Abstract
Purpose
To determine whether a selected set of mRNA biomarkers expressed in individual cumulus granulosa cell (CC) masses show association with oocyte developmental competence, embryo ploidy status, and embryo outcomes.
Methods
This prospective observational cohort pilot study assessed levels of mRNA biomarkers in 163 individual CC samples from 15 women stimulated in antagonist cycles. Nineteen mRNA biomarker levels were measured by real-time PCR and related to the development of their corresponding individually cultured oocytes and subsequent embryos, embryo ploidy status, and live birth outcomes.
Results
PAPPA mRNA levels were significantly higher in CC from oocytes that led to euploid embryos resulting in live births and aneuploid embryos compared to immature oocytes by ANOVA. LHCGR mRNA levels were significantly higher in CC of oocytes resulting in embryos associated with live birth compared to immature oocytes and oocytes resulting in arrested embryos by ANOVA. Using a general linearized mixed model to assess ploidy status, CC HSD3B mRNA levels in oocytes producing euploid embryos were significantly lower than other oocyte outcomes, collectively. When transferred euploid embryos outcomes were analyzed by ANOVA, AREG mRNA levels were significantly lower and PAPPA mRNA levels significantly higher in CC from oocytes that produced live births compared to transferred embryos that did not form a pregnancy.
Conclusions
Collectively, PAPPA, LHCGR, and AREG mRNA levels in CC may be able to identify oocytes with the best odds of resulting in a live birth, and HSD3B1 mRNA levels may be able to identify oocytes capable of producing euploid embryos.
Electronic supplementary material
The online version of this article (10.1007/s10815-019-01489-8) contains supplementary material, which is available to authorized users.
Keywords: Cumulus cells, Real-time PCR, mRNA levels, Oocyte developmental competence, Euploid embryo
Introduction
Successful in vitro fertilization (IVF) is highly dependent upon the quality of the oocytes harvested and the subsequent quality of embryos used for uterine transfer [1]. Several studies have shown that use of preimplantation genetic testing for aneuploidy (PGT-A) to select euploid embryos for uterine transfer increases implantation, ongoing pregnancy, and delivery rates [2–4], whereas other studies showed PGT-A decreased pregnancy rates [5, 6]. Conflicting PGT-A findings might indicate that other methods beyond genetic screening are needed to improve IVF pregnancy rates. The differing PGT-A study results may also reflect the invasive methodology of PGT-A, which requires the removal of 3–10 cells from the trophectoderm. Increased numbers of trophectoderm cells per biopsy have been correlated with decreased implantation rates [7]. Therefore, a method for identifying oocytes that will produce euploid embryos with the greatest chances of leading to a live birth that does not require embryo manipulation would be advantageous.
Some non-invasive methods for assessing oocyte and embryo live birth potential include preimplantation embryo metabolomics and proteomics of secreted products [8, 9] and granulosa cell (GC) mRNA biomarkers [10]. The use of GC biomarkers is based on knowledge that cumulus granulosa cells (CCs) and mural granulosa cells (MGCs) interact with oocytes and can reflect the health and maturational status of the oocyte [1, 11]. GCs are obtained as a by-product of oocyte retrieval and can be rapidly assessed for mRNA levels by reverse transcription and quantitative polymerase chain reaction (PCR). Several studies suggest that CC biomarker mRNA levels from individual oocytes can be used to generate models capable of predicting pregnancy and live birth [12–14].
Approaches to evaluate differentially expressed genes in CCs have employed large-scale microarray, transcriptome deep sequencing, PCR arrays, and quantitative PCR [10]. In most cases, the ploidy status of the embryos was not known, which could have altered the study outcomes. Only a handful of studies have examined CC mRNA levels in conjunction with oocyte or embryo ploidy status [15–17]. Taken together, a focused quantitative PCR analysis of CC target genes selected from prior studies combined with the ploidy status of the embryos should yield valuable information regarding mRNA biomarker usefulness in predicting live birth.
Many oocytes fail to fertilize, or when fertilized, arrest before reaching the blastocyst stage, or fail to develop into euploid blastocysts. We hypothesize that while many mature oocytes appear morphologically normal, the actual developmental competence of those oocytes is variable and will be reflected by varying mRNA levels of specific biomarkers in their associated CCs. Furthermore, we propose that a CC mRNA biomarker model could potentially indicate whether a mature oocyte, when fertilized, will result in a euploid embryo. In addition, we hypothesize that certain CC mRNA biomarkers and their levels will associate with oocytes that are capable of producing viable embryos which result in live births.
A comprehensive review of the human literature for potential CC candidate mRNA biomarkers was performed [10], and genes were selected based on their functions. In some cases, the genes selected were based on data in non-human animals [18–24]. Gene products associated with oocyte maturation included calmodulin 1 (CALM1), gap junction alpha-4 protein (GJA4), gremlin (GREM1), and inhibitor of growth protein 1 (ING1). Genes associated with cumulus mass expansion included amphiregulin (AREG), prostaglandin-endoperoxide synthase 2 (PTGS2), and versican (VCAN). Genes encoding proteins mediating de novo steroidogenesis, estrogen, and progesterone production included aromatase (CYP19A1), cholesterol side-chain cleavage enzyme (CYP11A1), hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 (HSD3B1), and steroidogenic acute regulatory protein (STARD1). Genes related to hormone modulation included insulin-like growth factor 2 (IGF2), insulin-like growth factor binding protein 5 (IGFBP5), and inhibin beta A subunit (INHBA), and pappalysin-1 (PAPPA). Gene encoding hormone receptors included follicle-stimulating hormone receptor (FSHR), luteinizing hormone/choriogonadotropin receptor (LHCGR), progesterone receptor (PGR), and progesterone receptor, membrane component 1 (PGRMC1). The primary goal of this study was to determine if a model of specific CC mRNA biomarker levels could identify a mature oocyte’s ability to produce euploid embryos. The secondary goal of this study was to determine if specific CC mRNA biomarker levels could indicate oocytes capable of leading to euploid embryos with live birth outcomes.
Materials and methods
Study population, participants, and CC isolation
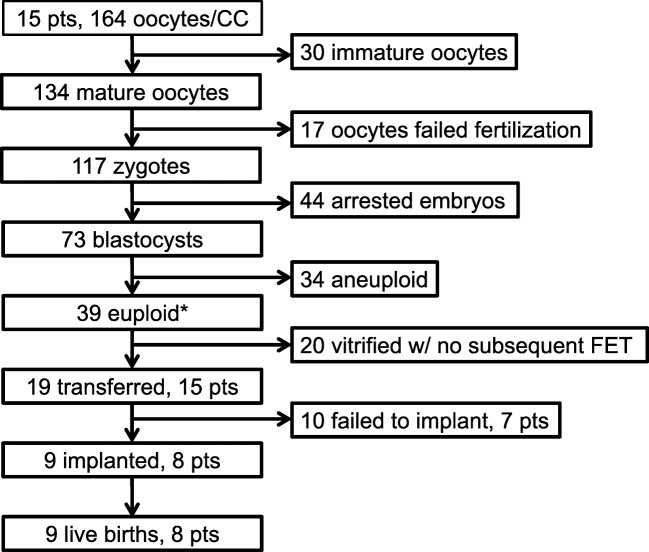
This study is a prospective observational cohort pilot study. In total, 164 individual CC masses were harvested from their corresponding oocytes from 15 patients undergoing infertility treatment with PGT-A at Advanced Fertility and Reproductive Endocrinology Institute (Columbia, SC, USA) from February 2015 through June 2016. Patients either electively desired PGT-A or it was recommended based on the patients’ diagnosis (Table 1). Patients with endometriosis or diminished ovarian reserve (DOR) were not included as prior work indicates these conditions alter GC gene expression [25–28]. Patient demographics and cycle information (Table 1), blastocyst development, PGT-A results, and live birth data (Supplemental Table 1), as well as CC mRNA levels were collected. Each patient’s ovaries were stimulated using an antagonist protocol until at least one follicle at 15 mm was present when a GnRH antagonist (Cetrotide, EMD-Serono, USA, or Ganirelix, EMD-Serono, USA) was administered. When two follicles reached 18 mm or 50% of the follicles were ≥ 15 mm by ultrasound, an ovulatory dose of human chorionic gonadotropin (hCG) (Pregnyl, EMD-Serono, USA) or Lupron (Leuprolide Acetate, Sandoz, USA) was delivered. Thirty-six hours later, patients underwent oocyte retrieval. Each CC mass was mechanically separated from its oocyte and rinsed in medium (HTF-HEPES, Irvine, USA) to remove debris. Each CC mass and its associated oocyte/embryo were kept separate throughout the entire process. A summary flowchart of the fate of the collected oocytes is shown in Fig. 1. CC masses were snap frozen and maintained in liquid nitrogen until they were transported to the University of South Carolina School of Medicine for further processing.
Table 1.
Patient demographics and cycle information
| Pt# | Age | BMI | Diagnosis/PGT-A reason | Cycle # | Day 3 FSH level (IU/ml) | E2 per oocyte (pg/ml) | # oocytes | # mature | # fertilized | # embryos biopsied | Trigger |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 27.8 | 25.3 | Sex selection | 3 | 5.4 | 141.9 | 8 | 8 | 8 | 4 | hCG |
| 2 | 36.4 | 26.4 | Male factor | 1 | 7.2 | 86.6 | 8 | 7 | 6 | 4 | hCG |
| 3 | 39.7 | 25 | AMA | 1 | 12 | 100.4 | 7 | 4 | 3 | 3 | hCG |
| 4 | 30 | 25.6 | Donor | 1 | 6 | 147.4 | 10 | 9 | 7 | 6 | hCG |
| 5 | 32.6 | 26.4 | Translocation w/PGT-A | 2 | 9.2 | 131.0 | 5 | 5 | 5 | 5 | Lupron |
| 6 | 35.9 | 21 | Idiopathic | 1 | 8.3 | 163.3 | 4 | 4 | 4 | 2 | Lupron |
| 7 | 39.7 | 38.8 | AMA | 1 | 11 | 145.5 | 9 | 9 | 6 | 5 | Lupron |
| 8 | 40 | 32 | AMA + male factor | 1 | 12 | 31.4 | 7 | 5 | 5 | 4 | Lupron |
| 9 | 33 | 18.8 | Male factor | 1 | 10 | 194.7 | 11 | 10 | 10 | 4 | Lupron |
| 10 | 42 | 20.3 | AMA | 1 | 6.6 | 108.2 | 5 | 5 | 5 | 3 | hCG |
| 11 | 36.1 | 29.2 | Tubal factor | 1 | 5.2 | 179.5 | 23 | 23 | 18 | 12 | Lupron |
| 12 | 37 | 20.7 | AMA | 1 | 11 | 142.2 | 16 | 15 | 14 | 8 | Lupron |
| 13 | 33.9 | 35 | Male factor | 2 | 10 | 183.1 | 11 | 10 | 8 | 2 | hCG |
| 14 | 28 | 28.4 | PCO | 1 | 5.9 | 184.2 | 24 | 6 | 5 | 4 | hCG |
| 15 | 39.7 | 31 | AMA | 1 | 7.5 | 132.6 | 16 | 14 | 14 | 7 | Lupron |
AMA advanced maternal age, PCO polycystic ovaries, hCG human chorionic gonadotropin
Fig. 1.
Flow diagram summarizing the study population from oocyte retrieval to final outcome. Oocytes (164) were retrieved from 15 patients and individually cultured. Individual CC masses from each oocyte were collected and mRNA was harvested from each mass. Of the 164 oocytes, 134 were mature and 30 were immature. All 134 were injected with sperm. Seventy-three of the fertilized oocytes became blastocysts and were biopsied for PGT-A testing and all were vitrified. PGT-A results indicated that 39 embryos were euploid and 34 were aneuploid. Nineteen of the euploid embryos were transferred and 20 remained in cryostorage. Ten embryos from 7 patients failed to implant while 9 embryos from 8 patients implanted and resulted in live births. *The control TBP mRNA did not amplify in one CC sample from an oocyte yielding a euploid embryo and was not included in the data analysis. RNA analyses were performed on n = 163 individual cumulus masses
Embryo culture and ploidy assessment
Mature oocytes were inseminated via intracytoplasmic sperm injection. Fertilization was confirmed by the presence of two pronuclei and two polar bodies 16–18 h after insemination. Oocytes and embryos were individually cultured in 20 μL media drops (Global total, Global, USA) under oil (Ovoil, Vitrolife, USA) at 37 °C, 6% CO2, 5% O2, 89% N2 in a humidified atmosphere until day 5 or day 6 post retrieval. Blastocyst morphologies were assessed using Gardner’s blastocyst grading scale [29], and 3–7 trophectoderm cells were biopsied and sent to Igenomix (Miami, FL, USA) for PGT-A using next-generation sequencing. Fifteen frozen embryo transfers (FETs) were performed using the highest quality euploid embryos (19 embryos total) for each patient. Live birth outcomes were obtained from the patient’s obstetrician including delivery dates and if there were any maternal or neonatal interventions or complications.
RNA isolation and cDNA synthesis
RNA was isolated from each CC mass using the Direct-zol MiniPrep Kit with DNase treatment (Zymo Research, Irvine, CA, USA). RNA concentration and purity were assessed at the wavelengths of 260 and 280 nm using a spectrophotometer with a 2 mm lid (NanoDrop 2000C, Thermo Scientific, USA). Sample amounts varied between 80 and 360 ng RNA per individual sample. Each sample was reverse transcribed into cDNA using the Bio-Rad iScript kit (Hercules, CA, USA). The reverse transcription reaction was carried out in an Eppendorf thermocycler, for 5 min at 25 °C, 30 min at 42 °C, and 5 min at 85 °C.
Quantitative real-time PCR
Gene primer sequences, annealing temperatures, and amplicon sizes can be found in Table 2. Synthesized primers were cartridge purified (Life Technologies, Carlsbad, CA, USA). PCR reactions were run with two or more wells per sample for 45 cycles and threshold cycle (Ct) values were averaged. The reaction included 2 μL of cDNA (6–27 ng starting RNA), 300 nM of each upstream and downstream primer, and 10 μL 2X SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA) and sterilized PCR-grade water to a final volume to 20 μL per well. PCR-grade water was substituted for the cDNA as a negative control. PCR amplification was performed using the iCycler iQ Real-Time PCR Detection System (Bio-Rad, USA). TATA-box binding protein (TBP) mRNA was used as an internal control [30]. Single amplicons of correct size were verified using agarose gels and by the presence of single melt-curve peaks. mRNA quantities were derived from a standard curve for each mRNA made using serial dilutions of its purified amplicon. TBP has extensively been tested as a control gene in our laboratory with human granulosa cells under multiple conditions or treatments and has been found not to be regulated under any circumstances. Target mRNA values were expressed relative to values of the TBP mRNA control.
Table 2.
Primer set information
| Gene | Primer sequences 5′–3′ | Annealing temperature (°C) | Amplicon size (base pairs) | Reference or NCBI no. |
|---|---|---|---|---|
| AREG |
For TGGACCTCAATGACACCTACTCTG Rev GGGCTTAACTACCTGTTCAACTCTG |
62° | 251 | [44] |
| CALM1 |
For TACTTCGTGTGCTCCGACCCAT Rev AGTCCACAGCCACAGCCTACTC |
62 | 231 | [51] |
| CYP11A1 |
For GCAACGTGGAGTCGGTTTATGTC Rev GTGCAGGACACTGACGAAGTC |
60 | 269 | [52] |
| CYP19A1 |
For GCACATCCTCAATACCAGGTC Rev TTTGAGGGATTCAGCACAGAC |
60 | 380 | [38] |
| FSHR |
For ACCAAGCTTCGAGTCATCC Rev CATCTGCCTCTATCACCTCC |
58 | 103 | [53] |
| GJA4 |
For CATCTCCCACATCCGCTACT Rev GAAGCCTGCCTCTAGCACAC |
58 | 295 | [54] |
| GREM1 |
For CGCCGCACTGACAGTATGAG Rev ACCTTGGGACCCTTTCTTTTTC |
62 | 108 | NM_013372.7 |
| HSD3B1 |
For TGTGCCAGTCTTCATCTACAC Rev TGTTTTCCAGAGGCTCTTCTTC |
60 | 101 | [38] |
| IGF2 |
For TTCCGCAGCTGTGACCTGGC Rev CCTCGAGCTCCTTGGCGAGC |
62 | 229 | NG_008849.1 |
| IGFBP5 |
For AAGAAGCTGACCCAGTCCAA Rev GAATCCTTTGCGGTCACAAT |
60 | 201 | [55] |
| ING1 |
For GCCTGGTGTGAGGAGGACAA Rev CCCTATGAAAGGAATGGTTCCTT |
62 | 124 | [56] |
| INHBA |
For AAGTCGGGGAGAACGGGTATGTGG Rev TCTTCCTGGCTGTTCCTGACTCG |
62 | 123 | [57] |
| LHCGR |
For TGGAGAAGATGCACAATGGA Rev GGCAATTAGCCTCTGAATGG |
60 | 122 | [58] |
| PAPPA |
For GTCATCTTTGCCTGGAAGGGAGAA Rev AGGGCTGTTCAACATCAGGATGAC |
62 | 129 | [61] |
| PGR |
For GTCATAGACCCCCGTTGCTA Rev GCTAAGCCAGCAAGAAATGG |
60 | 124 | [60] |
| PGRMC1 |
For TCTGGACTGCACTGTTGTCCTTG Rev GCAAACACCTGTTCCTATTCTG |
60 | 290 | [61] |
| PTGS2 |
For GCTTTATGCTGAAGCCCTATGA Rev TCCAACTCTGCAGACATTTCC |
60 | 70 | [45] |
| STARD1 |
For TACGTGGCTACTCAGCATCG Rev ACAGCAGGCTGGTCTTCAAC |
60 | 157 | [62] |
| TBP |
For CACGGCACTGATTTTCAGTTC Rev TCTTGCTGCCAGTCTGGACT |
62 | 79 | [63] |
| VCAN |
For GCACCTGTGTGCCAGGATA Rev CAGGGATTAGAGTGACATTCATCA |
60 | 70 | [45] |
Statistical analysis
The mRNA levels were normalized using log2 transformation. Principal component analysis (PCA) was conducted to explore the mRNA level correlation patterns between the CC samples. CC mRNA levels were compared between different oocyte and subsequent embryo developmental outcomes using repeated measures analysis of variance (ANOVA) to accommodate for the variable number of oocytes per patient followed by Tukey’s post hoc tests for pairwise comparisons. The CC samples used for these studies were divided into the following categories: immature oocytes, mature oocytes that failed fertilization, oocytes that produced embryos that arrested and were not appropriate for biopsy, oocytes that produced biopsied blastocysts that were aneuploid, and oocytes that produced biopsied blastocysts that were euploid. Oocytes that produced euploid embryos were further subdivided into embryos that resulted in live birth, embryos that resulted in a negative pregnancy, and embryos that were vitrified and not transferred. Each figure states which categories were used for the specific statistical analysis.
Statistical models
Models were estimated to predict multiple dichotomous oocyte developmental outcomes using generalized linear mixed models (GLMMs) fitted by maximum likelihood using the R package lme4 [31]. The GLMMs included the normalized gene expressions as variables and used patients’ demographics (age and body mass index (BMI) as controlling factors and adjusted for two ovulation induction or trigger medications). Unstructured correlation patterns were used in fitted GLMMs to capture the correlation among the repeated observations from same individual. The statistical models aimed to include as many patients as possible. However, some of the genes had high proportion of missing values for some patients because of limited RNA amounts that were obtained from CC masses. To ensure maximum utilization of the data, we started the modeling by including all the genes and proceeded backward dropping the genes one-by-one with highest proportion of missing observations until the fitted mixed-effects model attained computational convergence successfully. Since different patients had missing observations on different genes, exclusion of the missing values eventually reduced the number of patients and number of observations in the sample used to fit the model. The analysis sample size differed from model-to-model (for different response variables) as the maximum possible data points were used for a model to converge. Statistical significance in all analyses was determined at a 0.05 significance level. All the inference P values were reported after adjusting for multiple comparisons [32]. The predictive performances of the fitted models were assessed using receiver operating characteristics (ROC) curves and area under ROC curves (AUC). The AUC values of the models used in this study ranged from 0.75 to 1.00 implying the models exhibited good/very good to excellent predictive accuracy. Statistical analysis was performed using the statistical software R 3.4.4 (http://www.r.project.org/).
Results
In total, 164 CCs were harvested from 15 patients undergoing infertility treatment with PGT-A. Fifteen FETs were performed after PGT-A results were obtained. Eleven patients had single embryo transfers and four had double embryo transfers. In the four cases where two embryos were selected, one embryo of each gender was transferred. Eight patients gave birth to nine healthy children with no complications. One hundred sixty-three of 164 CC samples (n = 15 patients) analyzed contained a sufficient amount of RNA to detect the endogenous control house-keeping gene, TBP. Due to limited starting sample RNA amounts, not all samples were able to be tested for each biomarker and the variations in sample size are noted in figures.
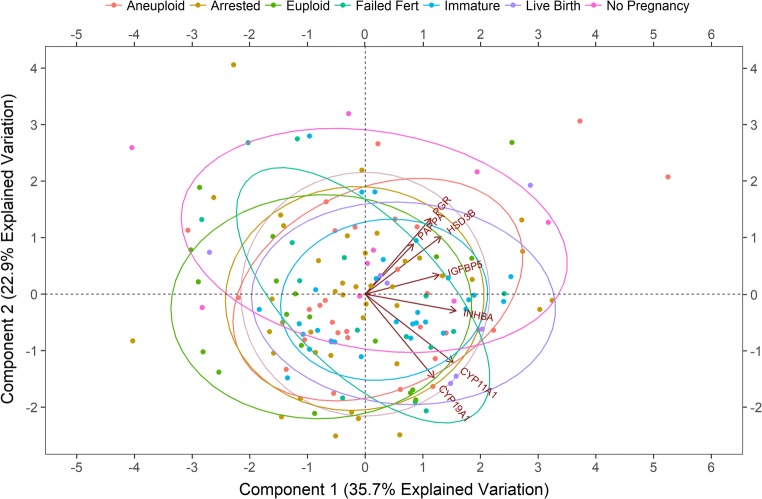
PCA was utilized to test the hypothesis that several of the CC genes we examined would be correlated with one another. The correlations of the CC mRNA levels of 151 observations from 14 patients were examined by creating a biplot within the multivariable dataset that contained the most biomarkers and individual patient observations that fit the model (Fig. 2). The biplot is based on the first and second principal components (PC1 and PC2) of the log2-transformed mRNA levels of each individual CC mass. PC1 and PC2 explain 35.7% and 22.9% of the variability of the model, respectively. All genes shown were positively correlated to PC1. HSD3B1, IGFBP5, PAPPA, and PGR were all positively correlated with PC2 while CYP11A1, CYP19A1, and INHBA are negatively correlated with PC2. In addition, a significant (P < 0.001) strong positive correlation was observed between CYP19A1 and CYP11A1 (r = .68), and significant (P < 0.001) moderate positive correlations were observed for PGR and HSD3B (r = .59), CYP11A1 and INHBA (r = .50), PGR and IGFBP5 (r = .41), and PAPPA and INHBA (r = .41).
Fig. 2.
Principal component analysis biplot for CC mRNA expression. Biplot of the log2-transformed mRNA levels for 7 genes from 14 patients along the first and second principal components (PC1 and PC2). PC1 accounted for 35.7% of the observed variance while PC2 accounted for 22.9% of the variance. Each data point represents an individual CC mass mRNA level associated with an individual oocyte (n = 151 oocytes). The biplot demonstrates the relationship between individual CC mRNA level patterns and the correlation between the different genes. The closer the data points are to each other, the more similar the normalized CC gene expression patterns. The closer the arrows are to each other, the higher the correlation between the normalized CC gene expressions. Groups represent CC mRNA from oocytes with the following descriptions: aneuploid = mature oocytes resulting in aneuploid embryos; arrested = mature oocytes resulting in embryos that did not reach the blastocyst stage; euploid = mature oocytes resulting in euploid blastocysts that were not transferred; failed fert = mature oocytes that did not fertilize; immature = immature oocytes that were not fertilized; live birth = oocytes that resulted in euploid embryos that resulted in live births; no pregnancy = oocytes that resulted in euploid embryos that did not result in a pregnancy
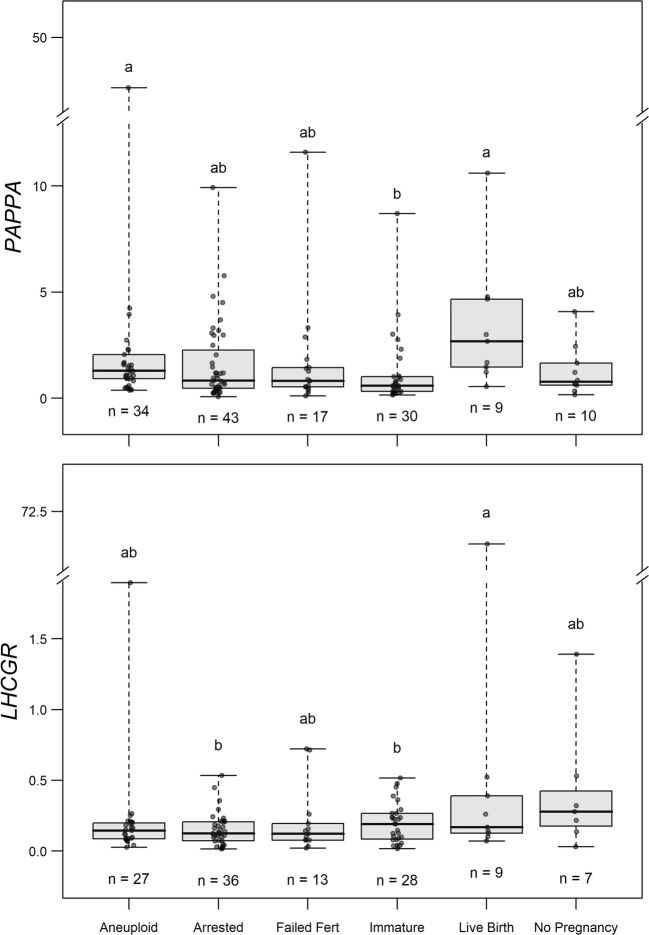
We next wanted to test the hypothesis that the CC mRNA levels of the selected genes would vary by oocyte developmental competence and resulting embryo endpoint. To test this hypothesis, we analyzed all the CC mRNA level data by the resulting developmental outcome for each group of oocytes by repeated measures ANOVA followed by Tukey’s post hoc test (adjusted P values). PAPPA mRNA levels in CCs were higher in oocytes that resulted in euploid embryos that led to a live birth compared to immature oocytes and in oocytes that resulted in aneuploid embryos compared to immature oocytes, when comparing all groups (P < 0.05) (Fig. 3). LHCGR mRNA levels in CCs were higher for oocytes that resulted in embryos which led to a live birth compared to immature oocytes and oocytes that resulted in arrested embryos compared to immature oocytes, when comparing all groups (P < 0.05). Other CC biomarkers showed no statistical differences between groups and their expression profiles are shown in Supplemental figure 1.
Fig. 3.
Biomarkers for CC mRNA expression associated with mature oocyte competence and embryo outcomes. To determine the differences in CC mRNA between groups where oocytes had different developmental and embryo outcomes, target mRNA levels were compared using repeated measures ANOVA followed by Tukey’s post hoc test (adjusted P values) for pairwise comparisons. Groups represent CC mRNA from oocytes with the following descriptions: aneuploid = mature oocytes resulting in aneuploid embryos; arrested = mature oocytes resulting in embryos that did not reach the blastocyst stage; failed fert = mature oocytes that did not fertilize; immature = immature oocytes that were not fertilized; live birth = oocytes that resulted in transferred euploid embryos that resulted in live births; no pregnancy = oocytes that resulted in transferred euploid embryos that did not result in a pregnancy. Data are presented as the median copy number (line inside box), first and third quartile (bottom and top of box), and highest and lowest data points (top and bottom of whiskers). Groups with different letters (a and b) exhibit significant differences (P < 0.05)
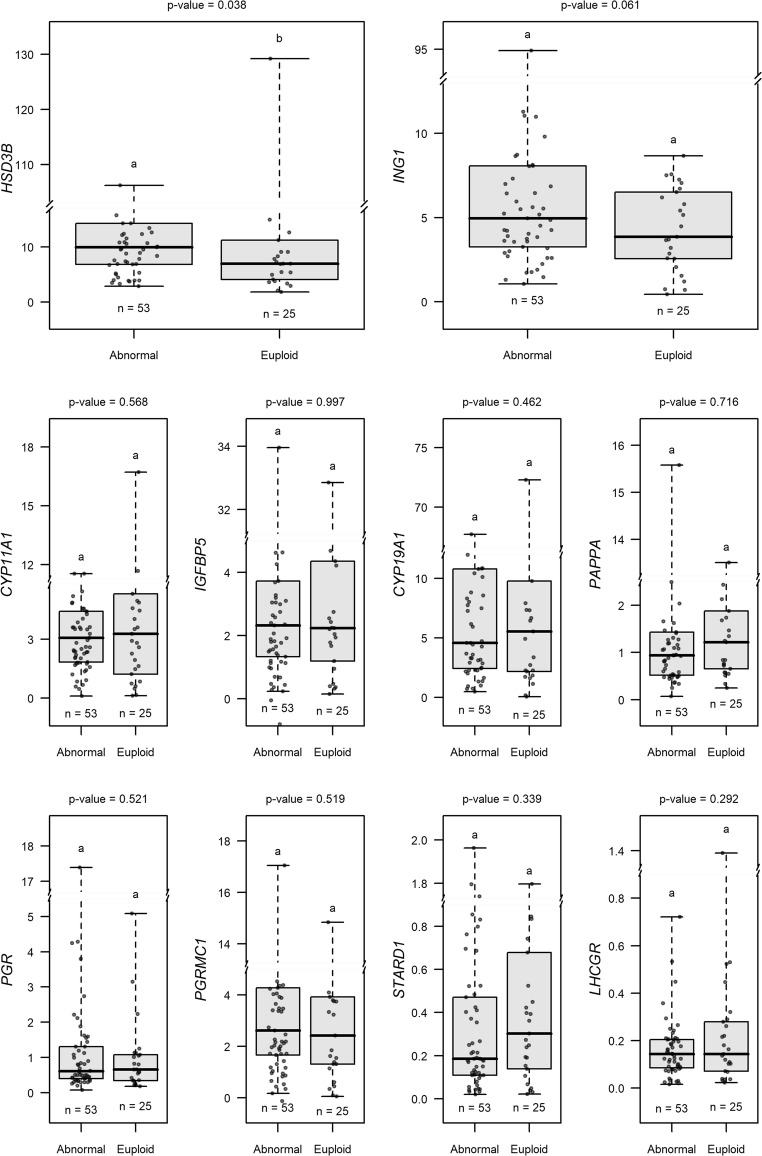
We also wanted to test the hypothesis that certain CC biomarker mRNA levels associated with oocytes giving rise to euploid embryos would be distinct from those giving rise to aneuploid embryos and those oocytes that resulted in arrested embryos or those that failed fertilization. Using a GLMM, the best fitting model examining whether CC biomarker mRNA expression associated with oocytes that became euploid embryos included 11 patients (n = 78 CC masses) and CYP11A1, CYP19A1, HSD3B1, IGFBP5, PAPPA, PGR, PGRMC1, ING1, LHCGR, and STARD1 (Fig. 4). CCs associated with oocytes that produced euploid embryos exhibited significantly (P < 0.05) lower CC HSD3B1 mRNA levels than those oocytes that led to mature oocytes resulting in aneuploid blastocysts, oocytes producing embryos that arrested and did not reach the blastocyst stage, and mature oocytes that failed to fertilize, collectively (abnormal group). CC ING1 mRNA levels associated with oocytes that produced euploid embryos trended toward being lower than CCs from oocytes that gave rise to the abnormal outcomes, collectively (P = 0.061).
Fig. 4.
Biomarkers from the GLMM model associated with oocytes producing euploid embryos versus mature oocytes with other outcomes. To evaluate CC biomarker mRNA level association with oocytes capable of producing euploid embryos, a model was fit with 78 CC samples from 11 patients and included the biomarkers: CYP11A1, CYP19A1, HSD3B, IGFBP5, PAPPA, PGR, PGRMC1, ING1, LHCGR, and STARD1. Higher HSD3B mRNA level significantly decreased the odds of an oocyte resulting in a euploid embryo (OR = 0.408, 95% CI 0.175 to 0.953). Higher ING1 mRNA levels marginally decreased the odds of an oocyte resulting in a euploid embryo (OR = 0.552, 95% CI 0.297 to 1.027). Groups represent CC mRNA from oocytes with the following descriptions: abnormal = mature oocytes resulting in aneuploid blastocysts, oocytes producing embryos that did not reach the blastocyst stage, and mature oocytes that failed to fertilize; euploid = mature oocytes that resulted in euploid embryos regardless of whether they were transferred or remained vitrified. Data are presented as stated in Fig. 3 legend
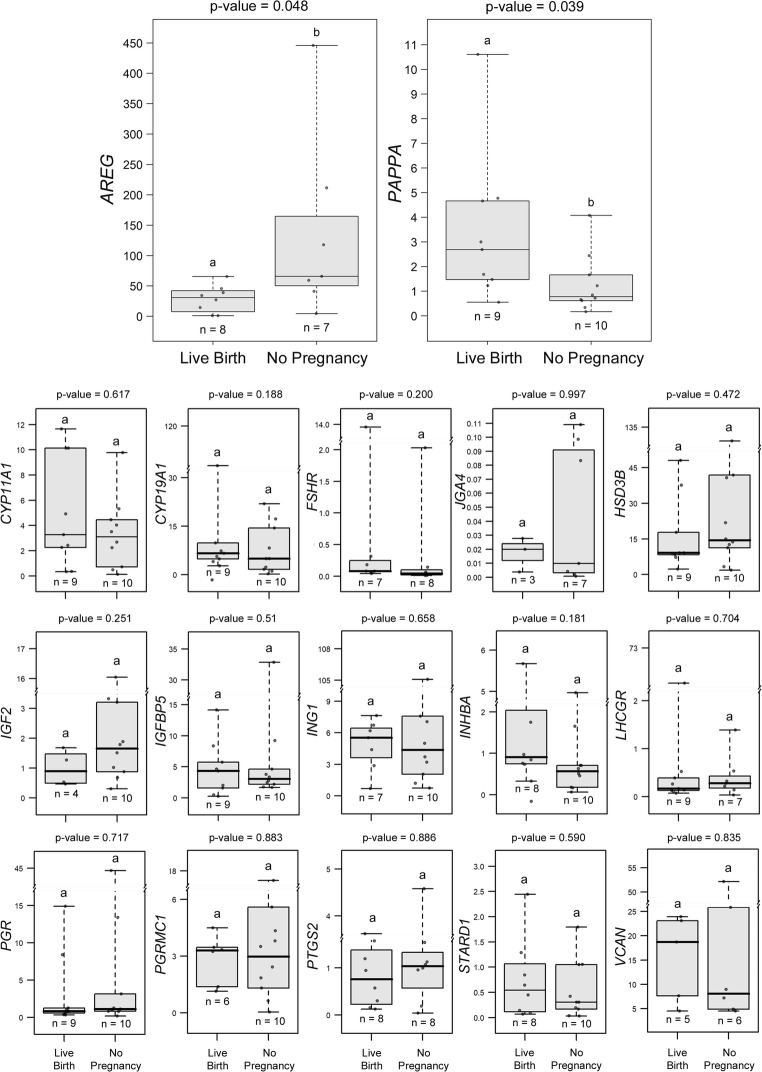
A critical hypothesis we tested was that certain CC mRNA biomarker levels of oocytes giving rise to euploid embryos would be associated with live birth outcomes. CC mRNA levels from all 15 patients (n = 19 CC masses) were assessed for differences between oocytes yielding transferred embryos that resulted in a live birth and those that did not form a pregnancy, using repeated measures ANOVA followed by Tukey’s post hoc test (adjusted P values). AREG mRNA levels were significantly lower in CCs from oocytes that resulted in live births (n = 8 from 8 patients) compared to the no pregnancy group (n = 7 from 5 patients) (P < 0.05) (Fig. 5). PAPPA mRNA levels were higher in CCs from oocytes producing euploid embryos that resulted in live births (n = 9 from 8 patients) compared to those that produced no pregnancy (n = 10 from 7 patients) (P < 0.05). The other biomarkers showed no statistical differences between groups.
Fig. 5.
CC biomarkers associated with oocytes giving rise to euploid embryos with live birth or no pregnancy. Transferred embryos (n = 19) from all 15 patients were assessed for mRNA level differences between those oocytes yielding embryos that resulted in a live birth and those that did not form a pregnancy using repeated measures ANOVA followed by Tukey’s post hoc test (adjusted P values) for pairwise comparisons. Of the transferred embryos, 9 resulted in live births. AREG mRNA levels were significantly lower in CCs from oocytes that resulted in live births (n = 8 from 8 patients) compared to the no pregnancy group (n = 7 from 5 patients) (P < 0.05). PAPPA mRNA expression was significantly increased in CCs from oocytes producing embryos that resulted in live births (n = 9 from 8 patients) compared to no pregnancy (n = 10 from 7 patients) (P < 0.05). GREM1 mRNA levels (n = 3/group) were not significantly different and not shown. Data are presented as in Fig. 3 legend
Using a GLMM, we also compared the CC mRNA biomarkers associated with oocytes that resulted in euploid embryos producing a live birth compared to oocyte outcomes not producing a live birth (non-viable group), collectively. The non-viable group included immature oocytes, mature oocytes resulting in aneuploid blastocysts, oocytes producing embryos that did not reach the blastocyst stage, and mature oocytes that failed to fertilize. The best fitting model for this comparison included 110 CC samples from 14 patients and included mRNA levels for CYP11A, CYP19A1, IGFBP5, PAPPA, PGRMC1, and STARD1 (Supplemental Figure 2). As revealed in the fitted model, higher CC PAPPA mRNA levels (P < 0.05) significantly increased the odds of an oocyte producing an embryo that resulted in a live birth (OR = 4.591, 95% CI 1.098 to 19.201). The other five CC mRNA biomarkers did not significantly associate with live births.
Discussion
The key findings of this research were that higher PAPPA, higher LHCGR, and lower AREG mRNA levels in CCs were associated with oocytes that produced embryos capable of yielding live birth outcomes. In addition, lower CC HSD3B1 mRNA was associated with oocytes that resulted in euploid embryos compared to other outcomes. Our premise was that although many retrieved oocytes in our study appeared mature, several oocytes would have mRNA expression profiles that were similar to those of immature oocytes. Aberrant mRNA levels would cause them to be incapable of forming euploid embryos and leading to live births. Therefore, an individual oocyte’s developmental competence would be reflected in specific mRNA levels of their CC masses. For example, we predicted that an immature oocyte CC profile would differ from that of a mature oocyte CC profile capable of giving rise to a euploid embryo resulting in a successful pregnancy. This was partly the case for CC PAPPA and LHCGR mRNA levels which were higher in CCs surrounding mature oocytes that gave rise to embryos resulting in live birth than immature oocytes (by ANOVA). However, by ANOVA, the levels of CC PAPPA and LHCGR could not distinguish between oocytes giving rise to morphologically normal embryos that were aneuploid and transferred euploid embryos that resulted in live birth or no pregnancy. When utilizing the best fit GLMM, higher CC PAPPA levels continued to associate with oocytes giving rise to embryos with live birth compared to transferred embryos that did not result in pregnancies as well as other oocyte outcomes, collectively. In transferred embryos, lower CC AREG associated with oocytes giving rise to embryos with live birth outcomes by ANOVA analysis. Combining these data with that for CC HSD3B1, where lower levels were associated with euploid embryos, one can envision using these findings to create a future practical model where the CC levels of all four mRNA biomarkers could be used to predict which of a patient’s cohort of oocytes and embryos will most likely be euploid and give rise to a live birth outcome. Although four biomarkers of the selected group exhibited significant differences in statistical models, this was a small pilot study and the data is limited by the small sample numbers for FETs. However, these results provide a starting point for a larger prospective future study.
Several of our analyses indicated PAPPA may be a reliable novel biomarker in CCs for identifying oocytes capable of leading to a live birth. PAPPA encodes a metalloproteinase that cleaves insulin-like growth factor binding protein 4 (IGFBP4) and IGFBP5 [33]. Increased levels of PAPPA mRNA and protein in peri-ovulatory follicles may be necessary to increase intrafollicular free IGFs through enhanced cleavage of IGFBPs [34]. IGFs are known to enhance gonadotropin signaling and FSH-induced LHCGR abundance [35]. We also found that immature oocytes possessed lower CC LHCGR mRNA levels compared to all the other groups except oocytes that resulted in arrested embryos. Similarly, others have shown that CC LHCGR mRNA levels were higher in mature compared to immature oocytes [36]. In addition, the ratio of CC LHCGR mRNA splice variants was previously reported to be higher in pregnant compared to non-pregnant patients [37].
Differential mRNA levels of steroidogenic genes in CCs may impact the fate of their associated oocytes. A previous study utilizing GCs pooled from multiple patients and follicles showed increased CYP19A1 and HSD3B1 mRNA levels in GCs from oocytes producing embryos that resulted in pregnancies compared to oocytes resulting in embryos that failed to develop [38]. In our study, lower CC HSD3B1 mRNA levels associated significantly with oocytes which yielded euploid embryos. Additionally, HSD3B1 and PAPPA both positively correlated with principal component 1 and 2 in our biplot analysis. The difference may have become apparent as we evaluated individual CC masses rather than pooling. Before including the trigger in our best fitting model, lower CC CYP19A1 mRNA level was associated with live birth (P = 0.048, not shown), but showed only a trend when trigger medication was added to the model (P = 0.052; Supplemental figure 2). Given that different trigger medications can alter steroidogenic gene expression [39], caution must be used in interpreting HSD3B1 and CYP19A1 mRNA data. Like LHCGR, CYP19A1 is induced through FSH signaling, and CYP19A1 is higher in dominant follicles of monovulatory species [40–42].
Huang and colleagues found that higher AREG levels in CCs and MGCs were more likely to be associated with pregnancy [43]. This differs from our study as we saw lower AREG levels in CCs from oocytes associated with live birth. As above, this discordant finding may be the result of pooling CCs and MGCs and/or the use four stimulation protocols in the Huang study. Our results are consistent with Feuerstein and colleagues who showed reduced AREG mRNA levels for CCs of oocytes that resulted in high-quality blastocysts [44].
Two strengths of our study were that all patients received the same stimulation regimen, albeit with two differing trigger medications, and we excluded patients with endometriosis or DOR. Differing stimulation protocols have been reported to alter mRNA expression and bias results [45–47]. In previous studies, CCs and MGCs of patients with DOR [27, 28] or endometriosis [25, 26] exhibited altered mRNA expression relative to their control groups. For these reasons, we excluded patients with these conditions.
As with any patient-driven clinical study, our study had limitations. Patient heterogeneity [45, 48], patient age [48, 49], and differing trigger medications have been previously demonstrated to alter gene expression [39]. A study design including younger patients with only male factor infertility using the same trigger medication would be a more ideal group to examine. Additionally, sample size may have influenced the findings. While we were able to analyze 163 samples, the samples were collected from 15 patients. Additionally, we were not able to analyze all genes for each of the samples as RNA amounts were a limiting factor with individual cumulus masses. With more patients, other mRNA expression level differences may have been apparent, and a larger prospective study is necessary to confirm the results we found and determine the predictive value of the CC mRNA levels for oocyte endpoints such as euploidy and live birth outcomes. Moreover, an argument can be made that looking at oocyte CC biomarkers and comparing the ploidy status of embryos disregards the potential that aneuploidy may have resulted from the paternal contribution to the embryos. We feel our study is still a valid assessment as aneuploidy is predominantly due to maternal origins [50]. Finally, we cannot overlook the impact of uterine receptivity. Although we have no evidence of patient uterine receptivity issues, it is possible some non-implanting embryos were capable of leading to a live birth but were not able to implant due to a suboptimal uterine environment. A way to account for this would be to perform double embryo transfers with euploid embryos and assess CC mRNA expression for those where one implants and one does not [16].
In conclusion, taken together the ideal oocyte developmental competence needed to produce live birth may be reflected by the interplay between AREG, HSD3B1, LHCGR, and PAPPA mRNA levels and likely their resulting proteins. The current data support that CC mRNA levels of these four biomarkers may be useful in assisted reproduction. A larger prospective study will be needed in to order to corroborate the results and to determine numerical ranges for levels of these biomarkers that could potentially have clinical value in selecting oocytes to fertilize or embryos for uterine transfer that will have the highest odds of yielding live birth.
Electronic supplementary material
Biomarkers for CC mRNA expression not associated with mature oocyte competence and embryo outcomes. To determine the differences in CC mRNA between groups where oocytes had different developmental and embryo outcomes target mRNA levels were compared using repeated measures ANOVA followed by Tukey’s post hoc test (adjusted P-values) for pairwise comparisons. Groups represent CC mRNA from oocytes with the following descriptions: Aneuploid = mature oocytes resulting in aneuploid embryos; Arrested = mature oocytes resulting in embryos that did not reach the blastocyst stage; Failed Fert = mature oocytes that did not fertilize; Immature = immature oocytes that were not fertilized; Live Birth = oocytes that resulted in transferred euploid embryos that resulted in live births; No Pregnancy = oocytes that resulted in transferred euploid embryos that did not result in a pregnancy. No differences were seen in these biomarkers (P > 0.05). Data are presented as stated in fig. 3 legend (PNG 732 kb)
GLMM model distinguishing CC from oocytes giving rise to live births and oocytes with outcomes not resulting in live birth, collectively. To evaluate the potential of CC biomarker mRNA levels to identify oocytes capable of producing euploid embryos resulting in a live birth versus groups not producing live births, the best fitting model included 110 CC samples from 14 patients and included biomarkers: CYP11A1, CYP19A1, IGFBP5, PAPPA, PGRMC1, and STARD1. Increased PAPPA mRNA expression (P < 0.05) significantly increased the odds of an oocytes producing an embryo resulting in a live birth (OR = 4.591, 95% CI: 1.098, to 19.201). All previously stated categories were included in this model except oocytes yielding euploid embryos that were not transferred. Groups represent CC mRNA from oocytes with the following descriptions: Live Birth = mature oocytes that resulted in euploid embryos that lead to live births; Non-Viable = immature oocytes, mature oocytes resulting in aneuploid blastocysts, oocytes producing embryos that did not reach the blastocyst stage, and mature oocytes that failed to fertilize, collectively. Data are presented as in fig. 3 legend (PNG 262 kb)
(DOCX 32 kb)
Funding
This study was supported by an ASPIRE-I grant from the University of South Carolina. MCC was supported by the University of South Carolina School of Medicine Research Program for Medical Students.
Compliance with ethical standards
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and in its later amendments or comparable ethical standards. This study was approved by the University of South Carolina Institution Review Board (IRB registration number: 00000240).
Informed consent
Informed consent was obtained from all individual participants included in the study.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Uyar A, Torrealday S, Seli E. Cumulus and granulosa cell markers of oocyte and embryo quality. Fertil Steril. 2013;99(4):979–997. doi: 10.1016/j.fertnstert.2013.01.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen M, Wei S, Hu J, Quan S. Can comprehensive chromosome screening technology improve IVF/ICSI outcomes? A meta-analysis. PLoS One. 2015;10(10):e0140779. doi: 10.1371/journal.pone.0140779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott RT, Jr, Upham KM, Forman EJ, Hong KH, Scott KL, Taylor D, et al. Blastocyst biopsy with comprehensive chromosome screening and fresh embryo transfer significantly increases in vitro fertilization implantation and delivery rates: a randomized controlled trial. Fertil Steril. 2013;100(3):697–703. doi: 10.1016/j.fertnstert.2013.04.035. [DOI] [PubMed] [Google Scholar]
- 4.Yang Z, Liu J, Collins GS, Salem SA, Liu X, Lyle SS, Peck AC, Sills E, Salem RD. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: results from a randomized pilot study. Mol Cytogenet. 2012;5(1):24. doi: 10.1186/1755-8166-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang J, Boulet SL, Jeng G, Flowers L, Kissin DM. Outcomes of in vitro fertilization with preimplantation genetic diagnosis: an analysis of the United States Assisted Reproductive Technology Surveillance Data, 2011–2012. Fertil Steril. 2016;105(2):394–400. doi: 10.1016/j.fertnstert.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kushnir VA, Darmon SK, Albertini DF, Barad DH, Gleicher N. Effectiveness of in vitro fertilization with preimplantation genetic screening: a reanalysis of United States assisted reproductive technology data 2011-2012. Fertil Steril. 2016;106(1):75–79. doi: 10.1016/j.fertnstert.2016.02.026. [DOI] [PubMed] [Google Scholar]
- 7.Zhang S, Luo K, Cheng D, Tan Y, Lu C, He H, Gu Y, Lu G, Gong F, Lin G. Number of biopsied trophectoderm cells is likely to affect the implantation potential of blastocysts with poor trophectoderm quality. Fertil Steril. 2016;105(5):1222–1227. doi: 10.1016/j.fertnstert.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 8.Nel-Themaat L, Nagy ZP. A review of the promises and pitfalls of oocyte and embryo metabolomics. Placenta. 2011;32(Suppl 3):S257–SS63. doi: 10.1016/j.placenta.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Uyar A, Seli E. Metabolomic assessment of embryo viability. Semin Reprod Med. 2014;32(2):141–152. doi: 10.1055/s-0033-1363556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kordus RJ, LaVoie HA. Granulosa cell biomarkers to predict pregnancy in ART: pieces to solve the puzzle. Reproduction. 2017;153(2):R69–R83. doi: 10.1530/REP-16-0500. [DOI] [PubMed] [Google Scholar]
- 11.Tanghe S, Van SA, Nauwynck H, Coryn M, de KA. Minireview: functions of the cumulus oophorus during oocyte maturation, ovulation, and fertilization. Mol Reprod Dev. 2002;61(3):414–424. doi: 10.1002/mrd.10102. [DOI] [PubMed] [Google Scholar]
- 12.Ekart J, McNatty K, Hutton J, Pitman J. Ranking and selection of MII oocytes in human ICSI cycles using gene expression levels from associated cumulus cells. Hum Reprod. 2013;28(11):2930–2942. doi: 10.1093/humrep/det357. [DOI] [PubMed] [Google Scholar]
- 13.Iager AE, Kocabas AM, Otu HH, Ruppel P, Langerveld A, Schnarr P, Suarez M, Jarrett JC, Conaghan J, Rosa GJM, Fernández E, Rawlins RG, Cibelli JB, Crosby JA. Identification of a novel gene set in human cumulus cells predictive of an oocyte’s pregnancy potential. Fertil Steril. 2013;99(3):745–752. doi: 10.1016/j.fertnstert.2012.10.041. [DOI] [PubMed] [Google Scholar]
- 14.Wathlet S, Adriaenssens T, Segers I, Verheyen G, Van d V, Coucke W, et al. Cumulus cell gene expression predicts better cleavage-stage embryo or blastocyst development and pregnancy for ICSI patients. Hum Reprod. 2011;26(5):1035–1051. doi: 10.1093/humrep/der036. [DOI] [PubMed] [Google Scholar]
- 15.Fragouli E, Wells D, Iager AE, Kayisli UA, Patrizio P. Alteration of gene expression in human cumulus cells as a potential indicator of oocyte aneuploidy. Hum Reprod. 2012;27(8):2559–2568. doi: 10.1093/humrep/des170. [DOI] [PubMed] [Google Scholar]
- 16.Green KA, Franasiak JM, Werner MD, Tao X, Landis JN, Scott RT, Jr, Treff NR. Cumulus cell transcriptome profiling is not predictive of live birth after in vitro fertilization: a paired analysis of euploid sibling blastocysts. Fertil Steril. 2018;109(3):460–466. doi: 10.1016/j.fertnstert.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Parks JC, Patton AL, McCallie BR, Griffin DK, Schoolcraft WB, Katz-Jaffe MG. Corona cell RNA sequencing from individual oocytes revealed transcripts and pathways linked to euploid oocyte competence and live birth. Reprod BioMed Online. 2016;32(5):518–526. doi: 10.1016/j.rbmo.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Assidi M, Dufort I, Ali A, Hamel M, Algriany O, Dielemann S, Sirard MA. Identification of potential markers of oocyte competence expressed in bovine cumulus cells matured with follicle-stimulating hormone and/or phorbol myristate acetate in vitro. Biol Reprod. 2008;79(2):209–222. doi: 10.1095/biolreprod.108.067686. [DOI] [PubMed] [Google Scholar]
- 19.Blaha M, Nemcova L, Kepkova KV, Vodicka P, Prochazka R. Gene expression analysis of pig cumulus-oocyte complexes stimulated in vitro with follicle stimulating hormone or epidermal growth factor-like peptides. Reprod Biol Endocrinol. 2015;13:113. doi: 10.1186/s12958-015-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christenson LK, Gunewardena S, Hong X, Spitschak M, Baufeld A, Vanselow J. Research resource: preovulatory LH surge effects on follicular theca and granulosa transcriptomes. Mol Endocrinol. 2013;27(7):1153–1171. doi: 10.1210/me.2013-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glister C, Satchell L, Knight PG. Changes in expression of bone morphogenetic proteins (BMPs), their receptors and inhibin co-receptor betaglycan during bovine antral follicle development: inhibin can antagonize the suppressive effect of BMPs on thecal androgen production. Reproduction. 2010;140(5):699–712. doi: 10.1530/REP-10-0216. [DOI] [PubMed] [Google Scholar]
- 22.Nivet AL, Vigneault C, Blondin P, Sirard MA. Changes in granulosa cells’ gene expression associated with increased oocyte competence in bovine. Reproduction. 2013;145(6):555–565. doi: 10.1530/REP-13-0032. [DOI] [PubMed] [Google Scholar]
- 23.Saini N, Singh MK, Shah SM, Singh KP, Kaushik R, Manik RS, Singla SK, Palta P, Chauhan MS. Developmental competence of different quality bovine oocytes retrieved through ovum pick-up following in vitro maturation and fertilization. Animal. 2015;9(12):1979–1985. doi: 10.1017/S1751731115001226. [DOI] [PubMed] [Google Scholar]
- 24.Vigone G, Merico V, Prigione A, Mulas F, Sacchi L, Gabetta M, Bellazzi R, Redi C, Mazzini G, Adjaye J, Garagna S, Zuccotti M. Transcriptome based identification of mouse cumulus cell markers that predict the developmental competence of their enclosed antral oocytes. BMC Genomics. 2013;14:380. doi: 10.1186/1471-2164-14-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allegra A, Raimondo S, Volpes A, Fanale D, Marino A, Cicero G, de Leo G, Sammartano F, Allegra G, Alessandro R. The gene expression profile of cumulus cells reveals altered pathways in patients with endometriosis. J Assist Reprod Genet. 2014;31(10):1277–1285. doi: 10.1007/s10815-014-0305-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barcelos ID, Donabella FC, Ribas CP, Meola J, Ferriani RA, de Paz CC, et al. Down-regulation of the CYP19A1 gene in cumulus cells of infertile women with endometriosis. Reprod BioMed Online. 2015;30(5):532–541. doi: 10.1016/j.rbmo.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Greenseid K, Jindal S, Hurwitz J, Santoro N, Pal L. Differential granulosa cell gene expression in young women with diminished ovarian reserve. Reprod Sci. 2011;18(9):892–899. doi: 10.1177/1933719111398502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.May-Panloup P, Ferre-L'Hotellier V, Moriniere C, Marcaillou C, Lemerle S, Malinge MC, et al. Molecular characterization of corona radiata cells from patients with diminished ovarian reserve using microarray and microfluidic-based gene expression profiling. Hum Reprod. 2012;27(3):829–843. doi: 10.1093/humrep/der431. [DOI] [PubMed] [Google Scholar]
- 29.Gardner DK, Lane M, Stevens J, Schlenker T, Schoolcraft WB. Blastocyst score affects implantation and pregnancy outcome: towards a single blastocyst transfer. Fertil Steril. 2000;73(6):1155–1158. doi: 10.1016/S0015-0282(00)00518-5. [DOI] [PubMed] [Google Scholar]
- 30.Nelson-Degrave VL, Wickenheisser JK, Hendricks KL, Asano T, Fujishiro M, Legro RS, Kimball SR, Strauss JF, III, McAllister JM. Alterations in mitogen-activated protein kinase kinase and extracellular regulated kinase signaling in theca cells contribute to excessive androgen production in polycystic ovary syndrome. Mol Endocrinol. 2005;19(2):379–390. doi: 10.1210/me.2004-0178. [DOI] [PubMed] [Google Scholar]
- 31.Bates D, Machler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67(1):1–48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 32.Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models. Biom J. 2008;50(3):346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 33.Laursen LS, Overgaard MT, Soe R, Boldt HB, Sottrup-Jensen L, Giudice LC, et al. Pregnancy-associated plasma protein-A (PAPP-A) cleaves insulin-like growth factor binding protein (IGFBP)-5 independent of IGF: implications for the mechanism of IGFBP-4 proteolysis by PAPP-A. FEBS Lett. 2001;504(1–2):36–40. doi: 10.1016/S0014-5793(01)02760-0. [DOI] [PubMed] [Google Scholar]
- 34.Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev. 2002;23(6):824–854. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- 35.Wang HS, Chard T. IGFs and IGF-binding proteins in the regulation of human ovarian and endometrial function. J Endocrinol. 1999;161(1):1–13. doi: 10.1677/joe.0.1610001. [DOI] [PubMed] [Google Scholar]
- 36.Maman E, Yung Y, Kedem A, Yerushalmi GM, Konopnicki S, Cohen B, Dor J, Hourvitz A. High expression of luteinizing hormone receptors messenger RNA by human cumulus granulosa cells is in correlation with decreased fertilization. Fertil Steril. 2012;97(3):592–598. doi: 10.1016/j.fertnstert.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 37.Papamentzelopoulou M, Mavrogianni D, Partsinevelos GA, Marinopoulos S, Dinopoulou V, Theofanakis C, Anagnostou E, Loutradis D. LH receptor gene expression in cumulus cells in women entering an ART program. J Assist Reprod Genet. 2012;29(5):409–416. doi: 10.1007/s10815-012-9729-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamel M, Dufort I, Robert C, Gravel C, Leveille MC, Leader A, Sirard MA. Identification of differentially expressed markers in human follicular cells associated with competent oocytes. Hum Reprod. 2008;23(5):1118–1127. doi: 10.1093/humrep/den048. [DOI] [PubMed] [Google Scholar]
- 39.Borgbo T, Povlsen BB, Andersen CY, Borup R, Humaidan P, Grondahl ML. Comparison of gene expression profiles in granulosa and cumulus cells after ovulation induction with either human chorionic gonadotropin or a gonadotropin-releasing hormone agonist trigger. Fertil Steril. 2013;100(4):994–1001. doi: 10.1016/j.fertnstert.2013.05.038. [DOI] [PubMed] [Google Scholar]
- 40.Hayashi KG, Ushizawa K, Hosoe M, Takahashi T. Differential genome-wide gene expression profiling of bovine largest and second-largest follicles: identification of genes associated with growth of dominant follicles. Reprod Biol Endocrinol. 2010;8:11. doi: 10.1186/1477-7827-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kristensen SG, Mamsen LS, Jeppesen JV, Botkjaer JA, Pors SE, Borgbo T, et al. Hallmarks of human small antral follicle development: implications for regulation of ovarian steroidogenesis and selection of the dominant follicle. Front Endocrinol. 2017;8:376. doi: 10.3389/fendo.2017.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sisco B, Hagemann LJ, Shelling AN, Pfeffer PL. Isolation of genes differentially expressed in dominant and subordinate bovine follicles. Endocrinology. 2003;144(9):3904–3913. doi: 10.1210/en.2003-0485. [DOI] [PubMed] [Google Scholar]
- 43.Huang Y, Zhao Y, Yu Y, Li R, Lin S, Zhang C, Liu P, Qiao J. Altered amphiregulin expression induced by diverse luteinizing hormone receptor reactivity in granulosa cells affects IVF outcomes. Reprod BioMed Online. 2015;30(6):593–601. doi: 10.1016/j.rbmo.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 44.Feuerstein P, Cadoret V, Dalbies-Tran R, Guerif F, Bidault R, Royere D. Gene expression in human cumulus cells: one approach to oocyte competence. Hum Reprod. 2007;22(12):3069–3077. doi: 10.1093/humrep/dem336. [DOI] [PubMed] [Google Scholar]
- 45.Adriaenssens T, Wathlet S, Segers I, Verheyen G, De VA, Van der Elst J, et al. Cumulus cell gene expression is associated with oocyte developmental quality and influenced by patient and treatment characteristics. Hum Reprod. 2010;25(5):1259–1270. doi: 10.1093/humrep/deq049. [DOI] [PubMed] [Google Scholar]
- 46.Assou S, Haouzi D, Dechaud H, Gala A, Ferrieres A, Hamamah S. Comparative gene expression profiling in human cumulus cells according to ovarian gonadotropin treatments. Biomed Res Int. 2013;2013:1–13. doi: 10.1155/2013/354582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grondahl ML, Borup R, Lee YB, Myrhoj V, Meinertz H, Sorensen S. Differences in gene expression of granulosa cells from women undergoing controlled ovarian hyperstimulation with either recombinant follicle-stimulating hormone or highly purified human menopausal gonadotropin. Fertil Steril. 2009;91(5):1820–1830. doi: 10.1016/j.fertnstert.2008.02.137. [DOI] [PubMed] [Google Scholar]
- 48.Hurwitz JM, Jindal S, Greenseid K, Berger D, Brooks A, Santoro N, Pal L. Reproductive aging is associated with altered gene expression in human luteinized granulosa cells. Reprod Sci. 2010;17(1):56–67. doi: 10.1177/1933719109348028. [DOI] [PubMed] [Google Scholar]
- 49.Al-Edani T, Assou S, Ferrieres A, Bringer DS, Gala A, Lecellier CH, et al. Female aging alters expression of human cumulus cells genes that are essential for oocyte quality. Biomed Res Int. 2014;2014:1–10. doi: 10.1155/2014/964614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rabinowitz M, Ryan A, Gemelos G, Hill M, Baner J, Cinnioglu C, Banjevic M, Potter D, Petrov DA, Demko Z. Origins and rates of aneuploidy in human blastomeres. Fertil Steril. 2012;97(2):395–401. doi: 10.1016/j.fertnstert.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 51.Assidi M, Montag M, Van d V, Sirard MA. Biomarkers of human oocyte developmental competence expressed in cumulus cells before ICSI: a preliminary study. J Assist Reprod Genet. 2011;28(2):173–188. doi: 10.1007/s10815-010-9491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsutsumi R, Hiroi H, Momoeda M, Hosokawa Y, Nakazawa F, Koizumi M, et al. Inhibitory effects of cholesterol sulfate on progesterone production in human granulosa-like tumor cell line, KGN. Endocr J. 2008;55(3):575–581. doi: 10.1507/endocrj.K07-097. [DOI] [PubMed] [Google Scholar]
- 53.Gonzalez-Fernandez R, Pena O, Hernandez J, Martin-Vasallo P, Palumbo A, Avila J. Patients with endometriosis and patients with poor ovarian reserve have abnormal follicle-stimulating hormone receptor signaling pathways. Fertil Steril. 2011;95(7):2373–2378. doi: 10.1016/j.fertnstert.2011.03.030. [DOI] [PubMed] [Google Scholar]
- 54.Wang HX, Tong D, El-Gehani F, Tekpetey FR, Kidder GM. Connexin expression and gap junctional coupling in human cumulus cells: contribution to embryo quality. J Cell Mol Med. 2009;13(5):972–984. doi: 10.1111/j.1582-4934.2008.00373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walker G, MacLeod K, Williams AR, Cameron DA, Smyth JF, Langdon SP. Insulin-like growth factor binding proteins IGFBP3, IGFBP4, and IGFBP5 predict endocrine responsiveness in patients with ovarian cancer. Clin Cancer Res. 2007;13(5):1438–1444. doi: 10.1158/1078-0432.CCR-06-2245. [DOI] [PubMed] [Google Scholar]
- 56.He YY, Huang JL, Sik RH, Liu J, Waalkes MP, Chignell CF. Expression profiling of human keratinocyte response to ultraviolet A: implications in apoptosis. J Invest Dermatol. 2004;122(2):533–543. doi: 10.1046/j.0022-202X.2003.22123.x. [DOI] [PubMed] [Google Scholar]
- 57.Seder CW, Hartojo W, Lin L, Silvers AL, Wang Z, Thomas DG, et al. Upregulated INHBA expression may promote cell proliferation and is associated with poor survival in lung adenocarcinoma. Neoplasia. 2009;11(4):388–396. doi: 10.1593/neo.81582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yung Y, Maman E, Ophir L, Rubinstein N, Barzilay E, Yerushalmi GM, et al. Progesterone antagonist, RU486, represses LHCGR expression and LH/hCG signaling in cultured luteinized human mural granulosa cells. Gynecol Endocrinol. 2014;30(1):42–47. doi: 10.3109/09513590.2013.848426. [DOI] [PubMed] [Google Scholar]
- 59.Wagner PK, Otomo A, Christians JK. Regulation of pregnancy-associated plasma protein A2 (PAPPA2) in a human placental trophoblast cell line (BeWo) Reprod Biol Endocrinol. 2011;9:48. doi: 10.1186/1477-7827-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guzman L, Adriaenssens T, Ortega-Hrepich C, Albuz FK, Mateizel I, Devroey P, et al. Human antral follicles <6 mm: a comparison between in vivo maturation and in vitro maturation in non-hCG primed cycles using cumulus cell gene expression. Mol Hum Reprod. 2013;19(1):7–16. doi: 10.1093/molehr/gas038. [DOI] [PubMed] [Google Scholar]
- 61.Zachariades E, Foster H, Goumenou A, Thomas P, Rand-Weaver M, Karteris E. Expression of membrane and nuclear progesterone receptors in two human placental choriocarcinoma cell lines (JEG-3 and BeWo): Effects of syncytialization. Int J Mol Med. 2011;27(6):767–774. doi: 10.3892/ijmm.2011.657. [DOI] [PubMed] [Google Scholar]
- 62.Chen Q, Sun X, Chen J, Cheng L, Wang J, Wang Y, et al. Direct rosiglitazone action on steroidogenesis and proinflammatory factor production in human granulosa-lutein cells. Reprod Biol Endocrinol. 2009;7:147. doi: 10.1186/1477-7827-7-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nelson-Degrave VL, Wickenheisser JK, Cockrell JE, Wood JR, Legro RS, Strauss JF, III, et al. Valproate potentiates androgen biosynthesis in human ovarian theca cells. Endocrinology. 2004;145(2):799–808. doi: 10.1210/en.2003-0940. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Biomarkers for CC mRNA expression not associated with mature oocyte competence and embryo outcomes. To determine the differences in CC mRNA between groups where oocytes had different developmental and embryo outcomes target mRNA levels were compared using repeated measures ANOVA followed by Tukey’s post hoc test (adjusted P-values) for pairwise comparisons. Groups represent CC mRNA from oocytes with the following descriptions: Aneuploid = mature oocytes resulting in aneuploid embryos; Arrested = mature oocytes resulting in embryos that did not reach the blastocyst stage; Failed Fert = mature oocytes that did not fertilize; Immature = immature oocytes that were not fertilized; Live Birth = oocytes that resulted in transferred euploid embryos that resulted in live births; No Pregnancy = oocytes that resulted in transferred euploid embryos that did not result in a pregnancy. No differences were seen in these biomarkers (P > 0.05). Data are presented as stated in fig. 3 legend (PNG 732 kb)
GLMM model distinguishing CC from oocytes giving rise to live births and oocytes with outcomes not resulting in live birth, collectively. To evaluate the potential of CC biomarker mRNA levels to identify oocytes capable of producing euploid embryos resulting in a live birth versus groups not producing live births, the best fitting model included 110 CC samples from 14 patients and included biomarkers: CYP11A1, CYP19A1, IGFBP5, PAPPA, PGRMC1, and STARD1. Increased PAPPA mRNA expression (P < 0.05) significantly increased the odds of an oocytes producing an embryo resulting in a live birth (OR = 4.591, 95% CI: 1.098, to 19.201). All previously stated categories were included in this model except oocytes yielding euploid embryos that were not transferred. Groups represent CC mRNA from oocytes with the following descriptions: Live Birth = mature oocytes that resulted in euploid embryos that lead to live births; Non-Viable = immature oocytes, mature oocytes resulting in aneuploid blastocysts, oocytes producing embryos that did not reach the blastocyst stage, and mature oocytes that failed to fertilize, collectively. Data are presented as in fig. 3 legend (PNG 262 kb)
(DOCX 32 kb)