Abstract
Significance: Nicotinamide adenine dinucleotide (NAD+) spans diverse roles in biology, serving as both an important redox cofactor in metabolism and a substrate for signaling enzymes that regulate protein post-translational modifications (PTMs).
Critical Issues: Although the interactions between these different roles of NAD+ (and its reduced form NADH) have been considered, little attention has been paid to the role of compartmentation in these processes. Specifically, the role of NAD+ in metabolism is compartment specific (e.g., mitochondrial vs. cytosolic), affording a very different redox landscape for PTM-modulating enzymes such as sirtuins and poly(ADP-ribose) polymerases in different cell compartments. In addition, the orders of magnitude differences in expression levels between NAD+-dependent enzymes are often not considered when assuming the effects of bulk changes in NAD+ levels on their relative activities.
Recent Advances: In this review, we discuss the metabolic, nonmetabolic, redox, and enzyme substrate roles of cellular NAD+, and the recent discoveries regarding the interplay between these roles in different cell compartments.
Future Directions: Therapeutic implications for the compartmentation and manipulation of NAD+ biology are discussed. Antioxid. Redox Signal. 31, 623–642.
Keywords: nicotinamide adenine dinucleotide, redox, mitochondria, metabolism, glycolysis, compartmentation
Introduction
Nicotinamide adenine dinucleotide (NAD+) functions both as a redox cofactor in metabolism and as a substrate for several families of enzymes that regulate protein post-translational modifications (PTMs), including the silent information regulator 2p homolog family of lysine deacylases (sirtuins, SIRTs) and the poly(ADP-ribose) polymerases (PARPs). Given that NAD+ availability in vivo is likely to be limited, the general concept of competition for NAD+ between these different roles (redox cofactor vs. substrate) has not been extensively explored. Herein, we address the role of compartmentation in regulating the balance between NAD+ redox versus substrate roles. We focus on the cytosolic, mitochondrial, and nuclear compartments, since these are the most studied, with NAD(H) in other compartments such as endoplasmic reticulum (ER) or peroxisomes being beyond the scope of this review [see (166) for review].
Although the redox couple between NAD+ and its reduced form NADH is often considered alongside the redox couple between phosphorylated NAD+ (i.e., NADP+) and its reduced form (NADPH), these reactions are normally exclusive—NAD(H)-dependent enzymes do not typically use NADP(H) and vice versa. While there are some exceptions [e.g., glutamate dehydrogenase in mammals (39)], most enzymes (e.g., isocitrate dehydrogenase [IDH]) exist as separate NAD(H)- or NADP(H)-dependent isoforms encoded by different genes. The focus of this review will be NAD+, with NADP+ only mentioned as it pertains to NAD+ biology.
First, we discuss the synthesis and biological sources of NAD+, followed by fates of NAD+ both metabolic and redox, the interplay between these fates, and how compartmentation facilitates the different cellular roles of NAD+. Throughout this review, therapeutic approaches to manipulate cellular levels of NAD+ and its precursors or metabolites are discussed in the context of various diseases.
NAD+ Synthesis and Compartmentation
Routes of NAD+ biosynthesis
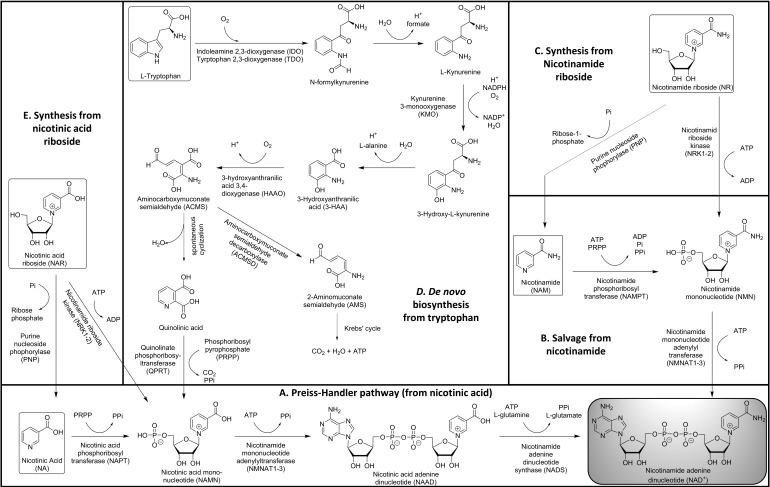
In mammalian cells, NAD+ can by synthesized using various forms of vitamin B3—nicotinic acid (NA), nicotinamide (NAM), or nicotinamide riboside (NR)—as the starting point. Alternatively, NAD+ can be synthesized de novo from tryptophan. NA, NAM, NR, and tryptophan are ingested through the diet and afford four main routes to NAD+ (Fig. 1A–D). Intermediates of NAD+ biosynthetic pathways such as nicotinamide mononucleotide (NMN) and alternative precursors such as nicotinic acid riboside (NAR) can also serve as sources for NAD+ synthesis (Fig. 1E) (81, 135). Besides dietary sources, products of NAD+-degrading enzymes such as SIRT, PARP, CD38, CD157 (all produce NAM), CD73 (NMN and NR), and NUDT13 (NMN) are also salvaged to replenish cellular NAD+ pools (note: these enzymes are discussed, and abbreviations are defined in the NAD+-Consuming Pathways and Compartmentalization section).
FIG. 1.
Biosynthesis of NAD+. Five biosynthetic routes to NAD+ in mammalian cells are shown, with NAD+ itself in the gray box at lower right. (A) The Priess–Handler pathway is the main synthetic route to NAD from precursors such as NA. (B) NAD+ can also be salvaged from NAM, the product of many NAD+-consuming enzymes. (C) NMN can be synthesized from NR, which is orally bioavailable. (D) NAMN in the Priess–Handler pathway (A) can be synthesized de novo from the amino acid tryptophan. (E) NA or NAMN can be synthesized from NAR. For full discussion, see text (the NAD+ Synthesis and Compartmentation section). NA, nicotinic acid; NAD+, nicotinamide adenine dinucleotide; NAM, nicotinamide; NAMN, nicotinic acid mononucleotide; NAR, nicotinic acid riboside; NMN, nicotinamide mononucleotide; NR, nicotinamide riboside.
The three-step pathway for NAD+ generation from NA is the Preiss–Handler pathway (Fig. 1A) (130, 131). The enzyme nicotinic acid phosphoribosyltransferase converts NA to nicotinic acid mononucleotide (NAMN) using the ribose-5-phosphate group of phosphoribosyl pyrophosphate (PRPP). NAMN is then adenylated by nicotinamide mononucleotide adenylyltransferase (NMNAT) using adenosine triphosphate (ATP) to yield nicotinic acid adenine dinucleotide (NAAD). Three isoforms of NMNAT have been reported—NMNAT1 is nuclear, NMNAT2 is in the cytosol and Golgi apparatus, and NMNAT3 is mitochondrial (12). In the final step, the cytosolic enzyme NAD+ synthase amidates NAAD using l-glutamine and ATP to give NAD+.
The two-step salvage pathway to recycle NAM into NAD+ is similar to the first two steps of the Preiss–Handler pathway (Fig. 1B). Using PRPP, nicotinamide phosphoribosyltransferase (NAMPT) transforms NAM to NMN. This is the rate-limiting step in the salvage of NAM (136). NAMPT is primarily located in the nucleus and cytosol. Early research reported the presence of NAMPT in mitochondria (172), although more recently advanced techniques have led to this finding being disputed (33, 128). NMNAT adenylates NMN in the presence of ATP to give NAD+. Since many NAD+-consuming enzymes produce NAM as a product, this pathway is a key mechanism for the maintenance of cellular NAD+ levels. Furthermore, NAM is a feedback inhibitor of NAD+-consuming enzymes such as SIRTs, so swift removal of NAM is required to maintain these enzymes' activities.
Synthesis of NAD+ from NR can proceed one of two ways (Fig. 1C). Predominantly, NR is phosphorylated by nicotinamide riboside kinases 1 and 2 (NRK1/NRK2) using ATP to give NMN (16), which is then converted to NAD+ by the NMNATs. The phosphorylation step is rate limiting for the use of exogenous NR in NAD+ synthesis (135). An alternate route involves the cleavage of the glycosidic linkage in NR by purine nucleoside phosphorylase (PNP) to make NAM, which subsequently enters the salvage pathway to give NAD+ (10, 46). Besides NR, both NRKs and PNP also accept NAR as a substrate to give NAMN and NA as respective products (10, 158). Both compounds then feed into the Preiss–Handler pathway.
The de novo synthesis of NAD+ from dietary tryptophan occurs via the kynurenine pathway (Fig. 1D) (11, 71). The first and rate-limiting step requires oxygen to convert l-tryptophan to N-formylkynurenine. This transformation is performed by two enzymes—indoleamine-2,3-dioxygenase and tryptophan-2,3-dioxygenase, the latter of which is liver-specific in mammals. Both these cytosolic enzymes are frequently overexpressed in a variety of cancers and confer immune resistance to cancer cells (68, 129, 177). The subsequent four steps include alternating hydrolysis and oxidation. Spontaneous hydrolytic removal of formate from N-formylkynurenine gives l-kynurenine, which is oxidized by kynurenine 3-monooxygenase to 3-hydroxy-l-kynurenine using oxygen and NADPH. This is followed by the formation of 3-hydroxyanthranilic acid (3-HAA) by removal of l-alanine. Subsequent oxidation of 3-HAA by 3-hydroxyanthranilic acid 3,4-dioxygenase gives an unstable intermediate aminocarboxymuconate semialdedhye (ACMS). ACMS can go two ways, only one of which yields NAD+: Normally, enzymatic conversion of ACMS to 2-aminomuconate semialdehyde (AMS) by aminocarboxymuconate semialdehyde decarboxylase (ACMSD) occurs, with AMS subsequently converted to acetyl-CoA. Alternatively, when ACMSD is saturated by high concentrations of ACMS, it spontaneously cyclizes, forming quinolinic acid, which is then adenylated and decarboxylated by quinolinate phosphoribosyltransferase, another rate-limiting enzyme that gives NAMN, which feeds into the Preiss–Handler pathway (11).
Cellular uptake of NAD+ precursors
Tryptophan is taken up into cells via carrier proteins that also transport large neutral amino acids (Fig. 2) (118). Among the different forms of vitamin B3, cellular uptake of NA is mediated by membrane carrier systems potentially including either a pH-dependent anion antiporter or a proton cotransporter (107, 144, 156). The exact mechanism of cellular NAM uptake, either as direct transport in intact form or conversion to salvage pathway metabolites, which are then taken up, is still unknown. NAMPT, the enzyme that converts NAM to NMN, is found both intracellularly and extracellularly, indicating that both routes of NAM uptake are possible. However, NAMPTs substrates ATP and PRPP were shown to be not available in sufficient quantities in the extracellular space (63). Studies in rodents have indicated that NAM is directly absorbed by the intestines (29, 55). The third form of vitamin B3, NR, is directly absorbed into cells with the help of equilibrative nucleoside transporters (ENTs) (Fig. 2) (111, 153).
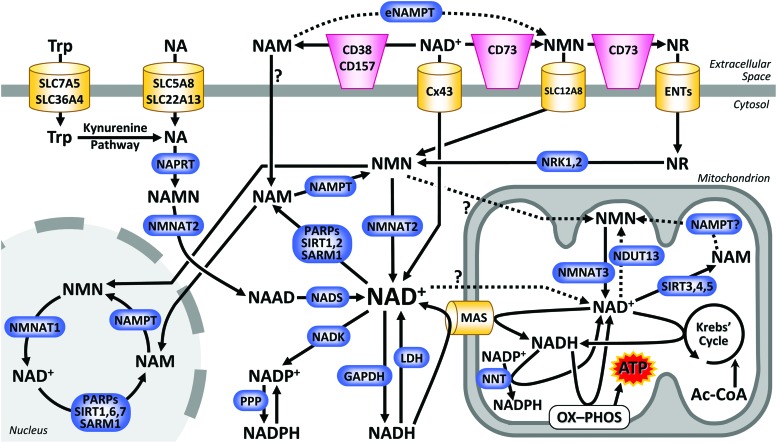
FIG. 2.
Compartmentation of NAD+ biology. The cellular compartments considered herein are cytosol, nucleus, mitochondrion, and extracellular space. The major routes for import, biosynthesis, and consumption of NAD+ in each of these compartments are shown. Dotted lines and question marks denote pathways for which limited evidence is available. ATP, adenosine triphosphate; ENT, equilibrative nucleoside transporter; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; LDH, lactate dehydrogenase; MAS, malate-aspartate shuttle; NAAD, nicotinic acid adenine; NADH, nicotinamide adenine dinucleotide reduced form; NADK, NAD+ kinase; NADP+, nicotinamide adenine; NADPH, nicotinamide adenine dinucleotide phosphate reduced form; NAMPT, nicotinamide phosphoribosyltransferase; NMNAT, nicotinamide mononucleotide adenylyltransferase; NNT, nicotinamide nucleotide transhydrogenase; NRK, nicotinamide riboside kinase; OX-PHOS, oxidative phosphorylation; PARP, poly(ADP-ribose) polymerase; PPP, pentose phosphate pathway; SARM1, sterile alpha and TIR motif-containing protein 1; SIRT, sirtuin. Color images are available online.
NMN, an intermediate in the NAD+ salvage pathway (Fig. 1B), can itself serve as an exogenous precursor for NAD+ and has been shown to increase NAD+ levels in mice (135). Until recently, there was debate regarding whether NMN is directly taken up by cells or must undergo metabolism before uptake. Several researchers proposed that NMN is extracellularly dephosphorylated to NR, which then enters the cell (Fig. 2) (9, 57, 111), whereas others suggested that the cell membrane is permeable to NMN (47). It has been argued that NMN dephosphorylation is likely a cell-type-specific phenomenon, and intact NMN can be rapidly transported into certain cell types such as hepatocytes (102, 176). However, recent studies showed that even in hepatocytes, NRK1 is necessary and rate limiting for the use of exogenous NMN for NAD+ synthesis (135). This suggests that if NMN was directly taken up into the cell, it would solely require the NMNAT activity for NAD+ synthesis. However, NRK1 deficiency, inhibition of CD73 (the cell surface protein thought to convert extracellular NMN to NR), and inhibition of ENTs all prevented NAD+ generation from exogenous NMN, providing evidence for conversion of NMN to NR before cellular uptake. The recent discovery of a specific NMN transporter, Slc12a8 (56), will require careful analysis of its cell type expression patterns to resolve this debate.
NAD(H) membrane transport
The transport of NAD(H) across biological membranes has been similarly controversial. The canonical view is that NAD+ is unable to cross lipid bilayers (34). The brush border intestinal lining has been shown to hydrolyze NAD+ to NMN, which is further hydrolyzed to NR and NAM (55). The cell surface protein CD73 has also been shown to catalyze the conversion of NAD+ to NR via NMN (48, 57). However, dedicated NAD+ transporters are thought to exist. One way of importing NAD+ is via connexin 43 (Cx43) channels, which are permeable to NAD+ (22) and are highly expressed on cardiomyocytes. Indeed, exogenous NAD+ was shown to block cardiac hypertrophy, and this effect was abolished in the presence of the Cx43 blocker carbenoxolone (126). However, despite considerable evidence that exogenous NAD+ (without prior hydrolysis) can be taken up into different types of mammalian cells (17, 127, 167, 175), besides Cx43 the transporters responsible for this are still to be discovered.
Source of mitochondrial NAD+
Whereas cytosolic and nuclear NAD+ pools are considered to be exchangeable via diffusion through the nuclear pore, the canonical view is that the mitochondrial NAD+ pool is sequestered due to the impermeability of the inner mitochondrial membrane. Whereas NMNAT3 is found to localize to the mitochondrial matrix, NAMPT is not (111). As such, cytosolic NMN (but not NAM) is widely believed to be the precursor of mitochondrial NAD+, although the exact mechanism of its mitochondrial transport is unknown. Recent evidence suggests that intact NAD+ is able to cross the inner mitochondrial membrane (33, 42, 141), and this was conclusively demonstrated using isotope labeling experiments (33). Whether it is NAD+ or NADH that crosses into mitochondria is still unknown, and the possibility that NMNAT3 still contributes to mitochondrial NAD+ via NMN import has not been ruled out. Indeed, experiments using a genetically encoded biosensor have shown that the mitochondrial NAD+ pool is maintained by both direct import of cytosolic NAD+ and synthesis from imported NMN (23). While an NAD+ transporter exists in bacteria (59) and has also been identified in mitochondria of yeast (159) and plants (119), the search for its mammalian counterpart continues.
NAD+-Consuming Pathways and Compartmentalization
Poly(ADP-ribose) polymerases
PARP is a family of enzymes that transfer adenosine diphosphate (ADP)-ribose (ADPR) onto target proteins (Fig. 3). There are 17 members of the PARP family in humans (69) including both poly-ADPR polymerases (PARP-1, 2, and 5) and mono-ADPR transferases (PARP-3, 4, 6, 7, 10, 12, and 14–16) (90). PARPs are predominantly located in the nucleus (PARPs 1–5, 9, and 14) but are also found in the cytoplasm (PARP-5, 10, 12, 13, and 15), cell membrane (PARP-4, 9, 14, and 16), and ER (PARP-16) (5, 90). Activity (poly vs. mono ADP-ribosylation) and location of certain PARP isoforms are still unknown (e.g., PARP-8 and 11).
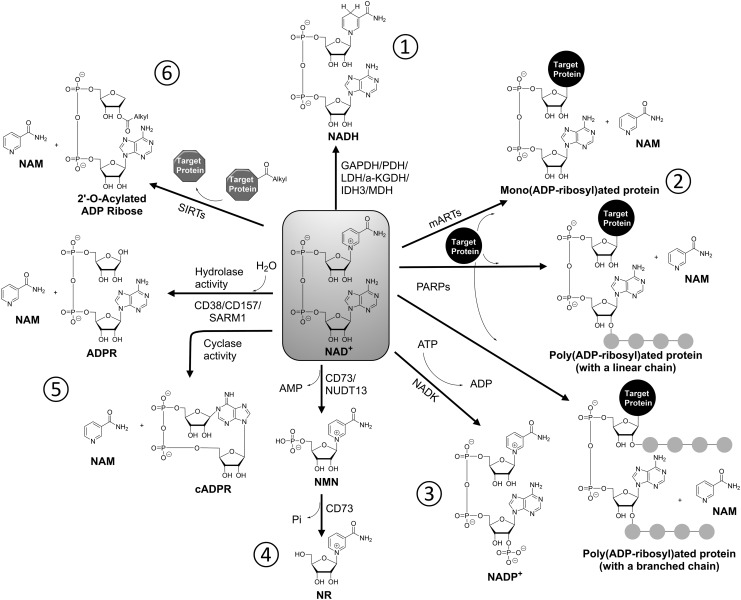
FIG. 3.
Cellular fates of NAD+. NAD+ has numerous fates in the cell; clockwise from top. (1) Numerous metabolic enzymes use NAD+ as redox cofactor, with the reduced form (NADH) being a cellular “reducing equivalent” used to drive biosynthetic and other reactions. (2) mARTs and PARPs catalyze the adduction of ADP-ribosyl groups onto proteins, either singly or in linear or branched chains. This is an important protein post-translational modification that regulates protein function. (3) NAD+ can be phosphorylated to make NADP+, which is used as a redox cofactor like NAD+. Its reduced form (NADPH) is used to drive many antioxidant enzymes important for defense against oxidative stress. (4) Cell surface CD73 and mitochondrial nudix hydrolase 13 (NUDT13) enzymes have the pyrophosphohydrolase activity, converting NAD+ to NMN. CD73 further converts NMN into NR and plays an important role in cellular uptake of NAD+ and NMN. (5) CD38/CD57/SARM1 enzymes can perform either (i) a cyclase activity that generates NAM and cyclic ADPR (cADPR), the latter being an important cell signaling second messenger, or (ii) a hydrolase activity that generates NAM and ADPR. CD hydrolase is considered an NAD+ degradation pathway. (6) The SIRT family of lysine deacylases use NAD+ as a substrate, generating NAM while removing an acyl group (e.g., acetyl, succinyl, malonyl) from a protein lysine residue to form 2′-O-acyl-ADPR. ADPR, adenosine diphosphate ribose; mARTs, mono-ADPR transferases.
PARPs bind NAD+ and cleave it into NAM and ADP-ribose. Depending on the activity of the particular PARP isoform, the target protein is either mono or poly(ADP-ribosyl)ated (PARylation). PARylation can either be linear or branched (Fig. 3). PAR polymers can reach lengths of 200 ADPR units (110). Typical amino acid residues of the target protein onto which ADPR is transferred include aspartate, glutamate, asparagine, arginine, lysine, serine, and cysteine (89). The majority of PARP substrates are nuclear proteins involved in DNA synthesis and repair, nucleic acid metabolism, and chromatin structure modulation (104). PARP-1 is also known to PARylate itself (auto-PARylation) (in addition to being PARylated by PARP-2) and is one of the main acceptors of poly-ADPR in vivo (80). PARylation is a dynamic process, and PAR polymer is rapidly degraded by poly(ADPR) glycohydrolase (PARG) and ADPR protein lyase (half-life <1 min) (160).
PARPs are thought to be responsible for the majority of NAD+ consumption both constitutively and upon hyperactivation during DNA damage (140). PARP-1 (85%–90%) and PARP-2 (10%–15%) contribute to most of the total PARP activity (155). The main role of PARPs is to detect DNA damage and initiate DNA repair (38). The extent of PARP activation following DNA damage determines the fate of the cell (117). If the damage is repairable, PARP activation contributes to cell survival by engaging DNA repair machinery. More severe damage engages apoptotic machinery in which caspases cleave PARP-1, thereby inactivating it and preventing the loss of cellular ATP, which is then used to dispose of the cell in a safe manner. In case of extensive DNA damage (e.g., in tissue ischemia and reperfusion injury), PARP hyperactivation causes severe depletion of cellular NAD+ stores. Salvage of NAD+ from the resulting NAM is an energy intensive process, which depletes cellular ATP levels. As a result, energy intensive apoptotic machinery is not engaged, and the cell undergoes necrosis. Besides DNA repair, PARPs are involved in regulating inflammation, metabolism, and cell proliferation [reviewed in detail in (5, 94, 104)].
Sirtuins
Sirtuins are a class of NAD+-dependent proteins with deacylase activity. The term “sirtuin” comes from the first member of the family, Sir2 (silent information regulator two), identified in yeast as a protein responsible for transcriptional silencing (137). In mammals, there are seven Sir2 homologs identified as SIRT1–SIRT7. SIRT1, SIRT6, and SIRT7 are predominantly nuclear, SIRT2 is cytosolic, and SIRT3–SIRT5 are mitochondrial (101). Under certain conditions, SIRTs may be present in subcellular compartments different from their primary location. For example, in cardiomyocytes, SIRT1 is located in the nucleus during development but is mostly cytosolic in adult (157). SIRT1 is known to shuttle between the nucleus and the cytosol in response to pathological conditions or stress (70, 171). SIRT3 is also known to localize and function in the nucleus in its full-length form, whereas the processed short form is mitochondrial (73).
The classical reaction catalyzed by sirtuins is the removal of an acetyl group from protein lysine residues, using NAD+ as an acetyl acceptor. In this process, NAM is released first and the ADPR moiety is covalently attached to the acetyl group on the protein lysine residue. The amide bond between acetyl carbonyl functionality and ɛ-amino group of lysine is then cleaved, thereby liberating the deacetylated lysine and 2′-O-acetyl-ADP ribose (61). While deacetylation is the main activity of sirtuins, they are also capable of removing other acyl groups in a target protein- and isoform-specific manner. SIRT1–SIRT3 and SIRT7 have the deacetylase activity. SIRT5 has demalonylase and desuccinylase activities in addition to deacetylase (36). SIRT4 has been shown to act as an ADPR transferase, catalyzing the release of NAM, but not the cleavage of the amide bond in the second step of the reaction (60). Like SIRT4, SIRT6 also has the ADPR transferase activity (88) and also removes long-chain fatty acid groups like myristoyl and palmitoyl from lysines (74). Because NAD+ is necessary for the sirtuin activity, sirtuins can act as a direct link between cellular metabolic status and protein PTMs, in effect acting as “metabolic sentinels,” in a manner similar to adenosine monophosphate (AMP)-dependent protein kinase (66). Additionally, NAM, which is a product of the reaction, is a noncompetitive inhibitor of sirtuins.
Sirtuins regulate a plethora of mammalian proteins involved in diverse processes, such as energy metabolism, genomic stability, inflammation, cellular stress resistance, circadian rhythms, tumorigenesis, autophagy, and apoptosis [reviewed in (43, 61)]. Notably, the NAD+ consumption potential of sirtuins (i.e., specific activity, combined with relative expression levels vs. other NAD+ consumers) is thought to be significantly lower than that of PARPs, as discussed in the Competition for NAD+ among SIRTs/PARPs/CDs section.
Glycohydrolases
Cluster of differentiation (CD) proteins CD38 and CD157 (Fig. 2) are cell surface glycohydrolases that cleave a glycosidic linkage within NAD+ to release NAM and ADPR (Fig. 3) (27, 132). ADPR is a product of the hydrolase activity (i.e., it uses water). Both CD38 and CD157 also have the cyclase activity to a smaller extent, in which the NAD+ molecule is broken down into NAM and cyclic ADPR (cADPR), without using water. The NAM generated in the reactions rapidly enters the salvage pathway to regenerate NAD+. Both ADPR and cADPR act as second messengers to regulate cytosolic Ca2+ flux. This in turn activates various signaling pathways critical for energy metabolism, muscle contraction, immune response, and cell adhesion (58, 121, 132). In addition, recent work has found that CD38 inhibitors can confer protection against cardiac ischemia-reperfusion injury, potentially by enhancing NAD+ availability for the SIRT activity (21) (see the Balancing NAD Consumption and Redox in a Compartment-Specific Manner section). Besides ADPR and cADPR, CD38 and CD157 also generate nicotinic acid adenine dinucleotide phosphate from NADP+, which also acts as a second messenger for Ca2+ mobilization (96).
Sterile alpha and toll-interleukin receptor (TIR) motif-containing 1 (SARM1) protein is the first member of recently discovered class of proteins, which contains two putative protein–protein interaction domains—sterile alpha and TIR—and was thought to play a role in innate immunity in Caenorhabditis elegans (31, 103). In recent years, a central role for NAD+ metabolism in neurodegeneration has emerged, where SARM1 initiates cell destruction by mediating NAD+ depletion while NMNAT1 protects neurons by maintaining NAD+ levels (41, 49, 114, 146) [reviewed in (30, 50)]. Interestingly, SARM1's NADase activity is mediated by the TIR domain, which usually is responsible for protein–protein interactions, and is the first example of a TIR domain exhibiting enzymatic activity. SARM1 is triggered by neuronal injury and catalyzes cleavage of NAD+ into ADPR, cADPR, and NAM (Fig. 3), with NAM acting as a feedback inhibitor of SARM1 (41). Overexpression of NAMPT or NMNAT or supplementation with NR increases NAD+ synthesis and counteracts axon destruction by SARM1 (49). As such, SARM1 inhibition is emerging as a promising therapeutic strategy in the treatment of acute neuronal injury as well as neurodegeneration and aging.
Pyrophosphohydrolases
CD73 is a cell surface enzyme (Fig. 2) with primary activity as 5′-nucleotidase acting on AMP. It was recently reported to also have the pyrophosphohydrolase activity to convert NAD+ to NMN, which is then further converted to NR by its 5′-nucleotidase activity (57). CD73 is required for use of NMN as extracellular NAD+ precursor (Fig. 3) (153).
Nudix hydrolases are a superfamily of hydrolytic “housecleaning” enzymes that catalyze the cleavage of nucleoside diphosphates linked to x (i.e., any moiety) (14). The human genome has 24 nudix hydrolase genes (99), and the enzymes hydrolyze a wide variety of substrates including organic pyrophosphates such as nucleoside di- and triphosphates and dinucleotides, nucleotide sugars, and RNA caps, with varying degrees substrate specificity (99). NUDT13, a member of this superfamily with the pyrophosphohydrolase activity, catalyzes the hydrolysis of NADH to NMNH and AMP, and NADPH to NMNH and 2′,5′-ADP (2). NUDT13 showed marked preference for the reduced form of pyridine nucleotides with its KM of 0.34 mM for NADH and was shown to localize exclusively to mitochondria. Related human NUDT12 protein hydrolyzes NADH and NADPH with 20-fold greater efficiency than NAD+ and localizes to peroxisomes (1). NUDT12 and NUDT13 are thought to regulate the NAD(P)+/NAD(P)H ratio within peroxisomes, mitochondria, and possibly also in the cytosol (99).
NADH/NAD+ Metabolic and Redox Roles
Glycolysis
The process by which glucose and other hexoses are lysed to yield the triose pyruvate consists of two halves (Fig. 4). The first or upper part of glycolysis comprises breakdown of glucose into two interchangeable trioses—dihydroxyacetone phosphate and glyceraldehyde-3-phosphate—and consumes two molecules of ATP, the first at hexokinase (65). In the lower half of glycolysis, glyceraldehyde-3-phosphate is converted to pyruvate, yielding two ATPs. The lower reactions occur twice for each glucose, yielding net two ATP per glucose for glycolysis as a whole (145). The first reaction of this lower part is catalyzed by glyceraldehyde-3-phosphate dehydrogenase (GAPDH), one of the most studied enzymes in all of biology. GAPDH is famously used as a “housekeeping” protein or gene in Western blotting or quantitative polymerase chain reaction experiments. GAPDH uses NAD+ and makes NADH. As such, despite the dogma that phosphofructokinase is the rate-limiting step of glycolysis (98), this requirement for NAD+ is such that without the oxidized cofactor, glycolysis cannot proceed. Indeed, in the absence of mitochondrial function, pyruvate is converted to lactate by lactate dehydrogenase (LDH), thus recycling NADH to NAD+ and permitting glycolysis to continue. It could be argued that LDH exists for the sole purpose of recycling NADH. Alternatively, NADH from glycolysis is recycled back to NAD+ via the malate-aspartate shuttle (MAS), transmitting NADH reducing equivalents into mitochondria to join the NADH pool generated by the Krebs' cycle (see the Krebs' Cycle section and Fig. 4). In some metabolically active tissues, cytosolic NADH reducing equivalency can also be shuttled into mitochondria via the glycerol-3-phosphate dehydrogenase system, linked to the reduction of coenzyme Q10 at the mitochondrial inner membrane (105). Furthermore, it was recently reported that the process of desaturating fatty acids is a significant pathway for recycling of NADH to NAD+ in the cytosol (78).
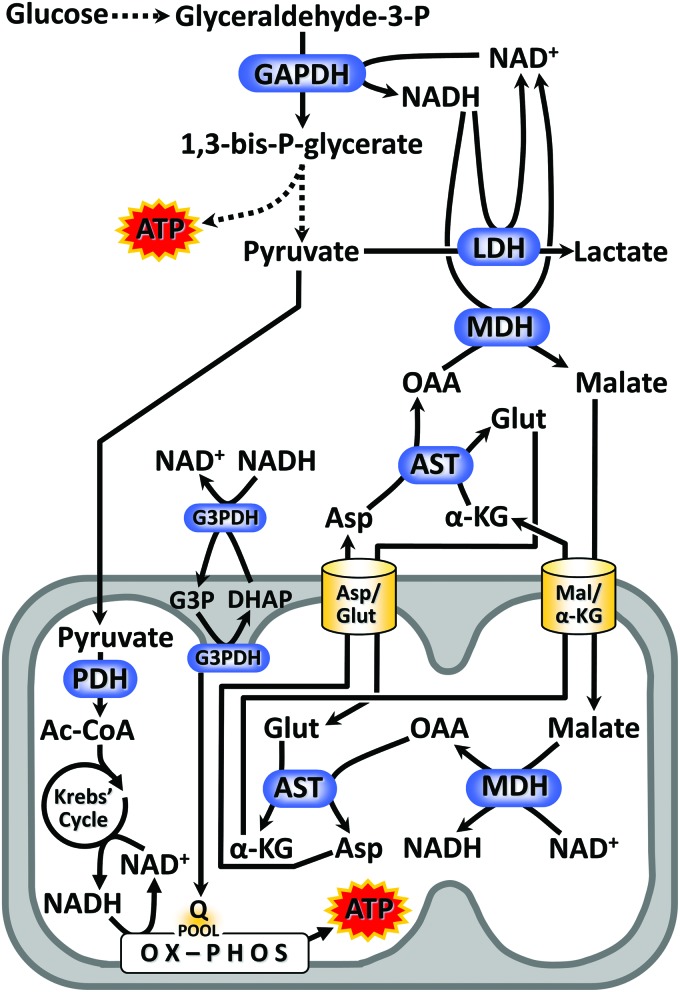
FIG. 4.
NADH/NAD+ redox in metabolism. The enzyme GAPDH, a key step in glycolysis, reduces NAD+ to NADH. Depending on the functionality of mitochondria, glycolytic NADH has two different routes for reoxidation. In the presence of oxygen (the so called “aerobic glycolysis”), the reducing equivalency of NADH is transmitted into mitochondria via the MAS, comprising cytosolic plus mitochondrial isoforms of MDH working in opposing directions, and cytosolic plus mitochondrial isoforms of AST working in opposing directions. In addition the G3PDH shuttle can transfer NADH reducing equivalents into mitochondria. The mitochondrial import/export of the relevant α-keto acids and amino acids is handled by a pair of electrophoretically driven membrane exchange proteins. The end product of glycolysis, pyruvate, is imported to mitochondria and further oxidized by the Krebs' cycle. In the absence of adequate mitochondrial function (e.g., hypoxia), glycolytic NADH is instead reoxidized by LDH, generating lactic acid as a terminal product (i.e., “anaerobic glycolysis”). In either scenario, the reoxidation of NADH is an absolute requirement for glycolysis to function. AST, aspartate aminotransferase; G3PDH, glycerol-3-phosphate dehydrogenase. Color images are available online.
Recently, we showed that acute dosing with bolus NAD+ precursors such as NMN induced a massive and transient cytosolic acidosis, attributable to a burst of glycolytic activity (109). The importance of this phenomenon to the wide-ranging effects of oral NMN supplementation (102) is underappreciated. Furthermore, it should be emphasized that the relative expression levels of GAPDH versus other NAD+-consuming enzymes (SIRTs, PARPs, etc.) are such that the bulk of NAD+ from bolus additions such as this is likely consumed by GAPDH (see the Competition for NAD+ between redox/metabolic cofactor enzymes versus NAD+ consumers section).
Krebs' cycle
The entry of the glycolytic end product pyruvate into the Krebs' cycle in the mitochondrial matrix requires its conversion to acetyl-CoA via pyruvate dehydrogenase (PDH), an NADH-generating enzyme (Figs. 4 and 5). The cycle contains three enzymes that reduce NAD+ to NADH, namely α-ketoglutarate dehydrogenase, malate dehydrogenase (MDH), and IDH3. There are three isoforms of IDH: IDH1 uses NADP+ and resides in the cytosol. IDH2 is mitochondrial and uses NADP+. IDH3 is the NAD+-dependent mitochondrial isoform that is largely responsible for maintaining Krebs' cycle flux for cellular bioenergetic purposes (145). The mitochondrial NADP+-dependent IDH2 is likely a source of NADPH within the organelle, with the latter required for antioxidant functions (e.g., mitochondrial glutathione reductase [GR]). Further discussion of NADPH-dependent antioxidants is beyond the scope of this review.
FIG. 5.
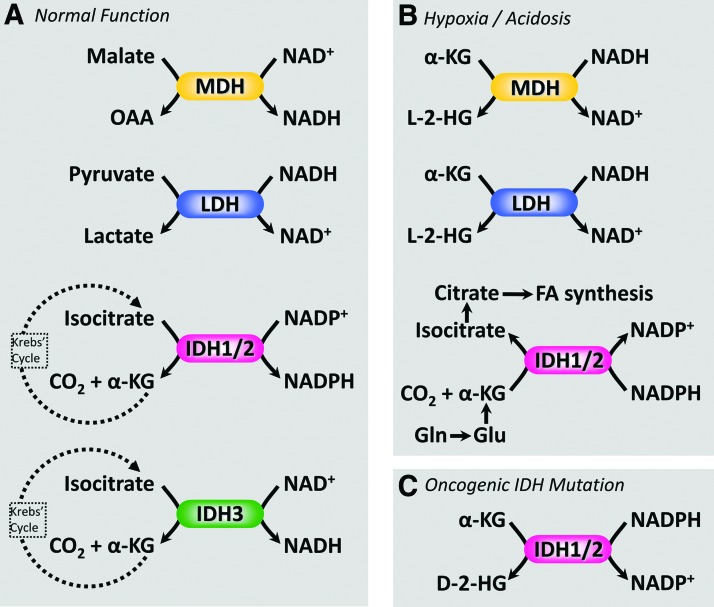
Noncanonical functions of NAD(P)-dependent dehydrogenases. Several pathophysiological conditions can manifest neomorphic enzyme activities from NAD(H)- and NADP(H)-dependent dehydrogenases such as those found in the Krebs' cycle. (A) Normal (canonical) function of dehydrogenases: MDH oxidizes malate to OAA, reducing NAD+ to NADH. LDH reduces pyruvate to lactate, oxidizing NADH to NAD+. IDH isoforms 1 and 2 decarboxylate isocitrate to α-KG in an NADP+ dependent manner, whereas IDH isoform 3 performs the same reaction in an NAD+-dependent manner. (B) Under conditions of hypoxia and/or acidosis, both MDH and LDH can use NADH to reduce α-KG to the l-enantiomer of 2-hydroxyglutarate (l-2-HG). In addition, IDH 1/2 can operate in reverse, using NADPH to perform reductive carboxylation of α-KG to form isocitrate. The latter is an important biosynthetic pathway in cancer cells, using α-KG derived from the amino acids glutamine or glutamate, to fuel fatty acid synthesis for biomembranes. (C) Mutations in IDH 1/2 are associated with aggressive forms of cancer such as glioma and result in a neomorphic activity, that is, the ability to use NADPH to reduce α-KG to the d-enantiomer of 2-hydroxyglutarate (d-2-HG). For this reason, d-2-HG is commonly referred to as an “oncometabolite.” α-KG, α-ketoglutarate; IDH, isocitrate dehydrogenase; OAA, oxaloacetate. Color images are available online.
While normally the canonical (clockwise) function of the Krebs' cycle results in the generation of NADH, there are a couple of scenarios in which noncanonical functions may exist (Fig. 5): (i) Mutant forms of IDH1 or IDH2 associated with cancer elicit a neomorphic activity that uses NADPH and α-ketoglutarate (α-KG) as substrates to generate NADP+ plus the “oncometabolite” d-2-hydroxyglutarate (d-2-HG) (32). In addition, under acidic or hypoxic conditions, either MDH or LDH can adopt a similar neomorphic activity to use NADH to reduce α-KG to the l-enantiomer of 2-HG (72, 108, 113). (ii) Under acidic or hypoxic conditions, mitochondrial IDH2 or IDH3 can run in the reverse direction to convert α-KG back to citrate (108, 169). Such reductive carboxylation of α-KG uses NADH (for IDH3) or NADPH (for IDH2) as reductants and also consumes CO2 and a proton. This reversal of IDH to eventually generate citrate is a component of cancer metabolism since citrate can be exported from mitochondria and used by ATP-citrate lyase to generate acetyl-CoA, which is then used for lipid biosynthesis (151). By sourcing α-KG from the amino acid glutamine (a preferred fuel for cancer cells), carbon is thus diverted from glutamine to building lipids, an important process for dividing cancer cells. As such, ATP-citrate lyase is an emerging cancer drug target (52).
The noncanonical NADH-consuming reactions of the Krebs' cycle have recently been claimed to be part of a redox sink that exists in hypoxia (92). Specifically, it has been hypothesized that by sinking electrons from NADH into metabolites such as l-2-HG, the cell may avoid “reductive stress” (92). However, it should be noted that the bulk flux afforded by such pathways is unlikely to be sufficient to impact global redox status. Specifically, for IDH2/MDH/LDH, their rates of conversion of α-KG to l-2-HG are at least three orders of magnitude slower than the canonical activities of these enzymes (108). Furthermore, it has been claimed that l-2-HG and similar metabolic dead-ends may be “redox reservoirs” because reverse enzymes exist to return 2-HG to α-KG (92). Again, however, it should be noted that the reverse reactions (catalyzed by l- or d-2-HG dehydrogenases) do not yield NADH (143). Rather, they rely on electron transfer to a flavin adenine dinucleotide (FAD) and then to the ubiquinone cycle embedded in the mitochondrial membrane. Thus, these reactions are not true reservoirs because the product yielded (FADH2) is not the same as the one initially deposited (NADH). Notably, electrons entering the ubiquinone cycle in this manner bypass complex I and thus converting NADH into a 2-HG reservoir and back again sacrifices the proton pumping that would have occurred at complex I.
While these issues do not detract from the importance of 2-HG as a signaling molecule (170, 173), beyond the scope of this review, they highlight that the magnitude and directionality of metabolic fluxes should consider all conditions including relative concentrations and rates for the reactions involved.
Mitochondrial electron transport chain
Complex I of the mitochondrial respiratory chain is the primary consumer of NADH in the organelle, passing electrons via a flavin mononucleotide (FMN) and along a “molecular wire” comprising at least nine Fe-S centers, eventually onto coenzyme Q10 (ubiquinone), and resulting in the pumping of protons from the matrix to the intermembrane space, yielding the energetic intermediate of oxidative phosphorylation (OX-PHOS), the transmembrane proton gradient (145).
Under conditions in which complex I is inhibited (e.g., electron transport chain [ETC] inhibition by lack of oxygen), the NADH/NAD+ ratio increases, and the convenient autofluorescence of NADH (at 340 nm excitation) is often used as a readout of this phenomenon (147). In this regard, it should be noted that a significant portion of the NADH/NAD+ pool in mitochondria is thought to be bound to either complex I or the dehydrogenases of the Krebs' cycle, which physically interact with it (20). A study using “ghost native” electrophoresis, enabling direct measurements of NADH autofluorescence bound to the complexes, found that the binding of NADH to complex I significantly enhances its fluorescent efficiency (20). As such, measurements of NADH fluorescence are thought to primarily report the bound pool of NADH, not free NADH in the mitochondrial matrix. Thus, it cannot be assumed that changes in NADH fluorescence alone represent meaningful changes in the concentration of free NAD(H) available for use by other enzymes, such as SIRTs. Recent technological advances permitting fluorescent distinction between NADH and NADPH (19) may provide further insight to the interplay of nucleotide pools in mitochondria (18).
Although beyond the scope of this review, it cannot be ignored that the mitochondrial respiratory chain, in particular complexes I and III, is a major source of cellular reactive oxygen species (ROS) (106). Complex I is thought to generate superoxide anion radical (O2•−) from either its matrix-located FMN site or its ubiquinone-binding site. Complex III has two ubiquinone-binding sites, with the outside-facing (QO) site (also known as the P-site) being the predominant ROS source. At these sites, the ubisemiquinone radical (Q•) can transfer an electron directly onto molecular oxygen to generate O2•−.
In the situation of reperfusion injury following tissue ischemia, a large burst of ROS generation occurs, and the primary source of these ROS is thought to be the reversal of electron transport through complex I, resulting in O2•− generation at the upstream FMN site (28). Such reverse electron transfer (RET) is thought to be driven by a hyperreduced ubiquinol pool during early reperfusion, which originates from an abundance of succinate, driving ubiquinone reduction by complex II. It is important to state that RET does not necessarily proceed all the way to the NAD(H)-binding site of complex I, and the formation of NADH is not typically observed with RET. Indeed, [NADH] is already very high at the beginning of reperfusion, and this high NADH level may in fact contribute to the diversion of electrons out of the FMN site to form ROS. Regardless the precise mechanisms, the mitochondrion is the proximal source for much of the ROS that the cellular antioxidant machinery has evolved to detoxify.
Cytosolic and mitochondrial NADPH
Although the focus of this review is NADH/NAD+, the relationship between NAD+ and its phosphorylated congener NADP+ is highly compartment specific. In the cytosol, NAD+ kinase (NADK, Fig. 3) phosphorylates NAD+ to NADP+. NADK was first isolated from Saccharomyces cerevisiae by Kornberg and Pricer in 1950 (79) and has since been purified from other organisms. Unlike prokaryotic NADKs, which use inorganic phosphates, human cytosolic NADK requires ATP and Mg2+ as cofactors for conversion of NAD+ to NADP+ (83). NADK is the only pathway for de novo NADP+/NADPH biosynthesis. Given the differences in the active site structure and cofactors between prokaryotic and mammalian NADKs, efforts to develop NADK inhibitors as antibacterial agents are underway (15, 120, 124, 150). In recent years, inhibition of human NADK for the treatment of inflammation and cancer has also been proposed (123).
Given the central role NADP(H) plays in regulating crucial processes such as reductive anabolic pathways, signal transduction, and fighting oxidative stress, NADK is critical for normal cellular functions. In the cytosol, the redox state of NADPH is primarily maintained by the initial oxidative stages of the pentose phosphate pathway (PPP) (Fig. 2) (145). NADPH is critical for the maintenance of cellular glutathione redox state (i.e., the balance between reduced [GSH] and oxidized [GSSG], maintained by GR). Indeed, a defect in the first PPP enzyme glucose-6-phosphate dehydrogenase (G6PDH) is one of the most common inherited enzymopathies (26) and results in an inability to maintain glutathione in the reduced form, such that xenobiotics consumption elicits hemolysis.
A mitochondrial isoform of NADK has also been recently reported (112, 179) and was shown to facilitate fatty acid oxidation (FAO), maintenance of mitochondrial SIRT activities, and protection of hepatocytes from oxidative stress (178). In addition, just as in the cytosol, there are numerous reactions in the mitochondrion that require NADPH, including the re-reduction of GSSG to GSH by mitochondrial GR. As such, specific pathways for the maintenance of NADPH redox status exist in the organelle.
As mentioned above, IDH2 can reduce NADP+ to NADPH (Fig. 5). Alternatively, embedded in the inner mitochondrial membrane, the nicotinamide nucleotide transhydrogenase (NNT) consumes the H+ gradient to drive the interconversion of NADH+NADP+ to NADPH+NAD+ (Fig. 2). Notably, the commonly used C57BL/6J mouse strain (from JAX Laboratories) has a polymorphism in the NNT, resulting in an inability to maintain reduced mitochondrial GSH, and an overall increased susceptibility to a wide variety of oxidative stress diseases (139). The C57BL/6N mouse (from EV Taconic) does not have this mutation, making it critical for all mitochondrial investigators to report substrain (J/N) usage.
Recently, it has been proposed that the mitochondrial NNT may exist in concert with ROS-generating dehydrogenases (such as PDH) to form an energy-wasting redox circuit. Specifically, the rate of H2O2 production by PDH is a direct function of its flux, and this H2O2 is detoxified by the NADPH-consuming GR. The subsequent regeneration of GSH by the NNT consumes the mitochondrial H+ gradient, thus providing an OX-PHOS uncoupling pathway that is only active at high rates of PDH flux. This overall PDH/NNT complex pathway and its energy-wasting function are thought to be important for whole-organism energetic balance, with implications for diseases such as obesity (45). In addition, it should be noted that NADPH is a critical requirement for the maintenance of other key antioxidants in both the cytosol and mitochondria, namely the thioredoxin, glutaredoxin, and peroxiredoxin systems [see (62) for review].
Transport of NADH reducing equivalents into mitochondria
As mentioned above (the Glycolysis section), glycolysis generates NADH, and this must be converted back to NAD+ to permit continued glycolytic flux. Under normoxic conditions, this is accomplished by the MAS (Fig. 4). The shuttle consists of a cytosolic isoform of MDH running in reverse to convert oxaloacetate (OAA) to malate, using NADH. Malate is then transported into the mitochondrial matrix, where the Krebs' cycle isoform of MDH converts it back to OAA, regenerating NADH for use by the ETC. To avoid accumulation of OAA in the matrix, OAA is converted to aspartate with the addition of an amino group from glutamate, catalyzed by aspartate aminotransferase (AST), and also yielding α-KG. This α-KG is then transported out of the mitochondrion in exchange for malate coming in. The removal of aspartate and import of glutamate are handled by the mitochondrial glutamate (Glu)/aspartate (Asp) carrier. A corollary isoform of AST in the cytosol then catalyzes the reverse reaction (Asp + α-KG → OAA + Glu). Thus, the MAS can be considered as consisting of two parts—one to carry reducing equivalents of NADH into the mitochondrion and the other to bring the carrier molecules back out again via an amino acid shuttle. It should be noted that NADH itself is not transported by the MAS, rather only its reducing equivalency. Also, the enzymes and transporters of the MAS are poised to operate only in one direction under normal conditions. The MAS does not transport NADPH equivalents.
Although the primary function of the MAS is transportation of NADH reducing equivalents for metabolic purposes to facilitate glycolytic flux, the role of this transport system in equilibrating redox power across the membrane (e.g., to provide reducing power for antioxidant systems or to modulate NADH/NAD+ ratios in the mitochondrion for the purpose of affecting the SIRT activity) is unclear. Furthermore, the existence of the MAS for the purpose of reducing mitochondrial NAD+ using cytosolic NADH reducing power implies that the import of NAD+ from the cytosol (see the Source of Mitochondrial NAD+ section) is likely in the form of NAD+ and not NADH.
Balancing NAD Consumption and Redox in a Compartment-Specific Manner
Compartment-specific levels of NAD+ and related metabolites
The estimated total intracellular NAD+ content in mammalian cells ranges between ∼200 and 500 μM (24, 152, 172). However, NAD+ levels can vary widely in response to diverse stimuli, including glucose deprivation, fasting, caloric restriction, exercise, and circadian cycles (24). In terms of subcellular compartmentation, NAD+ is typically found in the cytosol and nucleus at ∼100 μM and in mitochondria at ∼250 μM, although this varies with cell type (23). Generally, the cytosolic pool of NADH/NAD+ is heavily favored toward the oxidized state, such that NADH/NAD+ ratio is ∼1:1000 under normal conditions (i.e., ∼0.1% is in the reduced state) (164). The use of NADH as the major reducing equivalent between the Krebs' cycle and the ETC is such that NADH/NAD+ ratio in mitochondria is much higher (1:10) compared with cytosol, although the absolute value is highly dynamic depending on respiratory and metabolic activities (164).
In contrast to NAD+, the cytosolic NADP+ pool is primarily found in the reduced state (i.e., NADPH/NADP+ ∼200:1) (164). It should also be noted that in the ER, the redox state is generally more oxidized to facilitate the formation of disulfide bonds for protein folding. Surprisingly, however, indirect measurements have suggested that the ER pool of NAD(P)H is able to remain relatively reduced, despite a prevailing oxidative environment (125).
Even though at the cellular level NADP(H) pools are present in a generally more reduced state than NAD(H), the absolute cellular levels of NADH are reported to be higher than NADPH (53, 174), indicating that the total pool of NAD(H) is bigger than NADP(H).
Competition for NAD+ among SIRTs/PARPs/CDs
Some studies have suggested that NAD+-consuming enzymes can compete for NAD+ within a given cellular compartment. For example, hyperactivation of PARP-1 following ischemia-reperfusion injury in the heart was shown to result in a depression of the SIRT1 activity (7). In addition, PARP-1 and SIRT1 are thought to play reciprocal roles in regulating each other's expression—SIRT1 is able to negatively regulate the PARP-1 promoter and the SIRT1 promoter is also under the influence of PARP-2 (6, 7). Furthermore, SIRT1 is known to deacetylate PARP-1 directly resulting in its inhibition, suggesting that these enzymes may cross talk at both the genetic and activity levels to coordinate nuclear NAD+ availability (133).
It has also been shown that NMNAT1 can bind to ADPR polymers that result from the PARP activity. This raises the possibility that NMNAT1 may be recruited to sites of the PARP activity (including PARP auto-PARylation), thus providing more NAD+ for PARP to function (13). In addition, SIRT1 binds to NMNAT1, helping to recruit NMNAT1 to specific sites, which may help stimulate the SIRT1 activity (180). As such, PARP and SIRT1 may compete not only for NAD+ itself but also for NMNAT as a source of NAD+. The complexity of PARP-SIRT interactions is reviewed in Refs. (25, 93).
Similar to PARPs, CD38/CD157 also compete with SIRTs for NAD+, and these glycohydrolases have been shown to modulate the SIRT activity [reviewed in (97)]. Genetic ablation or pharmacological inhibition of CD38 in mice results in 10- to 30-fold increase in cellular NAD+ levels (3, 8, 40). These elevated NAD+ levels serve to activate SIRT1, thereby providing resistance to the development of diet-induced obesity and accompanying glucose intolerance and liver steatosis (8). Similarly, CD38 inhibition has been shown to confer protection against cardiac ischemia-reperfusion injury, potentially by enhancing NAD+ availability for the SIRT activity (21). The extracellular location of CD38 and its ability to modulate the SIRT activity suggest that cellular SIRT activity can respond dynamically to extracellular NAD+ availability.
Since many NAD+-consuming enzymes are inhibited by the by-product NAM, it is likely that enhanced activity of any one class of NAD+-consuming enzymes (SIRTs, PARPs) may influence the activity of the others not only by reducing the availability of NAD+ but also by increasing NAM levels.
Competition for NAD+ between redox/metabolic cofactor enzymes versus NAD+ consumers
A common observation in the cell death field is that PARP-1 hyperactivation leads to an “energy crisis” (37). However, this is not necessarily due to depletion of the bioenergetic pool of NAD+, which is localized in the mitochondrion, while PARP is primarily in the nucleus. Indeed, massive NAD+ depletion was shown to result in virtually no effect on mitochondrial function for up to 24 h. (128, 154, 172). Instead, the bioenergetic effects of PARP hyperactivation are likely due to the large amounts of ATP required for salvaging NAD+ from NAM (see the NAD+ Synthesis and Compartmentation section, Fig. 1). In this regard, an energetic crisis involving NAD+ depletion is worse than one due to ATP depletion alone. With ATP depletion (e.g., due to mitochondrial dysfunction), glycolysis can continue. However, when NAD+ depletion is the root cause of ATP depletion, glycolysis is also impaired, leading to a deeper energetic crisis.
While metabolism typically uses NAD+ as a redox cofactor, SIRTs/PARPs and so on use NAD+ as a substrate (yielding NAM and ADPR derivatives). A key factor that has received little attention in the competition between these processes is the relative levels of the enzymes concerned. Global analysis of protein expression levels (The Human Protein Atlas) (161) indicates that in most tissues the transcript levels of the key glycolytic enzyme GAPDH are several orders of magnitude greater than NAD+-consuming enzymes. For example, in human heart, the relative expression levels (in “TPM” units—transcripts per million) for Sirt1, Parp1, and Gapdh are 6.9, 32.9, and 2936, respectively. Even in the tissue with the highest levels of Sirt1 expression (testis), Sirt1 at 49 TPM is far lower than Gapdh at 891 TPM. For the highest Parp1 expressing tissue (lymph node), Parp1 at 150 TPM is 10-fold lower than Gapdh at 1635 TPM.
Although transcript levels may not be truly indicative of protein levels, in addition to expression, the affinity (KM) of these enzymes for NAD+ should also be considered. SIRT1 has a KM for NAD+ of ∼95 μM, whereas PARP-1 is ∼50–100 μM and CD38 is 15–25 μM (24). The KM for NAD+ of mammalian GAPDH is ∼35 μM (122). Given these similar KM values, it is suggested that competition for NAD+ between GAPDH and other consuming enzymes (e.g., SIRTs/PARPs) will be based largely on their relative concentrations, such that GAPDH will be the dominant NAD+ consumer. Indeed, in a recent study, we showed that acute administration of the NAD+ precursor NMN to cells or intact hearts led to a massive and rapid acidification of the cytosol, concurrent with stimulation of glycolysis (109). Notably, since SIRT1 in the heart is mostly cytosolic, no change in cytosolic lysine acetylation was observed, suggesting no stimulation of the SIRT1 activity by acute NMN.
Metabolism/NAD+ cross talk at genetic and metabolic levels
In addition NAD+ being both a metabolic redox cofactor and a substrate to enzymes such as PARPs and SIRTs, it has also been proposed that nuclear NAD+ regulates the expression of several genes that encode metabolic enzymes. Indeed, a recent study found that compartmentation of NAD+ synthesis and consumption machineries plays an important role in balancing glucose consumption for energy generation versus conversion of glucose into fat for energy storage, via regulation of gene transcription (142). Besides its role in DNA repair, PARP-1 regulates the adipogenic transcriptional program (95). The PARylation process converts NAD+ to NAM, and nuclear NMNAT1 helps to sustain the PARP-1 activity by regenerating NAD+ from NMN (which itself is generated from NAM by NAMPT, Fig. 2). However, NMNAT2 provides NAD+ for glycolysis. This competition between NMNAT1 and NMNAT2 for the common substrate NMN helps regulate the balance between energy consumption (glycolysis) and storage (adipocyte differentiation). Thus, NAD+ serves as a powerful tool to intimately link metabolism and gene regulation.
Another example of the complexity of NADs regulatory role in metabolism and biology is that NADH/NAD+ and their related metabolites often have allosteric regulatory interactions with the same metabolic enzymes that use the NADH/NAD+ redox couple as a cofactor. One example is mitochondrial Cx-I (NADH ubiquinone oxidoreductase), which is allosterically activated by ADPR (54). Another example is that NADH allosterically activates PDH kinase, which phosphorylates PDH resulting in its inhibition (145). As such, the accumulation of the product of the Krebs' cycle NADH serves to slowdown the entry of carbon into the cycle in the form of acetyl-CoA.
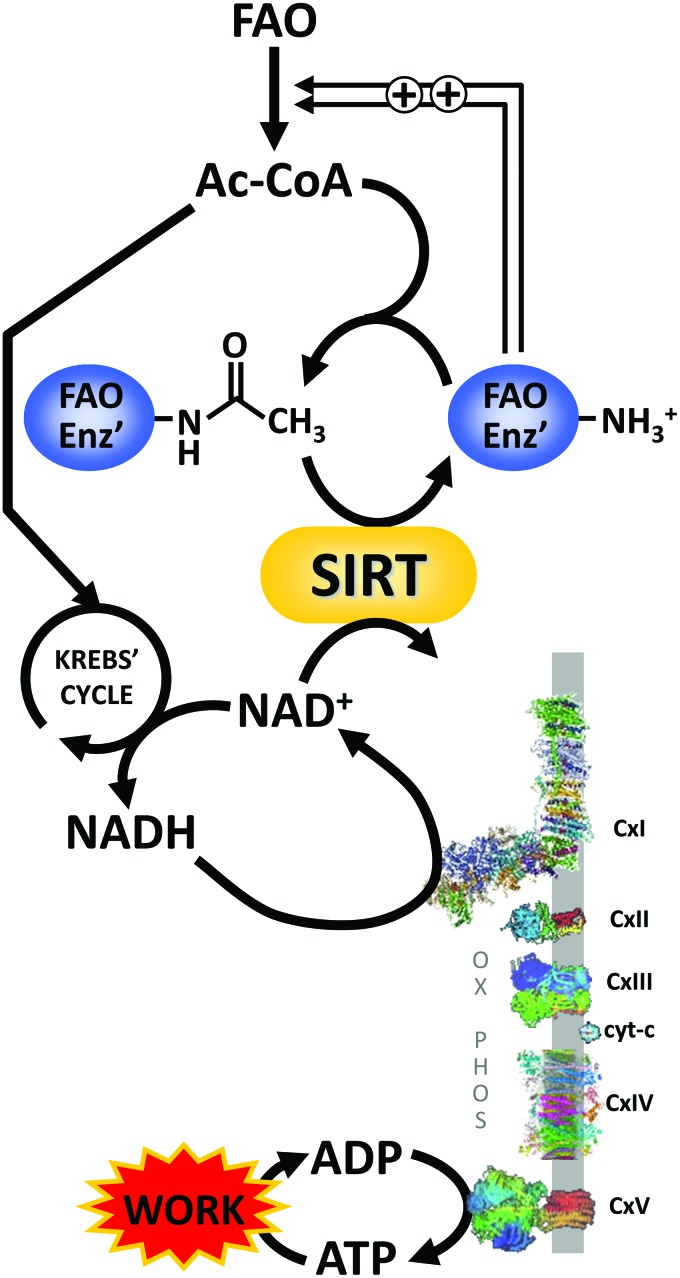
Furthermore, it should not be forgotten that the acetyl groups that are used to modify lysine residues (i.e., the same acetyl groups that NAD+-dependent SIRTs remove) originate from acetyl-CoA. Although lysine acetyltransferase enzymes exist (148), a significant portion of cellular acetylation may derive from nonenzymatic reactions of acetyl-CoA at slightly alkaline pH (such as found in the mitochondrion) (165). Thus, lysine acetylation can be viewed as reflecting the overall acetyl-CoA/NAD+ balance within a given cell compartment. Generally, deacetylation of metabolic enzymes involved in FAO leads to their stimulation (67). As such, under conditions of high workload in which respiratory complex I generates more NAD+, SIRT-mediated deacetylation may stimulate FAO to provide more ATP for the higher workload. Conversely, under conditions of low workload, the accumulation of acetyl-CoA (due to feedback inhibition of the Krebs' cycle) leads to acetylation and inhibition of FAO proteins. This is illustrated in Figure 6.
FIG. 6.
Lysine acetylation as a metabolic signal in the regulation of fatty acid β-oxidation. It is generally recognized that lysine acetylation of FAO enzymes (FAO Enz′) is inhibitory. In tissues such as the heart that rely heavily on FAO for bioenergetic needs, the ratio of acetyl-CoA (Ac-CoA) to NAD+ can serve as a metabolic signal for workload, to adjust FAO accordingly to provide ATP. Under conditions of low work, Krebs' cycle activity is low, leading to build up of Ac-CoA, which results in acetylation and inhibition of FAO enzymes. This may be an important feedback mechanism to prevent acyl-carbon-stress due to too much accumulation of acyl-CoA groups. Contrastingly, under conditions of high workload NADH will be rapidly oxidized by respiratory complex I (CxI) of OX-PHOS, yielding NAD+, which stimulates sirtuin activity to deacetylate (and thus activate) FAO enzymes. Resulting stimulation of FAO provides the ATP needed to drive the increased workload. FAO, fatty acid oxidation. Color images are available online.
On a related note, it is reported that complex I defects can lead to the inhibition of mitochondrial sirtuins (75). Specifically, the Ndufs4−/− mouse model of Cx-I deficiency exhibits hyperacetylation of mitochondrial proteins. This has been attributed to the inhibition of SIRT3 activity due to an increased NADH/NAD+ ratio. However, although SIRTs can be inhibited by NADH (87), this only occurs at very high NADH/NAD+ ratios that are not relevant in vivo. At physiological NADH/NAD+ ratios (1:10, see the Compartment-specific levels of NAD+ and related metabolites section), SIRTs are unaffected by NADH. Notably, in the Ndufs4−/− mouse, the driver of the higher NADH/NAD+ ratio was an increase in NADH, whereas NAD+ levels remained static. A more likely explanation for the hyperacetylation is that Cx-I inhibition and NADH buildup triggered feedback inhibition of the Krebs' cycle, leading to the accumulation of acetyl-CoA, which drives lysine acetylation. Clearly, when using surrogate markers such as global lysine acetylation to infer the SIRT activity, the acetylation side of the equation should not be overlooked.
An important caveat in the field of lysine acylation PTMs is the apparent discord between phenotypic effects and the stoichiometry of acylation (i.e., site occupancy on a given residue). Although reports of acetylation stoichiometry are generally low (∼1%), an emerging concept is that overall “lysine load” from various PTMs (acetylation, sumoylation, ubiquitination, succinylation, glutarylation, etc.) may be much higher (4, 100).
Further evidence for the intricate cross talk between sirtuin activity, redox state, and metabolic regulation is seen with the proposed role of SIRT5 as a regulator of two important NADPH-generating enzymes, namely IDH2 and G6PDH. Activation of SIRT5 by oxidative stress (H2O2) is thought to trigger the deacylation of these enzymes, activating them to generate NADPH to drive an antioxidant defense (181). In a similar manner, in addition to its role as a regulator of mitochondrial bioenergetics [e.g., deacetylation of ATP synthase (163)], SIRT3 is also reported to deacetylate and activate mitochondrial MnSOD as part of an antioxidant response to metabolic perturbation (35).
Conclusions and Therapeutic Considerations
In summary, there are numerous levels at which the enzymes that use NAD+ as a substrate, and those that use the NADH/NAD+ redox couple as a cofactor, compete for this essential nucleotide. While early studies in this area made bulk assumptions regarding the interchangeability of the cellular NADH/NAD+ pools, recent discoveries about the subcellular localization of these enzymes has led to a more nuanced understanding, and the notion of isolated NADH/NAD+ pools in specific cell compartments. As such, it cannot be assumed that these pools are interchangeable.
While a thorough review of the various pharmacological/therapeutic approaches to manipulation of NAD+ and its consuming enzymes is beyond the scope of this review [reviewed in (134)], the compartmentation and pleiotropic roles of NAD+ in the cell outlined herein suggest that the manipulation of NAD+ and its consuming enzymes en masse may carry unforeseen risks, due to impacts on other parts of the NAD+ cellular network. This is important because NAD+ supplementation is now receiving considerable attention in the field of aging.
NAD+ levels are known to decline with age (51), and indeed, the sirtuin family of NAD+-dependent proteins was initially discovered within the context of the anti-aging effects of caloric restriction. Several anti-aging therapeutic approaches including caloric restriction are thought to involve in the effects on NAD+ homeostasis (51). However, the complexities of NAD+ homeostasis should be appreciated during the design of therapeutic interventions for in vivo manipulation of NAD+. This is perhaps best illustrated by discussing one of the primary targets of such interventions—SIRT1.
SIRT1 has been shown to play a key role in the regulation of autoimmunity. Specifically, the SIRT1 activity is involved both in the suppression of regulatory T cells, which protect against autoimmunity (82, 162), and in the activation of T helper 17 CD4 cells, which contribute to autoimmunity (86). As such, global SIRT1 activation might confer an increased risk of autoimmune diseases.
In cancer, evidence exists for both tumor prevention (44) and promotion by SIRT1 (84). In addition, SIRT3 has been shown to function as a tumor suppressor (77). Notably, some tumors show increased NAMPT expression, such that interventions that increase NAD+ might enhance or promote tumor development (149). Similarly, given the importance of glycolysis for cancer cell metabolism (the Warburg effect), our results showing that acute supplementation of NMN results in bulk stimulation of glycolysis (109) suggest that the use of such compounds to boost NAD+ availability should be considered carefully, to avoid a pro-cancer metabolic environment.
Both SIRT1 and SIRT2 play a critical role in regulating neurodegeneration but act in opposite directions (116, 168). SIRT1 overexpression has been shown to be beneficial for neuronal survival in tissue culture and animal models of Alzheimer's disease, amyotrophic lateral sclerosis, and polyglutamine toxicity (76, 85). However, SIRT2 has been implicated in neuronal death, and genetic and pharmacological inhibition of SIRT2 has been shown to rescue α-synuclein toxicity models of Parkinson's disease (115). These observations underline the importance of tailoring therapeutic interventions that specifically stimulate NAD+ levels in the desired subcellular compartment (in this case, the nucleus) versus stimulating global NAD+ levels to avoid undesired side effects.
While the above examples highlight the importance of stimulating NAD+ levels in a target cell type- and subcellular compartment-specific manner, to stimulate only the desired enzyme isoform, in some instances doing so might not be enough. For example, in the liver, SIRT1 regulates gluconeogenesis and fat homeostasis by deacetylating various proteins involved in these processes, but these deacetylation events can have opposing effects. SIRT1 deacetylation of PGC-1α activates gluconeogenesis (138), whereas deacetylation of CRTC2, another coactivator of gluconeogenesis, triggers its ubiquitination and degradation (91).
SIRT1 function also changes depending on the pathological state in certain tissues. SIRT1 has been shown to protect kidney and suppress diabetic albuminuria (64). On the contrary, SIRT1 is implicated in the pathophysiology of autosomal-dominant polycystic kidney disease (ADPKD) (182). ADPKD is marked by the development of multiple bilateral cysts that replace normal kidney tissue, and genetic or pharmacological ablation of the SIRT1 activity delays cyst growth in animal model of this disease. As such, SIRT1 activation is beneficial under normal conditions, whereas its inhibition is beneficial in the disease state.
NAD+ precursor supplementation for the purpose of increasing cellular NAD+ levels is already undergoing clinical trials (NCT NCT03151239, NCT03432871, NCT03423342, NCT03501433, NCT02835664). However, large scale trials of nicotinic acid (NA, niacin) for cardiovascular disease (NCT00461630 and NCT00880178) showed efficacy but with considerably more adverse side effects. As such, the aforementioned complex roles of NAD+ in disease biology suggest that simple nutraceutical approaches to alter NAD+ levels may not represent a clinical panacea.
Acknowledgments
Work in the laboratory of P.S.B. is funded by a grant from the U.S. National Institutes of Health (R01-HL071158). C.A.K. is funded by an American Heart Association Post-Doctoral Fellowship (19POST34380212).
Abbreviations Used
- α-KG
α-ketoglutarate
- 2-HG
2-hydroxyglutarate
- 3-HAA
3-hydroxyanthranilic acid
- ACMS
aminocarboxymuconate semialdedhye
- ACMSD
aminocarboxymuconate semialdehyde decarboxylase
- ADP
adenosine diphosphate
- ADPKD
autosomal-dominant polycystic kidney disease
- ADPR
adenosine diphosphate-ribose
- AMP
adenosine monophosphate
- AMS
2-aminomuconate semialdehyde
- Asp
aspartate
- AST
aspartate aminotransferase
- ATP
adenosine triphosphate
- Cx43
connexin 43
- ENT
equilibrative nucleoside transporter
- ER
endoplasmic reticulum
- ETC
electron transport chain
- FAD
flavin adenine dinucleotide
- FAO
fatty acid oxidation
- FMN
flavin mononucleotide
- G6PDH
glucose-6-phosphate dehydrogenase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- Glu
glutamate
- GR
glutathione reductase
- GSH
glutathione reduced form
- GSSG
glutathione oxidized form
- IDH
isocitrate dehydrogenase
- LDH
lactate dehydrogenase
- MAS
malate-aspartate shuttle
- MDH
malate dehydrogenase
- NA
nicotinic acid
- NAAD
nicotinic acid adenine dinucleotide
- NAD+
nicotinamide adenine dinucleotide
- NADH
nicotinamide adenine dinucleotide reduced form
- NADK
NAD+ kinase
- NADP+
nicotinamide adenine dinucleotide phosphate
- NADPH
nicotinamide adenine dinucleotide phosphate reduced form
- NAM
nicotinamide
- NAMPT
nicotinamide phosphoribosyltransferase
- NAR
nicotinic acid riboside
- NAMN
nicotinic acid mononucleotide
- NMN
nicotinamide mononucleotide
- NMNAT
nicotinamide mononucleotide adenylyltransferase
- NNT
nicotinamide nucleotide transhydrogenase
- NR
nicotinamide riboside
- NRK
nicotinamide riboside kinase
- OAA
oxaloacetate
- OX-PHOS
oxidative phosphorylation
- PARP
poly(ADP-ribose) polymerase
- PDH
pyruvate dehydrogenase
- PNP
purine nucleoside phosphorylase
- PPP
pentose phosphate pathway
- PRPP
phosphoribosyl pyrophosphate
- PTMs
post-translational modifications
- RET
reverse electron transfer
- ROS
reactive oxygen species
- SARM1
sterile alpha and TIR motif-containing protein 1
- SIRT
sirtuin
- TIR
toll-interleukin receptor
- TPM
transcripts per million
References
- 1. Abdelraheim SR, Spiller DG, and McLennan AG. Mammalian NADH diphosphatases of the Nudix family: cloning and characterization of the human peroxisomal NUDT12 protein. Biochem J 374: 329–335, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abdelraheim SR, Spiller DG, and McLennan AG. Mouse Nudt13 is a mitochondrial Nudix hydrolase with NAD(P)H pyrophosphohydrolase activity. Protein J 36: 425–432, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aksoy P, White TA, Thompson M, and Chini EN. Regulation of intracellular levels of NAD: a novel role for CD38. Biochem Biophys Res Commun 345: 1386–1392, 2006 [DOI] [PubMed] [Google Scholar]
- 4. Ali HR, Assiri MA, Harris PS, Michel CR, Yun Y, Marentette JO, Huynh FK, Orlicky DJ, Shearn CT, Saba LM, Reisdorph R, Reisdorph N, Hirschey MD, and Fritz KS. Quantifying competition among mitochondrial protein acylation events induced by ethanol metabolism. J Proteome Res 2019. [Epub ahead of print]; DOI: 10.1021/acs.jproteome.8b00800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bai P. Biology of poly(ADP-Ribose) polymerases: the factotums of cell maintenance. Mol Cell 58: 947–958, 2015 [DOI] [PubMed] [Google Scholar]
- 6. Bai P, Canto C, Brunyanszki A, Huber A, Szanto M, Cen Y, Yamamoto H, Houten SM, Kiss B, Oudart H, Gergely P, Menissier-de Murcia J, Schreiber V, Sauve AA, and Auwerx J. PARP-2 regulates SIRT1 expression and whole-body energy expenditure. Cell Metab 13: 450–460, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, Schreiber V, Sauve AA, Menissier-de Murcia J, and Auwerx J. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab 13: 461–468, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barbosa MT, Soares SM, Novak CM, Sinclair D, Levine JA, Aksoy P, and Chini EN. The enzyme CD38 (a NAD glycohydrolase, EC 3.2.2.5) is necessary for the development of diet-induced obesity. FASEB J 21: 3629–3639, 2007 [DOI] [PubMed] [Google Scholar]
- 9. Belenky P, Bogan KL, and Brenner C. NAD+ metabolism in health and disease. Trends Biochem Sci 32: 12–19, 2007 [DOI] [PubMed] [Google Scholar]
- 10. Belenky P, Christensen KC, Gazzaniga F, Pletnev AA, and Brenner C. Nicotinamide riboside and nicotinic acid riboside salvage in fungi and mammals. Quantitative basis for Urh1 and purine nucleoside phosphorylase function in NAD+ metabolism. J Biol Chem 284: 158–164, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bender DA. Biochemistry of tryptophan in health and disease. Mol Aspects Med 6: 101–197, 1983 [DOI] [PubMed] [Google Scholar]
- 12. Berger F, Lau C, Dahlmann M, and Ziegler M. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J Biol Chem 280: 36334–36341, 2005 [DOI] [PubMed] [Google Scholar]
- 13. Berger F, Lau C, and Ziegler M. Regulation of poly(ADP-ribose) polymerase 1 activity by the phosphorylation state of the nuclear NAD biosynthetic enzyme NMN adenylyl transferase 1. Proc Natl Acad Sci U S A 104: 3765–3770, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bessman MJ, Frick DN, and O'Handley SF. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem 271: 25059–25062, 1996 [DOI] [PubMed] [Google Scholar]
- 15. Bi J, Wang H, and Xie J. Comparative genomics of NAD(P) biosynthesis and novel antibiotic drug targets. J Cell Physiol 226: 331–340, 2011 [DOI] [PubMed] [Google Scholar]
- 16. Bieganowski P. and Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 117: 495–502, 2004 [DOI] [PubMed] [Google Scholar]
- 17. Billington RA, Travelli C, Ercolano E, Galli U, Roman CB, Grolla AA, Canonico PL, Condorelli F, and Genazzani AA. Characterization of NAD uptake in mammalian cells. J Biol Chem 283: 6367–6374, 2008 [DOI] [PubMed] [Google Scholar]
- 18. Blacker TS. and Duchen MR. Investigating mitochondrial redox state using NADH and NADPH autofluorescence. Free Radic Biol Med 100: 53–65, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blacker TS, Mann ZF, Gale JE, Ziegler M, Bain AJ, Szabadkai G, and Duchen MR. Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat Commun 5: 3936, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blinova K, Levine RL, Boja ES, Griffiths GL, Shi ZD, Ruddy B, and Balaban RS. Mitochondrial NADH fluorescence is enhanced by complex I binding. Biochemistry 47: 9636–9645, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boslett J, Reddy N, Alzarie YA, and Zweier JL. Inhibition of CD38 with the thiazoloquin(az)olin(on)e 78c protects the heart against post-ischemic injury. J Pharmacol Exp Ther 369: 55–64, 2019. Epub ahead of publication. DOI: 10.1124/jpet.118.254557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bruzzone S, Guida L, Zocchi E, Franco L, and De Flora A. Connexin 43 hemi channels mediate Ca2+-regulated transmembrane NAD+ fluxes in intact cells. FASEB J 15: 10–12, 2001 [DOI] [PubMed] [Google Scholar]
- 23. Cambronne XA, Stewart ML, Kim D, Jones-Brunette AM, Morgan RK, Farrens DL, Cohen MS, and Goodman RH. Biosensor reveals multiple sources for mitochondrial NAD+. Science 352: 1474–1477, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Canto C, Menzies KJ, and Auwerx J. NAD+ Metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab 22: 31–53, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Canto C, Sauve AA, and Bai P. Crosstalk between poly(ADP-ribose) polymerase and sirtuin enzymes. Mol Aspects Med 34: 1168–1201, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cappellini MD. and Fiorelli G. Glucose-6-phosphate dehydrogenase deficiency. Lancet 371: 64–74, 2008 [DOI] [PubMed] [Google Scholar]
- 27. Chini EN, Chini CCS, Espindola Netto JM, de Oliveira GC, and van Schooten W. The pharmacology of CD38/NADase: an emerging target in cancer and diseases of aging. Trends Pharmacol Sci 39: 424–436, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chouchani ET, Pell VR, Gaude E, Aksentijevic D, Sundier SY, Robb EL, Logan A, Nadtochiy SM, Ord ENJ, Smith AC, Eyassu F, Shirley R, Hu CH, Dare AJ, James AM, Rogatti S, Hartley RC, Eaton S, Costa ASH, Brookes PS, Davidson SM, Duchen MR, Saeb-Parsy K, Shattock MJ, Robinson AJ, Work LM, Frezza C, Krieg T, and Murphy MP. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 515: 431–435, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Collins PB. and Chaykin S. The management of nicotinamide and nicotinic acid in the mouse. J Biol Chem 247: 778–783, 1972 [PubMed] [Google Scholar]
- 30. Conforti L, Gilley J, and Coleman MP. Wallerian degeneration: an emerging axon death pathway linking injury and disease. Nat Rev Neurosci 15: 394–409, 2014 [DOI] [PubMed] [Google Scholar]
- 31. Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, and Ewbank JJ. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol 5: 488–494, 2004 [DOI] [PubMed] [Google Scholar]
- 32. Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, and Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 465: 966, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Davila A, Liu L, Chellappa K, Redpath P, Nakamaru-Ogiso E, Paolella LM, Zhang Z, Migaud ME, Rabinowitz JD, and Baur JA. Nicotinamide adenine dinucleotide is transported into mammalian mitochondria. Elife 7: e33246, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Di Lisa F. and Ziegler M. Pathophysiological relevance of mitochondria in NAD+ metabolism. FEBS Lett 492: 4–8, 2001 [DOI] [PubMed] [Google Scholar]
- 35. Dikalova AE, Itani HA, Nazarewicz RR, McMaster WG, Flynn CR, Uzhachenko R, Fessel JP, Gamboa JL, Harrison DG, and Dikalov SI. Sirt3 impairment and SOD2 hyperacetylation in vascular oxidative stress and hypertension. Circ Res 121: 564–574, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, and Lin H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334: 806–809, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duan Y, Gross RA, and Sheu SS. Ca2+-dependent generation of mitochondrial reactive oxygen species serves as a signal for poly(ADP-ribose) polymerase-1 activation during glutamate excitotoxicity. J Physiol 585: 741–758, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Durkacz BW, Omidiji O, Gray DA, and Shall S. (ADP-ribose)n participates in DNA excision repair. Nature 283: 593–596, 1980 [DOI] [PubMed] [Google Scholar]
- 39. Engel PC. Glutamate dehydrogenases: the why and how of coenzyme specificity. Neurochem Res 39: 426–432, 2014 [DOI] [PubMed] [Google Scholar]
- 40. Escande C, Nin V, Price NL, Capellini V, Gomes AP, Barbosa MT, O'Neil L, White TA, Sinclair DA, and Chini EN. Flavonoid apigenin is an inhibitor of the NAD+ ase CD38: implications for cellular NAD+ metabolism, protein acetylation, and treatment of metabolic syndrome. Diabetes 62: 1084–1093, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Essuman K, Summers DW, Sasaki Y, Mao X, DiAntonio A, and Milbrandt J. The SARM1 Toll/interleukin-1 receptor domain possesses intrinsic NAD+ cleavage activity that promotes pathological axonal degeneration. Neuron 93: 1334–1343 e5, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Felici R, Lapucci A, Ramazzotti M, and Chiarugi A. Insight into molecular and functional properties of NMNAT3 reveals new hints of NAD homeostasis within human mitochondria. PLoS One 8: e76938, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Finkel T, Deng CX, and Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature 460: 587–591, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S, de Cabo R, Fuchs C, Hahn WC, Guarente LP, and Sinclair DA. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One 3: e2020, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fisher-Wellman KH, Lin CT, Ryan TE, Reese LR, Gilliam LA, Cathey BL, Lark DS, Smith CD, Muoio DM, and Neufer PD. Pyruvate dehydrogenase complex and nicotinamide nucleotide transhydrogenase constitute an energy-consuming redox circuit. Biochem J 467: 271–280, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fletcher RS, Ratajczak J, Doig CL, Oakey LA, Callingham R, Da Silva Xavier G, Garten A, Elhassan YS, Redpath P, Migaud ME, Philp A, Brenner C, Canto C, and Lavery GG. Nicotinamide riboside kinases display redundancy in mediating nicotinamide mononucleotide and nicotinamide riboside metabolism in skeletal muscle cells. Mol Metab 6: 819–832, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Formentini L, Moroni F, and Chiarugi A. Detection and pharmacological modulation of nicotinamide mononucleotide (NMN) in vitro and in vivo. Biochem Pharmacol 77: 1612–1620, 2009 [DOI] [PubMed] [Google Scholar]
- 48. Garavaglia S, Bruzzone S, Cassani C, Canella L, Allegrone G, Sturla L, Mannino E, Millo E, De Flora A, and Rizzi M. The high-resolution crystal structure of periplasmic Haemophilus influenzae NAD nucleotidase reveals a novel enzymatic function of human CD73 related to NAD metabolism. Biochem J 441: 131–141, 2012 [DOI] [PubMed] [Google Scholar]
- 49. Gerdts J, Brace EJ, Sasaki Y, DiAntonio A, and Milbrandt J. SARM1 activation triggers axon degeneration locally via NAD+ destruction. Science 348: 453–457, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gerdts J, Summers DW, Milbrandt J, and DiAntonio A. Axon Self-Destruction: new Links among SARM1, MAPKs, and NAD+ Metabolism. Neuron 89: 449–460, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, Mercken EM, Palmeira CM, de Cabo R, Rolo AP, Turner N, Bell EL, and Sinclair DA. Declining NAD+ induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell 155: 1624–1638, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Granchi C. ATP citrate lyase (ACLY) inhibitors: an anti-cancer strategy at the crossroads of glucose and lipid metabolism. Eur J Med Chem 157: 1276–1291, 2018 [DOI] [PubMed] [Google Scholar]
- 53. Griffiths MH, Thomas DP, Xipell JM, and Hope RN. Thorotrast-induced bilateral carcinoma of the kidney. Pathology 9: 43–48, 1977 [DOI] [PubMed] [Google Scholar]
- 54. Grivennikova VG, Gladyshev GV, and Vinogradov AD. Allosteric nucleotide-binding site in the mitochondrial NADH:ubiquinone oxidoreductase (respiratory complex I). FEBS Lett 585: 2212–2216, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gross CJ. and Henderson LM. Digestion and absorption of NAD by the small intestine of the rat. J Nutr 113: 412–420, 1983 [DOI] [PubMed] [Google Scholar]
- 56. Grozio A, Mills KF, Yoshino J, Bruzzone S, Sociali G, Tokizane K, Lei HC, Cunningham R, Sasaki Y, Migaud ME, and Imai S-i. Slc12a8 is a nicotinamide mononucleotide transporter. Nat Metab 1: 47–57, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Grozio A, Sociali G, Sturla L, Caffa I, Soncini D, Salis A, Raffaelli N, De Flora A, Nencioni A, and Bruzzone S. CD73 protein as a source of extracellular precursors for sustained NAD+ biosynthesis in FK866-treated tumor cells. J Biol Chem 288: 25938–25949, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Guse AH. Calcium mobilizing second messengers derived from NAD. Biochim Biophys Acta 1854: 1132–1137, 2015 [DOI] [PubMed] [Google Scholar]
- 59. Haferkamp I, Schmitz-Esser S, Linka N, Urbany C, Collingro A, Wagner M, Horn M, and Neuhaus HE. A candidate NAD+ transporter in an intracellular bacterial symbiont related to Chlamydiae. Nature 432: 622–625, 2004 [DOI] [PubMed] [Google Scholar]
- 60. Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, and Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 126: 941–954, 2006 [DOI] [PubMed] [Google Scholar]
- 61. Haigis MC. and Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol 5: 253–295, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hanschmann EM, Godoy JR, Berndt C, Hudemann C, and Lillig CH. Thioredoxins, glutaredoxins, and peroxiredoxins—molecular mechanisms and health significance: from cofactors to antioxidants to redox signaling. Antioxid Redox Signal 19: 1539–1605, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hara N, Yamada K, Shibata T, Osago H, and Tsuchiya M. Nicotinamide phosphoribosyltransferase/visfatin does not catalyze nicotinamide mononucleotide formation in blood plasma. PLoS One 6: e22781, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hasegawa K, Wakino S, Simic P, Sakamaki Y, Minakuchi H, Fujimura K, Hosoya K, Komatsu M, Kaneko Y, Kanda T, Kubota E, Tokuyama H, Hayashi K, Guarente L, and Itoh H. Renal tubular Sirt1 attenuates diabetic albuminuria by epigenetically suppressing Claudin-1 overexpression in podocytes. Nat Med 19: 1496–1504, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Heneberg P. Redox regulation of hexokinases. Antioxid Redox Signal 30: 415–442, 2019 [DOI] [PubMed] [Google Scholar]
- 66. Herzig S. and Shaw RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol 19: 121–135, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr., Alt FW, Kahn CR, and Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464: 121–125, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hornyak L, Dobos N, Koncz G, Karanyi Z, Pall D, Szabo Z, Halmos G, and Szekvolgyi L. The role of indoleamine-2,3-dioxygenase in cancer development, diagnostics, and therapy. Front Immunol 9: 151, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hottiger MO, Hassa PO, Luscher B, Schuler H, and Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci 35: 208–219, 2010 [DOI] [PubMed] [Google Scholar]
- 70. Hou J, Chong ZZ, Shang YC, and Maiese K. Early apoptotic vascular signaling is determined by Sirt1 through nuclear shuttling, forkhead trafficking, bad, and mitochondrial caspase activation. Curr Neurovasc Res 7: 95–112, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Houtkooper RH, Canto C, Wanders RJ, and Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev 31: 194–223, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Intlekofer AM, Dematteo RG, Venneti S, Finley LW, Lu C, Judkins AR, Rustenburg AS, Grinaway PB, Chodera JD, Cross JR, and Thompson CB. Hypoxia induces production of L-2-hydroxyglutarate. Cell Metab 22: 304–311, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Iwahara T, Bonasio R, Narendra V, and Reinberg D. SIRT3 functions in the nucleus in the control of stress-related gene expression. Mol Cell Biol 32: 5022–5034, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, Hang HC, Hao Q, and Lin H. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496: 110–113, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Karamanlidis G, Lee CF, Garcia-Menendez L, Kolwicz SC, Jr., Suthammarak W, Gong G, Sedensky MM, Morgan PG, Wang W, and Tian R. Mitochondrial complex I deficiency increases protein acetylation and accelerates heart failure. Cell Metab 18: 239–250, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, Puigserver P, Sinclair DA, and Tsai LH. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO J 26: 3169–3179, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, and Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 17: 41–52, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kim W, Deik A, Gonzalez C, Gonzalez ME, Fu F, Ferrari M, Churchhouse CL, Florez JC, Jacobs SBR, Clish CB, and Rhee EP. Polyunsaturated fatty acid desaturation is a mechanism for glycolytic NAD+ recycling. Cell Metab 2019. [Epub ahead of print]; DOI: 10.1016/j.cmet.2018.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kornberg A. and Pricer WE., Jr On the structure of triphosphopyridine nucleotide. J Biol Chem 186: 557–567, 1950 [PubMed] [Google Scholar]
- 80. Krishnakumar R. and Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell 39: 8–24, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kulikova V, Shabalin K, Nerinovski K, Dolle C, Niere M, Yakimov A, Redpath P, Khodorkovskiy M, Migaud ME, Ziegler M, and Nikiforov A. Generation, release, and uptake of the NAD precursor nicotinic acid riboside by human cells. J Biol Chem 290: 27124–27137, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kwon HS, Lim HW, Wu J, Schnolzer M, Verdin E, and Ott M. Three novel acetylation sites in the Foxp3 transcription factor regulate the suppressive activity of regulatory T cells. J Immunol 188: 2712–2721, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lerner F, Niere M, Ludwig A, and Ziegler M. Structural and functional characterization of human NAD kinase. Biochem Biophys Res Commun 288: 69–74, 2001 [DOI] [PubMed] [Google Scholar]
- 84. Li HJ, Che XM, Zhao W, He SC, Zhang ZL, Chen R, Fan L, and Jia ZL. Diet-induced obesity promotes murine gastric cancer growth through a nampt/sirt1/c-myc positive feedback loop. Oncol Rep 30: 2153–2160, 2013 [DOI] [PubMed] [Google Scholar]
- 85. Li Y, Yokota T, Gama V, Yoshida T, Gomez JA, Ishikawa K, Sasaguri H, Cohen HY, Sinclair DA, Mizusawa H, and Matsuyama S. Bax-inhibiting peptide protects cells from polyglutamine toxicity caused by Ku70 acetylation. Cell Death Differ 14: 2058–2067, 2007 [DOI] [PubMed] [Google Scholar]
- 86. Lim HW, Kang SG, Ryu JK, Schilling B, Fei M, Lee IS, Kehasse A, Shirakawa K, Yokoyama M, Schnolzer M, Kasler HG, Kwon HS, Gibson BW, Sato H, Akassoglou K, Xiao C, Littman DR, Ott M, and Verdin E. SIRT1 deacetylates RORgammat and enhances Th17 cell generation. J Exp Med 212: 607–617, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lin SJ, Ford E, Haigis M, Liszt G, and Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev 18: 12–16, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Liszt G, Ford E, Kurtev M, and Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem 280: 21313–21320, 2005 [DOI] [PubMed] [Google Scholar]
- 89. Liu C, Vyas A, Kassab MA, Singh AK, and Yu X. The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res 45: 8129–8141, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Liu C. and Yu X. ADP-ribosyltransferases and poly ADP-ribosylation. Curr Protein Pept Sci 16: 491–501, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, 3rd, Olefsky J, Guarente L, and Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature 456: 269–273, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Loscalzo J. Adaptions to hypoxia and redox stress: essential concepts confounded by misleading terminology. Circ Res 119: 511–513, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Luna A, Aladjem MI, and Kohn KW. SIRT1/PARP1 crosstalk: connecting DNA damage and metabolism. Genome Integr 4: 6, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Luo X. and Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev 26: 417–432, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Luo X, Ryu KW, Kim DS, Nandu T, Medina CJ, Gupte R, Gibson BA, Soccio RE, Yu Y, Gupta RK, and Kraus WL. PARP-1 controls the adipogenic transcriptional program by PARylating C/EBPbeta and modulating its transcriptional activity. Mol Cell 65: 260–271, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Malavasi F, Deaglio S, Funaro A, Ferrero E, Horenstein AL, Ortolan E, Vaisitti T, and Aydin S. Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol Rev 88: 841–886, 2008 [DOI] [PubMed] [Google Scholar]
- 97. Malavasi F, Deaglio S, Zaccarello G, Horenstein AL, Chillemi A, Audrito V, Serra S, Gandione M, Zitella A, and Tizzani A. The hidden life of NAD+-consuming ectoenzymes in the endocrine system. J Mol Endocrinol 45: 183–191, 2010 [DOI] [PubMed] [Google Scholar]
- 98. Mansour TE. Phosphofructokinase. Curr Top Cell Regul 5: 1–46, 1972 [PubMed] [Google Scholar]
- 99. McLennan AG. The Nudix hydrolase superfamily. Cell Mol Life Sci 63: 123–143, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Meyer JG, D'Souza AK, Sorensen DJ, Rardin MJ, Wolfe AJ, Gibson BW, and Schilling B. Quantification of lysine acetylation and succinylation stoichiometry in proteins using mass spectrometric data-independent acquisitions (SWATH). J Am Soc Mass Spectrom 27: 1758–1771, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Michishita E, Park JY, Burneskis JM, Barrett JC, and Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 16: 4623–4635, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Mills KF, Yoshida S, Stein LR, Grozio A, Kubota S, Sasaki Y, Redpath P, Migaud ME, Apte RS, Uchida K, Yoshino J, and Imai SI. Long-term administration of nicotinamide mononucleotide mitigates age-associated physiological decline in mice. Cell Metab 24: 795–806, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Mink M, Fogelgren B, Olszewski K, Maroy P, and Csiszar K. A novel human gene (SARM) at chromosome 17q11 encodes a protein with a SAM motif and structural similarity to Armadillo/beta-catenin that is conserved in mouse, Drosophila, and Caenorhabditis elegans. Genomics 74: 234–244, 2001 [DOI] [PubMed] [Google Scholar]
- 104. Morales J, Li L, Fattah FJ, Dong Y, Bey EA, Patel M, Gao J, and Boothman DA. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit Rev Eukaryot Gene Expr 24: 15–28, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Mracek T, Drahota Z, and Houstek J. The function and the role of the mitochondrial glycerol-3-phosphate dehydrogenase in mammalian tissues. Biochim Biophys Acta 1827: 401–410, 2013 [DOI] [PubMed] [Google Scholar]
- 106. Murphy MP. How mitochondria produce reactive oxygen species. Biochem J 417: 1–13, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Nabokina SM, Kashyap ML, and Said HM. Mechanism and regulation of human intestinal niacin uptake. Am J Physiol Cell Physiol 289: C97–C103, 2005 [DOI] [PubMed] [Google Scholar]
- 108. Nadtochiy SM, Schafer X, Fu D, Nehrke K, Munger J, and Brookes PS. Acidic pH is a metabolic switch for 2-hydroxyglutarate generation and signaling. J Biol Chem 291: 20188–20197, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Nadtochiy SM, Wang YT, Nehrke K, Munger J, and Brookes PS. Cardioprotection by nicotinamide mononucleotide (NMN): involvement of glycolysis and acidic pH. J Mol Cell Cardiol 121: 155–162, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Niere M, Kernstock S, Koch-Nolte F, and Ziegler M. Functional localization of two poly(ADP-ribose)-degrading enzymes to the mitochondrial matrix. Mol Cell Biol 28: 814–824, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Nikiforov A, Dolle C, Niere M, and Ziegler M. Pathways and subcellular compartmentation of NAD biosynthesis in human cells: from entry of extracellular precursors to mitochondrial NAD generation. J Biol Chem 286: 21767–21778, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ohashi K, Kawai S, and Murata K. Identification and characterization of a human mitochondrial NAD kinase. Nat Commun 3: 1248, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Oldham WM, Clish CB, Yang Y, and Loscalzo J. Hypoxia-mediated increases in L-2-hydroxyglutarate coordinate the metabolic response to reductive stress. Cell Metab 22: 291–303, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Osterloh JM, Yang J, Rooney TM, Fox AN, Adalbert R, Powell EH, Sheehan AE, Avery MA, Hackett R, Logan MA, MacDonald JM, Ziegenfuss JS, Milde S, Hou YJ, Nathan C, Ding A, Brown RH, Jr., Conforti L, Coleman M, Tessier-Lavigne M, Zuchner S, and Freeman MR. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science 337: 481–484, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ, Young AB, Abagyan R, Feany MB, Hyman BT, and Kazantsev AG. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science 317: 516–519, 2007 [DOI] [PubMed] [Google Scholar]
- 116. Outeiro TF, Marques O, and Kazantsev A. Therapeutic role of sirtuins in neurodegenerative disease. Biochim Biophys Acta 1782: 363–369, 2008 [DOI] [PubMed] [Google Scholar]
- 117. Pacher P. and Szabo C. Role of poly(ADP-ribose) polymerase 1 (PARP-1) in cardiovascular diseases: the therapeutic potential of PARP inhibitors. Cardiovasc Drug Rev 25: 235–260, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Palego L, Betti L, Rossi A, and Giannaccini G. Tryptophan biochemistry: structural, nutritional, metabolic, and medical aspects in humans. J Amino Acids 2016: 8952520, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Palmieri F, Rieder B, Ventrella A, Blanco E, Do PT, Nunes-Nesi A, Trauth AU, Fiermonte G, Tjaden J, Agrimi G, Kirchberger S, Paradies E, Fernie AR, and Neuhaus HE. Molecular identification and functional characterization of Arabidopsis thaliana mitochondrial and chloroplastic NAD+ carrier proteins. J Biol Chem 284: 31249–31259, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Paoletti J, Assairi L, Gelin M, Huteau V, Nahori MA, Dussurget O, Labesse G, and Pochet S. 8-Thioalkyl-adenosine derivatives inhibit Listeria monocytogenes NAD kinase through a novel binding mode. Eur J Med Chem 124: 1041–1056, 2016 [DOI] [PubMed] [Google Scholar]
- 121. Partida-Sanchez S, Rivero-Nava L, Shi G, and Lund FE. CD38: an ecto-enzyme at the crossroads of innate and adaptive immune responses. Adv Exp Med Biol 590: 171–183, 2007 [DOI] [PubMed] [Google Scholar]
- 122. Patra S, Ghosh S, Bera S, Roy A, Ray S, and Ray M. Molecular characterization of tumor associated glyceraldehyde-3-phosphate dehydrogenase. Biochemistry (Mosc) 74: 717–727, 2009 [DOI] [PubMed] [Google Scholar]
- 123. Petrelli R, Felczak K, and Cappellacci L. NMN/NaMN adenylyltransferase (NMNAT) and NAD kinase (NADK) inhibitors: chemistry and potential therapeutic applications. Curr Med Chem 18: 1973–1992, 2011 [DOI] [PubMed] [Google Scholar]
- 124. Petrelli R, Sham YY, Chen L, Felczak K, Bennett E, Wilson D, Aldrich C, Yu JS, Cappellacci L, Franchetti P, Grifantini M, Mazzola F, Di Stefano M, Magni G, and Pankiewicz KW. Selective inhibition of nicotinamide adenine dinucleotide kinases by dinucleoside disulfide mimics of nicotinamide adenine dinucleotide analogues. Bioorg Med Chem 17: 5656–5664, 2009 [DOI] [PubMed] [Google Scholar]
- 125. Piccirella S, Czegle I, Lizak B, Margittai E, Senesi S, Papp E, Csala M, Fulceri R, Csermely P, Mandl J, Benedetti A, and Banhegyi G. Uncoupled redox systems in the lumen of the endoplasmic reticulum. Pyridine nucleotides stay reduced in an oxidative environment. J Biol Chem 281: 4671–4677, 2006 [DOI] [PubMed] [Google Scholar]
- 126. Pillai VB, Sundaresan NR, Kim G, Gupta M, Rajamohan SB, Pillai JB, Samant S, Ravindra PV, Isbatan A, and Gupta MP. Exogenous NAD blocks cardiac hypertrophic response via activation of the SIRT3-LKB1-AMP-activated kinase pathway. J Biol Chem 285: 3133–3144, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Pittelli M, Felici R, Pitozzi V, Giovannelli L, Bigagli E, Cialdai F, Romano G, Moroni F, and Chiarugi A. Pharmacological effects of exogenous NAD on mitochondrial bioenergetics, DNA repair, and apoptosis. Mol Pharmacol 80: 1136–1146, 2011 [DOI] [PubMed] [Google Scholar]
- 128. Pittelli M, Formentini L, Faraco G, Lapucci A, Rapizzi E, Cialdai F, Romano G, Moneti G, Moroni F, and Chiarugi A. Inhibition of nicotinamide phosphoribosyltransferase: cellular bioenergetics reveals a mitochondrial insensitive NAD pool. J Biol Chem 285: 34106–34114, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Platten M, von Knebel Doeberitz N, Oezen I, Wick W, and Ochs K. Cancer immunotherapy by targeting IDO1/TDO and their downstream effectors. Front Immunol 5: 673, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Preiss J. and Handler P. Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J Biol Chem 233: 488–492, 1958 [PubMed] [Google Scholar]
- 131. Preiss J. and Handler P. Biosynthesis of diphosphopyridine nucleotide. II. Enzymatic aspects. J Biol Chem 233: 493–500, 1958 [PubMed] [Google Scholar]
- 132. Quarona V, Zaccarello G, Chillemi A, Brunetti E, Singh VK, Ferrero E, Funaro A, Horenstein AL, and Malavasi F. CD38 and CD157: a long journey from activation markers to multifunctional molecules. Cytometry B Clin Cytom 84: 207–217, 2013 [DOI] [PubMed] [Google Scholar]
- 133. Rajamohan SB, Pillai VB, Gupta M, Sundaresan NR, Birukov KG, Samant S, Hottiger MO, and Gupta MP. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol Cell Biol 29: 4116–4129, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Rajman L, Chwalek K, and Sinclair DA. Therapeutic potential of NAD-boosting molecules: the in vivo evidence. Cell Metab 27: 529–547, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Ratajczak J, Joffraud M, Trammell SA, Ras R, Canela N, Boutant M, Kulkarni SS, Rodrigues M, Redpath P, Migaud ME, Auwerx J, Yanes O, Brenner C, and Canto C. NRK1 controls nicotinamide mononucleotide and nicotinamide riboside metabolism in mammalian cells. Nat Commun 7: 13103, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Revollo JR, Grimm AA, and Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem 279: 50754–50763, 2004 [DOI] [PubMed] [Google Scholar]
- 137. Rine J. and Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 116: 9–22, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, and Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434: 113–118, 2005 [DOI] [PubMed] [Google Scholar]
- 139. Ronchi JA, Figueira TR, Ravagnani FG, Oliveira HC, Vercesi AE, and Castilho RF. A spontaneous mutation in the nicotinamide nucleotide transhydrogenase gene of C57BL/6J mice results in mitochondrial redox abnormalities. Free Radic Biol Med 63: 446–456, 2013 [DOI] [PubMed] [Google Scholar]
- 140. Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, and Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer 10: 293–301, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Rustin P, Chretien D, Parfait B, Rotig A, and Munnich A. Nicotinamide adenine dinucleotides permeate through mitochondrial membranes in human Epstein-Barr virus-transformed lymphocytes. Mol Cell Biochem 174: 115–119, 1997 [PubMed] [Google Scholar]
- 142. Ryu KW, Nandu T, Kim J, Challa S, DeBerardinis RJ, and Kraus WL. Metabolic regulation of transcription through compartmentalized NAD+ biosynthesis. Science 360: eaan5780, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Rzem R, Veiga-da-Cunha M, Noel G, Goffette S, Nassogne MC, Tabarki B, Scholler C, Marquardt T, Vikkula M, and Van Schaftingen E. A gene encoding a putative FAD-dependent L-2-hydroxyglutarate dehydrogenase is mutated in L-2-hydroxyglutaric aciduria. Proc Natl Acad Sci U S A 101: 16849–16854, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Said HM, Nabokina SM, Balamurugan K, Mohammed ZM, Urbina C, and Kashyap ML. Mechanism of nicotinic acid transport in human liver cells: experiments with HepG2 cells and primary hepatocytes. Am J Physiol Cell Physiol 293: C1773–C1778, 2007 [DOI] [PubMed] [Google Scholar]
- 145. Salway JG. Metabolism at a Glance. Chichester, United Kingdom: Wiley, 2017 [Google Scholar]
- 146. Sasaki Y, Nakagawa T, Mao X, DiAntonio A, and Milbrandt J. NMNAT1 inhibits axon degeneration via blockade of SARM1-mediated NAD+ depletion. Elife 5: e19749, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Scholz TD, Laughlin MR, Balaban RS, Kupriyanov VV, and Heineman FW. Effect of substrate on mitochondrial NADH, cytosolic redox state, and phosphorylated compounds in isolated hearts. Am J Physiol 268: H82–H91, 1995 [DOI] [PubMed] [Google Scholar]
- 148. Scott I, Webster BR, Li JH, and Sack MN. Identification of a molecular component of the mitochondrial acetyltransferase programme: a novel role for GCN5L1. Biochem J 443: 655–661, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Shackelford RE, Mayhall K, Maxwell NM, Kandil E, and Coppola D. Nicotinamide phosphoribosyltransferase in malignancy: a review. Genes Cancer 4: 447–456, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Shi F, Li Y, Li Y, and Wang X. Molecular properties, functions, and potential applications of NAD kinases. Acta Biochim Biophys Sin (Shanghai) 41: 352–361, 2009 [DOI] [PubMed] [Google Scholar]
- 151. Smith B, Schafer XL, Ambeskovic A, Spencer CM, Land H, and Munger J. Addiction to coupling of the Warburg effect with glutamine catabolism in cancer cells. Cell Rep 17: 821–836, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Smith WC. and Harland RM. Injected Xwnt-8 RNA acts early in Xenopus embryos to promote formation of a vegetal dorsalizing center. Cell 67: 753–765, 1991 [DOI] [PubMed] [Google Scholar]
- 153. Sociali G, Raffaghello L, Magnone M, Zamporlini F, Emionite L, Sturla L, Bianchi G, Vigliarolo T, Nahimana A, Nencioni A, Raffaelli N, and Bruzzone S. Antitumor effect of combined NAMPT and CD73 inhibition in an ovarian cancer model. Oncotarget 7: 2968–2984, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Stein LR. and Imai S. The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol Metab 23: 420–428, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Szanto M, Brunyanszki A, Kiss B, Nagy L, Gergely P, Virag L, and Bai P. Poly(ADP-ribose) polymerase-2: emerging transcriptional roles of a DNA-repair protein. Cell Mol Life Sci 69: 4079–4092, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Takanaga H, Maeda H, Yabuuchi H, Tamai I, Higashida H, and Tsuji A. Nicotinic acid transport mediated by pH-dependent anion antiporter and proton cotransporter in rabbit intestinal brush-border membrane. J Pharm Pharmacol 48: 1073–1077, 1996 [DOI] [PubMed] [Google Scholar]
- 157. Tanno M, Kuno A, Yano T, Miura T, Hisahara S, Ishikawa S, Shimamoto K, and Horio Y. Induction of manganese superoxide dismutase by nuclear translocation and activation of SIRT1 promotes cell survival in chronic heart failure. J Biol Chem 285: 8375–8382, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Tempel W, Rabeh WM, Bogan KL, Belenky P, Wojcik M, Seidle HF, Nedyalkova L, Yang T, Sauve AA, Park HW, and Brenner C. Nicotinamide riboside kinase structures reveal new pathways to NAD+. PLoS Biol 5: e263, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Todisco S, Agrimi G, Castegna A, and Palmieri F. Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae. J Biol Chem 281: 1524–1531, 2006 [DOI] [PubMed] [Google Scholar]
- 160. Ueda K, Oka J, Naruniya S, Miyakawa N, and Hayaishi O. Poly ADP-ribose glycohydrolase from rat liver nuclei, a novel enzyme degrading the polymer. Biochem Biophys Res Commun 46: 516–523, 1972 [DOI] [PubMed] [Google Scholar]
- 161. Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, and Ponten F. Proteomics. Tissue-based map of the human proteome. Science 347: 1260419, 2015 [DOI] [PubMed] [Google Scholar]
- 162. van Loosdregt J, Vercoulen Y, Guichelaar T, Gent YY, Beekman JM, van Beekum O, Brenkman AB, Hijnen DJ, Mutis T, Kalkhoven E, Prakken BJ, and Coffer PJ. Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood 115: 965–974, 2010 [DOI] [PubMed] [Google Scholar]
- 163. Vassilopoulos A, Pennington JD, Andresson T, Rees DM, Bosley AD, Fearnley IM, Ham A, Flynn CR, Hill S, Rose KL, Kim HS, Deng CX, Walker JE, and Gius D. SIRT3 deacetylates ATP synthase F1 complex proteins in response to nutrient- and exercise-induced stress. Antioxid Redox Signal 21: 551–564, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Veech RL, Eggleston LV, and Krebs HA. The redox state of free nicotinamide-adenine dinucleotide phosphate in the cytoplasm of rat liver. Biochem J 115: 609–619, 1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Wagner GR. and Hirschey MD. Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol Cell 54: 5–16, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Wanders RJ. and Waterham HR, Ferdinandusse S. Metabolic interplay between peroxisomes and other subcellular organelles including mitochondria and the endoplasmic reticulum. Front Cell Dev Biol 3: 83, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Wang S, Xing Z, Vosler PS, Yin H, Li W, Zhang F, Signore AP, Stetler RA, Gao Y, and Chen J. Cellular NAD replenishment confers marked neuroprotection against ischemic cell death: role of enhanced DNA repair. Stroke 39: 2587–2595, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Westphal CH, Dipp MA, and Guarente L. A therapeutic role for sirtuins in diseases of aging? Trends Biochem Sci 32: 555–560, 2007 [DOI] [PubMed] [Google Scholar]
- 169. Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, and Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A 108: 19611–19616, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, and Xiong Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Yanagisawa S, Baker JR, Vuppusetty C, Koga T, Colley T, Fenwick P, Donnelly LE, Barnes PJ, and Ito K. The dynamic shuttling of SIRT1 between cytoplasm and nuclei in bronchial epithelial cells by single and repeated cigarette smoke exposure. PLoS One 13: e0193921, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, and Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell 130: 1095–1107, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Ye D, Guan KL, and Xiong Y. Metabolism, activity, and targeting of D- and L-2-hydroxyglutarates. Trends Cancer 4: 151–165, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Signal 10: 179–206, 2008 [DOI] [PubMed] [Google Scholar]
- 175. Ying W, Wei G, Wang D, Wang Q, Tang X, Shi J, Zhang P, and Lu H. Intranasal administration with NAD+ profoundly decreases brain injury in a rat model of transient focal ischemia. Front Biosci 12: 2728–2734, 2007 [DOI] [PubMed] [Google Scholar]
- 176. Yoshino J, Mills KF, Yoon MJ, and Imai S. Nicotinamide mononucleotide, a key NAD+ intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab 14: 528–536, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Yu CP, Song YL, Zhu ZM, Huang B, Xiao YQ, and Luo DY. Targeting TDO in cancer immunotherapy. Med Oncol 34: 73, 2017 [DOI] [PubMed] [Google Scholar]
- 178. Zhang K, Kim H, Fu Z, Qiu Y, Yang Z, Wang J, Zhang D, Tong X, Yin L, Li J, Wu J, Qi NR, Houten SM, and Zhang R. Deficiency of the mitochondrial NAD kinase causes stress-induced hepatic steatosis in mice. Gastroenterology 154: 224–237, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Zhang R. MNADK, a novel liver-enriched mitochondrion-localized NAD kinase. Biol Open 2: 432–438, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Zhang T, Berrocal JG, Frizzell KM, Gamble MJ, DuMond ME, Krishnakumar R, Yang T, Sauve AA, and Kraus WL. Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters. J Biol Chem 284: 20408–20417, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Zhou L, Wang F, Sun R, Chen X, Zhang M, Xu Q, Wang Y, Wang S, Xiong Y, Guan KL, Yang P, Yu H, and Ye D. SIRT5 promotes IDH2 desuccinylation and G6PD deglutarylation to enhance cellular antioxidant defense. EMBO Rep 17: 811–822, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Zhou X, Fan LX, Sweeney WE, Jr., Denu JM, Avner ED, and Li X. Sirtuin 1 inhibition delays cyst formation in autosomal-dominant polycystic kidney disease. J Clin Invest 123: 3084–3098, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]