Abstract
Purpose
To investigate the impact of chronically elevated androgens in the presence and absence of an obesogenic diet on oocyte quality in the naturally selected primate periovulatory follicle.
Methods
Rhesus macaques were treated using a 2-by-2 factorial design (n = 10/treatment) near the onset of menarche with implants containing either cholesterol (C) or testosterone (T, 4–5-fold increase above C) and a standard or “Western-style” diet alone (WSD) or in combination (T+WSD). Following ~ 3.5 years of treatment, females underwent controlled ovulation (COv, n = 7–10/treatment) cycles, and contents of the naturally selected periovulatory follicle were aspirated. Follicular fluid (FF) was analyzed for cytokines, chemokines, growth factors, and steroids. RNA was extracted from luteinizing granulosa cells (LGCs) and assessed by RNA-seq.
Results
Only healthy, metaphase (M) I/II-stage oocytes (100%) were retrieved in the C group, whereas several degenerated oocytes were recovered in other groups (33–43% of T, WSD, and T+WSD samples). Levels of two chemokines and one growth factor were reduced (p < 0.04) in FF of follicles with a MI/MII oocyte in WSD+T (CCL11) or T and WSD+T groups (CCL2 and FGF2) compared to C and/or WSD. Intrafollicular cortisol was elevated in T compared to C follicles (p < 0.02). Changes in the expression pattern of 640+ gene products were detected in LGC samples from follicles with degenerated versus MI/MII-stage oocytes. Pathway analysis on RNAs altered by T and/or WSD found enrichment of genes mapping to steroidogenic and immune cell pathways.
Conclusions
Female primates experiencing hyperandrogenemia and/or consuming a WSD exhibit an altered intrafollicular microenvironment and reduced oocyte quality/competency, despite displaying menstrual cyclicity.
Electronic supplementary material
The online version of this article (10.1007/s10815-019-01497-8) contains supplementary material, which is available to authorized users.
Keywords: Androgen, Follicular cytokines, Nonhuman primate, Oocyte quality, Periovulatory follicle, Western-style diet
Background
Polycystic ovary syndrome (PCOS) is a multifaceted disorder in ~ 5–10% of reproductive-age women associated with elevated androgens and infertility/subfertility due to oligo- or anovulation and altered folliculogenesis. While this is a multifaceted disorder, a high proportion of PCOS women are at risk for metabolic dysfunction or are obese compared to non-affected women [1, 2]. Further studies revealed up to four distinct phenotypes of PCOS, with women experiencing both obesity and elevated androgens having worse metabolic function compared to those with hyperandrogenemia or obesity alone [3].
Since ovaries containing large numbers of atretic, small antral follicles are a defining characteristic of PCOS, researchers have focused on the effects of elevated androgen on follicular function. Women with PCOS experience higher rates of pregnancy loss compared to non-PCOS women in populations seeking infertility therapy [4], which may be related to poor oocyte quality due to impaired folliculogenesis [5, 6]. Studies on nonprimate [7] and primate species [8, 9] identified stage-specific effects of excess androgen on follicular development and function. In vitro studies employing three-dimensional culture of macaque follicles suggest that androgen actions are critical for pre-antral folliculogenesis in primates, but once follicles reach the antral stage, continued exposure to elevated androgens results in smaller antral follicles producing less local factors (VEGF, AMH) and steroid (estradiol, E2) compared to controls [9]. However, some of these androgen-exposed, less-differentiated antral follicles retained the ability to produce a mature metaphase II (MII) oocyte upon exposure to human chorionic gonadotropin (hCG), but preimplantation embryogenesis was not evaluated in these studies.
Murine studies also indicated that diet-induced obesity negatively affects oocyte quality [10]. Similarly, several studies suggest that oocyte quality and fertilization rates are lower after ovarian stimulation in women with high BMIs (as reviewed in references [4, 11]). Obese patients seeking infertility therapies may also require higher doses of gonadotropins to stimulate follicular development, but these women also tended to produce less E2 during ovarian stimulation and had fewer oocytes retrieved [11]. Nevertheless, a PCOS diagnosis is associated with fewer days of gonadotropin stimulation, regardless of BMI [11], and many PCOS women recruit more follicles during stimulation compared to BMI-matched non-PCOS women [12]. Despite these findings, rates of fertilization are reduced in PCOS women compared to BMI-matched controls, suggesting their oocytes are of lower quality [12]. While there are very few studies of women during spontaneous menstrual cycles, one study of women undergoing intrauterine insemination found those with increased BMI and increased waist-to-hip ratio (a measurement of android fat, possibly linked to increased androgen levels) required a greater number of menstrual cycles to achieve pregnancy [13]. However, it is unclear whether oocyte or uterine factors, or both, contributed to subfertility in this cohort. Furthermore, many studies on the influence of BMI in the absence and presence of PCOS on follicle/oocyte quality in women are retrospective, and several use different criteria for PCOS diagnosis and/or BMI cutoff (as reviewed in reference [14]). Therefore, further studies that evaluate the effects of androgen, in the presence and absence of a high-fat diet, on oocyte quality and the microenvironment in the maturing follicle are needed, particularly in primates.
Recently, our research group developed a nonhuman primate model to determine if hyperandrogenemia beginning at puberty, in the absence and presence of an obesogenic diet, results in the onset of PCOS symptoms and metabolic/reproductive dysfunction [15]. Rhesus macaque females were randomly selected near menarche for exposure to mildly elevated testosterone (T) levels (4–5-fold) via silastic implant, and/or consumption of a Western-style diet (WSD). This cohort also included age-matched, non-treated female monkeys, resulting in experimental groups of controls (C) versus chronic exposure to T alone, WSD alone, or T plus WSD. We recently reported that those young adult macaque females treated with T+WSD for 3 years gain more weight and fat depots, with greater impairment of insulin sensitivity compared to the other groups [15]. While the majority of these macaque females had menstrual cycles, there was evidence of ovarian dysfunction including high incidence of PCO morphology, reduced E2 and elevated LH levels in the early follicular phase, plus lower progesterone (P4) levels and ovarian blood flow during the luteal phase with T and/or T+WSD treatment compared to controls [16]. Therefore, it is likely that many of these rhesus females recapitulate the ovulatory PCOS phenotype in women. However, the effects of T, WSD, and T+WSD on the maturing periovulatory follicle and oocyte quality had not yet been evaluated. Thus, studies were designed to use this primate cohort to investigate the effects of chronic T exposure, consumption of a WSD, or T+WSD on the function of the naturally selected ovulatory follicle and its oocyte.
Methods
Young, adult reproductive-aged (5.5–6 years of age, equivalent to ~ 18 years old human) rhesus females (Macaca mulatta) were utilized in this study. Details of ongoing androgen (T) and WSD treatments of these macaque females were reported previously [15]. Briefly, silastic implants containing either cholesterol (control) or T (serum T level ~ 1.4 ng/mL) were placed subcutaneously near menarche (~ 2.5 years old, n = 20/group). These females were also randomized to consume either a standard chow diet, or a WSD, resulting in a 2-by-2 factorial design of treatments. Groups were denoted controls (C), T, WSD, and T+WSD (n = 10/group). Animal care was provided by the Division of Comparative Medicine (DCM) at the Oregon National Primate Research Center (ONPRC) and all procedures were reviewed and approved by the ONPRC/Oregon Health and Science University (OHSU) Institutional Animal Care and Use Committee (IACUC) in accordance with the U.S. Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals. All surgical procedures were performed by trained laboratory animal veterinarians and veterinary technicians of the ONPRC DCM Surgical Services Unit.
Controlled ovulation cycles
At 5.5–6.0 years of age (3.0–3.5 years on treatment), a controlled ovulation protocol (COv, n = 7–10/group; Table 1) was performed similar to previous studies in rhesus monkeys [17]. Serum E2 levels were monitored daily beginning 4–6 days after onset of menses. Once E2 levels rose above 100 pg/mL, indicating the selection of a dominant antral follicle, females received a sc injection of a gonadotropin-releasing hormone antagonist, Antide (1–3 mg/kg; Salk Institute for Biological Studies, La Jolla, CA, USA), along with an im injection of follicle stimulating hormone and luteinizing hormone (FSH:LH, 30 IU each; Menopur, Ferring Pharmaceuticals Inc. Parsippany, NJ, USA) at 1600 h. The next morning at 0800 h, females received Antide (0.5 mg/kg) and FSH:LH (30 IU each), followed by a second FSH:LH injection (30 IU each) that afternoon at 1600. Human chorionic gonadotropin (hCG, 1000 IU; Ovidrel, EMD Serono, Inc. Rockland, MA, USA) was injected im the following morning. Serum P4 levels were monitored daily from the onset of Antide treatment through the day of follicle aspiration. The contents of the single periovulatory follicle were aspirated 26–30 h post-hCG under anesthesia 16: [16] during laparoscopy using a hand syringe. Unsuccessful oocyte retrievals due to failure to block the LH surge/follicle rupture and ovulation unexpectedly occurred in 50% (5/10) of C, 14% (1/7) of T, 25% (2/8) of WSD, and 13% (1/8) of T+WSD cycles (see “Discussion”).
Table 1.
Results of COv cycles to aspirate COC’s from rhesus females by treatment group
| C | T | WSD | T+WSD | |
|---|---|---|---|---|
| # Cycles | 10 | 7 | 8 | 8 |
| # Oocytes | 5 | 6 | 6 | 7 |
| Degenerating (%) | 0 (0%) | 2 (33%) | 2 (33%) | 3 (43%) |
| MI/MII (%) | 5 (100%) | 3 (50%) | 4 (67%) | 4 (57%) |
| GV | 0 | 1 | 0 | 0 |
| MI | 3 | 1 | 3 | 2 |
| MII | 2 | 2 | 1 | 2 |
Oocyte quality assessment
The aspirate from each individual follicle was centrifuged at 67×g for 1 min at ambient temperature to separate the fluid and cellular components. The follicular fluid was removed and stored at − 80 °C until assayed, while the pelleted cellular contents (granulosa cells and oocyte) were dispersed by repeated pipetting in 500 μL TALP-HEPES to isolate and recover the oocyte. After transfer to a 30-mm petri dish, cumulus cells (when present) were removed from the oocyte by gentle pipetting through a fine-bore pipette (135-μm internal diameter) and each ovum was evaluated for stage of meiosis as follows: germinal vesicle (GV) = meiotic arrest and immature oocyte; metaphase I (MI) = meiosis resumed and oocyte maturing; and metaphase II (MII) = meiosis complete; oocyte is mature with an extruded first polar body visible in the peri-vitelline space. Recovered MII ova were subjected to intracytoplasmic sperm injection (ICSI) with fresh sperm from a rhesus male of proven fertility [18] and cultured in 500 μL Global + 10% protein supplement (Life Global Group, Guilford, CT, USA) covered with oil in a humidified proportional mixture of CO2/O2/N2 gas (6/5/89%, respectively); medium was replenished every 2 days. Any oocytes retrieved at the MI stage that did not complete cytoplasmic maturation within a few hours following aspiration were in vitro fertilized by conventional method overnight as previously described by Hanna et al. [19] and cultured as noted above. Fertilization procedures were not performed on GV oocytes recovered or those that were grossly degenerated. Fertilization was noted by the presence of two polar bodies, the formation of two pronuclei, and/or the first mitotic cleavage of a presumed zygote. Fertilized oocytes were cultured under dark field time-lapse imaging until undergoing embryonic arrest or blastocyst formation as previously reported by Daughtry et al. [20].
Follicular fluid cytokine/chemokine/growth factor protein analysis
Intrafollicular levels of chemokine/cytokine/growth factors were assayed using a multiplex immunoassay platform (cytokine/chemokine/growth factor 37-Plex NHP ProcartaPlex™ Panel, Life Technologies Co, ThermoFisher Scientific, Waltham, MA USA). All samples were processed in the same assay, and for each protein intra-assay, coefficients of variation (CV) are provided as Supplemental Table 1A.
FF liquid chromatography-tandem mass spectrometry analysis of selected steroids
Not all follicles yielded a large enough volume of FF for extraction after cytokine analyses (10 μL, n = 22 follicles). Standard, quality control (QC), or FF samples (350 μL) were placed into microtiter plates, along with 200 μL of ultrapure water containing 7.5 ng/mL cortisol (CSL)-d4, 3.75 ng/mL, E2-d5, 2.5 ng/mL cortisone (CSN)-d8, 1.5 ng/mL estrone (E1)-13C3, 0.8 ng/mL each P4-d9, and androstenedione (A4)-13C3 as internal standards. After mixing, the contents of each well were transferred to a Stata DE SLE plate (Phenomenex, Torrance, CA) and incubated for 5 min at room temperature. Analytes were eluted into an advantage 2 mL polypropylene 96-well deep-well plate (Analytical Sales and Services, Flanders, NJ) with 3 × 600 μL dichloromethane (DCM; Millipore Sigma, Burlington, MA) and dried under forced nitrogen at 40 °C in a TurboVap 96 automated evaporation system (Biotage, Uppsala, Sweden). After reconstitution in 50 μL of 25:75 methanol:water (v:v), the contents of each well were transferred to a V-bottom 96-well microtiter plate (Shimadzu, Kyoto, Japan) and analyzed by LC-MS/MS.
Calibration curves were created using a mixture of unlabeled standards for CSL, E2, CSN, E1, P4, and A4 prepared in charcoal-stripped human serum. Serial dilutions of this mixture were made in charcoal-stripped human serum to yield the final nine-point calibration curve, which ranged from 6.5 pg/mL to 26.67 ng/mL. Serum QC samples were prepared by either (a) spiking a pool of macaque serum with a mixture of unlabeled hormone standards or (b) spiking a mixture of unlabeled hormone standards into the charcoal-stripped human serum. These QCs were prepared and assayed in triplicate at the beginning and end of each assay. A double blank containing no isotope-labeled standards or unlabeled standards in charcoal-stripped human serum and ultrapure water was also prepared and analyzed in duplicate.
Microtiter plates were prepared as described above and loaded onto a SIL-30ACMP autosampler (Shimadzu) set at 10 °C. Twenty-five microliters of each standard, QC or sample, was injected onto a Raptor 2.7 μm Biphenyl 100 mm × 2.1 mm column (Restek, Bellefonte, PA) with an in-line guard column (2.7 μm Biphenyl 5 mm × 2.1 mm, Restek) at 35 °C using reversed phase chromatography. Mobile phase A was 0.15 mM ammonium fluoride in water; mobile phase B was 100% methanol. The LC time gradient was created using two Nexera LC-30AD pumps (Shimadzu) as follows: 0.00–4.00 min, 65–97% B; 4.01–5.10 min, 97–100% B; 5.11–6.90 min, 100% B; 6.91–7.00 min, return to 65% B and hold until 10.40 min for re-equilibration. The entire gradient was run at a flow rate of 0.4 mL/min. Heated electrospray injection in both positive (CSL, CSN, P4, A4) and negative (E1, E2) modes with ultra-fast polarity switching and scheduled multiple reaction monitoring (MRM) on a Shimadzu LCMS-8050 was used for detection of steroids. The MS/MS conditions for each target were optimized using the automated MRM optimization procedure in LabSolutions software (Shimadzu). The MS/MS conditions for each target were optimized using the automated MRM optimization procedure in LabSolutions software (Shimadzu) and can be found in Supplemental Table 1B.
LC-MS/MS trace data were quantified using LabSolutions version 5.72 (Shimadzu). Internal standardized linear calibration curves using 1/C weighting were used for each hormone. Target reference ion ratios for the qualifying ions were set based on the area ratio observed in the highest standard between the quantifying ion and the qualifying ions.
Intra-and inter-assay precisions were less than 10% and less than 12% for all assays within the method, respectively. Accuracies for assays within the method ranged from 86%–101% and extraction efficiencies ranged from 60%–90%. Matrix effects ranged from 50%–200%, and were compensated by using appropriate internal standards for each target. Matrix effects did not vary by different concentrations of analyte and were stable across replicates in each analysis.
Oocyte, fertilization data, and FF statistics
Due to low numbers of mature oocytes/group available, statistics were not performed on fertilization rate or other measures of embryogenesis. Oocyte morphological data (MI/MII or degenerated) were analyzed by a generalized linear model, while intrafollicular steroid hormones and cytokine/chemokine/growth factor protein levels were analyzed by a linear model of SAS (SAS v4.6, SAS Institute Inc. Cary, NC, USA). Serum E2 and P4 levels during COv protocols were analyzed by mixed model/repeated measures function of SAS. For all models, when overall effects of WSD, T, time, or interactions (T by WSD, etc.) were statistically significant (p < 0.05), pairwise comparisons between relevant treatments/time points were performed by least squared means function of SAS with Tukey-Kramer adjustment for multiple comparisons.
Mural granulosa cell RNA-seq analysis and statistics
Luteinizing granulosa cells (LGCs) of individual follicular aspirates were recovered from 30-mm plates and pelleted by centrifugation at 300×g for 5 min at 4 °C. Total RNA was then isolated from LGCs by Relia-Prep RNA cell MiniPrep system (Promega, Madison, WI, USA). Purified RNA was stored at − 80 °C until RNA quality was analyzed by NanoChip. The RNA of samples which passed quality control (QC) assessment with RIN scores > 5 n = 4 C, n = 4 T, n = 5 WSD, and n = 6 T+WSD were sequenced by a HiSeq 2500 using 8-lane high-throughput flow cells (provided by OHSU Massively Parallel Sequencing Shared Resource). All sequence data are archived at NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/gds/; GEO Dataset Series ID: GSE128569). The quality of the raw sequencing files was evaluated using FastQC [21] combined with MultiQC [22] (http://multiqc.info/). The files were then imported into ONPRC’s DISCVR-Seq [23], LabKey server-based system, PRIMe-Seq [24]. Trimmomatic was used to remove any remaining Illumina adapters [25]. Reads were aligned to the Mmul_8.0.1 genome in Ensembl along with its corresponding annotation, release 87. The program STAR (v020201; two-pass mode) was used to align the reads to the genome [26]. STAR has been shown to perform well compared to other RNA-seq aligners [27]. Since STAR utilizes the gene annotation file, it calculated the number of reads aligned to each gene. The total unstranded counts were selected. RNA-SeQC (v1.1.8.1) was then utilized to ensure alignments were of sufficient quality [28].
RNA-seq data yielded summarized raw counts of 32,386 RNAs from 19 samples. RNA-seq statistical analyses were performed in open-source software R [29]. Gene-level raw counts were filtered to remove genes with extremely low counts (sequences) across all samples, normalized using the trimmed mean of M values method (TMM) [30], and transformed to log counts per million with associated sample-wise quality weight and observational precision weights using the voom method [31, 32]. After filtering, 15,349 RNAs were retained. Gene-wise linear models accommodating 2 × 2 factorial design between WSD and Androgen (T) exposure were employed for differential expression analyses using limma with empirical Bayes moderation [33]. Contrasts of interest interrogated were T vs C, C vs WSD, C vs T+WSD, T vs T+WSD, and T+WSD vs WSD. A separate gene-wise linear model analysis was performed contrasting RNAs of LGCs from follicles yielding MI/MII oocytes versus those from follicles with degenerating oocytes (MI/MII vs Deg.). Those RNAs with at least a 2-fold change in transcript levels (upregulated or downregulated) and p ≤ 0.05 between each contrast were considered to significantly change in expression. Ingenuity Pathway Analyses (IPA) software (QIAGEN Sciences, Germantown, MD, USA) was used to mine the database of all significantly altered RNAs in the contrasts (comparisons). Gene ontologies mapping to cellular components were analyzed by PANTHER software (Overrepresentation Test release 20170413, GeneOntology (GO) database Annotation version Released 2017-08-14; PANTHER Software Version 12.0, Gene Ontology Phylogenetic Annotation Project, http://pantherdb.org/) GeneOntology (GO) pathways were also assessed using the PANTHER classification software and Ensembl gene ID’s (www.ensembl.org) were mined using all LGC RNAs in contrasts.
Results
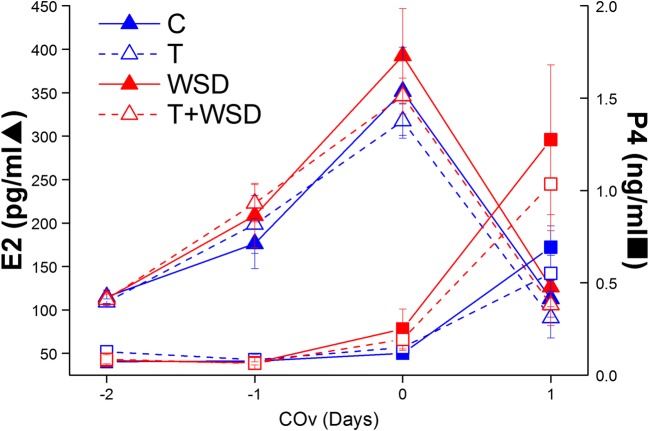
There were no differences by treatment group in levels of E2 or P4 produced during COv cycles where the oocyte was successfully recovered (all p > 0.16; Fig. 1). Serum E2 levels rose to > 250 pg/mL in all groups and declined to < 150 pg/mL following hCG administration. Serum P4 levels remained below 0.3 ng/mL until after exposure to hCG bolus. Different E2 and P4 patterns were not associated with oocyte stage or fertilization success; however, a rise of P4 > 0.5 ng/mL before hCG administration was associated with premature follicle rupture (ovulated; examples T group, Supplemental Fig. 1).
Fig. 1.
Serum estradiol (E2, triangles) and progesterone (P4, squares) levels in rhesus females during controlled ovulation (COv) protocols in C (solid blue lines), T (dashed blue lines), WSD (solid red lines), and T+WSD (dashed red lines) treatment groups which resulted in successful oocyte retrieval (i.e., without premature follicle rupture). There were no significant differences in E2 and P4 profiles between groups during COv (all p > 0.05)
Results of successful COv cycles are depicted in Table 1. Only MI/II-stage oocytes were retrieved from control (C) animals (100%), whereas only 50% of T, 67% of WSD, and 57% of T+WSD follicles yielded MI/MII oocytes. One immature, GV-stage ovum was recovered from a follicle in the T group (17%). Degenerated oocytes were recovered from 33% of T, 33% of WSD, and 43% of T+WSD follicles. Of the 11MI/MII-stage oocytes recovered from T, WSD, and T+WSD groups, 7 fertilized, leading to five embryos that progressed to the morula or blastocyst stage. Time-lapse imaging revealed that one of the WSD embryos underwent an unusual 1:4 multi-polar division at the zygote stage and exhibited two inner cell masses upon blastocyst formation, along with a large excluded blastomere (Fig. 2a–c, Supplemental File 1).
Fig. 2.
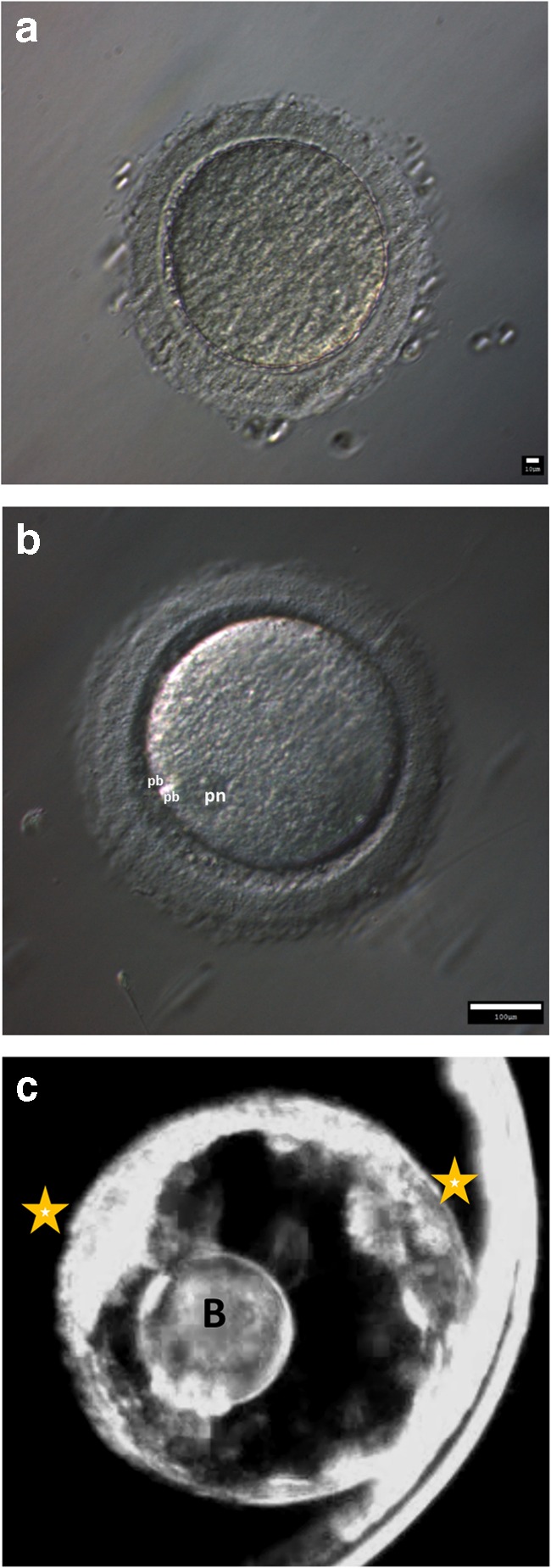
Fertilization and embryogenesis of oocyte recovered from WSD-treated female resulting in abnormal cleavage of 1 cell to 4 cells, bypassing 2 and 3 cell stages (time-lapse video: Supplemental File 1). a Oocyte recovered at metaphase I (MI)-stage, underwent subsequent IVF. b Fertilized oocyte with 2 polar bodies (denoted pb on image) and pronucleus (pn). c Blastocyst-stage embryo with 2 apparent inner-cell masses (denoted by stars) and a large excluded blastomere
Twenty-two chemokine/cytokine/growth factors were detected in FF (Supplemental Table 1A). In FF from follicles yielding an MI/MII-stage oocyte (n = 3–5/group), levels of three chemokines and one growth factor were reduced by exposure to T (overall effect of T, p < 0.04) and WSD (overall effect of WSD, p < 0.03; Table 2). Two chemokines CXCL8 and CCL2, as well one growth factor, FGF2, were reduced in FF by T alone (7.5-, 2.6-, and 2.4-fold, respectively), and by T+WSD (2.2-, 2.2-, and 3-fold, respectively) compared to C group follicles. The chemokine CCL11 was reduced 1.8-fold in follicles of T+WSD ovaries compared to C. Additionally, there was a trend for the chemokine CXCL12 to be reduced by T-exposure (overall effect of androgen p < 0.06). Because of the large number of degenerated oocytes recovered, an additional analysis was performed comparing cytokine/chemokine/growth factor levels in follicles containing MII/MI-stage oocytes (n = 16) and those with degenerating ovum (n = 7, Table 3). This analysis found increased levels (p ≤ 0.05) of the chemokines, CCL11 (1.5-fold) and CXCL12 (1.4-fold), the interleukin (IL)-1 receptor antagonist, IL1RN (2.9-fold), and IL18 (5.2-fold) in FF containing poor quality/degraded oocytes compared to those with MI/MII-stage ova (Table 3).
Table 2.
Cytokine/chemokine/growth factors in the follicular fluid milieu within follicles yielding MI/MII-stage oocytes altered by T and/or WSD exposure
| Protein (pg/mL) | Main effect androgen | Main effect diet | C (n = 5) | T (n = 3) | WSD (n = 4) | T+WSD (n = 4) |
|---|---|---|---|---|---|---|
| CCL11 | 0.2 | 0.03 | 686 ± 109* | 687 ± 119* | 600 ± 59 | 374 ± 41** |
| FGF2 | 0.02 | 0.4 | 3540 ± 628* | 1503 ± 514 | 2629 ± 714 | 1200 ± 572** |
| CXCL8 | 0.04 | 0.6 | 227 ± 87 | 30 ± 13 | 235 ± 89 | 103 ± 42 |
| CCL2 | 0.03 | 0.9 | 335 ± 124 | 128 ± 71 | 326 ± 63 | 154 ± 53 |
| CXCL12 | 0.06 | 0.5 | 675 ± 121 | 503 ± 62 | 757 ± 69 | 546 ± 104 |
| VEGFA | 0.11 | 0.2 | 7260 ± 2196 | 3880 ± 517 | 8444 ± 1673 | 6437 ± 983 |
Values = (pg/mL); mean of each group ± standard error of the mean (SEM)
Bold indicates significance (p ≤ 0.05); italics indicate trend (p = 0.06–0.11) towards significance by either androgen, diet, or androgen by diet main factors. When main effects are significant, different lower case letters indicate significant differences between individual groups; asterisks (* vs **) denote trend towards differences. When no symbols are present, there were no differences detected by pairwise comparisons. No significant or trends for interaction between diet and androgen were detected
Table 3.
Alterations to cytokine/chemokine/growth factor follicular fluid milieu by the presence of degenerating oocytes
| Protein | p value | MI/MII-stage oocytes (n = 16) | Degenerating oocytes (n = 7) |
|---|---|---|---|
| CCL11 | 0.03 | 572 ± 50 | 857 ± 150 |
| FGF2 | 0.1 | 2175 ± 375 | 9966 ± 7286 |
| IL18 | 0.05 | 7 ± 3 | 35 ± 20 |
| IL1RN | 0.02 | 720 ± 170 | 2064 ± 724 |
| IL6 | 0.06 | 5 ± 1.7 | 10 ± 1.8 |
| CXCL8 | 0.08 | 152 ± 38 | 352 ± 140 |
| CXCL12 | 0.05 | 625 ± 49 | 905 ± 172 |
Values = (pg/mL); mean of each group ± standard error of the mean (SEM)
Bold indicates significance (p ≤ 0.05); italics indicate trend (p = 0.06–0.11) towards significance between follicular fluid of MI/MII-stage versus degenerating oocytes
Intrafollicular steroid hormone levels were measured by LC-MS/MS in a single extraction. In FF of follicles yielding a MI/MII-stage oocyte, only intrafollicular cortisol was significantly elevated over 4-fold by T treatment alone compared to control follicles (Table 4, p < 0.02). Because of the low number of samples/treatment group, an additional analysis of all follicles was performed regardless of oocyte health, and cortisol was still significantly elevated by T exposure 3.3-fold relative to C (Table 5, p < 0.01). Intrafollicular cortisone, A4, E2, estrone, and P4 were not affected by T and/or WSD treatments (Tables 4 and 5). Follicles containing degenerating oocytes tended to maintain elevated levels of intrafollicular E2 26–30 h post-hCG treatment (Table 6, p = 0.09).
Table 4.
Steroids in the follicular fluid milieu of follicles yielding MI/MII-stage oocytes: impact of T and/or WSD exposure
| Steroid | Main effect androgen | Main effect diet | Interaction T by WSD | C (n = 5) | T (n = 2) | WSD (n = 4) | T+WSD (n = 4) |
|---|---|---|---|---|---|---|---|
| Cortisol | 0.02 | 0.6 | 0.03 | 26 ± 8 a | 103 ± 5 b | 54 ± 19a,b | 59 ± 12a,b |
| Cortisone | 0.6 | 0.6 | 0.7 | 67 ± 20 | 88 ± 17 | 66 ± 14 | 68 ± 21 |
| E2 | 0.5 | 0.2 | 0.3 | 322 ± 147 | 554 ± 216 | 288 ± 86 | 250 ± 48 |
| Estrone | 0.4 | 0.18 | 0.15 | 31 ± 13 | 70 ± 48 | 33 ± 12 | 21 ± 5 |
| P4 | 0.9 | 0.4 | 0.9 | 4588 ± 1854 | 4246 ± 391 | 6146 ± 1194 | 6104 ± 2031 |
| A4$ | 0.18 | 0.5 | 0.9 | 517 ± 284 | 1037 | 360 ± 107 | 763 ± 165 |
Values = (ng/mL); mean of each group ± standard error of the mean (SEM)
Bold indicates significance (p ≤ 0.05); italics indicate trend (p = 0.06–0.11) towards significance by either androgen, diet, or interaction T by WSD main factors. When main effects are significant, different lower case letters indicate significant differences between individual groups; asterisks (* vs **) denote trend towards differences. When no symbols are present, there were no differences detected by pairwise comparisons. $ = A4 values were below the limit of detected in some samples, therefore, n = 4 C, 1 T, 3 WSD, and 2 T+WSD
Table 5.
Steroids in the follicular fluid milieu of all follicles aspirated: impact of T and/or WSD exposure
| Steroid | Main effect androgen | Main effect diet | Interaction T by WSD | C (n = 5) | T (n = 5) | WSD (n = 5) | T+WSD (n = 7) |
|---|---|---|---|---|---|---|---|
| Cortisol | 0.01 | 0.6 | 0.07 | 26 ± 8 a | 83 ± 13 b | 44 ± 18a,b | 54 ± 8a,b |
| Cortisone | 0.5 | 0.9 | 0.5 | 67 ± 20 | 81 ± 11 | 67 ± 11 | 59 ± 14 |
| E2 | 0.3 | 1 | 0.8 | 322 ± 147 | 341 ± 115 | 237 ± 85 | 209 ± 40 |
| Estrone | 0.8 | 0.7 | 0.15 | 31 ± 13 | 45 ± 19 | 49 ± 18 | 22 ± 6 |
| P4 | 0.5 | 0.8 | 0.8 | 4588 ± 1854 | 5144 ± 597 | 5803 ± 911 | 5768 ± 1305 |
| A4 | 0.5 | 0.9 | 0.9 | 517 ± 284 | 534 ± 252 | 700 ± 349 | 760 ± 95 |
Values = (ng/mL); mean of each group ± standard error of the mean (SEM)
Bold indicates significance (p ≤ 0.05); italics indicate trend (p = 0.06–0.11) towards significance by either androgen, diet, or interaction T by WSD main factors. When main effects are significant, different lower case letters indicate significant differences between individual groups; asterisks (* vs **) denote trend towards differences. When no symbols are present, there were no differences detected by pairwise comparisons
Table 6.
Steroids in the follicular fluid milieu: impact of oocyte health
| Steroid | p value | MI/MII-stage oocytes (n = 15) | Degenerating oocytes (n = 6) |
|---|---|---|---|
| Cortisol | 0.9 | 50 ± 14 | 52 ± 9 |
| Cortisone | 0.4 | 56 ± 10 | 70 ± 9 |
| E2 | 0.09 | 144 ± 43 | 325 ± 61 |
| Estrone | 0.8 | 38 ± 16 | 34 ± 8 |
| P4 | 0.9 | 5045 ± 331 | 5306 ± 866 |
| A4 | 0.5 | 760 ± 339 | 571 ± 130 |
Values = (ng/mL); mean of each group ± standard error of the mean (SEM)
Italics indicates significance (p ≤ 0.05); italics indicate trend (p = 0.06–0.11) towards significance between follicular fluid of healthy versus degenerating oocytes
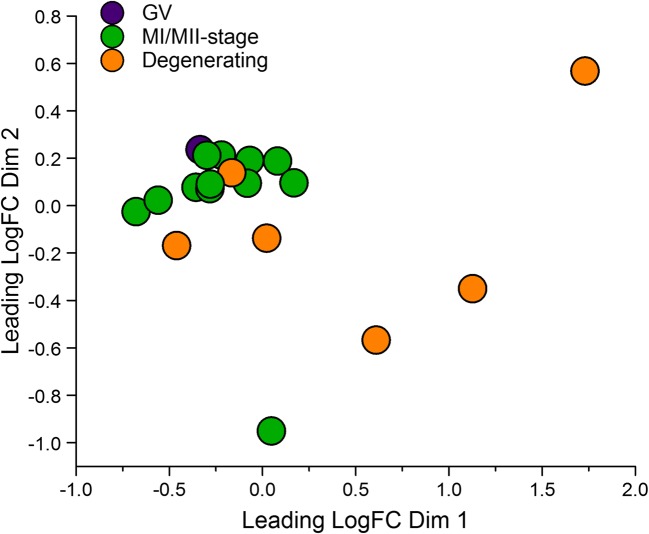
Principle component analyses of luteinizing granulosa cell (LGC) transcriptomes (n = 4–6/group) show those from follicles containing MI/MII-stage oocytes were mostly distinct from those containing poor quality/degenerating oocytes (Fig. 3). Since the latter were potentially confounding outliers, only LGC samples from follicles with MI/MII-stage oocytes were included in contrast/comparison analyses (n = 4 C, n = 2 T, n = 3 WSD, n = 3 T+WSD). RNAs which changed in expression ≥ ± 2-fold (p < 0.05) were considered significantly altered (Supplemental File 2). Those RNAs with the greatest fold-change in expression (both upregulated and downregulated) in each pairwise comparison are listed in Table 7. The pairwise comparisons of differentially expressed RNA transcripts were then visualized through a Venn diagram [34]. In comparisons of C versus treated groups, 44 RNAs were also altered in expression by T and/or WSD exposure (Fig. 4a, center of diagram). Of the 974 RNAs altered by T exposure, 835 (91%) were unique to T alone. A total of 503 RNAs were altered by WSD exposure, and 342 (68%) were uniquely altered by WSD alone. While T+WSD altered expression of 358 RNAs, 52 (14.5%) were commonly altered by T alone and 74 (21%) by WSD alone. Comparing the effects of T in the presence and absence of WSD (T vs T+WSD), 942 RNAs were identified as differentially expressed. Furthermore, analysis of the effects of WSD in the presence and absence of T (WSD vs T+WSD) identified 247 differentially expressed RNAs. When these two pairwise contrasts were compared by Venn diagram, only a small number of RNAs [34] were altered similarly (Fig. 4b, center of diagram). Additional Venn diagram analyses (Supplemental Fig. 2) show that a large percentage of RNAs (46%) were altered in both pairwise comparisons of C vs T and T vs T+WSD.
Fig. 3.
Principle component analyses of luteinizing granulosa cell (LGC) RNA transcriptomes yielding MI/MII-stage oocytes (green) versus those containing degenerating (orange) and germinal vesicle intact (GV-stage, purple) oocytes. See text for details
Table 7.
Changes in LGC RNA expression by T and/or WSD identified by RNA-seq
| Pairwise comparison | Total # RNAs | # Upregulated | # Downregulated | Most upregulated RNAa Ensembl gene ID| gene product | Fold change | p value | Most downregulated RNAa Ensembl gene ID| gene product | Fold change | p value |
|---|---|---|---|---|---|---|---|---|---|
| C vs T | 974 | 540 | 434 | 47114| CCL18 | 256 | 0.02 | 23363| AQP10 | − 181 | 0.04 |
| C vs WSD | 503 | 322 | 181 | 46863| SLC26A2 | 27 | 0.03 | 28703| predicted MT-RNR1 | − 134 | 0.01 |
| C vs T+WSD | 358 | 200 | 158 | 28703| predicted MT-RNR1 | 43 | 0.03 | 32525| predicted TCF4 | − 25 | 0.03 |
| T vs T+WSD | 942 | 517 | 425 | 39933| Novel lincRNA | 247 | 0.02 | 23363| AQP10 | − 3029 | 0.005 |
| WSD vs T+WSD | 247 | 188 | 59 | 20035| uncharacterized ribosomal protein | 118 | 0.04 | 47184| predicted MUC4 | − 13 | 0.025 |
aEnsembl Gene ID: ENSMMUG000000XXXXX
Predicted RNAs identified by Ensembl BLAST/TBLASTN against human genome/proteome
In follicles containing MI/MII-stage ovum
Fig. 4.
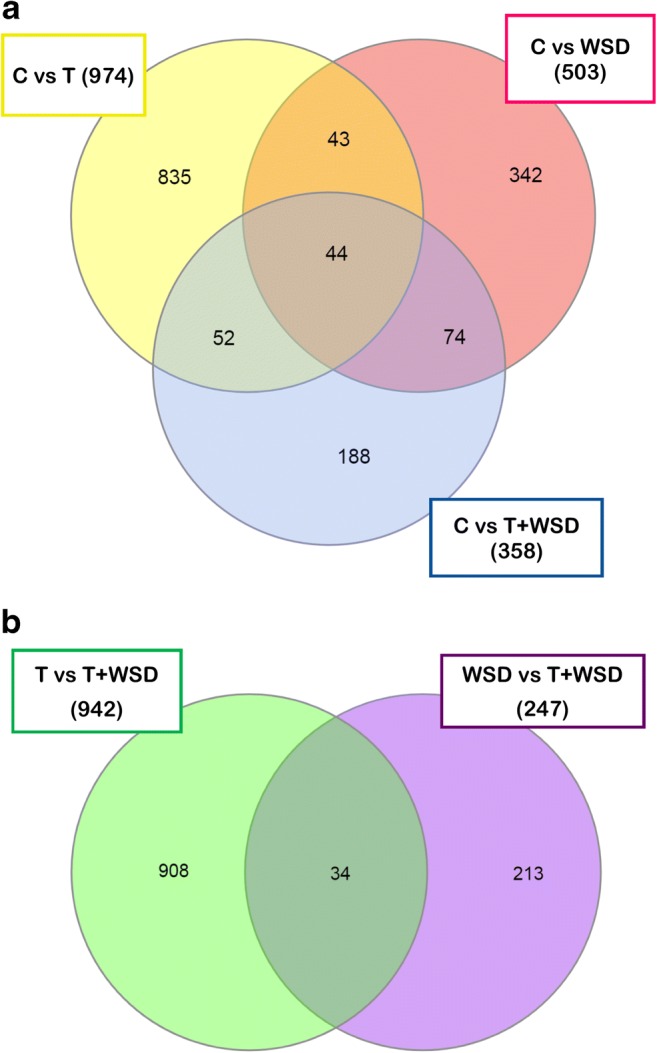
Venn diagram showing differences and similarities between different pairwise comparisons performed to identify changes to LGC RNA transcriptome induced by chronic androgen and/or WSD treatment. a Numbers of RNAs in each contrast and overlap between C vs T (yellow ellipse), C vs WSD (red ellipse), and C vs T+WSD (purple ellipse). b Numbers of RNAs in each contrast and overlap between T vs T+WSD (green ellipse) and WSD vs T+WSD (blue ellipse). See “Methods” for details of statistical analyses and Table 7 for total RNAs significantly upregulated and downregulated identified by each contrast
Pathway analyses were performed on all significantly altered RNAs to determine the most impacted canonical pathways (Supplemental Table 3). Top canonical pathways impacted by T compared to C included several that suggested alterations to gene processes controlling steroidogenesis (pregnenolone biosynthesis, Supplemental Table 3). In contrast, WSD compared to C included mostly immune cell-related pathways, and C compared to T+WSD groups demonstrated changes in several pathways associated with immune function. The contrast of T vs T+WSD exhibited divergence of pathways associated with cholesterol biosynthetic processes, while WSD vs T+WSD included pathways associated with the immune system (Supplemental Table 3). To further investigate the impact of T and WSD on the LGC transcriptome, pairwise comparisons were further analyzed by IPA for regulatory networks which could lead to the changes in RNA expression observed [35]. Examples of “Top Regulator Effect Networks” (Supplemental Table 4) identified by IPA for each pairwise comparison are depicted in Supplemental Figs. 3–5. Notably, gene networks that regulate changes in cellular viability, invasion, and proliferation were associated with the RNA expression changes identified in the pairwise comparison C vs T.
A separate analysis was performed to compare the LGC transcriptome from all follicles (regardless of treatment) containing MI/MII-stage (n = 12) versus those containing degenerating oocytes (n = 7; Supplemental File 3). A total of 646 RNAs were significantly different in the expression: 440 with increased and 206 with decreased expression in LGCs of follicles with degenerating oocytes (Table 8). The most upregulated and downregulated RNAs were associated with mitochondrial genes: MT-RNR1 (mitochondrially encoded 12S RNA) and MT-ND1 (mitochondrially encoded NADH dehydrogenase 1). Significantly (p < 0.05) overrepresented gene ontologies were analyzed by PANTHER and expression changes to RNAs mapping to ontologies associated with the extracellular matrix and plasma membrane components were enriched in LGCs of follicles containing degenerating oocytes (Table 9). The list of significantly altered RNAs in LGCs by the presence of degenerating oocytes was also analyzed for GO pathways (Supplemental Table 5). From this analysis, the most impacted pathway was the “Gonadotropin-releasing hormone receptor pathway”, which includes upregulated RNA transcripts mapping to the genes, PTGER4 (3.6-fold), ALOX15 (2.4-fold), COX2 (2.2-fold), MAPK12 (2.1-fold), BMP2 (3.7-fold), NR3C1 (3-fold), MAPK11 (2.1-fold), and a putative heat shock protein (2.1-fold), while the RNA encoding SLC2A1 is downregulated in degenerating oocytes (− 2.1-fold; Supplemental Table 5).
Table 8.
Changes in LGC RNA expression by the presence of degenerating oocytes identified by RNA-seq
| Total # RNAs | # Upregulated | # Downregulated | Most upregulated RNAa Ensembl gene ID| gene product | Fold change | p value | Most downregulated RNAa Ensembl gene ID| gene product | Fold change | p value | |
|---|---|---|---|---|---|---|---|---|---|
| Degenerating vs MI/MII-stage oocytes | 646 | 440 | 206 | 28703| predicted MT-RNR1 | 27 | 0.03 | 45979| predicted MT-ND1 | − 483 | 0.02 |
aEnsembl Gene ID: ENSMMUG000000XXXXX
Predicted RNAs identified by Ensembl BLAST/TBLASTN against human genome/proteome
Table 9.
GeneOntology (GO) terms altered in LGCs by the presence of degenerated oocytes
| GO cellular component complete | # In reference genome (Macaca mulatta) | # In list | # expected | Fold enrichment | Enriched (+)/depleted (−) | p value* |
|---|---|---|---|---|---|---|
| Extracellular matrix | 311 | 20 | 6.1 | 3.3 | + | 0.005 |
| -Extracellular region | 2607 | 82 | 50.7 | 1.6 | + | 0.009 |
| Integral component of plasma membrane | 811 | 35 | 15.8 | 2.2 | + | 0.015 |
| -Integral component of membrane | 4519 | 145 | 87.9 | 1.7 | + | 9.72E-08 |
| --Intrinsic component of membrane | 4562 | 150 | 88.7 | 1.7 | + | 5.48E-09 |
| ---Membrane part | 5181 | 168 | 100.7 | 1.7 | + | 3.47E-10 |
| ----Membrane | 6603 | 194 | 128.4 | 1.5 | + | 1.70E-08 |
| Intrinsic component of plasma membrane | 848 | 38 | 16.5 | 2.3 | + | 0.003 |
| -Plasma membrane part | 1397 | 64 | 27.2 | 2.4 | + | 2.54E-07 |
| --Plasma membrane | 2638 | 102 | 51.3 | 2.0 | + | 8.94E-09 |
| ---Cell periphery | 2717 | 105 | 52.8 | 2.0 | + | 4.05E-09 |
| Unclassified | 7501 | 80 | 145.8 | 0.6 | − | 0.00E+00 |
PANTHER overrepresentation test of RNAs mapping to cellular components
*Bonferroni correction applied for multiple testing, all results p < 0.05
All LGC RNAs in comparisons of C vs T, WSD, and T+WSD, and MI/MII-stage vs degenerating oocytes (Supplemental Files 2 and 3) were mined in Ensembl to identify gene changes associated with ovarian and metabolic processes (Supplemental Table 6). Treatment with T alone significantly (p < 0.05) upregulated expression of AR and XBP1, while expressions of CYP11A1, HSD3B2, IRS1, FADS1, and PRKAA2 were downregulated. Consumption of WSD significantly upregulated RNA encoding CYP17A1, while downregulating IGF2 and SCD5. Because RNA samples were isolated from crude aspirates of the entire periovulatory follicle, some luteinizing theca cells are likely to present as demonstrated by expression of CYP17A1 [36]. The combination of T+WSD upregulated expression of the only DDIT3 in this analysis. The expressions of IGF2, FADS1, and PRKAA2 were similarly downregulated in LGCs of follicles with degenerating oocytes. In contrast, upregulation of AREG, VEGFA, and IGF1R, and downregulation of CDKN1A, CYP19A1, DENND1A, ATF6, and HACD1 were observed in LGCs of follicles with degenerating oocytes in this dataset.
Discussion
Unlike human studies that evaluated the effects of diet and elevated androgens on human oocyte/follicle quality using multiple heterogeneous antral follicles from ovarian stimulation protocols [5, 6], this nonhuman primate study is unique in that it leveraged a standardized COv protocol to examine the contents of the single, dominant periovulatory follicle. In addition, the experimental design allowed for strict comparison of a similar level of chronic androgen exposure (4- to 5-fold elevation above controls) in the presence and absence of an obesogenic diet in a primate model [15]. The WSD utilized here provides 36% of total caloric intake from fat, while the standard chow diet contains 14% of calories from fat [15]. This is in contrast to some high-fat diets in studies with rodent models, in which up to 60% of caloric intake is from fat [37]. Thus, the current WSD diet closely mimics the typical human Western-style diet of 30–40% of caloric intake from fat [38]. Our data demonstrate that unlike in control animals, the oocyte from the dominant follicle of the natural menstrual cycle in chronically T-, WSD-, and T+WSD-treated monkeys was often degenerated or immature, despite the persistence of ovarian steroid patterns from the COv protocol. Due to focus on the single pre-ovulatory follicle, only small numbers of oocytes were collected from each group, but it is noteworthy that degenerated oocytes were not collected in any of our previous studies in control groups or other efforts to identify local regulatory factors important for ovarian physiology (e.g., P4 or prostaglandin E2 [17, 39]). Here, degenerated oocytes were only found previously if the treatment profoundly altered follicle health as well as vascularity and steroid patterns (e.g., angiopoietin 2 [40] or PGRMC1; unpublished data). However, the current data does complement our earlier studies comparing small pre-antral and antral follicles in young, adult rhesus females after short term (~ 14 months) exposure to a WSD in the absence and presence of chronically elevated androgen (~ 6 years) [41, 42]. In this previous study, we also observed increased numbers of atretic small antral follicles in vivo and increased numbers of degenerating oocytes following follicle culture after WSD and T+WSD treatments. Thus, the negative effects of T and/or WSD may occur during folliculogenesis and manifest later in the antral follicle during ovulation.
Unexpectedly, a number of follicles, especially in the control group, ovulated prematurely during the COv protocol before follicular contents could be aspirated. The macaque COv protocol developed by our research group [43] has been successfully used in rhesus monkeys for 15+ years [17, 39, 44] without early ovulation unless the COv protocol is initiated too late in the follicular phase to block the resulting endogenous surge of LH [43]. Serum E2 levels at the time of COv initiation did not differ between control females with premature follicle rupture and those without (no rupture 115.1 ± 7.6 pg/mL; ruptured 115.5 ± 8.1 pg/mL), and these levels are not near peak E2 levels that elicit an LH surge. In addition, daily serum LH levels in several females who ovulated prematurely demonstrated a 5 ± 1-fold reduction in serum LH levels (data not shown) indicating that GnRH antagonist Antide was successful in blunting the LH surge. These suggest the COv protocols of all control females were initiated correctly, timed in a similar fashion. If the COv protocol was biased towards the retrieval of either MI/MII or poor-quality oocytes, this would be reflected in the 5 successful control retrievals as well as the treated groups. As the data stand, only MI/MII-stage oocytes were recovered in controls, while several degenerated or immature oocytes were recovered in the other groups. There were also 1–2 unsuccessful attempts in the treated females as well. All of these were a result of premature follicle rupture, similar to the controls, precluding recovery of these oocytes of unknown quality. The age of the female may be significant, however, since these females are substantially younger than previous rhesus females used for similar COv studies (8–10 years old, selected for proven fertility, vs 5.5–6 years of age). The pituitary and/or ovaries/follicles of these females may be more sensitive to lower levels of GnRH/LH compared to their older counterparts. Protocols employing controlled ovarian stimulation to induce multi-follicular development are underway in this cohort of females to retrieve greater numbers of oocytes for analyses. These studies should be able to determine the impact of premature follicle rupture and retrieval success rates/oocyte quality in the T- and/or WSD-treated cohorts.
Despite the occurrence of degenerated and immature oocytes, the current data also demonstrated that fertilization and preimplantation embryogenesis is possible in the presence of chronic T and/or WSD if the oocyte retrieved is healthy and meiotically competent. Fertilization occurred in all groups, and blastocyst formation was observed in a few T- and WSD-exposed embryos. Time-lapse imaging revealed one WSD embryo that underwent an unusual 1:4 multi-polar division at the zygote stage and exhibited two possible inner cell masses upon blastocyst formation, as well as a largely excluded blastomere. While we did not perform chromosomal analyses on this embryo, recent studies suggest that multipolar divisions and the exclusion of non-dividing aneuploid blastomeres might provide a means to overcome aneuploidy during preimplantation development [45]. Because so few MI/MII oocytes were recovered in T, WSD, and T+WSD groups, further studies are needed to determine changes to embryonic developmental potential induced by exposure to chronic T and/or WSD in primates.
Previous studies of women with PCOS reported increased levels of ovarian immune cells [46] and enhanced cytokine production by follicular lymphocytes [47]. In addition, several studies demonstrated an association between ovulation, ovarian dysfunction, inflammation, and obesity [48]. Because of this, the FF obtained from aspirated follicles was analyzed for cytokine/chemokine and growth factor milieu. In the follicles containing healthy oocytes, significant declines in production of several factors were associated with chronic androgen exposure. Lower levels of one factor, CXCL8/IL8, were also observed in FF of women with PCOS [49]. While a decline in cytokine/chemokine production seems counter-intuitive to the hypothesis that these ovaries are exposed to chronic inflammation, CXCL8 levels were also shown to be lower in smaller antral follicles of non-PCOS women undergoing ovarian stimulation due to male factor infertility [50]. Therefore, reduced levels of these proteins may be indicative of overall follicle maturity, but this premise remains to be investigated. However, increased production of CCL11, IL18, ILRN, and CXCL12 were associated with follicles containing degenerated oocytes, which were only recovered in T, WSD, and T+WSD groups. One of these, IL18, was shown to be elevated in the serum of women with PCOS independent of BMI [51]. Further studies of these factors and steroid/metabolic regulation are needed to understand the contribution of cytokines/chemokines and growth factors to overall effects of T and/or WSD on primate folliculogenesis/oocyte quality.
Tandem LC-MS/MS analyses were performed on FF samples to detect any impact of chronic T and/or WSD on local androgenic or estrogenic steroidogenic pathways during ovulatory processes. Intrafollicular androgens (A4) and estrogens (E2, E1) were not altered by chronic androgen exposure in the naturally selected dominant follicle of our rhesus females. These data are contradictory to a previous study of prenatally androgenized rhesus females, which display elevated androgens in adult life [52], whereby androgen exposure was associated with reduced intrafollicular A4 and E2, which correlated with reductions in intrafollicular testosterone. In this previous study, however, intrafollicular steroid hormones were measured by RIA and not LC-MS/MS, and therefore increased the sensitivity of the current assay may explain differences between these two studies. This previous study also measured FF following FSH-induced ovarian stimulation, unlike the data reported here in macaques which only sampled the naturally selected periovulatory follicle. In women with PCOS, elevated intrafollicular androgens and declines in intrafollicular estrogens were detected by LC-MS/MS in non-stimulated follicles of the natural menstrual cycle [53]. These follicles displayed elevated A4, and decreased E2 and estrone, suggesting impairments to local aromatase activity. But, this study sampled the only medium- to large-sized antral follicle present during the mid-follicular phase of the menstrual cycle and pooled the fluid for analysis [53]. These previous reports of intrafollicular steroids in women (mid-follicular phase) and prenatally androgenized monkeys (multi-follicular ovaries) may not be directly comparable to the data from the single periovulatory follicle reported here. It is important to note the female macaques in our study were presumably still undergoing ovulatory cycles [16], evidenced by the mid-cycle rise in E2, and many studies of women with PCOS are performed on those seeking infertility treatment for oligo- and/or anovulation.
Elevated intrafollicular cortisol was detected in FF of T-treated females compared to controls. This effect appears to be independent of oocyte health in this study. Follicles of women with PCOS are also reported to have elevated cortisol [53]; this is postulated to be a result of suppressed HSD11B1 activity [54]. In women, this was previously associated with an anovulatory PCO phenotype: granulosa cells isolated from these women had reduced HSD11B1 activity [54]. RNA-seq evaluation of LGC transcriptomes found HSD11B1 tended to be reduced as a function of WSD exposure; however, this analysis also identified changes in expression for RNAs for several enzymes in ovarian steroidogenic pathways in LGCs of chronic T- and/or WSD-exposed follicles. In particular, CYP11A1 and HSD3B2 RNAs are significantly reduced in T versus control follicles. Alterations to local steroidogenesis by reduced HSD3B2 and/or CYP11A1 may impair the activities of HSD11B1. Additional experiments are underway which should help clarify alterations to local follicular steroidogenesis following exposure to chronically elevated T.
Although the rhesus females in this study were experiencing menstrual cycles [16], we hypothesized that ovarian and/or uterine defects resulting from 3+ years of treatment reduce their fertility. The presence of immature or degenerating oocytes in periovulatory follicles (current study), the reduction in ovarian blood flow and P4 levels during the luteal phase [16], and evidence of P4 resistance in the uterine endometrium [16] could disrupt onset or viable pregnancy. Indeed, in a recently reported fertility trial on these animals, chronic T treatment (T or T+WSD) increased the time to achieve pregnancy, WSD treatment (WSD or T+WSD) decreased the number of pregnancies by 30%, and combined T+WSD treatment was associated with pregnancy loss (2 of 6 before the third trimester) [55]. Fertility in this cohort was significantly associated with insulin resistance and elevated fasting glucose and not body fat or BMI. In this cohort, the presence of metabolic dysfunction preceded significant increases in BMI or body fat. Women with elevated BMI have higher rates of pregnancy loss during infertility therapy, and PCOS diagnosis combined with elevated BMI further increases these rates [4]. This same meta-analysis found poor outcomes were also noted in obese women seeking infertility therapy due to male factor. The data shown here corroborate these studies and suggest an alteration in preimplantation embryogenesis by T and/or WSD may contribute to reduced fecundity in primates with hyperandrogenemia/obesity.
As part of this fertility trial, placental structure function was analyzed in vivo (imaging) and in vitro (histology) [56]. Placentas obtained from the WSD groups displayed greater vascular impedance and accelerated villous maturation; a higher portion of these tissues displayed infarcts and calcification. Placentas of the T groups displayed decreased blood flux (flow) and a striking decrease in fetal capillary volume compared to controls at mid-third trimester. These data are similar to those of Palomba et al. [57], where these researchers demonstrated altered vascularization in placentas of hyperandrogenic/high BMI, normandrogenic/high BMI, and hyperandrogenic/normal BMI women with a PCOS diagnosis. However, our rhesus data are complicated by the small sample size due to the lower fertility rate in the T+WSD group [55]. Additional fertility trials are underway in this cohort to increase the number of offspring available for analyses. Nevertheless, these data support the concept that placentas of women with PCOS display lesions which may be associated with detrimental impacts on the health of offspring [58]. Whether these lesions form as a result of altered embryogenesis remains to be determined, studies on preimplantation embryos and third-trimester fetuses are ongoing.
Conclusions
This study demonstrates that the oocyte within the dominant follicle selected during the natural menstrual cycles in chronically T-, WSD-, and T+WSD-treated monkeys was often degenerated or immature, whereas only healthy MI/MII oocytes were present in follicles of controls. However, if the oocyte retrieved is healthy and meiotically competent, fertilization and preimplantation embryogenesis are possible despite chronic T and/or WSD treatment. While changes in the production of some cytokines/chemokines/growth factors were found in FF of T-and/or WSD-treated females with MI/MII-stage oocytes, increased production of a few of these factors was associated with follicles containing degenerated ova. Divergence in LGC mRNA expression occurred in samples from follicles with these degenerated ova, compared to LGCs of MI/MII-stage oocytes. Alterations to the LGC transcriptome associated with T and WSD exposure were also detected. These data provide insight into the processes contributing to lower fertility in females experiencing chronic hyperandrogenemia in the presence and absence of a high-fat diet. This “puberty-onset” model could be useful for investigating whether insulin-sensitizing agents [59, 60] or anti-androgen therapies [59] prevent or ameliorate the metabolic and/or reproductive effects of chronic hyperandrogenemia and high-fat diet in primates.
Electronic Supplementary Material
Examples of estradiol (E2) and progesterone (P4) patterns and oocyte/ fertilization outcomes from several T-treated females. E2 and P4 patterns were not associated with oocyte stage or fertilization success. A premature rise of P4 >0.5ng/ml before hCG administration (orange arrow, bottom graph and image) was associated with follicle rupture (ovulated). (PNG 2249 kb)
Additional Venn Diagram Analyses comparing all pairwise comparisons from LGC transcriptome analyses. Numbers of RNAs in each contrast and overlap between C vs T (yellow ellipse), C vs WSD (red ellipse) C vs T+WSD (purple ellipse), T vs T+WSD (green ellipse) and WSD vs T+WSD (blue ellipse). Panel A) All RNAs in dataset. Panel B) Only up-regulated RNAs in pairwise comparisons. Panel C) Only down-regulated RNAs in pairwise comparisons. See Methods for details of statistical analyses and Table 7 for total RNAs significantly up-regulated and down-regulated identified by each contrast. (PNG 807 kb)
The Top Regulator Effect Networks identified by IPA for pairwise comparison C vs T (Supplemental Table 4). Genes in top row are transcription factors by which changes in activity could lead to the observed changes in gene product expression in the center row. Processes identified in the bottom row are the predicted cellular outcomes of the observed changes to gene expression. Red colors indicate activated and green indicate inhibited molecules, while orange/yellow lines indicate direction of predicted activation and blue lines indicate directed of predicted inhibition (dashed lines indicate lower level of confidence for predictions, grey lines indicate those predictions that do not reach statistical significance). (PNG 1862 kb)
The Top Regulator Effect Networks identified by IPA for pairwise comparison C vs WSD (Supplemental Table 4). Genes in top row are transcription factors by which changes in activity could lead to the observed changes in gene product expression in the center row. Processes identified in the bottom row are the predicted cellular outcomes of the observed changes to gene expression. Red colors indicate activated and green indicate inhibited molecules, while orange/yellow lines indicate direction of predicted activation and blue lines indicate directed of predicted inhibition (dashed lines indicate lower level of confidence for predictions, grey lines indicate those predictions that do not reach statistical significance). (PNG 3593 kb)
The Top Regulator Effect Networks identified by IPA for pairwise comparisons C vs T+WSD (Panel A) and T vs T+WSD (Panel B; Supplemental Table 4). Genes in top row are transcription factors by which changes in activity could lead to the observed changes in gene product expression in the center row. Processes identified in the bottom row are the predicted cellular outcomes of the observed changes to gene expression. Red colors indicate activated and green indicate inhibited molecules, while orange/yellow lines indicate direction of predicted activation and blue lines indicate directed of predicted inhibition (dashed lines indicate lower level of confidence for predictions, grey lines indicate those predictions that do not reach statistical significance). (PNG 1220 kb)
(MP4 748 kb)
(CSV 2199 kb)
(CSV 744 kb)
(XLSX 17 kb)
(XLSX 18 kb)
(XLSX 13 kb)
(XLSX 13 kb)
(XLSX 15 kb)
(XLSX 19 kb)
Acknowledgments
The outstanding efforts of the NCTRI NHP Core members Emily Mishler, M.S., Corrine Wilcox, B.S., Kise Bond, P.S.M., and Andrea Calhoun, M.S. under the direction of Ov Slayden, Ph.D. as well as Diana Takahashi, M.S., were critical for the successful execution of all protocols and are greatly appreciated. In addition, the efforts of the staff of the ONPRC Endocrine Technology Support Core contributed to the success of this project. RNA sequencing was performed by the OHSU Massively Parallel Sequencing Shared Resource. Analysis of the RNA-seq data was performed by the ONPRC’s Biostatistics & Bioinformatics Core under the direction of Lucia Carbone, Ph.D. and Suzanne Fei, Ph.D. with statistical analyses performed by Lina Gao, Ph.D. and Byung Park, Ph.D. We greatly appreciate the core’s analysis pipelines that utilize OHSU’s Exacloud compute cluster.
Funding
Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development (NICHD) of the National Institutes of Health (NIH) under Award Number P50HD071836 (to RLS). Additional funding was provided by NIH Award Number P51OD011092 (Support for National Primate Research Center and Cores).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures involving rhesus monkeys were reviewed and approved by the ONPRC/Oregon Health and Science University (OHSU) Institutional Animal Care and Use Committee (IACUC) in accordance with the U.S. Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rosenfield RL, Ehrmann DA. The pathogenesis of polycystic ovary syndrome (PCOS): the hypothesis of PCOS as functional ovarian hyperandrogenism revisited. Endocr Rev. 2016;37(5):467–520. doi: 10.1210/er.2015-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Leo V, Musacchio MC, Cappelli V, Massaro MG, Morgante G, Petraglia F. Genetic, hormonal and metabolic aspects of PCOS: an update. Reprod Biol Endocrinol. 2016;14(1):38. doi: 10.1186/s12958-016-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang CC, Tien YJ, Chen MJ, Chen CH, Ho HN, Yang YS. Symptom patterns and phenotypic subgrouping of women with polycystic ovary syndrome: association between endocrine characteristics and metabolic aberrations. Hum Reprod. 2015;30(4):937–946. doi: 10.1093/humrep/dev010. [DOI] [PubMed] [Google Scholar]
- 4.Provost MP, Acharya KS, Acharya CR, Yeh JS, Steward RG, Eaton JL, et al. Pregnancy outcomes decline with increasing body mass index: analysis of 239,127 fresh autologous in vitro fertilization cycles from the 2008-2010 Society for Assisted Reproductive Technology registry. Fertil Steril. 2016;105(3):663–9. 10.1016/j.fertnstert.2015.11.008. [DOI] [PubMed]
- 5.Wood JR, Dumesic DA, Abbott DH, Strauss JF., 3rd Molecular abnormalities in oocytes from women with polycystic ovary syndrome revealed by microarray analysis. J Clin Endocrinol Metab. 2007;92(2):705–713. doi: 10.1210/jc.2006-2123. [DOI] [PubMed] [Google Scholar]
- 6.Palomba S, Daolio J, La Sala GB. Oocyte competence in women with polycystic ovary syndrome. Trends Endocrinol Metab. 2017;28(3):186–198. doi: 10.1016/j.tem.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 7.Murray AA, Swales AKE, Smith RE, Molinek MD, Hillier SG, Spears N. Follicular growth and oocyte competence in the in vitro cultured mouse follicle: effects of gonadotrophins and steroids. MHR: Basic Science of Reproductive Medicine" with: Mol Hum Reprod. 2008;14(2):75–83. doi: 10.1093/molehr/gam092. [DOI] [PubMed] [Google Scholar]
- 8.Franks S, Hardy K. Androgen action in the ovary. Front Endocrinol. 2018;9:452. doi: 10.3389/fendo.2018.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodrigues JK, Navarro PA, Zelinski MB, Stouffer RL, Xu J. Direct actions of androgens on the survival, growth and secretion of steroids and anti-Mullerian hormone by individual macaque follicles during three-dimensional culture. Hum Reprod. 2015;30(3):664–674. doi: 10.1093/humrep/deu335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynolds KA, Boudoures AL, Chi MM, Wang Q, Moley KH. Adverse effects of obesity and/or high-fat diet on oocyte quality and metabolism are not reversible with resumption of regular diet in mice. Reprod Fertil Dev. 2015;27(4):716–724. doi: 10.1071/RD14251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamer Erel C, Senturk LM. The impact of body mass index on assisted reproduction. Curr Opin Obstet Gynecol. 2009;21(3):228–235. doi: 10.1097/GCO.0b013e32832aee96. [DOI] [PubMed] [Google Scholar]
- 12.Vembu R, Reddy NS. Serum AMH level to predict the hyper response in women with PCOS and non-PCOS undergoing controlled ovarian stimulation in ART. J Hum Reprod Sci. 2017;10(2):91–4. 10.4103/jhrs.JHRS_15_16. [DOI] [PMC free article] [PubMed]
- 13.Zaadstra BM, Seidell JC, Van Noord PA, te Velde ER, Habbema JD, Vrieswijk B, et al. Fat and female fecundity: prospective study of effect of body fat distribution on conception rates. BMJ. 1993;306(6876):484–487. doi: 10.1136/bmj.306.6876.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pantasri T, Norman RJ. The effects of being overweight and obese on female reproduction: a review. Gynecol Endocrinol. 2014;30(2):90–94. doi: 10.3109/09513590.2013.850660. [DOI] [PubMed] [Google Scholar]
- 15.True CA, Takahashi DL, Burns SE, Mishler EC, Bond KR, Wilcox MC, et al. Chronic combined hyperandrogenemia and western-style diet in young female rhesus macaques causes greater metabolic impairments compared to either treatment alone. Hum Reprod. 2017;32(9):1880–91. 10.1093/humrep/dex246. [DOI] [PMC free article] [PubMed]
- 16.Bishop CV, Mishler EC, Takahashi DL, Reiter TE, Bond KR, True CA, et al. Chronic hyperandrogenemia in the presence and absence of a western-style diet impairs ovarian and uterine structure/function in young adult rhesus monkeys. Hum Reprod. 2018;33(1):128–39. 10.1093/humrep/dex338. [DOI] [PMC free article] [PubMed]
- 17.Bishop CV, Hennebold JD, Kahl CA, Stouffer RL. Knockdown of progesterone receptor (PGR) in macaque granulosa cells disrupts ovulation and progesterone production. Biol Reprod. 2016;94(5):109. doi: 10.1095/biolreprod.115.134981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf DP, Thormahlen S, Ramsey C, Yeoman RR, Fanton J, Mitalipov S. Use of assisted reproductive technologies in the propagation of rhesus macaque offspring. Biol Reprod. 2004;71(2):486–493. doi: 10.1095/biolreprod.103.025932. [DOI] [PubMed] [Google Scholar]
- 19.Hanna CB, Yao S, Ramsey CM, Hennebold JD, Zelinski MB, Jensen JT. Phosphodiesterase 3 (PDE3) inhibition with cilostazol does not block in vivo oocyte maturation in rhesus macaques (Macaca mulatta) Contraception. 2015;91:418–422. doi: 10.1016/j.contraception.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daughtry BL, Chavez SL. Time-lapse imaging for the detection of chromosomal abnormalities in primate preimplantation embryos. Methods Mol Biol (Clifton, NJ) 2018;1769:293–317. doi: 10.1007/978-1-4939-7780-2_19. [DOI] [PubMed] [Google Scholar]
- 21.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010.
- 22.Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bimber B. DISCVR-Seq: LabKey Server Extensions for Management and Analysis of Sequencing Data., at <https://github.com/bbimber/discvr-seq/wiki>. 2015. at <https://github.com/bbimber/discvr-seq/wiki>.
- 24.Nelson EK, Piehler B, Eckels J, Rauch A, Bellew M, Hussey P, et al. LabKey server: an open source platform for scientific data integration, analysis and collaboration. BMC Bioinformatics. 2011;12(1):71. 10.1186/1471-2105-12-71. [DOI] [PMC free article] [PubMed]
- 25.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed]
- 27.Engstrom PG, Steijger T, Sipos B, Grant GR, Kahles A, Ratsch G, et al. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat Methods. 2013;10(12):1185–1191. doi: 10.1038/nmeth.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28(11):1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Team RC. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 30.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu R, Holik AZ, Su S, Jansz N, Chen K, Leong HS, et al. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015;43(15):e97. 10.1093/nar/gkv412. [DOI] [PMC free article] [PubMed]
- 33.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed]
- 34.Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanders SL, Stouffer RL. Localization of steroidogenic enzymes in macaque luteal tissue during the menstrual cycle and simulated early pregnancy: immunohistochemical evidence supporting the two-cell model for estrogen production in the primate corpus luteum. Biol Reprod. 1997;56(5):1077–1087. doi: 10.1095/biolreprod56.5.1077. [DOI] [PubMed] [Google Scholar]
- 37.Hariri N, Thibault L. High-fat diet-induced obesity in animal models. Nutr Res Rev. 2010;23(2):270–299. doi: 10.1017/S0954422410000168. [DOI] [PubMed] [Google Scholar]
- 38.Odermatt A. The Western-style diet: a major risk factor for impaired kidney function and chronic kidney disease. Am J Physiol Renal Physiol. 2011;301(5):F919–F931. doi: 10.1152/ajprenal.00068.2011. [DOI] [PubMed] [Google Scholar]
- 39.Xu F, Stouffer RL, Muller J, Hennebold JD, Wright JW, Bahar A, et al. Dynamics of the transcriptome in the primate ovulatory follicle. Mol Hum Reprod. 2011;17(3):152–165. doi: 10.1093/molehr/gaq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu F, Stouffer RL. Local delivery of angiopoietin-2 into the preovulatory follicle terminates the menstrual cycle in rhesus monkeys. Biol Reprod. 2005;72(6):1352–1358. doi: 10.1095/biolreprod.104.037143. [DOI] [PubMed] [Google Scholar]
- 41.Bishop CV, Xu F, Xu J, Ting AY, Galbreath E, McGee WK, et al. Western-style diet, with and without chronic androgen treatment, alters the number, structure, and function of small antral follicles in ovaries of young adult monkeys. Fertil Steril. 2016;105(4):1023–1034. doi: 10.1016/j.fertnstert.2015.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ting AY, Xu J, Stouffer RL. Differential effects of estrogen and progesterone on development of primate secondary follicles in a steroid-depleted milieu in vitro. Hum Reprod. 2015;30(8):1907–1917. doi: 10.1093/humrep/dev119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young KA, Chaffin CL, Molskness TA, Stouffer RL. Controlled ovulation of the dominant follicle: a critical role for LH in the late follicular phase of the menstrual cycle. Hum Reprod. 2003;18(11):2257–2263. doi: 10.1093/humrep/deg467. [DOI] [PubMed] [Google Scholar]
- 44.Stouffer RL, Xu F, Duffy DM. Molecular control of ovulation and luteinization in the primate follicle. Front Biosci. 2007;12:297–307. doi: 10.2741/2065. [DOI] [PubMed] [Google Scholar]
- 45.Daughtry BL, Rosenkrantz JL, Lazar NH, Fei SS, Redmayne N, Torkenczy KA, et al. Single-cell sequencing of primate preimplantation embryos reveals chromosome elimination via cellular fragmentation and blastomere exclusion. Genome Res. 2019;29:367–82. 10.1101/gr.239830.118. [DOI] [PMC free article] [PubMed]
- 46.Xiong YL, Liang XY, Yang X, Li Y, Wei LN. Low-grade chronic inflammation in the peripheral blood and ovaries of women with polycystic ovarian syndrome. Eur J Obstet Gynecol Reprod Biol. 2011;159(1):148–150. doi: 10.1016/j.ejogrb.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 47.Qin L, Xu W, Li X, Meng W, Hu L, Luo Z, et al. Differential expression profile of immunological cytokines in local ovary in patients with polycystic ovarian syndrome: analysis by flow cytometry. Eur J Obstet Gynecol Reprod Biol. 2016;197:136–41. 10.1016/j.ejogrb.2015.12.003. [DOI] [PubMed]
- 48.Robker RL, Wu LL, Yang X. Inflammatory pathways linking obesity and ovarian dysfunction. J Reprod Immunol. 2011;88(2):142–148. doi: 10.1016/j.jri.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 49.Roth LW, McCallie B, Alvero R, Schoolcraft WB, Minjarez D, Katz-Jaffe MG. Altered microRNA and gene expression in the follicular fluid of women with polycystic ovary syndrome. J Assist Reprod Genet. 2014;31(3):355–362. doi: 10.1007/s10815-013-0161-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malizia BA, Wook YS, Penzias AS, Usheva A. The human ovarian follicular fluid level of interleukin-8 is associated with follicular size and patient age. Fertil Steril. 2010;93(2):537–543. doi: 10.1016/j.fertnstert.2008.11.033. [DOI] [PubMed] [Google Scholar]
- 51.Ebejer K, Calleja-Agius J. The role of cytokines in polycystic ovarian syndrome. Gynecol Endocrinol. 2013;29(6):536–540. doi: 10.3109/09513590.2012.760195. [DOI] [PubMed] [Google Scholar]
- 52.Dumesic DA, Schramm RD, Bird IM, Peterson E, Paprocki AM, Zhou R, et al. Reduced intrafollicular androstenedione and estradiol levels in early-treated prenatally androgenized female rhesus monkeys receiving follicle-stimulating hormone therapy for in vitro fertilization. Biol Reprod. 2003;69(4):1213–1219. doi: 10.1095/biolreprod.102.015164. [DOI] [PubMed] [Google Scholar]
- 53.Naessen T, Kushnir MM, Chaika A, Nosenko J, Mogilevkina I, Rockwood AL, et al. Steroid profiles in ovarian follicular fluid in women with and without polycystic ovary syndrome, analyzed by liquid chromatography-tandem mass spectrometry. Fertil Steril. 2010;94(6):2228–33. 10.1016/j.fertnstert.2009.12.081. [DOI] [PubMed]
- 54.Michael AE, Glenn C, Wood PJ, Webb RJ, Pellatt L, Mason HD. Ovarian 11beta-hydroxysteroid dehydrogenase (11betaHSD) activity is suppressed in women with anovulatory polycystic ovary syndrome (PCOS): apparent role for ovarian androgens. J Clin Endocrinol Metab. 2013;98(8):3375–3383. doi: 10.1210/jc.2013-1385. [DOI] [PubMed] [Google Scholar]
- 55.Bishop CV, Stouffer RL, Takahashi DL, Mishler EC, Wilcox MC, Slayden OD, et al. Chronic hyperandrogenemia and western-style diet beginning at puberty reduces fertility and increases metabolic dysfunction during pregnancy in young adult, female macaques. Hum Reprod. 2018;33(4):694–705. 10.1093/humrep/dey013. [DOI] [PMC free article] [PubMed]
- 56.Kuo K, Roberts VHJ, Gaffney J, Takahashi DL, Morgan T, Lo JO, et al. Maternal high fat diet and chronic hyperandrogenemia are associated with placental dysfunction in female rhesus macaques. Endocrinology. 2019; In Press. [DOI] [PMC free article] [PubMed]
- 57.Palomba S, Falbo A, Chiossi G, Tolino A, Tucci L, La Sala GB, et al. Early trophoblast invasion and placentation in women with different PCOS phenotypes. Reprod BioMed Online. 2014;29(3):370–381. doi: 10.1016/j.rbmo.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 58.Cesta CE, Oberg AS, Ibrahimson A, Yusuf I, Larsson H, Almqvist C, et al. Maternal polycystic ovary syndrome and risk of neuropsychiatric disorders in offspring: prenatal androgen exposure or genetic confounding? Psychol Med. 2019:1–9. 10.1017/s0033291719000424. [DOI] [PMC free article] [PubMed]
- 59.Mehrabian F, Ghasemi-Tehrani H, Mohamadkhani M, Moeinoddini M, Karimzadeh P. Comparison of the effects of metformin, flutamide plus oral contraceptives, and simvastatin on the metabolic consequences of polycystic ovary syndrome. J Res Med Sci. 2016;21:7. doi: 10.4103/1735-1995.177354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morgante G, Massaro MG, Di Sabatino A, Cappelli V, De Leo V. Therapeutic approach for metabolic disorders and infertility in women with PCOS. Gynecol Endocrinol. 2018;34(1):4–9. doi: 10.1080/09513590.2017.1370644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Examples of estradiol (E2) and progesterone (P4) patterns and oocyte/ fertilization outcomes from several T-treated females. E2 and P4 patterns were not associated with oocyte stage or fertilization success. A premature rise of P4 >0.5ng/ml before hCG administration (orange arrow, bottom graph and image) was associated with follicle rupture (ovulated). (PNG 2249 kb)
Additional Venn Diagram Analyses comparing all pairwise comparisons from LGC transcriptome analyses. Numbers of RNAs in each contrast and overlap between C vs T (yellow ellipse), C vs WSD (red ellipse) C vs T+WSD (purple ellipse), T vs T+WSD (green ellipse) and WSD vs T+WSD (blue ellipse). Panel A) All RNAs in dataset. Panel B) Only up-regulated RNAs in pairwise comparisons. Panel C) Only down-regulated RNAs in pairwise comparisons. See Methods for details of statistical analyses and Table 7 for total RNAs significantly up-regulated and down-regulated identified by each contrast. (PNG 807 kb)
The Top Regulator Effect Networks identified by IPA for pairwise comparison C vs T (Supplemental Table 4). Genes in top row are transcription factors by which changes in activity could lead to the observed changes in gene product expression in the center row. Processes identified in the bottom row are the predicted cellular outcomes of the observed changes to gene expression. Red colors indicate activated and green indicate inhibited molecules, while orange/yellow lines indicate direction of predicted activation and blue lines indicate directed of predicted inhibition (dashed lines indicate lower level of confidence for predictions, grey lines indicate those predictions that do not reach statistical significance). (PNG 1862 kb)
The Top Regulator Effect Networks identified by IPA for pairwise comparison C vs WSD (Supplemental Table 4). Genes in top row are transcription factors by which changes in activity could lead to the observed changes in gene product expression in the center row. Processes identified in the bottom row are the predicted cellular outcomes of the observed changes to gene expression. Red colors indicate activated and green indicate inhibited molecules, while orange/yellow lines indicate direction of predicted activation and blue lines indicate directed of predicted inhibition (dashed lines indicate lower level of confidence for predictions, grey lines indicate those predictions that do not reach statistical significance). (PNG 3593 kb)
The Top Regulator Effect Networks identified by IPA for pairwise comparisons C vs T+WSD (Panel A) and T vs T+WSD (Panel B; Supplemental Table 4). Genes in top row are transcription factors by which changes in activity could lead to the observed changes in gene product expression in the center row. Processes identified in the bottom row are the predicted cellular outcomes of the observed changes to gene expression. Red colors indicate activated and green indicate inhibited molecules, while orange/yellow lines indicate direction of predicted activation and blue lines indicate directed of predicted inhibition (dashed lines indicate lower level of confidence for predictions, grey lines indicate those predictions that do not reach statistical significance). (PNG 1220 kb)
(MP4 748 kb)
(CSV 2199 kb)
(CSV 744 kb)
(XLSX 17 kb)
(XLSX 18 kb)
(XLSX 13 kb)
(XLSX 13 kb)
(XLSX 15 kb)
(XLSX 19 kb)