Abstract
Most studies on amino acids are focused on the proteinogenic amino acids given their essential roles in protein synthesis among other pathways. In addition to 20 ubiquitous amino acids used in protein synthesis, plants synthesize over 250 non-proteinogenic amino acids that are involved in the synthesis of compounds that are anti-herbivory, anti-microbial, response to abiotic stresses, nitrogen storage, toxins against both vertebrates/invertebrates, and plant hormones among others. One such non-proteinogenic acid is β-alanine, which is known mainly for studies on humans. β-Alanine forms a part of pantothenate (vitamin B5), which is incorporated into the universal carbon shuttling compounds Coenzyme A and acyl carrier protein, in all organisms including plants. The focus of this review, however, is on the biosynthesis, metabolism, and the role of β-alanine in plants. There are several functions of β-alanine unique to plants. It is accumulated as a generic stress response molecule involved in protecting plants from temperature extremes, hypoxia, drought, heavy metal shock, and some biotic stresses. There is evidence of its participation in lignin biosynthesis and ethylene production in some species. It is further converted to the osmoprotective compound β-alanine betaine in some species and converted to the antioxidant homoglutathione in others. The polyamines spermine/spermidine, propionate and uracil have been shown to be precursors of β-alanine in plants. However, plants vary in terms of their biosynthetic pathways, and the primary metabolism of β-alanine is far from settled.
Keywords: β-alanine, L-alanine, non-proteinogenic, amino acid, secondary metabolites
Introduction
β-Alanine is a non-proteinogenic amino acid, where the amino group is at the β-position from the carboxylate group (IUPAC name = 3-aminopropanoic acid). In contrast to L-alanine, which is a proteinogenic amino acid, β-alanine has no stereo center (Figure 1). β-Alanine is incorporated into pantothenate (Vitamin B5), and therefore, is a precursor of Coenzyme A (CoA) and acyl-carrier protein, which shuttle carbon within the cell (Voet et al., 2006). β-Alanine is a component of carnosine, a dipeptide concentrated in muscle and brain tissue, which underlies the wide use of β-alanine in humans as a strength enhancing supplement. The vast majority of scientific articles about β-alanine deal with the exercise supplement aspect, please see for example Blancquaert et al. (2017). Much less is known about the role of β-alanine in plants. The focus of this manuscript is the current state of knowledge on the metabolism and role of β-alanine in plants.
Figure 1.
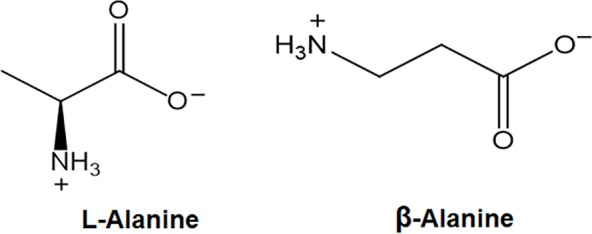
The chemical structures of L-alanine and β-alanine.
Apart from the universal importance of β-alanine (as the precursor of CoA) in the synthesis of phospholipids, synthesis and degradation of fatty acids, and the operation of the tricarboxylic acid cycle, plants also employ β-alanine in secondary metabolism, including lignin biosynthesis (Broeckling et al., 2005). Moreover, β-alanine is involved in multiple stress responses in plants. LeSvels of β-alanine have been shown to be elevated significantly following heat shock in Vigna unguiculata cell cultures (Mayer et al., 1990). Various biotic and abiotic stresses on Medicago truncatula cell cultures resulted in elevated β-alanine levels, suggesting changes in the metabolism of CoA and its thioesters, which are essential in secondary metabolism (Broeckling et al., 2005). Both drought and heat stress were found to induce increased β-alanine levels in the model plant Arabidopsis thaliana (Kaplan et al., 2004; Rizhsky et al., 2004). Metabolomics studies reveal that in response to cadmium ions (heavy metal stress); A. thaliana accumulates multiple compounds, including L-alanine, β-alanine, and the polyamine putrescine (Sun et al., 2010).
β-Alanine aminotransferases from plants have been known since at least the late 1960s (Stinson and Spencer, 1969a,b). Work on enzymes from Phaseolus vulgaris (wax bean) cotyledons showed that β-alanine could be converted into the well-known plant signaling molecule ethylene (Stinson and Spencer, 1969a,b). In some plants, β-alanine is additionally converted to β-alanine betaine, an important quaternary ammonium osmoprotective compound that is involved in tolerance to both high salt concentration and low oxygen (Hanson et al., 1991, 1994; van Dongen et al., 2009; Rocha et al., 2010a,b). A role for β-alanine in recovery from waterlogging was suggested in an earlier study employing a hydroponic system (Drakeford et al., 1985). In vitro protection against high temperature stress for the enzyme lactate dehydrogenase (LDH) is afforded by β-alanine (Mehta and Seidler, 2005).
In some leguminous plants, β-alanine forms a part of the thiol tripeptide homoglutathione, which is involved in protection against heavy metal toxicity and free radicals (reactive oxygen species) (Klapheck et al., 1988; Moran et al., 2000). For this reason, pathways involving β-alanine are considered attractive targets for the metabolic engineering of plants, potentially including crop plants in a world facing a growing human population and increasing environmental stresses due to climate change. Work that is more recent suggests that β-alanine metabolism in A. thaliana involved in a variety of pathways and roles such as nitrogen utilization efficiency, response to hypoxia, osmoprotection, vitamin B5 and CoA metabolism (Parthasarathy et al., 2019).
The Synthesis of β-Alanine In Plants
The biosynthesis of β-alanine in plants may be initiated from at least four different precursors, namely (1) the polyamines spermine/spermidine, (2) the carboxylic acid propionate, (3) the nucleotide base uracil, and (4) the proteinogenic amino acid L-aspartate. Among these, the first three are thoroughly characterized, while the fourth has long been postulated without conclusive experimental evidence. The physiological implications of each pathway are discussed below.
The Synthesis of β-Alanine in Plants: The Polyamine Pathway
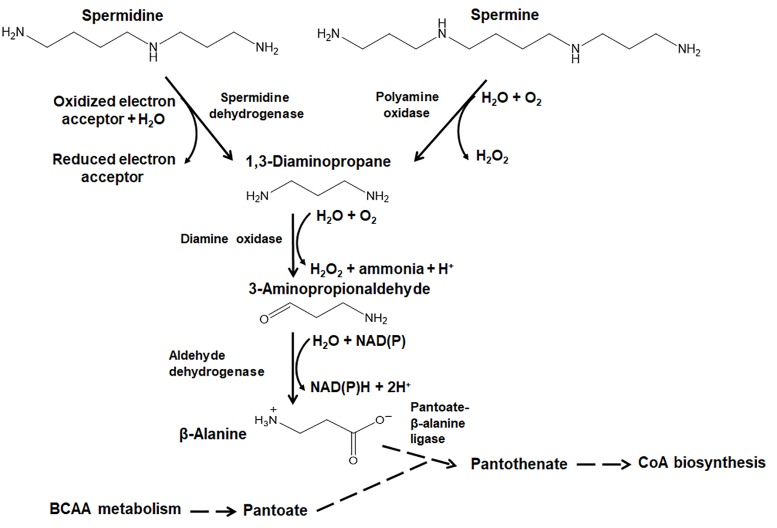
The polyamines spermine and spermidine are converted via 1,3-diaminopropane to β-alanine (Figure 2) in many plants and demonstrated in maize shoots (Terano and Suzuki, 1978; Galston and Sawhney, 1990). Pericarp discs of tomato fruits were also shown to degrade spermidine to putrescine and β-alanine (Rastogi and Davies, 1989). Spermidine is cleaved by the FAD- and heme-containing enzyme spermidine dehydrogenase [EC 1.5.99.6] into 1,3-diaminopropane and 4-aminobutyraldehyde. Spermine degradation is facilitated by the incorporation of water and molecular oxygen by the enzyme polyamine oxidase [EC 1.5.3.14], yielding 1,3-diaminopropane, 4-aminobutyraldehyde, and hydrogen peroxide. The common intermediate 1,3-diaminopropane is then deaminated with the addition of water and molecular oxygen into 3-aminopropionaldehyde, hydrogen peroxide, and ammonia by the enzyme diamine oxidase [EC 1.4.3.22]. Finally, oxidation of 3-aminopropionaldehyde with the addition of water by the NAD(P)-dependent aldehyde dehydrogenase [EC 1.2.1.3] yields β-alanine. Spermidine oxidation via these enzymes is induced by abiotic stresses (Kamada-Nobusada et al., 2008; Moschou et al., 2008a,b).
Figure 2.
Anabolism of β-Alanine via the spermine and spermidine pathways. The enzyme class (EC) numbers shown in figure correspond to the following enzymes: spermidine dehydrogenase [EC 1.5.99.6], polyamine oxidase [EC 1.5.3.14], diamine oxidase [EC 1.4.3.22], aldehyde dehydrogenase [EC 1.2.1.3], and pantoate-β-alanine ligase (AMP-forming) [EC 6.3.2.1]. The dotted lines show the pantothenate and CoA biosynthesis pathway into which β-alanine can be fed. BCAA, branched chain amino acid.
AtALDH10A8, an NAD+-dependent aminoaldehyde dehydrogenase [EC 1.2.1.19] in A. thaliana was shown to convert 3-aminopropionaldehyde into β-alanine in vitro, and similar enzymes were studied in apple fruit (Zarei et al., 2015, 2016). However, unlike gamma-aminobutyric acid (GABA), which was specifically accumulated in response to salinity, β-alanine was not, suggesting that β-alanine is accumulated as part of a non-specific rather than a specific stress response (Zarei et al., 2016). A. thaliana has been shown to survive without the ability to make spermine, but spermidine synthesis is essential to survival. This suggests that spermidine (but not spermine) could be linked to pantothenate biosynthesis via β-alanine (Imai et al., 2004). The condensation of β-alanine with pantoate in plants (Figure 2) yields pantothenate (vitamin B5), which is essential for all organisms as a precursor to the 4′-phosphopantetheine moiety of coenzyme A (CoA) and acyl carrier protein (Webb et al., 2004; Webb and Smith, 2011).
As seen in Figure 2, there is an intriguing link between the metabolism of the branched chain amino acids (BCAA) and β-alanine. It appears that there is a feedback loop whereby a change in the levels of β-alanine may be effected via BCAA degradation. Since valine and isoleucine may serve as precursors for propionyl-CoA, recent studies in this regard are discussed in the next section.
The Synthesis of β-Alanine in Plants: The Propionate Pathway
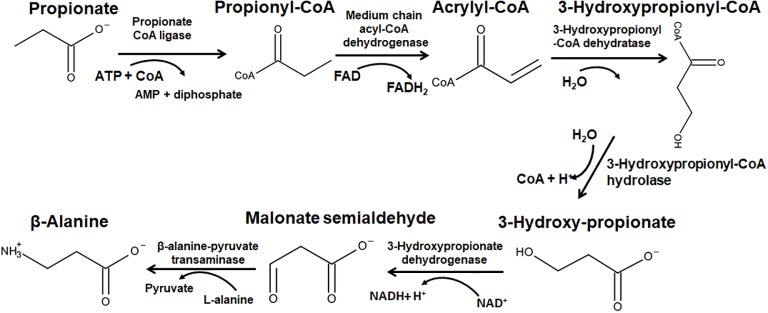
Propionate may act as the precursor for β-alanine in the pathway involving a β-alanine aminotransferase (Figure 3). Propionate is activated with the hydrolysis of ATP to propionyl-CoA A by propionate CoA ligase [EC 6.2.1.17]. Propionyl-CoA is then oxidized to acrylyl-CoA by the FAD-dependent medium chain acyl-CoA dehydrogenase [EC 1.3.8.7]. 3-hydroxypropionyl-CoA dehydratase [EC 4.2.1.116], then hydrates acrylyl-CoA yielding 3-hydroxypropionyl-CoA. Hydrolysis of the CoA ester by 3-hydroxypropionyl-CoA hydrolase [EC 3.1.2.4] generates 3-hydroxyproiponate, which is further oxidized to malonate semialdehyde by the NAD-dependent 3-hydroxypropionate dehydrogenase [EC 1.1.1.59]. The final step in this pathway is the transamination catalyzed by the pydridoxal-5′-phosphate (PLP)-dependent β-alanine-pyruvate transaminase [EC 2.6.1.18], whereby an amino group is transferred from L-alanine to malonate semialdehyde, yielding β-alanine and pyruvate (Hayaishi et al., 1961; Stinson and Spencer, 1969a,b). After 2001 study identified a β-alanine aminotransferase in A. thaliana (Liepman and Olsen, 2001), it was hypothesized that this pathway may be involved in pantothenate production. In A. thaliana, there is a gene encoding for the pantothenate synthase (PtS) enzyme, which condenses pantoate with β-alanine to yield pantothenate (Genschel et al., 1999; Ottenhof et al., 2004). It should be noted that homologous gene/s encoding aspartate decarboxylase (ADC), the enzyme that generates β-alanine in the bacterial pantothenate pathway were not identified in the plant (Ottenhof et al., 2004).
Figure 3.
The anabolism of β-alanine via the propionate pathway. The enzymes shown in the figure correspond to the following EC numbers: propionate CoA ligase = [EC 6.2.1.17]; medium chain acyl-CoA dehydrogenase = [EC 1.3.8.7]; 3-hydroxypropionyl-CoA dehydratase = [EC 4.2.1.116]; 3-hydroxypropionyl-CoA hydrolase = [EC 3.1.2.4]; 3-hydroxypropionate dehydrogenase = [EC 1.1.1.59], and β-alanine-pyruvate transaminase = [EC 2.6.1.18].
The propionate pathway connects the metabolism of L-alanine and β-alanine in plants, since the final step requires the former amino acid as the amino donor (Figure 3). Therefore, a change in the levels of one or the other of the two amino acids may have wide-ranging physiological effects. Gene regulation studies of alanine aminotransferase has been studied in many plants in response to low-oxygen stress, light, and nitrogen, and it was demonstrated that hypoxia induced the expression of two distinct alanine aminotransferase genes in A. thaliana (Miyashita et al., 2007). The function of the gene product from the locus tag At3g08860 was unknown for a long time, but was recently shown to encode a β-alanine/L-alanine aminotransferase using an in vivo functional complementation approach (Parthasarathy et al., 2019). The same study reported that a definitive preference of the corresponding enzymatic activity toward the synthesis of β-alanine. The gene corresponding to the β-alanine/L-alanine aminotransferase was shown to be most highly expressed in A. thaliana roots (Schmid et al., 2005). The enrichment of the β-alanine aminotransferase enzyme in the roots suggests that β-alanine possibly has a specific protective function in the root tissue. Earlier, hydroponics studies in Helianthus annuus L. revealed that roots which were excised and flooded for 24 h had reduced uptake of radioactive β-alanine than non-flooded roots and also that they exuded more radioactive β-alanine into distilled water (Drakeford et al., 1985). This suggests, first, that there is a regulation of β-alanine uptake (if available in the medium), and second, that flooded roots may exude β-alanine for protective purposes. Therefore, it is likely that plants require enhanced expression of the β-alanine aminotransferase in the roots to facilitate a β-alanine based defense response when faced with hypoxia or flooding.
Additional studies have addressed the links between branched-chain amino acids and β-alanine, since valine and isoleucine degradation can produce propionyl-CoA. Using radiolabeled precursors of both isoleucine and valine, it was shown in A. thaliana that only isoleucine directly generated β-alanine. However, if seedlings were treated with valine, an increase in the levels of β-alanine resulted, suggesting an indirect effect via amino acid homeostasis (Perrett et al., 2017). Seedlings harboring a mutated methylmalonate semialdehyde dehydrogenase (MMSD, At2g14170) gene cannot convert valine into propionyl-CoA (since the Mmsd-1 protein catalyzes the final step leading to propionyl-CoA); but the equilibrium is shifted toward β-alanine, presumably by the amino acid homeostatic mechanism (Perrett et al., 2017). More recently, feeding of 13C-labeled isoleucine and propionate were shown to result in the production of C13-labeled 3-hydroxypropionate and β-alanine in A. thaliana seedlings, offering direct evidence for the metabolic link between isoleucine and β-alanine via propionyl-CoA (Goldfarb and Rouhier, 2019). Further studies in transgenic A. thaliana and wheat seedlings confirmed that isoleucine degradation could initiate β-alanine synthesis (Rouhier et al., 2019). This suggests that isoleucine may serve as an additional precursor for β-alanine in plants (apart from those known already) and since valine is a precursor of pantoate, pantothenate production could depend on BCAA catabolism.
The Synthesis of β-Alanine in Plants: The Uracil Pathway
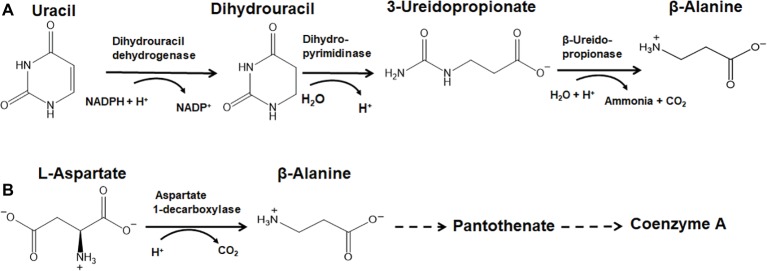
The nucleotide base uracil is reduced by NADPH-dependent dihydrouracil dehydrogenase [EC 1.3.1.2] into dihydrouracil (Figure 4A); in A. thaliana, this gene is identified with the At3g17810 locus. The hydrolysis of dihydrouracil into 3-ureidopropionate is catalyzed by dihydropyrimidinase [EC 3.5.2.2] (Figure 4A); the accession IDs in A. thaliana are At5g12200 and At5g12200.1. Further hydrolysis by β-ureidopropionase [EC 3.5.1.6] eliminates ammonia and carbon dioxide, yielding β-alanine as shown in Figure 4A (Campbell, 1960; Traut and Loechel, 1984); the A. thaliana accession IDs are At5g64370 and At5g64370.1. The degradation of uracil and thymine to produce carbon dioxide, ammonia, β-alanine, and γ-aminoisobutyrate has been shown to occur in seedlings of Brassica napus (rapeseed), while the β-ureidopropionase enzyme of maize was characterized by its overexpression in Escherichia coli (Tsai and Axelrod, 1965; Walsh et al., 2001). The existence of this pathway in pine trees was inferred already in the 1960s (Barnes and Naylor, 1962). In the halotolerant members of the leadwort or plumbago family (Plumbaginaceae) of perennial plants, uracil is only one of many sources for β-alanine (Duhazé et al., 2003). Thus, the distribution of this pathway in higher plants varies widely.
Figure 4.
The anabolism of β-Alanine biosynthesis via uracil and L-aspartate. (A) The nucleotide base uracil can be degraded to β-alanine. The enzymes in this pathway are dihydrouracil dehydrogenase [EC 1.3.1.2], dihydropyrimidinase [EC 3.5.2.2] and β-ureidopropionase [EC 3.5.1.6]. (B) Aspartate 1-decarboxylase [EC 4.1.1.11] can directly decarboxylate L-aspartate into β-alanine, which may feed into Coenzyme A biosynthesis via pantothenate.
The Synthesis of β-Alanine in Plants: The Aspartate Pathway
L-aspartate may be decarboxylated by aspartate 1-decarboxylase [EC 4.1.1.11] also yielding β-alanine, which may enter Coenzyme A biosynthesis (Figure 4B). This enzyme abbreviated ADC and encoded by the panD gene has been well characterized in prokaryotes (Williamson and Brown, 1979; Merkel and Nichols, 1996). However, its presence in plants has never been identified and or confirmed. Bioinformatics analysis based on sequence homology using the FUGUE tool failed to find homologs in either A. thaliana or the yeast Saccharomyces cerevisiae genomes, casting doubt on whether this pathway crossed the divide between prokaryotes and eukaryotes (Shi et al., 2001; Ottenhof et al., 2004). With the availability of numerous genome sequences of plants and fungi available since 2004, further inquiry of panD orthologs using bioinformatics tools is warranted.
Since only plants and microbes synthesize pantothenate de novo, there is significant interest in engineering this pathway in plants in order to fortify food crops with pantothenate, which is essential for animals since they lack the enzymes required to biosynthesize the compound (Coxon et al., 2005; Chakauya et al., 2008). Also, since only plants and microbes contain the pathway for pantothenate, it is also considered an attractive target for herbicides, fungicides and antibiotic development (Coxon et al., 2005). Increased heat tolerance via overproduction of β-alanine and pantothenate content could be achieved in transgenic tobacco plants harboring the gene for the E. coli ADC enzyme (Fouad and Rathinasabapathi, 2006). Later, the same system was also shown to enhance photosynthesis and to augment biomass production in response to higher temperatures (Fouad and Altpeter, 2009). These are important practical advances that could pave the way for heat resistant transgenic food crops that are better suited in warmer climate zones and in changing climates, where loss in agricultural productivity with increasing temperatures might be a concern.
The Utilization of β-Alanine in Other Plant Pathways
β-Alanine Betaine
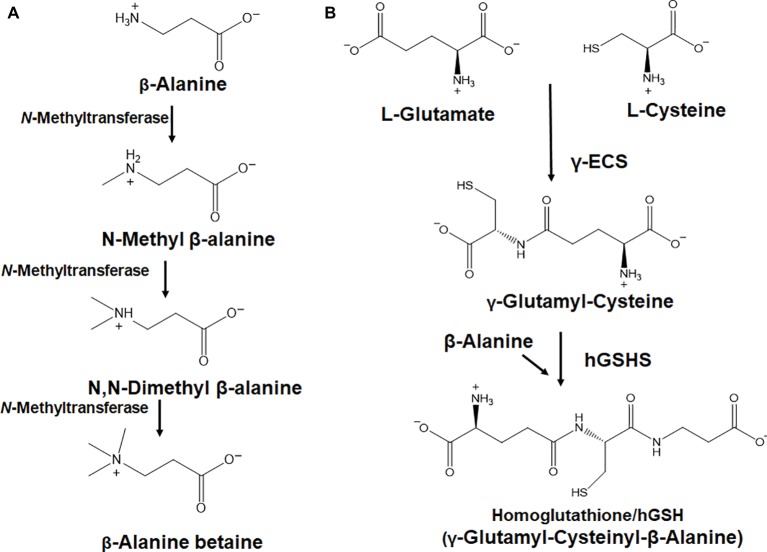
β-Alanine betaine is an osmoprotective compound accumulated by most members of the highly stress-tolerant leadwort or plumbago family (Plumbaginaceae). β-Alanine betaine is synthesized by S-adenosyl-L-methionine (SAM)-dependent enzymatic N-methylation of β-alanine (Figure 5A) via N-methyl β-alanine and N,N-dimethyl β-alanine (Raman and Rathinasabapathi, 2003). Most members of the highly stress-tolerant Plumbaginaceae accumulate β-alanine betaine instead of glycine betaine (Hanson et al., 1991, 1994). It was proposed that β-alanine betaine is a more suitable osmoprotectant than glycine betaine under saline hypoxic conditions because the first step in glycine betaine synthesis catalyzed by choline monooxygenase [EC 1.14.15.7] requires molecular oxygen (Hanson et al., 1991, 1994). Further, β-alanine betaine accumulation was proposed to be an evolutionary strategy to avoid the metabolic limitations existing for choline oxidation under hypoxic conditions, because β-alanine betaine is synthesized from the ubiquitous primary metabolite β-alanine (Hanson et al., 1994).
Figure 5.
The conversion of β-alanine to important secondary metabolites. (A) A trifunctional, S-adenosyl-L-methionine (SAM)-dependent N-methyltransferase [EC 2.1.1.49], performs iterative N-methylation of β-alanine to β-alanine betaine. (B) Homoglutathione biosynthesis involves the enzymes, glutamate cysteine ligase (GCL) or γ-ECS (γ-glutamyl-cysteine synthetase) [EC 6.3.2.2] and a β-alanine specific homoglutathione synthase (hGSHS) [EC 6.3.2.23], both of which perform specific amino acid condensations at the expense of ATP.
Homoglutathione
In many legumes, the thiol tripeptide homoglutathione (hGSH; γGlu-Cys-βAla) (Figure 5B) can partially or fully replace the better-known thiol, glutathione. γ-Glu-Cys is formed from L-glutamate and L-cysteine by glutamate cysteine ligase (γ-Glutamylcysteine synthetase or γ-ECS) at the expense of ATP, and γ-Glu-Cys and β-alanine are condensed at the expense of ATP by a specific hGSH synthetase (hGSHS), an enzyme which has affinity for β-alanine and low affinity for glycine (Macnicol, 1987; Klapheck, 1988; Klapheck et al., 1988). Multiple cellular compartments are involved in the biosynthesis of hGSH and fractionation of the root nodules demonstrated that the bacteroids contain high hGSH concentrations and the highest specific activities of GSHS, suggesting a critical role in nitrogen fixation processes in the root nodules of legume plants (Moran et al., 2000). More recent work in M. truncatula also showed that hGSH is essential for the growth of parasitic nematode worms, which infect plant roots and force the differentiation of root cells into giant cells. In addition, it was also shown that hGSH-depleted roots did not suffer similar damage by nematodes (Baldacci-Cresp et al., 2012). Thus, hGSH may play key roles in plant defense and could be the target of future interventions into improving plant resistance to nematodes.
Conclusions
Although underappreciated, the non-proteinogenic amino acid β-alanine has important roles in plant physiology and metabolism, directly as a defense compound that enables plants to withstand various stresses such as hypoxia, waterlogging and drought, and indirectly as a precursor to the compounds pantothenate and CoA, which are involved in a variety of functions. Furthermore, the amino acid is converted into β-alanine betaine, which has additional protective functions such as salt tolerance, and homoglutathione, which may be critical for nitrogen fixation.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge the College of Science and the Thomas H. Gosnell School of Life Sciences at the Rochester Institute of Technology for ongoing support.
Footnotes
Funding. This research was supported by a United States National Science Foundation (NSF) award (MCB-#1120541) to AH.
References
- Baldacci-Cresp F., Chang C., Maucourt M., Deborde C., Hopkins J., Lecomte P., et al. (2012). (Homo)glutathione deficiency impairs root-knot nematode development in Medicago truncatula. PLoS Pathog. 8:e1002471. 10.1371/journal.ppat.1002471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes R. L., Naylor A. W. (1962). Formation of beta-alanine by pine tissues supplied with intermediates in uracil & orotic acid metabolism. Plant Physiol. 37, 171–175. 10.1104/pp.37.2.171, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancquaert L., Everaert I., Missinne M., Baguet A., Stegen S., Volkaert A., et al. (2017). Effects of histidine and β-alanine supplementation on human muscle carnosine storage. Med. Sci. Sports Exerc. 49, 602–609. 10.1249/MSS.0000000000001213, PMID: [DOI] [PubMed] [Google Scholar]
- Broeckling C. D., Huhman D. V., Farag M. A., Smith J. T., May G. D., Mendes P., et al. (2005). Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism. J. Exp. Bot. 56, 323–336. 10.1093/jxb/eri058, PMID: [DOI] [PubMed] [Google Scholar]
- Campbell L. L. (1960). Reductive degradation of pyrimidines 5. Enzymatic conversion of N-carbamyl-beta-alanine to beta-alanine, carbon dioxide, and ammonia. J. Biol. Chem. 235, 2375–2378. PMID: [PubMed] [Google Scholar]
- Chakauya E., Coxon K. M., Wei M., Macdonald M. V., Barsby T., Abell C., et al. (2008). Towards engineering increased pantothenate (vitamin B5) levels in plants. Plant Mol. Biol. 68, 493–503. 10.1007/s11103-008-9386-5, PMID: [DOI] [PubMed] [Google Scholar]
- Coxon K. M., Chakauya E., Ottenhof H. H., Whitney H. M., Blundell T. L., Abell C., et al. (2005). Pantothenate biosynthesis in higher plants. Biochem. Soc. Trans. 33, 743–746. 10.1042/BST0330743, PMID: [DOI] [PubMed] [Google Scholar]
- Drakeford D. R., Mukherjee I., Reid D. M. (1985). Some early responses of Helianthus annuus L. to flooding: I. The effects of flooding on the uptake and leakage of ‘non-electrolytes’ by roots. J. Exp. Bot. 36, 1705–1715. 10.1093/jxb/36.11.1705 [DOI] [Google Scholar]
- Duhazé C., Gagneul D., Leport L., Larher F., Bouchereau A. (2003). Uracil as one of the multiple sources of β-alanine in Limonium latifolium a halotolerant β-alanine betaine accumulating Plumbaginaceae. Plant Physiol. Biochem. 41, 993–998. 10.1016/j.plaphy.2003.06.002 [DOI] [Google Scholar]
- Fouad W. M., Altpeter F. (2009). Transplastomic expression of bacterial L-aspartate-alpha-decarboxylase enhances photosynthesis and biomass production in response to high temperature stress. Transgenic Res. 18, 707–718. 10.1007/s11248-009-9258-z, PMID: [DOI] [PubMed] [Google Scholar]
- Fouad W. M., Rathinasabapathi B. (2006). Expression of bacterial L-aspartate-alpha-decarboxylase in tobacco increases beta-alanine and pantothenate levels and improves thermotolerance. Plant Mol. Biol. 60, 495–505. 10.1007/s11103-005-4844-9, PMID: [DOI] [PubMed] [Google Scholar]
- Galston A. W., Sawhney R. K. (1990). Polyamines in plant physiology. Plant Physiol. 94, 406–410. 10.1104/pp.94.2.406, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschel U., Powell C. A., Abell C., Smith A. G. (1999). The final step of pantothenate biosynthesis in higher plants: cloning and characterization of pantothenate synthetase from Lotus japonicus and Oryza sativum (rice). Biochem. J. 341, 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb M. H., Rouhier K. A. (2019). Evidence for the accumulation of 3-hydroxypropionate and β-alanine in Arabidopsis thaliana seedlings via isoleucine degradation. The FASEB J. Biochem. Mol. Biol. 33(Suppl. 1). 10.1096/fasebj.2019.33.1_supplement.486.3 [DOI] [Google Scholar]
- Hanson A. D., Rathinasabapathi B., Chamberlin B., Gage D. A. (1991). Comparative physiological evidence that beta-alanine betaine and choline-o-sulfate act as compatible osmolytes in halophytic limonium species. Plant Physiol. 97, 1199–1205. 10.1104/pp.97.3.1199, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson A. D., Rathinasabapathi B., Rivoal J., Burnet M., Dillon M. O., Gage D. A. (1994). Osmoprotective compounds in the Plumbaginaceae: a natural experiment in metabolic engineering of stress tolerance. Proc. Natl. Acad. Sci. USA 91, 306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayaishi O., Nishizuka Y., Tatibana M., Takeshita M., Kuno S. (1961). Enzymatic studies on the metabolism of beta-alanine. J. Biol. Chem. 236, 781–790. PMID: [PubMed] [Google Scholar]
- Imai A., Matsuyama T., Hanzawa Y., Akiyama T., Tamaoki M., Saji H., et al. (2004). Spermidine synthase genes are essential for survival of Arabidopsis. Plant Physiol. 135, 1565–1573. 10.1104/pp.104.041699, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada-Nobusada T., Hayashi M., Fukazawa M., Sakakibara H., Nishimura M. (2008). A putative peroxisomal polyamine oxidase, AtPAO4, is involved in polyamine catabolism in Arabidopsis thaliana. Plant Cell Physiol. 49, 1272–1282. 10.1093/pcp/pcn114, PMID: [DOI] [PubMed] [Google Scholar]
- Kaplan F., Kopka J., Haskell D. W., Zhao W., Schiller K. C., Gatzke N., et al. (2004). Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 136, 4159–4168. 10.1104/pp.104.052142, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapheck S. (1988). Homoglutathione: isolation, quantification, and occurrence in legumes. Physiol. Plant. 74, 727–732. 10.1111/j.1399-3054.1988.tb02044.x [DOI] [Google Scholar]
- Klapheck S., Zopes H., Levels H., Bergmann L. (1988). Properties and localization of the homoglutathione synthetase from Phaseolus coccineus leaves. Physiol. Plant. 74, 733–739. 10.1111/j.1399-3054.1988.tb02045.x [DOI] [Google Scholar]
- Liepman A. H., Olsen L. J. (2001). Peroxisomal alanine: glyoxylate aminotransferase (AGT1) is a photorespiratory enzyme with multiple substrates in Arabidopsis thaliana. Plant J. 25, 487–498. 10.1046/j.1365-313x.2001.00961.x, PMID: [DOI] [PubMed] [Google Scholar]
- Macnicol P. (1987). Homoglutathione and glutathione synthetases of legume seedlings: partial purification and substrate specificity. Plant Sci. 53, 229–235. 10.1016/0168-9452(87)90159-2 [DOI] [Google Scholar]
- Mayer R. R., Cherry J. H., Rhodes D. (1990). Effects of heat shock on amino acid metabolism of cowpea cells. Plant Physiol. 94, 796–810. 10.1104/pp.94.2.796, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A. D., Seidler N. W. (2005). Beta-alanine suppresses heat inactivation of lactate dehydrogenase. J. Enzyme Inhib. Med. Chem. 20, 199–203. 10.1080/14756360400020538 [DOI] [PubMed] [Google Scholar]
- Merkel W. K., Nichols B. P. (1996). Characterization and sequence of the Escherichia coli panBCD gene cluster. FEMS Microbiol. Lett. 143, 247–252. 10.1111/j.1574-6968.1996.tb08488.x, PMID: [DOI] [PubMed] [Google Scholar]
- Miyashita Y., Dolferus R., Ismond K. P., Good A. G. (2007). Alanine aminotransferase catalyses the breakdown of alanine after hypoxia in Arabidopsis thaliana. Plant J. 49, 1108–1121. 10.1111/j.1365-313X.2006.03023.x, PMID: [DOI] [PubMed] [Google Scholar]
- Moran J. F., Iturbe-Ormaetxe I., Matamoros M. A., Rubio M. C., Clemente M. R., Brewin N. J., et al. (2000). Glutathione and homoglutathione synthetases of legume nodules. Cloning, expression, and subcellular localization. Plant Physiol. 124, 1381–1392. 10.1104/pp.124.3.1381, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschou P. N., Paschalidis K. A., Delis I. D., Andriopoulou A. H., Lagiotis G. D., Yakoumakis D. I., et al. (2008a). Spermidine exodus and oxidation in the apoplast induced by abiotic stress is responsible for H2O2 signatures that direct tolerance responses in tobacco. Plant Cell 20, 1708–1724. 10.1105/tpc.108.059733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschou P. N., Sanmartin M., Andriopoulou A. H., Rojo E., Sanchez-Serrano J. J., Roubelakis-Angelakis K. A. (2008b). Bridging the gap between plant and mammalian polyamine catabolism: a novel peroxisomal polyamine oxidase responsible for a full back-conversion pathway in Arabidopsis. Plant Physiol. 147, 1845–1857. 10.1104/pp.108.123802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenhof H. H., Ashurst J., Whitney H., Saldanha S., Schmitzberger F., Gweon H., et al. (2004). Organisation of the pantothenate (vitamin B5) biosynthesis pathway in higher plants. Plant J. 37, 61–72. 10.1046/j.1365-313X.2003.01940.x, PMID: [DOI] [PubMed] [Google Scholar]
- Parthasarathy A., Savka F. C., Hudson A. O. (2019). The gene annotated by the locus tag At3g08860 encodes a β-alanine/L-alanine aminotransferase in Arabidopsis thaliana. Bioarxiv [Preprint]. 576041. 10.1101/576041. (Accessed March 21, 2019). [DOI] [PMC free article] [PubMed]
- Perrett M., Gothard M., Ludwig A., Rouhier K. A. (2017). Identifying a source of beta-alanine and its broader implications in Arabidopsis thaliana by GC/MS. The FASEB J. Biochem. Mol. Biol. 31(Suppl. 1). 10.1096/fasebj.31.1_supplement.626.1 [DOI] [Google Scholar]
- Raman S. B., Rathinasabapathi B. (2003). beta-Alanine N-methyltransferase of Limonium latifolium. cDNA cloning and functional expression of a novel N-methyltransferase implicated in the synthesis of the osmoprotectant beta-alanine betaine. Plant Physiol. 132, 1642–1651. 10.1104/pp.103.020453, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi R., Davies P. (1989). Polyamine metabolism in ripening tomato fruit. Plant Physiol. 94, 1449–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L., Liang H., Shuman J., Shulaev V., Davletova S., Mittler R. (2004). When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 134, 1683–1696. 10.1104/pp.103.033431, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha M., Licausi F., Araújo W. L., Nunes-Nesi A., Sodek L., Fernie A. R., et al. (2010a). Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol. 152, 1501–1513. 10.1104/pp.109.150045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha M., Sodek L., Licausi F., Hameed M. W., Dornelas M. C., van Dongen J. T. (2010b). Analysis of alanine aminotransferase in various organs of soybean (Glycine max) and in dependence of different nitrogen fertilisers during hypoxic stress. Amino Acids 39, 1043–1053. 10.1007/s00726-010-0596-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier K. A., Perrett M. E., Goldfarb M. H., Reinhart P. E., Wilczewski-Shirai K. C., Raymond-Schmidt L., et al. (2019). Isoleucine serves as a precursor for the synthesis of β-alanine in plants. The FASEB J. Biochem. Mol. Biol. 33(Suppl. 1). 10.1096/fasebj.2019.33.1_supplement.486.10 [DOI] [Google Scholar]
- Schmid M., Davison T. S., Henz S. R., Pape U. J., Demar M., Vingron M., et al. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506. 10.1038/ng1543, PMID: [DOI] [PubMed] [Google Scholar]
- Shi J., Blundell T. L., Mizuguchi K. (2001). FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310, 243–257. 10.1006/jmbi.2001.4762, PMID: [DOI] [PubMed] [Google Scholar]
- Stinson R. A., Spencer M. S. (1969a). beta-Alanine as an ethylene precursor. Investigations towards preparation, and properties, of a soluble enzyme system from a subcellular particulate fraction of bean cotyledons. Plant Physiol. 44, 1217–1226. 10.1104/pp.44.9.1217, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinson R. A., Spencer M. S. (1969b). beta Alanine aminotransferase(s) from a plant source. Biochem. Biophys. Res. Commun. 34, 120–127. 10.1016/0006-291X(69)90537-3, PMID: [DOI] [PubMed] [Google Scholar]
- Sun X., Zhang J., Zhang H., Ni Y., Zhang Q., Chen J., et al. (2010). The responses of Arabidopsis thaliana to cadmium exposure explored via metabolite profiling. Chemosphere 78, 840–845. 10.1016/j.chemosphere.2009.11.045, PMID: [DOI] [PubMed] [Google Scholar]
- Terano S., Suzuki Y. (1978). Formation of β-alanine from spermine and spermidine in maize shoots. Phytochemistry 17, 148–149. 10.1016/S0031-9422(00)89700-9 [DOI] [Google Scholar]
- Traut T. W., Loechel S. (1984). Pyrimidine catabolism: individual characterization of the three sequential enzymes with a new assay. Biochemistry 23, 2533–2539. [DOI] [PubMed] [Google Scholar]
- Tsai C. S., Axelrod B. (1965). Catabolism of pyrimidines in rape seedlings. Plant Physiol. 40, 39–44. 10.1104/pp.40.1.39, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen J. T., Fröhlich A., Ramírez-Aguilar S. J., Schauer N., Fernie A. R., Erban A., et al. (2009). Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of Arabidopsis plants. Ann. Bot. 103, 269–280. 10.1093/aob/mcn126, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voet D., Voet J., Pratt C. (2006). Fundamentals of biochemistry: Life at the molecular level. 2nd Edn New York: Wiley, ISBN 9780471214953 471214957. [Google Scholar]
- Walsh T. A., Green S. B., Larrinua I. M., Schmitzer P. R. (2001). Characterization of plant beta-ureidopropionase and functional overexpression in Escherichia coli. Plant Physiol. 125, 1001–1011. 10.1104/pp.125.2.1001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb M. E., Smith A. G. (2011). Pantothenate biosynthesis in higher plants. Adv. Bot. Res. 58, 203–255. 10.1042/BST0330743 [DOI] [Google Scholar]
- Webb M. E., Smith A. G., Abell C. (2004). Biosynthesis of pantothenate. Nat. Prod. Rep. 21, 695–721. 10.1039/b316419p, PMID: [DOI] [PubMed] [Google Scholar]
- Williamson J. M., Brown G. M. (1979). Purification and properties of L-Aspartate-alpha-decarboxylase, an enzyme that catalyzes the formation of beta-alanine in Escherichia coli. J. Biol. Chem. 254, 8074–8082. PMID: [PubMed] [Google Scholar]
- Zarei A., Trobacher C. P., Shelp B. J. (2015). NAD(+)-aminoaldehyde dehydrogenase candidates for 4-aminobutyrate (GABA) and β-alanine production during terminal oxidation of polyamines in apple fruit. FEBS Lett. 589, 2695–2700. 10.1016/j.febslet.2015.08.005, PMID: [DOI] [PubMed] [Google Scholar]
- Zarei A., Trobacher C. P., Shelp B. J. (2016). Arabidopsis aldehyde dehydrogenase 10 family members confer salt tolerance through putrescine-derived 4-aminobutyrate (GABA) production. Sci. Rep. 6:35115. 10.1038/srep35115, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]