 2G, CL82198 or the PI3K inhibitor LY294002 failed to alter BACE1 mRNA levels. Similarly, Bace1 mRNA was not altered by Mmp13 knockdown (Fig. 2H). It was also unlikely that the BACE1 reduction by CL82198 was mediated by protein degradation machinery because neither the proteasomal inhibitor MG132 (Zhan et al., 2016) nor the lysosomal inhibitor chloroquine (CQ) (Yang et al., 2011) blocked the effect of CL82198 on BACE1 (Fig. 2I and J). To test whether protein synthesis might be involved, we assessed the effect of the protein synthesis inhibitor cycloheximide (CHX) on BACE1 protein expression, and the transcriptional inhibitor actinomycin D (ActD) was used as a control. As shown in Fig. 2K, although basal protein levels of BACE1 were significantly decreased in cells treated with ActD or CHX, the effect of CL82198 on BACE1 was diminished in only the presence of CHX. These results suggested that MMP13 might not regulate BACE1 transcription or protein degradation. Instead, BACE1 protein synthesis was affected by MMP13.
eIF4B phosphorylation at S422 mediates regulation of BACE1 by MMP13
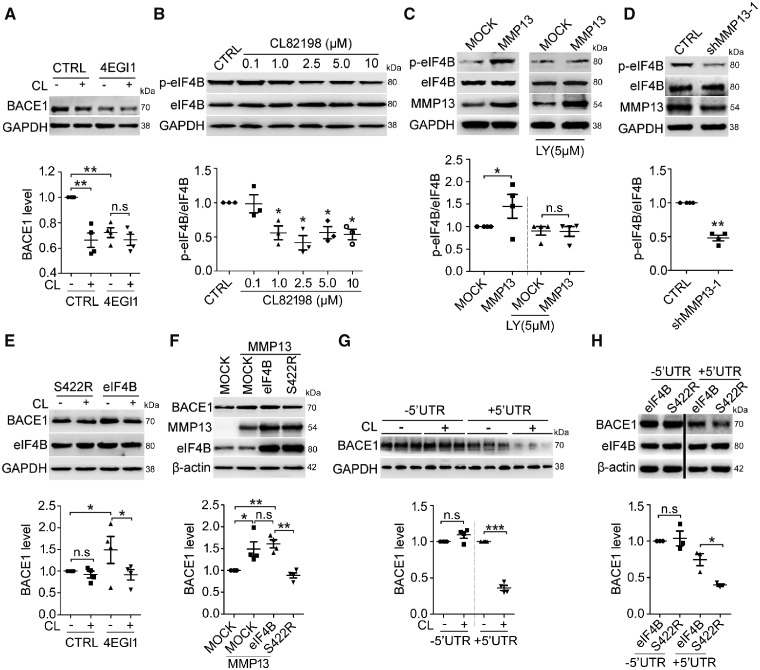
It has been reported that translation initiation is a rate-limiting step in protein synthesis, which involves a variety of eukaryotic translation initiation factors (Sonenberg and Hinnebusch, 2009). Among these, eIF4E and eIF4G join the eIF4F complex and facilitate cap-binding (Sonenberg and Hinnebusch, 2009; Bitterman and Polunovsky, 2012). Interestingly, eIF4B at serine 422 (S422) can be phosphorylated by PI3K (Raught et al., 2004; Shahbazian et al., 2010; Parsyan et al., 2011), suggesting an important role of eIF4B phosphorylation in PI3K signalling. Thus, we first tested the effect of the translation inhibitor, 4EGI1, which disrupts the eIF4E-eIF4G interaction (Moerke et al., 2007; Bitterman and Polunovsky, 2012), on BACE1 protein expression. As shown in Fig. 3A, the administration of 4EGI1 significantly reduced BACE1 protein levels in SH-SY5Y cells, which further diminished the effect of CL82198 on BACE1. We then tested whether eIF4B activity was controlled by MMP13. As shown in Fig. 3B, CL82198 at 1–10 μM inhibited eIF4B phosphorylation at S422 (p-eIF4B). Consistently, MMP13 overexpression significantly increased p-eIF4B levels, which were significantly attenuated in the presence of the PI3K inhibitor LY294002 (Fig. 3C). Conversely, Mmp13 knockdown led to a significant decrease in p-eIF4B levels (Fig. 3D). These results suggested that eIF4B activity might link MMP13 with PI3K signalling.
Figure 3.
eIF4B phosphorylation at S422 is involved in MMP13-mediated BACE1 regulation. (A) BACE1 protein levels in SH-SY5Y cells treated with 5 μM CL82198 (CL) for 48 h in the absence (CTRL) or presence of 25 μM 4EGI1 (a competitive inhibitor of eIF4E/eIF4G). (B) Dose-response of phosphorylated eIF4B (S422, p-eIF4B) in SH-SY5Y cells treated with 0.1, 1, 2.5, 5 and 10 μM CL82198 for 48 h. (C) p-eIF4B levels in HEK293 cells transfected with either control or MMP13 vector for 48 h in the absence or presence of LY294002 (LY, 5 μM). (D) p-eIF4B levels in HEK293 cells transfected with shRNA control (CTRL) or Mmp13 shRNA-1 vector (shMMP13-1). (E) BACE1 proteins in HEK293 cells transfected with eIF4B S422R mutant (S422R) or wild-type eIF4B vector (eIF4B) in the absence or presence of 5 μM CL82198 (CL) for 48 h. (F) BACE1 protein levels in HEK293 cells transfected with control vector (MOCK), MMP13 or MMP13 and co-transfected with S422R or eIF4B. (G) BACE1 protein levels in HEK293 cells transfected with truncated human BACE1 with a 5′UTR deletion (−5′UTR) or with the entire human BACE1 sequence containing the 5′UTR (+5′UTR) in the absence or presence of 5 μM CL82198 (CL) for 48 h. (H) BACE1 protein levels in HEK293 cells expressing the −5′UTR (left two lanes) or +5′UTR construct (right two lanes) in which eIF4B or S422R was co-transfected. S422R decreased BACE1 protein levels in only the +5′UTR transfected cells. All values were normalized to CTRL or MOCK (1.0) within each experiment. The error bars are the SEM. n.s. = no significant difference; *P < 0.05, **P < 0.01, ***P < 0.001 (ANOVA, n = 3 or 4).
To determine whether eIF4B phosphorylation is involved in CL82198/MMP13-mediated BACE1 regulation, we compared the effect of CL82198/MMP13 in cells overexpressing wild-type eIF4B and mutated eIF4B at 422 (S422R). As shown in Fig. 3E, eIF4B overexpression significantly increased BACE1 protein levels compared with S422R overexpression. S422R but not eIF4B attenuated the effect of CL82198 on BACE1 protein. We also found that in cells overexpressing MMP13, the presence of S422R, but not eIF4B, resulted in significantly reduced BACE1 levels (Fig. 3F). It has been reported that BACE1 translation is dependent on the 5′UTR (Pestova and Kolupaeva, 2002; Dmitriev et al., 2003; Rossner et al., 2006). To validate whether the function of eIF4B was also dependent on the BACE1 5′UTR, we assessed the effect of CL82198 and eIF4B mutants on cells transfected with BACE1 constructs with 5′UTR either included (+5′UTR) or deleted (−5′UTR). As shown in Fig. 3G, basal BACE1 protein levels were dramatically lower in cells overexpressing the +5′UTR construct than in cells overexpressing the −5′UTR construct, consistent with previous findings (Lammich et al., 2004; Mihailovich et al., 2007). In the presence of the +5′UTR but not the −5′UTR, CL82198 significantly decreased BACE1 protein levels, indicating that the CL82198 regulation of BACE1 was mediated by the 5′UTR (Fig. 3H). Similarly, eIF4B’s regulation of BACE1 was also dependent on the 5′UTR in BACE1 mRNA, as S422R reduced BACE1 protein levels in only the +5′UTR-expressing cells (Fig. 3H). These results indicated that eIF4B phosphorylation at S422 mediated the regulation of BACE1 by MMP13, and the 5′UTR-dependence further supported a translational mechanism.
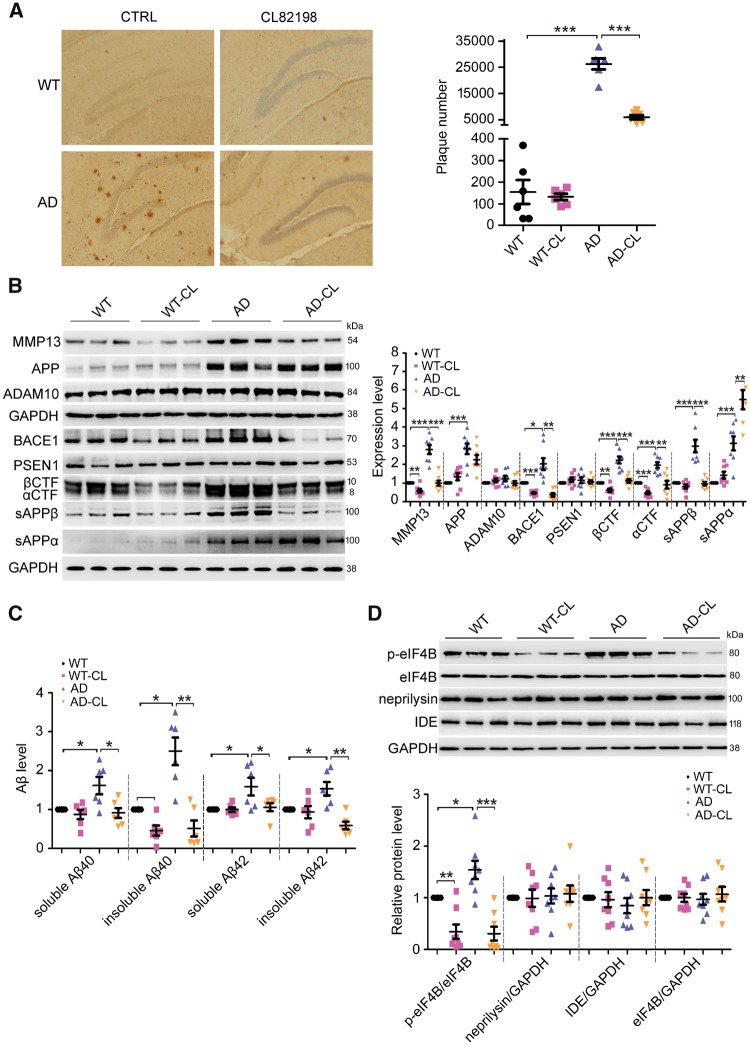
Mmp13 knockdown or CL82198 reduces Alzheimer’s disease- associated pathology in APP/PS1 mice
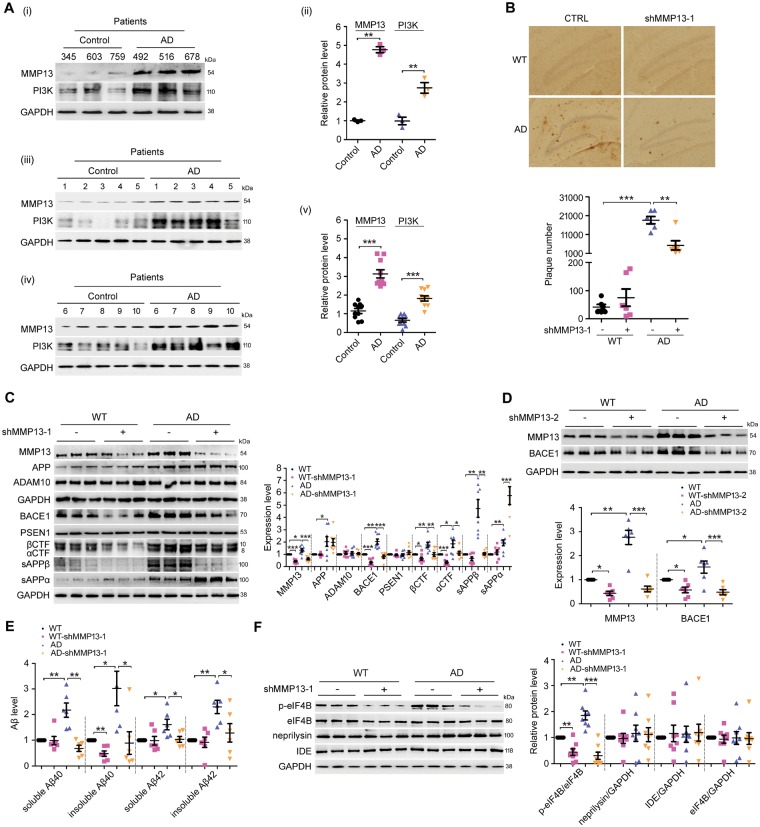
We found that MMP13 and PI3K expression levels were significantly increased in the brains of patients with Alzheimer’s disease (Fig. 4A). Based on the finding that MMP13 inhibition by CL82198 reduced BACE1 protein levels in cultured cells, we speculated that Mmp13 knockdown or CL82198 treatment would reduce Alzheimer’s disease-associated pathology in the brains of APP/PS1 mice. Thus, we first assessed the effect of Mmp13 knockdown on hippocampal amyloid-β deposition using a 6E10 antibody that recognizes all amyloid-β species (Sarsoza et al., 2009; Kim et al., 2015). As shown in Fig. 4B, Mmp13 knockdown significantly decreased the number of plaques in APP/PS1 mice. Interestingly, MMP13 protein levels were also significantly increased in the hippocampi of APP/PS1 mice (Fig. 4C and D), suggesting that MMP13 is involved in Alzheimer’s disease-associated pathology.
Figure 4.
Mmp13 knockdown ameliorates Alzheimer’s disease-associated pathology in APP/PS1 mice. (A) Representative western blots and bar plot summary of MMP13 and PI3K expression in the prefrontal cortex of control (n = 13) and Alzheimer’s disease patients (n = 13). Data in (i and ii) and in (iii–v) were from samples provided by Sydney Brain Bank and NIH NeuroBiobank, respectively. (B) Immunohistochemical images and bar plot summary of neuritic plaques in the hippocampi of wild-type (WT) and APP/PS1 mice (AD) treated with vehicle (CTRL) and Mmp13 shRNA-1 (shMMP13-1). Neuritic plaques were probed with the amyloid-β-specific monoclonal antibody 6E10 (n = 6). (C) Representative western blots and bar plot summary of MMP13, APP, ADAM10, BACE1, PSEN1, βCTF, αCTF, sAPPβ and sAPPα expression in the hippocampi of wild-type (n = 8), wild-type with Mmp13 shRNA-1 (WT-shMMP13-1, n = 8), Alzheimer’s disease (n = 8) and Alzheimer’s disease with Mmp13 shRNA-1 mice (AD-shMMP13-1, n = 8). The soluble fractions were used to detect sAPPβ and sAPPα with sAPPβ and 6E10 antibodies. (D) Representative western blots and bar plot summary of MMP13 and BACE1 expression in the hippocampi of wild-type (n = 6), wild-type with Mmp13 shRNA-2 (WT-shMMP13-2, n = 6), Alzheimer’s disease (n = 6) and Alzheimer’s disease with Mmp13 shRNA-2 mice (AD-shMMP13-2, n = 6). (E) ELISAs were used to measure soluble and insoluble amyloid-β40/42 levels in the brain homogenates of wild-type (n = 6), WT-shMMP13-1 (n = 6), Alzheimer’s disease (n = 6) and AD-shMMP13-1 (n = 6) mice. (F) Representative western blots and bar plot summary of p-eIF4B, eIF4B, neprilysin and insulin-degrading enzyme (IDE) levels in the hippocampi of wild-type (n = 8), WT-shMMP13-1 (n = 8), Alzheimer’s disease (n = 8) and AD-shMMP13-1 (n = 8) mice. All values were normalized to CTRL or wild-type (1.0) within each experiment. The error bars are the SEM. n.s. = no significant difference; *P < 0.05, **P < 0.01, ***P < 0.001 (ANOVA).
To investigate whether the reduced amyloid-β load was associated with BACE1 and APP processing, we assessed the effect of Mmp13 knockdown on APP and BACE1, as well as ADAM10 (a disintegrin and metalloproteinase domain 10) and presenilin 1 (PSEN1), which are also involved in the regulation of amyloid-β generation (Vincent and Checler, 2012). As shown in Fig. 4C, hippocampal Mmp13 knockdown caused significant reductions in BACE1 levels in wild-type and APP/PS1 mice, while APP, ADAM10 and PSEN1 levels were not altered; these data are consistent with the in vitro findings that BACE1 was selectively regulated by MMP13. As BACE1 is involved in APP processing and amyloid-β generation, we then assessed the levels of soluble APPβ (sAPPβ), sAPPα and the CTF of APP. Similar to BACE1 changes, sAPPβ and β-CTF levels were significantly reduced in wild-type and APP/PS1 mice, while sAPPα levels were significantly increased (Fig. 4C). The effect of Mmp13 knockdown was further validated by the second Mmp13 shRNA (shMMP13-2), which similarly reduced BACE1 protein levels in wild-type and APP/PS1 mice (Fig. 4D). The effect of Mmp13 knockdown on amyloid-β generation was further assessed. As shown in Fig. 4E, shMMP13 did not change soluble amyloid-β40/42 or insoluble amyloid-β42 levels in wild-type mice, whereas insoluble amyloid-β40 levels were significantly reduced. In APP/PS1 mice treated with shMMP13, the soluble and insoluble amyloid-β40/42 levels were all significantly decreased. To determine whether BACE1 alterations caused by Mmp13 knockdown were associated with eIF4B activity, we assessed p-eIF4B levels. As shown in Fig. 4F, the basal levels of p-eIF4B were significantly higher in APP/PS1 mice than in wild-type mice, and shMMP13 significantly decreased p-eIF4B levels in wild-type and APP/PS1 mice. On the other hand, the protein levels of the amyloid-β degrading enzymes neprilysin and insulin-degrading enzyme (IDE) were not altered by shMMP13 in wild-type or APP/PS1 mice (Fig. 4E). Interestingly, the levels of Iba1, a marker of inflammation, were also decreased in shMMP13-treated mice (Supplementary Fig. 1). These results indicated that shMMP13 effectively reduced hippocampal BACE1 levels and amyloid-β generation in wild-type and APP/PS1 mice, which was consistent with the altered levels of p-eIF4B.
Next, we assessed the effect of CL82198 on Alzheimer’s disease-associated pathology. As shown in Fig. 5A, the administration of CL82198 for 2 months significantly decreased the number of amyloid-β plaques in APP/PS1 mice. Similar to Mmp13 knockdown, CL82198 reduced BACE1, sAPPβ and β-CTF levels and increased sAPPα levels in wild-type and APP/PS1 mice without affecting APP, ADAM10 or PSEN1 levels (Fig. 5B). CL98198 also mimicked the effect of shMMP13 on amyloid-β generation. Insoluble amyloid-β40 levels were significantly decreased in wild-type mice, and soluble and insoluble amyloid-β40/42 levels were significantly reduced in APP/PS1 mice (Fig. 5C). Moreover, CL82198 similarly decreased p-eIF4B levels in wild-type and APP/PS1 mice without affecting neprilysin or IDE levels (Fig. 5D). CL82198 also reduced Iba1 levels in APP/PS1 mice (Supplementary Fig. 1). These results indicated that CL82198 mimicked the effect of MMP13 knockdown on Alzheimer’s disease-associated pathology.
Figure 5.
CL82198 ameliorates Alzheimer’s disease-associated pathology in APP/PS1 mice. (A) Immunohistochemical images and bar plot summary of neuritic plaques in the hippocampi of wild-type, wild-type with CL82198 (WT-CL), APP/PS1 (AD) and APP/PS1 mice treated with CL82198 (AD-CL) (n = 6). (B) Representative western blots and bar plot summary of MMP13, APP, ADAM10, BACE1, PSEN1, βCTF, αCTF, sAPPβ and sAPPα levels in the hippocampi of wild-type (n = 8), WT-CL (n = 8), AD (n = 8) and AD-CL (n = 8) mice. Soluble fractions were used to detect sAPPβ and sAPPα with sAPPβ and 6E10 antibodies. (C) ELISAs were used to measure soluble and insoluble amyloid-β40/42 levels in the brain homogenates of wild-type (n = 6), WT-CL (n = 6), AD (n = 6) and AD-CL (n = 6) mice. (D) Representative western blots and bar plot summary of p-eIF4B, eIF4B, neprilysin and IDE levels in the hippocampi of wild-type (n = 8), WT-CL (n = 8), AD (n = 8) and AD-CL (n = 8) mice. All values were normalized to CTRL or wild-type (1.0) within each experiment. The error bars are the SEM. n.s. = no significant difference; *P < 0.05, **P < 0.01, ***P < 0.001 (ANOVA). Aβ = amyloid-β.
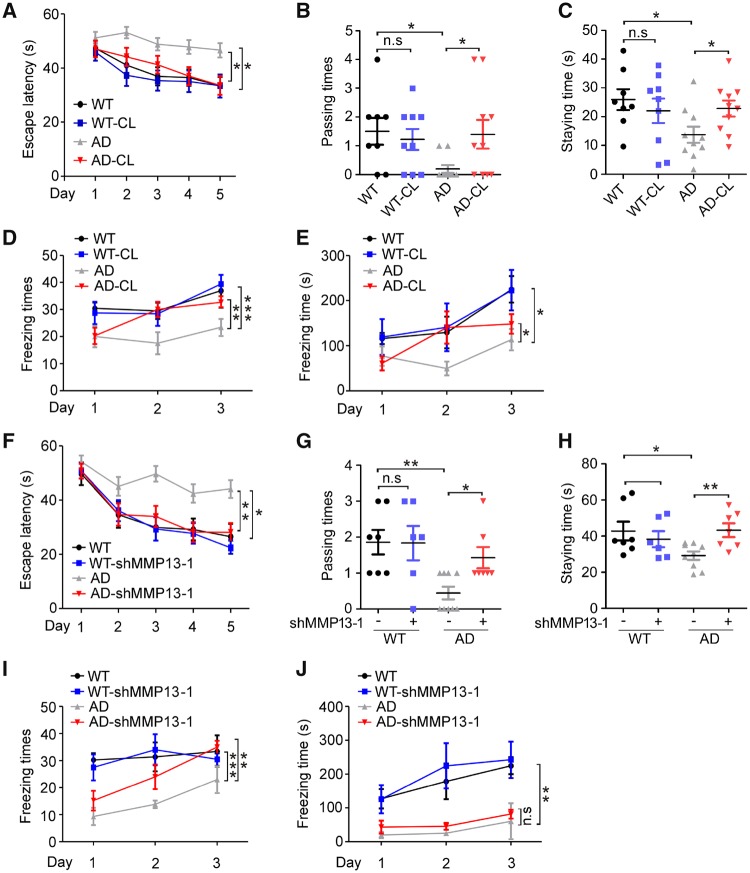
CL82198 treatment or Mmp13 knockdown improves cognitive function in APP/PS1 mice
To determine whether CL82198 or MMP13 may affect cognitive functions, we assessed spatial and associative learning memory using water maze tests and context fear conditioning tests in animals with Mmp13 knockdown or CL82198 treatment (Ly et al., 2013; Webster et al., 2014). In the hidden platform test, the escape latency was significantly shorter in CL82198-treated APP/PS1 [Alzheimer’s disease (AD)-CL] mice than in saline-treated APP/PS1 (Alzheimer’s disease) mice beginning on the third day (Fig. 6A). When the platform was removed during the probe trial, the passing time for crossing over the target site (Fig. 6B) and the staying time in the target quadrant (Fig. 6C) were significantly longer in AD-CL mice than in Alzheimer’s disease mice. No significant differences were observed between AD-CL and wild-type mice receiving CL82198 (WT-CL) or saline (wild-type, WT) for escape latency (Fig. 6A), passing times (Fig. 6B) or staying times (Fig. 6C). The subsequent context fear conditioning tests revealed that AD-CL mice exhibited more and longer freezing times than Alzheimer’s disease mice (Fig. 6D and E). No significant differences were observed between WT-CL and wild-type mice for their number of freezing times (Fig. 6D) or length of freezing times (Fig. 6E). These results indicated that CL82198 significantly improved spatial and associative learning memory in APP/PS1 mice.
Figure 6.
CL82198 treatment or Mmp13 knockdown improves learning and memory in APP/PS1 mice. Behavioural performance was assessed by Morris water maze tests followed by fear conditioning tests. (A) In the hidden platform tests, the time spent on reaching the platform (latency) was recorded by ANY-maze tracking software. Compared with vehicle-treated APP/PS1 mice (AD, n = 10), CL82198 treated APP/PS1 mice (AD-CL, n = 10) showed a significantly shorter latency on the third, fourth and fifth days. No significant differences were found for the AD-CL mice or wild-type mice treated with vehicle (WT, n = 8) or CL82198 (WT-CL, n = 9). (B and C) In the probe trial on the sixth day, AD-CL mice spent significantly more time travelling (B) and a significantly longer time staying (C) in the place where the hidden platform was previously placed than Alzheimer’s disease mice. (D and E) In the context fear conditioning tests, the freezing times and the total freezing times of the four groups of mice in the chamber were recorded and analysed after drug injection. (F) In the hidden platform tests, shMMP13-treated APP/PS1 mice (AD-shMMP13-1, n = 7) exhibited a significantly shorter latency on the third, fourth and fifth days than the vehicle-treated APP/PS1 mice (AD, n = 9). No significant differences were found between AD-shMMP13-1 and wild-type mice treated with vehicle (WT, n = 7) or shMMP13 (WT-shMMP13-1, n = 6). (G and H) On the sixth day of the probe trial, AD-shMMP13-1 mice spent significantly more time travelling (G) and a significantly longer time staying (H) in the place where the hidden platform was previously placed than the Alzheimer’s disease mice. (I and J) In the context fear conditioning tests, the freezing times and the total freezing times of the four groups of mice in the chamber were recorded and analysed for mice treated with shMMP13 or vehicle. All values were normalized to wild-type (1.0) within each experiment. The error bars are the SEM. n.s. = no significant difference; *P < 0.05, **P < 0.01, ***P < 0.001 (ANOVA).
Hippocampal Mmp13 knockdown had effects similar to CL82198 for the water maze test (Fig. 6F–H). In the fear conditioning test, while the freezing times were significantly greater in AD-shMMP13-1 mice than in Alzheimer’s disease mice, the difference was not significantly different (Fig. 6I and J). These results supported that MMP13 mediated the effect of CL82198 on behavioural activities in an animal model.
Discussion
The function of MMP13 in the CNS is not well understood. Studies suggest that MMP13 is involved in tumour progression (Kobayashi et al., 2012; Guan et al., 2015; Zheng et al., 2015) and membrane permeability of brain endothelial cells (Lu et al., 2009). Our study provides evidence that MMP13 controls BACE1 protein levels in multiple cell lines and primary cultured neurons (Fig. 1).
MMP13 is known to degrade ECM components, such as type I–IV collagen and perlecan (Leeman et al., 2002; Yurchenco, 2011). Perlecan is a heparin sulphate proteoglycan that is capable of binding a variety of growth factors (Iozzo et al., 2009). It has been suggested that heparin plays an important role in receptor dimerization, which enables the autophosphorylation of the receptor cytoplasmic tail (Schlessinger et al., 2000). The involvement of RTK-PI3K signalling in MMP13 function has been documented. For example, RTK/PI3K mediates MMP13’s regulation of prostate cancer invasion (Lin et al., 2016) and myeloma metastasis (Xu et al., 2015). Mmp13 knockdown inhibits the growth factor-induced proliferation of melanocytes (Meierjohann et al., 2010). In line with these data, our study provides evidence that MMP13 increases PI3K activity and p-Akt levels, and these effects are prevented by an RTK inhibitor. As RTK or PI3K inhibitors reduce basal BACE1 protein levels and further diminish the effect of MMP13/CL82198 on BACE1 (Fig. 2), we speculate that PI3K signalling is essential for the MMP13 regulation of BACE1.
PI3K signalling plays an important role in Alzheimer’s disease pathology (O’ Neill, 2013). Enhanced PI3K activity is associated with disrupted autophagic clearance of amyloid-β and tau (Heras-Sandoval et al., 2014). However, how PI3K might regulate BACE1 processing is currently unclear. It is unlikely that PI3K might regulate BACE1 through transcriptional regulation and protein degradation (Fig. 2). Alternatively, PI3K might achieve translational control of BACE1 through eIF4B. PI3K is known to increase eIF4B phosphorylation at S422 through the S6 kinase (S6K) (Raught et al., 2004; Holz et al., 2005). S6K and the p90 ribosomal protein S6 kinase (RSK) phosphorylate eIF4B at the same residue, which promotes eIF4B interaction with eIF3 and translation (Shahbazian et al., 2006, 2010). In our study, the overexpression or knockdown of MMP13 enhanced or reduced eIF4B phosphorylation at S422, respectively (Fig. 3), supporting that eIF4B activity might bridge PI3K signalling with the MMP13-mediated regulation of BACE1.
At the translational level, BACE1 can be regulated by eIF2α, but conflicting results have been reported. While one study shows that the phosphorylation of eIF2α increases BACE1 protein levels and amyloid-β production (O’Connor et al., 2008), another reveals that eIF2α deficiency does not result in BACE1 inhibition in mice (Sadleir et al., 2014). To the best of our knowledge, the involvement of eIF4B in BACE1 translation has not been previously reported. eIF4B phosphorylation at S422 is essential for cap-dependent translation (Shahbazian et al., 2006; van Gorp et al., 2009). In addition, eIF4B is unique in that it selectively regulates long and structured 5′UTR function (Dmitriev et al., 2003), while being insufficient for ribosomal movement on unstructured 5′UTRs (Pestova and Kolupaeva, 2002). Interestingly, the BACE1 5′UTR is GC-rich and structured (Lammich et al., 2004; Rossner et al., 2006). Thus, it is plausible that eIF4B phosphorylation might play a role in BACE1 translation. In our study, the following findings indicate that eIF4B is critical in MMP13′s regulation of BACE1: (i) in cells overexpressing the eIF4B mutant S422R, CL82198-induced inhibition or MMP13-induced increases in BACE1 protein levels were diminished (Fig. 3E and F); and (ii) the inhibition of BACE1 protein expression by S422R or CL82198 occurred only when the BACE1 5′UTR was present (Fig. 3G and H).
MMP13 is upregulated in the brains of rats and humans after cerebral ischaemia (Cuadrado et al., 2009). In our study, MMP13 expression was increased in patients with Alzheimer’s disease and APP/PS1 mice, suggesting that MMP13 also plays an important role in the pathology of Alzheimer’s disease (Graham et al., 2017). In line with these data, Mmp13 knockdown reduces BACE1, sAPPβ and β-CTF protein levels, with a concomitant increase in sAPPα. The latter might be a secondary effect of BACE1 inhibition because sAPPα can be increased after BACE1 inhibition (Fukumoto et al., 2010). Notably, Mmp13 knockdown significantly reduced insoluble amyloid-β40, but not amyloid-β42 levels in wild-type mice, while soluble and insoluble amyloid-β40/42 levels were decreased in APP/PS1 mice. Although it has been reported that BACE1 deficiency reduces both amyloid-β40 and amyloid-β42 levels (Ohno et al., 2004), there is evidence showing that BACE1 inhibition preferentially reduces amyloid-β40 levels in the mouse brain or in mouse cell lines (Ohno et al., 2004; Herzig et al., 2007; Fukumoto et al., 2010). In a mouse model of Alzheimer’s disease with an APP mutation, the partial inhibition of BACE1 results in reduced total amyloid-β levels, whereas amyloid-β42 levels remain unchanged (McConlogue et al., 2007). We speculate that the slight but significant inhibition of BACE1 might be responsible for the selective decrease in amyloid-β40 levels in wild-type mice. On the other hand, APP/PS1 mice differ from wild-type mice in APP and PSEN1 mutations (Kurt et al., 2001). While APP mutations dramatically increase (∼50%) amyloid-β40 levels without altering amyloid-β42 levels (Morel et al., 2013), mutations of γ-secretase favour the generation of amyloid-β42 over amyloid-β40 (Jankowsky et al., 2004; Mastrangelo et al., 2005). Thus, combined mutations in APP and PS1 lead to enhanced basal levels of amyloid-β40 and amyloid-β42 in APP/PS1 mice, in which inhibiting BACE1 reduces both amyloid-β40 and amyloid-β42 levels (Ly et al., 2013). Interestingly, Iba1 staining was also decreased in shMMP13/CL82198-treated mice (Supplementary Fig. 1). This effect might have been a result of the reduced amyloid-β load, since amyloid-β is neurotoxic and promotes inflammation (Sengupta et al., 2016).
The role of MMP13 in Alzheimer’s disease pathology was further supported by the results of behavioural tests in our study, as Mmp13 knockdown or CL82198 treatment improved spatial and associative learning memory (Fig. 6). It was noted that Mmp13 knockdown and CL82198 treatment had different effects on freezing times (Fig. 6E versus Fig. 6J). shMMP13-1 was introduced into only the hippocampus, while CL82198 was administered systematically. It has been reported that spatial learning (water maze) is critically dependent on the hippocampus (Neves et al., 2008), whereas associative learning (fear conditioning) also involves the amygdala (Pape and Pare, 2010). We speculate that one of the mechanisms contributing to the difference is that the amygdala was affected by CL82198 but was relatively spared from shMMP13-1.
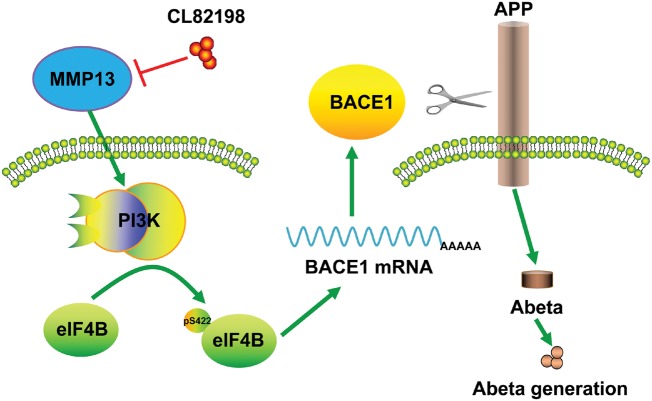
We propose the following possible mechanisms by which MMP13 regulates BACE1 protein expression. The activation of PI3K signalling promotes eIF4B phosphorylation at S422, which leads to the 5′UTR-dependent translation of BACE1. According to this mechanism, CL82198 reduces BACE1 protein levels and amyloid-β accumulation, which contributes to the improved learning and memory in APP/PS1 mice (Fig. 7). It is worth noting that although MMP13-mediated regulation of BACE1 is dependent on PI3K signalling, the cognitive improvement by MMP13 inhibition in APP/PS1 mice may involve mechanisms beyond BACE1 and PI3K. It is suggested that ECM molecules play an important role in synaptic plasticity and control dendritic spines through the interaction with their receptors (Dansie and Ethell, 2011; Wlodarczyk et al., 2011). ECM also mediates the crosstalk between neurons and glia in remodelling brain network function (Song and Dityatev, 2018). In pathological conditions, accumulation of ECM components coincides with the synaptic dysfunction in APP/PS1 mice (Vegh et al., 2014). Moreover, inhibition of MMP9 attenuates traumatic injury and behavioural impairment (Jia et al., 2014). In line with this, the elevated MMP13 level in the ischaemic brain may suggest a role in neuronal damage (Cuadrado et al., 2009). Therefore, the potential role of MMP13 in synaptic dysfunction or cellular stress requires further study.
Figure 7.
Schematic diagram depicting the possible mechanisms through which MMP13 regulates BACE1 translation. PI3K signalling downstream of MMP13 promotes eIF4B phosphorylation, which in turn facilitates the 5′UTR-dependent BACE1 translation. Consequently, increased BACE1 protein levels enhance amyloid-β production and impair cognitive function. The inhibition of MMP13 by CL82198 results in reduced BACE1 and amyloid-β accumulation in the brain, which contributes to the improved learning and memory functions in APP/PS1 mice. AD = Alzheimer;s disease; WT = wild-type.
The clinical relevance of the current study may include the following: (i) the study design provides an alternative strategy for BACE1 inhibitors: reducing BACE1 protein levels while allowing residual BACE1 to exert its function. To the best of our knowledge, luciferase-based drug screening for BACE1 in living cells has not been previously reported; (ii) the translational mechanism associated with MMP13 and PI3K signalling in BACE1 regulation adds to the understanding of why BACE1 protein levels rather than BACE1 mRNA levels are significantly increased in Alzheimer’s disease patients; and (iii) furthermore, we found that MMP13 levels are significantly increased in both Alzheimer’s disease patients and APP/PS1 mice. By inhibiting MMP13, which is closely associated with BACE1 regulation, CL82198 might have therapeutic potential for Alzheimer’s disease.
Funding
This work was supported by NSFC (The National Natural Science Foundation of China) grant (81220108010), Chongqing Science and Technology Commission grant (cstc2018jcyjAX0067) and Chongqing Yu-Zhong Science and Technology Project (001) to G.-J.C., and by National Key R&D Program of China (2018YFC1314700) to L.-J.J. and G.-J.C.
Competing interests
The authors report no competing interests.
Supplementary Material
Glossary
Abbreviations
- ECM
extracellular matrix
- eIF4B
eukaryotic translation initiation factor 4B
- PI3K
phosphatidylinositide 3-kinase
References
- Bitterman PB, Polunovsky VA. Attacking a nexus of the oncogenic circuitry by reversing aberrant eIF4F-mediated translation. Mol Cancer Ther 2012; 11: 1051–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chami L, Checler F. BACE1 is at the crossroad of a toxic vicious cycle involving cellular stress and beta-amyloid production in Alzheimer’s disease. Mol Neurodegener 2012; 7: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, Nelson FC, Levin JI, Mobilio D, Moy FJ, Nilakantan R et al. Structure-based design of a novel, potent, and selective inhibitor for MMP-13 utilizing NMR spectroscopy and computer-aided molecular design. J Am Chem Soc 2000; 122: 9648–54. [Google Scholar]
- Cheng JS, Craft R, Yu GQ, Ho K, Wang X, Mohan G et al. Tau reduction diminishes spatial learning and memory deficits after mild repetitive traumatic brain injury in mice. PLoS One 2014; 9: e115765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coimbra JRM, Marques DFF, Baptista SJ, Pereira CMF, Moreira PI, Dinis TCP et al. Highlights in BACE1 inhibitors for Alzheimer’s disease treatment. Front Chem 2018; 6: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado E, Rosell A, Borrell-Pages M, Garcia-Bonilla L, Hernandez-Guillamon M, Ortega-Aznar A et al. Matrix metalloproteinase-13 is activated and is found in the nucleus of neural cells after cerebral ischemia. J Cereb Blood Flow Metab 2009; 29: 398–410. [DOI] [PubMed] [Google Scholar]
- Cummings J, Lee G, Mortsdorf T, Ritter A, Zhong K. Alzheimer’s disease drug development pipeline: 2017. Alzheimers Dement (N Y) 2017; 3: 367–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansie LE, Ethell IM. Casting a net on dendritic spines: the extracellular matrix and its receptors. Dev Neurobiol 2011; 71: 956–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev SE, Terenin IM, Dunaevsky YE, Merrick WC, Shatsky IN. Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5’ untranslated regions. Mol Cell Biol 2003; 23: 8925–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan HF, Smith AJ, Fleming GJ, Partridge NC, Shimizu E, Moran GP et al. The histone-deacetylase-inhibitor suberoylanilide hydroxamic acid promotes dental pulp repair mechanisms through modulation of matrix metalloproteinase-13 activity. J Cell Physiol 2016; 231: 798–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Long Y, Lai Y, Wang X, Chen G, Zhu B. NPAS4 facilitates the autophagic clearance of endogenous Tau in rat cortical neurons. J Mol Neurosci 2016; 58: 401–10. [DOI] [PubMed] [Google Scholar]
- Filser S, Ovsepian SV, Masana M, Blazquez-Llorca L, Brandt Elvang A, Volbracht C et al. Pharmacological inhibition of BACE1 impairs synaptic plasticity and cognitive functions. Biol Psychiatry 2015; 77: 729–39. [DOI] [PubMed] [Google Scholar]
- Frost G, Li Y. The role of astrocytes in amyloid production and Alzheimer’s disease. Open Biol 2017; 7: 170228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto H, Takahashi H, Tarui N, Matsui J, Tomita T, Hirode M et al. A noncompetitive BACE1 inhibitor TAK-070 ameliorates Abeta pathology and behavioral deficits in a mouse model of Alzheimer’s disease. J Neurosci 2010; 30: 11157–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham WV, Bonito-Oliva A, Sakmar TP. Update on Alzheimer’s disease therapy and prevention strategies. Annu Rev Med 2017; 68: 413–30. [DOI] [PubMed] [Google Scholar]
- Guan N, Huo X, Zhang Z, Zhang S, Luo J, Guo W. Ginsenoside Rh2 inhibits metastasis of glioblastoma multiforme through Akt-regulated MMP13. Tumour Biol 2015; 36: 6789–95. [DOI] [PubMed] [Google Scholar]
- Hawkes N. Merck ends trial of potential Alzheimer’s drug verubecestat. BMJ 2017; 356: j845. [DOI] [PubMed] [Google Scholar]
- Heras-Sandoval D, Perez-Rojas JM, Hernandez-Damian J, Pedraza-Chaverri J. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cell Signal 2014; 26: 2694–701. [DOI] [PubMed] [Google Scholar]
- Herzig MC, Paganetti P, Staufenbiel M, Jucker M. BACE1 and mutated presenilin-1 differently modulate Abeta40 and Abeta42 levels and cerebral amyloidosis in APPDutch transgenic mice. Neurodegener Dis 2007; 4: 127–35. [DOI] [PubMed] [Google Scholar]
- Hevner RF. Brain overgrowth in disorders of RTK-PI3K-AKT signaling: a mosaic of malformations. Semin Perinatol 2015; 39: 36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger RM, McLean CA, Beyreuther K, Masters CL, Evin G. Increased expression of the amyloid precursor beta-secretase in Alzheimer’s disease. Ann Neurol 2002; 51: 783–6. [DOI] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 2005; 123: 569–80. [DOI] [PubMed] [Google Scholar]
- Hsiao YH, Hung HC, Chen SH, Gean PW. Social interaction rescues memory deficit in an animal model of Alzheimer’s disease by increasing BDNF-dependent hippocampal neurogenesis. J Neurosci 2014; 34: 16207–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Das B, Hou H, He W, Yan R. BACE1 deletion in the adult mouse reverses preformed amyloid deposition and improves cognitive functions. J Exp Med 2018; 215: 927–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley GW. Synaptic circuit remodelling by matrix metalloproteinases in health and disease. Nat Rev Neurosci 2012; 13: 743–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain I, Hawkins J, Harrison D, Hille C, Wayne G, Cutler L et al. Oral administration of a potent and selective non-peptidic BACE-1 inhibitor decreases beta-cleavage of amyloid precursor protein and amyloid-beta production in vivo. J Neurochem 2007; 100: 802–9. [DOI] [PubMed] [Google Scholar]
- Iozzo RV, Zoeller JJ, Nystrom A. Basement membrane proteoglycans: modulators Par Excellence of cancer growth and angiogenesis. Mol Cells 2009; 27: 503–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Kimura K, Haneda M, Ishida Y, Sawada M, Isobe K. Induction of matrix metalloproteinases (MMP3, MMP12 and MMP13) expression in the microglia by amyloid-beta stimulation via the PI3K/Akt pathway. Exp Gerontol 2007; 42: 532–7. [DOI] [PubMed] [Google Scholar]
- Jack CR, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB et al. NIA-AA research framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement 2018; 14: 535–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky JL, Fadale DJ, Anderson J, Xu GM, Gonzales V, Jenkins NA et al. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum Mol Genet 2004; 13: 159–70. [DOI] [PubMed] [Google Scholar]
- Jia F, Yin YH, Gao GY, Wang Y, Cen L, Jiang JY. MMP-9 inhibitor SB-3CT attenuates behavioral impairments and hippocampal loss after traumatic brain injury in rat. J Neurotrauma 2014; 31: 1225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Kim HV, Jo S, Lee CJ, Choi SY, Kim DJ et al. EPPS rescues hippocampus-dependent cognitive deficits in APP/PS1 mice by disaggregation of amyloid-beta oligomers and plaques. Nat Commun 2015; 6: 8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Takahashi H, Inoue A, Harada H, Toshimori S, Kobayashi Y et al. Oct-3/4 promotes migration and invasion of glioblastoma cells. J Cell Biochem 2012; 113: 508–17. [DOI] [PubMed] [Google Scholar]
- Kurt M, Davies D, Kidd M, Duff K, Rolph S, Jennings K et al. Neurodegenerative changes associated with beta-amyloid deposition in the brains of mice carrying mutant amyloid precursor protein and mutant presenilin-1 transgenes. Exp Neurol 2001; 171: 59–71. [DOI] [PubMed] [Google Scholar]
- Kusakawa G-i, Saito T, Onuki R, Ishiguro K, Kishimoto T, Hisanaga S-i. Calpain-dependent proteolytic cleavage of the p35 cyclin-dependent kinase 5 activator to p25. J Biol Chem 2000; 275: 17166–72. [DOI] [PubMed] [Google Scholar]
- Lammich S, Schobel S, Zimmer AK, Lichtenthaler SF, Haass C. Expression of the Alzheimer protease BACE1 is suppressed via its 5’-untranslated region. EMBO Rep 2004; 5: 620–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeman MF, Curran S, Murray GI. The structure, regulation, and function of human matrix metalloproteinase-13. Crit Rev Biochem Mol Biol 2002; 37: 149–66. [DOI] [PubMed] [Google Scholar]
- Lepelletier FX, Mann DM, Robinson AC, Pinteaux E, Boutin H. Early changes in extracellular matrix in Alzheimer’s disease. Neuropathol Appl Neurobiol 2017; 43: 167–82. [DOI] [PubMed] [Google Scholar]
- Li JJ, Johnson AR. Selective MMP13 inhibitors. Med Res Rev 2011; 31: 863–94. [DOI] [PubMed] [Google Scholar]
- Lin YH, Tian Y, Wang JS, Jiang YG, Luo Y, Chen YT. Pituitary tumor-transforming gene 1 regulates invasion of prostate cancer cells through MMP13. Tumour Biol 2016; 37: 15495–500. [DOI] [PubMed] [Google Scholar]
- Lu DY, Yu WH, Yeh WL, Tang CH, Leung YM, Wong KL et al. Hypoxia-induced matrix metalloproteinase-13 expression in astrocytes enhances permeability of brain endothelial cells. J Cell Physiol 2009; 220: 163–73. [DOI] [PubMed] [Google Scholar]
- Luo J, Lee SH, VandeVrede L, Qin Z, Ben Aissa M, Larson J et al. A multifunctional therapeutic approach to disease modification in multiple familial mouse models and a novel sporadic model of Alzheimer’s disease. Mol Neurodegener 2016; 11: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly PT, Wu Y, Zou H, Wang R, Zhou W, Kinoshita A et al. Inhibition of GSK3beta-mediated BACE1 expression reduces Alzheimer-associated phenotypes. J Clin Invest 2013; 123: 224–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo P, Mathews PM, Chishti MA, Schmidt SD, Gu Y, Yang J et al. Dissociated phenotypes in presenilin transgenic mice define functionally distinct gamma-secretases. Proc Natl Acad Sci USA 2005; 102: 8972–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConlogue L, Buttini M, Anderson JP, Brigham EF, Chen KS, Freedman SB et al. Partial reduction of BACE1 has dramatic effects on Alzheimer plaque and synaptic pathology in APP Transgenic Mice. J Biol Chem 2007; 282: 26326–34. [DOI] [PubMed] [Google Scholar]
- Meierjohann S, Hufnagel A, Wende E, Kleinschmidt MA, Wolf K, Friedl P et al. MMP13 mediates cell cycle progression in melanocytes and melanoma cells: in vitro studies of migration and proliferation. Mol Cancer 2010; 9: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menting KW, Claassen JA. beta-secretase inhibitor; a promising novel therapeutic drug in Alzheimer’s disease. Front Aging Neurosci 2014; 6: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihailovich M, Thermann R, Grohovaz F, Hentze MW, Zacchetti D. Complex translational regulation of BACE1 involves upstream AUGs and stimulatory elements within the 5’ untranslated region. Nucleic Acids Res 2007; 35: 2975–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 2007; 128: 257–67. [DOI] [PubMed] [Google Scholar]
- Morel E, Chamoun Z, Lasiecka Z, Chan R, Williamson R, Vetanovetz C et al. Phosphatidylinositol-3-phosphate regulates sorting and processing of amyloid precursor protein through the endosomal system. Nat Commun 2013; 4: 2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada M, Kita D, Teng L, Pyko IV, Watanabe T, Hayashi Y et al. Receptor tyrosine kinases: principles and functions in glioma invasion. Adv Exp Med Biol 2013; 986: 143–70. [DOI] [PubMed] [Google Scholar]
- Neves G, Cooke SF, Bliss TV. Synaptic plasticity, memory and the hippocampus: a neural network approach to causality. Nat Rev Neurosci 2008; 9: 65–75. [DOI] [PubMed] [Google Scholar]
- Nishimura R, Wakabayashi M, Hata K, Matsubara T, Honma S, Wakisaka S et al. Osterix regulates calcification and degradation of chondrogenic matrices through matrix metalloproteinase 13 (MMP13) expression in association with transcription factor Runx2 during endochondral ossification. J Biol Chem 2012; 287: 33179–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obregon D, Hou H, Deng J, Giunta B, Tian J, Darlington D et al. Soluble amyloid precursor protein-alpha modulates beta-secretase activity and amyloid-beta generation. Nat Commun 2012; 3: 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor T, Sadleir KR, Maus E, Velliquette RA, Zhao J, Cole SL et al. Phosphorylation of the translation initiation factor eIF2alpha increases BACE1 levels and promotes amyloidogenesis. Neuron 2008; 60: 988–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M, Sametsky EA, Younkin LH, Oakley H, Younkin SG, Citron M et al. BACE1 deficiency rescues memory deficits and cholinergic dysfunction in a mouse model of Alzheimer’s disease. Neuron 2004; 41: 27–33. [DOI] [PubMed] [Google Scholar]
- O’ Neill C. PI3-kinase/Akt/mTOR signaling: impaired on/off switches in aging, cognitive decline and Alzheimer’s disease. Exp Gerontol 2013; 48: 647–53. [DOI] [PubMed] [Google Scholar]
- Pape HC, Pare D. Plastic synaptic networks of the amygdala for the acquisition, expression, and extinction of conditioned fear. Physiol Rev 2010; 90: 419–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC et al. mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol 2011; 12: 235–45. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev 2002; 16: 2906–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preece P, Virley DJ, Costandi M, Coombes R, Moss SJ, Mudge AW et al. Beta-secretase (BACE) and GSK-3 mRNA levels in Alzheimer’s disease. Brain Res Mol Brain Res 2003; 116: 155–8. [DOI] [PubMed] [Google Scholar]
- Querfurth HW, LaFerla FM. Alzheimer’s disease. N Engl J Med 2010; 362: 329–44. [DOI] [PubMed] [Google Scholar]
- Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL et al. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J 2004; 23: 1761–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad T. Targeting RTK signaling pathways in cancer. Cancers (Basel) 2015; 7: 1758–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renhowe PA, Pecchi S, Shafer CM, Machajewski TD, Jazan EM, Taylor C et al. Design, structure-activity relationships and in vivo characterization of 4-amino-3-benzimidazol-2-ylhydroquinolin-2-ones: a novel class of receptor tyrosine kinase inhibitors. J Med Chem 2009; 52: 278–92. [DOI] [PubMed] [Google Scholar]
- Roland J-R, Helmut J. Alzheimer’s disease: from pathology to therapeutic approaches. Angew Chem Int Ed Engl 2009; 48: 3030–59. [DOI] [PubMed] [Google Scholar]
- Rossner S, Sastre M, Bourne K, Lichtenthaler SF. Transcriptional and translational regulation of BACE1 expression—implications for Alzheimer’s disease. Prog Neurobiol 2006; 79: 95–111. [DOI] [PubMed] [Google Scholar]
- Sadleir KR, Eimer WA, Kaufman RJ, Osten P, Vassar R. Genetic inhibition of phosphorylation of the translation initiation factor eIF2alpha does not block Abeta-dependent elevation of BACE1 and APP levels or reduce amyloid pathology in a mouse model of Alzheimer’s disease. PLoS One 2014; 9: e101643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarsoza F, Saing T, Kayed R, Dahlin R, Dick M, Broadwater-Hollifield C et al. A fibril-specific, conformation-dependent antibody recognizes a subset of Abeta plaques in Alzheimer disease, Down syndrome and Tg2576 transgenic mouse brain. Acta Neuropathol 2009; 118: 505–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheltens P, Blennow K, Breteler MM, de Strooper B, Frisoni GB, Salloway S et al. Alzheimer’s disease. Lancet 2016; 388: 505–17. [DOI] [PubMed] [Google Scholar]
- Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A et al. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell 2000; 6: 743–50. [DOI] [PubMed] [Google Scholar]
- Sengupta U, Nilson AN, Kayed R. The role of amyloid-beta oligomers in toxicity, propagation, and immunotherapy. EBioMedicine 2016; 6: 42–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian D, Parsyan A, Petroulakis E, Hershey J, Sonenberg N. eIF4B controls survival and proliferation and is regulated by proto-oncogenic signaling pathways. Cell Cycle 2010; 9: 4106–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J et al. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J 2006; 25: 2781–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer O, Marr RA, Rockenstein E, Crews L, Coufal NG, Gage FH et al. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat Neurosci 2005; 8: 1343–9. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009; 136: 731–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song I, Dityatev A. Crosstalk between glia, extracellular matrix and neurons. Brain Res Bull 2018; 136: 101–8. [DOI] [PubMed] [Google Scholar]
- Sun X, Bromley-Brits K, Song W. Regulation of beta-site APP-cleaving enzyme 1 gene expression and its role in Alzheimer’s disease. J Neurochem 2012; 120 (Suppl 1): 62–70. [DOI] [PubMed] [Google Scholar]
- Tamagno E, Bardini P, Obbili A, Vitali A, Borghi R, Zaccheo D et al. Oxidative stress increases expression and activity of BACE in NT2 neurons. Neurobiol Dis 2002; 10: 279–88. [DOI] [PubMed] [Google Scholar]
- van Gorp AG, van der Vos KE, Brenkman AB, Bremer A, van den Broek N, Zwartkruis F et al. AGC kinases regulate phosphorylation and activation of eukaryotic translation initiation factor 4B. Oncogene 2009; 28: 95–106. [DOI] [PubMed] [Google Scholar]
- Vassar R, Kovacs DM, Yan R, Wong PC. The beta-secretase enzyme BACE in health and Alzheimer’s disease: regulation, cell biology, function, and therapeutic potential. J Neurosci 2009; 29: 12787–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegh MJ, Heldring CM, Kamphuis W, Hijazi S, Timmerman AJ, Li KW et al. Reducing hippocampal extracellular matrix reverses early memory deficits in a mouse model of Alzheimer’s disease. Acta Neuropathol Commun 2014; 2: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent B, Checler F. alpha-Secretase in Alzheimer’s disease and beyond: mechanistic, regulation and function in the shedding of membrane proteins. Curr Alzheimer Res 2012; 9: 140–56. [DOI] [PubMed] [Google Scholar]
- Wang M, Sampson ER, Jin H, Li J, Ke QH, Im HJ et al. MMP13 is a critical target gene during the progression of osteoarthritis. Arthritis Res Ther 2013; 15: R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster SJ, Bachstetter AD, Nelson PT, Schmitt FA, Van Eldik LJ. Using mice to model Alzheimer’s dementia: an overview of the clinical disease and the preclinical behavioral changes in 10 mouse models. Front Genet 2014; 5: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Yu WH, Maloney B, Bailey J, Ma J, Marie I et al. Transcriptional regulation of beta-secretase by p25/cdk5 leads to enhanced amyloidogenic processing. Neuron 2008; 57: 680–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarczyk J, Mukhina I, Kaczmarek L, Dityatev A. Extracellular matrix molecules, their receptors, and secreted proteases in synaptic plasticity. Dev Neurobiol 2011; 71: 1040–53. [DOI] [PubMed] [Google Scholar]
- Xu L, Sun K, Xia M, Li X, Lu Y. MMP13 regulates aggressiveness of pediatric multiple myeloma through VEGF-C. Cell Physiol Biochem 2015; 36: 509–16. [DOI] [PubMed] [Google Scholar]
- Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther 2011; 10: 1533–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LB, Lindholm K, Yan R, Citron M, Xia W, Yang XL et al. Elevated beta-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat Med 2003; 9: 3–4. [DOI] [PubMed] [Google Scholar]
- Yurchenco PD. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol 2011; 3: a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha JS, Zhu BL, Liu L, Lai YJ, Long Y, Hu XT et al. Phorbol esters dPPA/dPA promote furin expression involving transcription factor CEBPbeta in neuronal cells. Oncotarget 2017; 8: 60159–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan J, He J, Zhou Y, Wu M, Liu Y, Shang F et al. Crosstalk between the autophagy-lysosome pathway and the ubiquitin-proteasome pathway in retinal pigment epithelial cells. Curr Mol Med 2016; 16: 487–95. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Li X, Qian X, Wang Y, Lee JH, Xia Y et al. Secreted and O-GlcNAcylated MIF binds to the human EGF receptor and inhibits its activation. Nat Cell Biol 2015; 17: 1348–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Hou H, Rezai-Zadeh K, Giunta B, Ruscin A, Gemma C et al. CD45 deficiency drives amyloid-beta peptide oligomers and neuronal loss in Alzheimer’s disease mice. J Neurosci 2011; 31: 1355–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, Peters F, Filser S, Herms J. Consequences of pharmacological BACE inhibition on synaptic structure and function. Biol Psychiatry 2018; 84: 478–87. [DOI] [PubMed] [Google Scholar]
- Zhu B, Zhou Y, Xu F, Shuai J, Li X, Fang W. Porcine circovirus type 2 induces autophagy via the AMPK/ERK/TSC2/mTOR signaling pathway in PK-15 cells. J Virol 2012; 86: 12003–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of present study are available from the corresponding author on reasonable request.