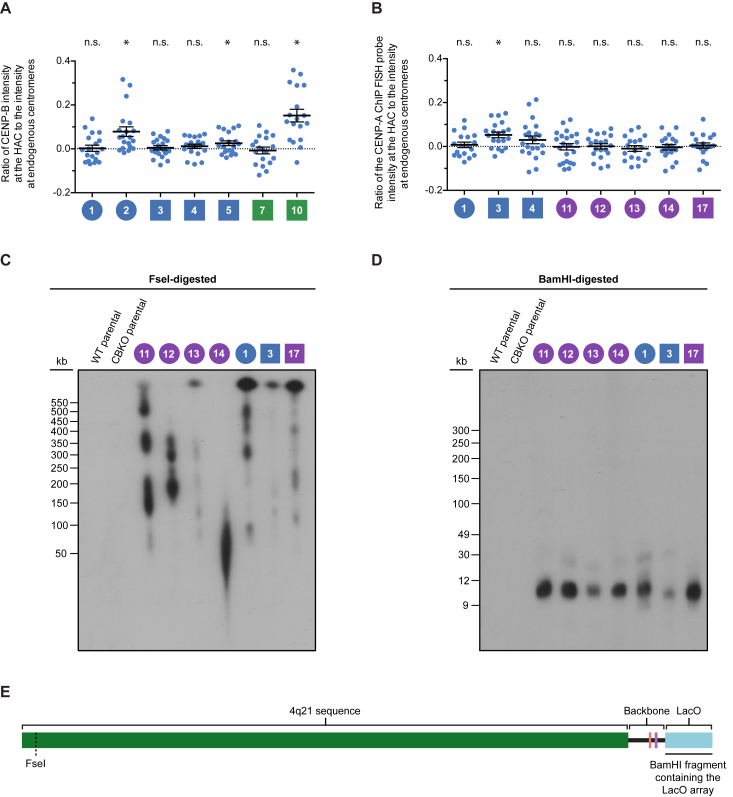
Figure S3.
Analysis of the Centromeric Protein and Sequence Abundance, as well as the Organization, of Various 4q21 BACLacO HAC Clones, Related to Figure 4
A,B) Plot with an expanded y axis of the ratio of CENP-B (A) and CENP-A ChIP FISH probe (B) intensity at the HAC relative to endogenous centromeres for clones with a mean below 0.2; related to Figures 4B and 4D. Clones with a p value < 0.05 are marked with an asterisk; clones with a p value ≥ 0.05 are marked as not significant (n.s.).
C,D) Southern blot analysis of the indicated cell lines showing variable sequence organization within the 4q21 HACs. Genomic DNA from each cell line was digested with the indicated restriction enzyme, separated by pulsed-field gel electrophoresis, transferred to a membrane, and hybridized with a LacO-specific probe. The FseI restriction enzyme digests the 4q21 BACLacO sequence one time; therefore, if the HAC had undergone a simple amplification of the 4q21 BACLacO sequence, multiples of a 203 kb band should be observed. However, we observed varying band sizes (C), indicating that each HAC had undergone structural rearrangements during HAC formation, which has been previously observed with α-satellite HACs (Kouprina et al., 2012). In all HACs assessed, the LacO array was largely intact (D), indicating that the rearrangements occurred in the 4q21 and backbone sequences within each HAC and not within the LacO array.
(E) Restriction enzyme map of the FseI cut site and the fragment produced by BamHI enzyme digestion of the 4q21 BACLacO construct. BamHI cuts 26 other times throughout the 4q21 sequence and backbone (not shown), but these fragments are largely not detected by the LacO-specific probe (as shown in Panel D).