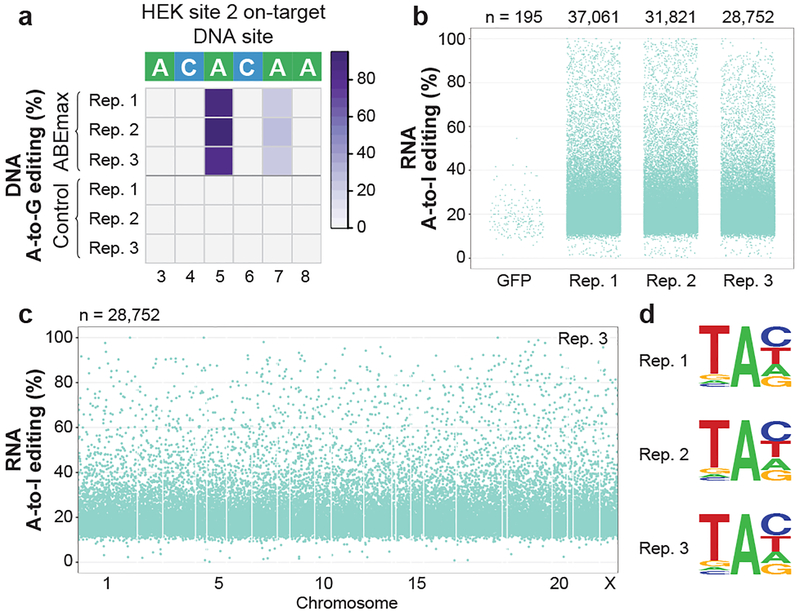
Figure 3. Adenine base editor ABEmax induces transcriptome-wide off-target A-to-I RNA editing in HEK293T cells.
(a) Heat map of on-target DNA base editing efficiencies of ABEmax and NLS-nCas9-NLS (Control) in HEK293T cells with the HEK site 2 gRNA. Bases shown are within the editing window of the on-target site (numbered with 1 as the most PAM distal position). (b) Jitter plots derived from RNA-seq experiments showing RNA adenines modified by ABEmax expression with the HEK site 2 gRNA or a GFP control (Methods). Y-axis represents the efficiencies of A-to-I RNA editing. n = total number of modified adenines observed. (c) Manhattan plot showing the distribution of modified adenines across the transcriptome for replicate 3 from (b). n = total number of modified adenines observed. (d) Sequence logos derived from edited adenines in each RNA-seq replicate. Analysis done using RNA-seq data generated from cDNA, thus every T depicted should be considered a U in RNA.