Abstract
Proactive identification of chemicals with skin sensitizing properties is a key toxicological endpoint within chemical safety assessment, as required by legislation for registration of chemicals. In order to meet demands of increased animal welfare and facilitate increased testing efficiency also in nonregulatory settings, considerable efforts have been made to develop nonanimal approaches to replace current animal testing. Genomic Allergen Rapid Detection (GARD™) is a state-of-the-art technology platform, the most advanced application of which is the assay for assessment of skin sensitizing chemicals, GARD™skin. The methodology is based on a dendritic cell (DC)-like cell line, thus mimicking the mechanistic events leading to initiation and modulation of downstream immunological responses. Induced transcriptional changes are measured following exposure to test chemicals, providing a detailed evaluation of cell activation. These changes are associated with the immunological decision-making role of DCs in vivo and include among other phenotypic modifications, up-regulation of co-stimulatory molecules, induction of cellular and oxidative stress pathways and xenobiotic responses, and provide a holistic readout of substance-induced DC activation. Here, results from an inter-laboratory ring trial of GARD™skin, conducted in compliance with OECD guidance documents and comprising a blinded chemical test set of 28 chemicals, are summarized. The assay was found to be transferable to naïve laboratories, with an inter-laboratory reproducibility of 92.0%. The within-laboratory reproducibility ranged between 82.1% and 88.9%, whereas the cumulative predictive accuracy across the 3 laboratories was 93.8%. It was concluded that GARD™skin is a robust and reliable method for the identification of skin sensitizing chemicals and suitable for stand-alone use or as a constituent of integrated testing. These data form the basis for the regulatory validation of GARD™skin.
Keywords: GARD™, GARD™skin, in vitro, sensitization, chemical sensitizers
Skin sensitization resulting in allergic contact dermatitis (ACD) is a common occupational and environmental health problem. Topical exposure to chemical allergens triggers an adaptive immune response that causes immunological priming resulting in the acquisition of skin sensitization. The development of sensitization is dependent upon the stimulation of T lymphocyte responses. If the sensitized subject is exposed again to the same chemical allergen, a heightened response will be triggered resulting in ACD.
There is a general agreement regarding the key biological events underlying skin sensitization, which have been continuously and extensively reviewed (Adler et al., 2011; Ainscough et al., 2013; Kimber et al., 2011; Martin, 2015; Martin et al., 2011). The existing knowledge of the chemical and biological mechanisms associated with skin sensitization has been summarized in the form of an Adverse Outcome Pathway (AOP) (OECD, 2014). The AOP describes the sensitization process by the definition of 4 key events (KE). In order, KEs include: (1) the covalent binding of a sensitizing chemical to endogenous proteins, a process also referred to as haptenization, (2) the induction of inflammatory responses in keratinocytes, (3) the activation of dendritic cells (DC), allowing for cell migration, antigen presentation, and attenuation of downstream immunological pathways, thus bridging the innate and specific immunity of the host, and (4) the priming and subsequent proliferation of specific T cells.
Assessment of skin sensitizing activity of chemicals initially required the use of laboratory animals. The classical methods based on guinea pigs, the Guinea Pig Maximization Test (GPMT) (Magnusson and Kligman, 1969) and the Buehler Occluded Patch Test (Test Guideline [TG] 406) (OECD, 1992), study both the induction and elicitation phases of skin sensitization. A murine test, the Local Lymph Node Assay (LLNA, TG 429) (OECD, 2010) has gained acceptance as gold standard, because it provide advantages over the guinea pig tests in terms of animal welfare and predictive performance, whereas also having the ability to evaluate the relative potency of skin sensitizing chemicals, measured as a function of the lowest concentration of chemical required to trigger a certain level of T cell activation. However, driven by legislation (EC, 2006; EU, 1976), public opinion, and economic interests, nonanimal alternatives have become mandatory in predictive toxicology in general, and for skin sensitization testing in particular.
Recently, mechanistically based in chemico and in vitro test methods have been described and validated. These methods include the Direct Peptide Reactivity Assay (DPRA) (Gerberick et al., 2004), the Keratinosens assay (Natsch, 2010), and the h-CLAT assay (Ashikaga et al., 2006), addressing the first, second, and third KE of the AOP, respectively. These methods are currently available as OECD test guidelines TG 442C (OECD, 2015), TG 442D, which also include the LuSens assay (OECD, 2018a) and TG 442E, which also include the U-SENS and IL-8 Luc assays (OECD, 2018b). In addition, a TG is currently being drafted for the Sens-IS assay (Cottrez et al., 2015).
However, information generated with the above-mentioned methods are not regarded as being sufficiently predictive for use as stand-alone tests for the identification of skin sensitization hazards and the current view is that more than 1 KE has to be monitored for safe hazard assessment. Rather, Defined Approaches (DA) in the context of Integrated Approaches to Testing and Assessment (IATA) are proposed as currently being the best paradigm for nonanimal testing (Casati et al., 2018; Hartung et al., 2013). Furthermore, none of the methods presently embraced by OECD guidelines are considered to allow potency-associated subclassification of sensitizers into categories 1A and 1B as defined by UN GHS (UN, 2009). For these reasons, the need for complementary in vitro assays to assist in these predictions remains unfulfilled, and the room for improvement in terms of predictive accuracy in next-generation methodologies remains substantial.
The Genomic Allergen Rapid Detection (GARD™) assay for assessment of chemical skin sensitizers (GARD™skin), was conceived by whole genome analysis of human DC-like cells following chemical exposure. A predictive genomic biomarker signature, comprising genes associated with, eg, xenobiotic recognition, antigen presentation and co-stimulation, and induction of cellular and oxidative stress pathways, was established (Johansson et al., 2011). The methodology has since been transferred from a whole genome technological platform to the Nanostring nCounter system (Geiss et al., 2008), a resource-effective gene expression readout system well suited for streamlined standard operating procedures (SOP) and widespread implementation (Forreryd et al., 2014, 2016). Furthermore, the GARD™skin assay has been shown, in a series of publications and industry-sponsored collaborations, to provide high levels of predictive accuracy (Forreryd et al., 2016; Johansson et al., 2014, 2017).
Here, we describe the validation study of GARD™skin, based on an inter-laboratory ring trial, and report performance parameters of the method, including transferability, reproducibility, and predictive capacity. This publication constitutes a summary of the validation report that has been submitted to ECVAM and is currently in review of validating bodies.
MATERIALS AND METHODS
Participating organizations and structure of the study
The study was initiated by SenzaGen AB (Lund, Sweden), which acted as lead laboratory throughout the validation process. 3RsMC (Lyngby, Denmark) acted as validation manager, and formed a validation management group (VMG) with Adriaens Consulting (Aalter, Belgium) and Triskelion (Zeist, Netherlands). The overall purpose of the VMG was to define, guide, facilitate, and evaluate the validation process, although ensuring compliance with the EURL-ECVAM modular approach to validation (Hartung et al., 2004) and with the OECD Guidance documents (GD) on the validation and international acceptance of new or updated test methods for hazard assessment, as defined in OECD GD Nos 1 and 34 (OECD, 2005, 2009). Two naïve laboratories, Burleson Research Technologies (BRT, Morrisville, North Carolina), and Eurofins BioPharma Product Testing Munich GmbH (Eurofins, Planegg, Germany), were recruited. In addition, the 2 naïve laboratories were assisted in application of the technical readout system by Covance Genomics Laboratory (Redmond, Western Australia), and KIGene (Karolinska University Hospital, Stockholm, Sweden), respectively.
The study was composed of a training phase, a transfer phase, and a validation phase. During the training phase, naïve laboratories were given hands-on training in all aspects of the GARD™skin SOP. Initial training was followed by an opportunity to provide feedback on the SOP, which was incorporated in a revised document. Following training, the transfer phase was performed using a set of nonblinded chemicals, giving the naïve laboratories the opportunity to demonstrate proficiency in all aspects of the protocols. Once the validation phase commenced, the revised SOP was locked for any further changes, all communication between the lead and naïve laboratories ceased and each participating laboratory individually assessed a larger set of blinded chemicals. Each laboratory performed 3 independent experiments, in which all blinded test chemicals were assayed. The coding of test chemicals was unique for each participating laboratory, and for every independent experiment within each laboratory. All data were reported to the VMG, which evaluated and summarized the results which form the basis of this report. The true identities of the test chemicals were not decoded until after all data had been generated and submitted to the VMG.
Test chemicals
The set of chemicals used during the transfer phase is listed in Table 1. Chemicals used during training and transfer phases were chosen by the lead laboratory. For the validation phase, all chemical selection was done by the VMG, without any knowledge or influence from the participating laboratories. Chemical selection was based on predefined criteria relating to chemical reactivity, sensitizing potency diversity and solubility diversity, and the goal to minimize inclusion of chemicals previously assayed in GARD™skin, while maintaining a diverse and balanced chemical subset with enough data availability from clinical, in vivo and in vitro sources. The chemicals selected for inclusion in the blinded validation phase are presented in Table 2. All chemicals used during training, transfer, and validation phases were purchased from Sigma Aldrich (St. Louis, Missouri) and in the case of the validation phase, distributed and randomly coded by the VMG.
Table 1.
Transfer Phase Chemical Set
| Chemical Name | CAS No. | True Group |
|---|---|---|
| 2,4-Dinitrochlorobenzene | 97-00-7 | Sensitizer |
| Resorcinol | 108-46-3 | Sensitizer |
| Geraniol | 106-24-1 | Sensitizer |
| 1-Butanol | 71-36-3 | Nonsensitizer |
| Chlorobenzene | 108-90-7 | Nonsensitizer |
Table 2.
Validation Phase Chemical Set
| Chemical Name | CAS No. | True Group |
|---|---|---|
| 4-Nitrobenzyl bromide | 100-11-8 | Sensitizer |
| 2-Bromo-2-glutaronitrile | 35691-65-7 | Sensitizer |
| Cinnamal | 104-55-2 | Sensitizer |
| Formaldehyde | 50-00-0 | Sensitizer |
| Lauryl gallate | 1166-52-5 | Sensitizer |
| 4-(Methylamino)phenol sulfate | 55-55-0 | Sensitizer |
| Methylisothiazolinone | 2682-20-4 | Sensitizer |
| Propyl gallate | 121-79-9 | Sensitizer |
| Tolouene diamine sulfate | 615-50-9 | Sensitizer |
| Diethyl maleate | 141-05-9 | Sensitizer |
| 3-Dimethylaminopropylamine | 109-55-7 | Sensitizer |
| Ethylene diamine | 107-15-3 | Sensitizer |
| Isoeugenol | 97-54-1 | Sensitizer |
| 2-Mercaptobenzothiazole | 149-30-4 | Sensitizer |
| Benzyl benzoate | 120-51-4 | Sensitizer |
| Cinnamyl alcohol | 104-54-1 | Sensitizer |
| Citral | 5392-40-5 | Sensitizer |
| Ethylene glycol dimethacrylate | 97-90-5 | Sensitizer |
| Eugenol | 97-53-0 | Sensitizer |
| Dextran | 9004-54-0 | Nonsensitizer |
| Glycerol | 56-81-5 | Nonsensitizer |
| Hexane | 110-54-3 | Nonsensitizer |
| Isopropanol | 67-63-0 | Nonsensitizer |
| Kanamycin | 70560-51-9 | non-sensitizer |
| Lactic acid | 50-21-5 | Nonsensitizer |
| Propylene glycol | 57-55-6 | Nonsensitizer |
| Salicylic acid | 69-72-7 | Nonsensitizer |
| Vanillin | 121-33-5 | Nonsensitizer |
GARD™ protocols
All protocols associated with the GARD™skin assay have been previously published (Forreryd et al., 2016; Johansson et al., 2013) and summarized in SOPs following the standard recommended format of the EURL ECVAM Database Service on Alternative Methods to Animal Experimentation (DB-ALM), with appropriate incremental version control. Following feedback from training and transfer phases and inclusion of clarifying revisions, the updated GARD™ assay SOP v.05.01 was finalized and distributed prior to the commencement of the blinded validation phase. Hence, the GARD™ assay SOP v.05.01 was used as the sole GD available to participating laboratories during the validation phase. The GARD™ assay SOP v.05.01 is attached in its entirety to this publication as Supplementary Material S1.
In short, cultivated SenzaCells (ATCC Depository PTA-123875), are exposed in vitro to (the) test chemical(s) of interest for 24 h. Following dose-response measurements of induced cell toxicity, an appropriate and test chemical specific input concentration is defined at non- to low-toxic levels. Genetic material (ie mRNA) is harvested from cells exposed to the appropriate input concentration of (the) test chemical(s) in 3 biological replicates, and the transcriptional levels of the GARD™skin prediction signature (Johansson et al., 2011) are quantified using the Nanostring nCounter system. The data are analyzed by a Support Vector Machine (SVM) (Cortes and Vapnik, 1995), which has been appropriately trained on samples generated during technology platform transfer, consisting of expressional profiles generated by the reference panel of chemicals, as described in Forreryd et al. (2016). Final prediction calls are derived from the mean SVM decision value (GARD™ DV) generated by the biological triplicate samples; any test chemical inducing a positive (≥0) mean GARD™ DV is classified as a skin sensitizer. Consequently, any test chemical inducing a negative (<0) mean GARD DV is classified as a nonsensitizer.
Statistical analysis
After data submission, the within-laboratory reproducibility (WLR), between-laboratory reproducibility (BLR), and predictive performance were calculated. WLR was assessed based on concordance between classifications in the 3 repeated experiments in each laboratory. BLR was assessed based on concordance between the majority classification of each chemical in the 3 different laboratories. Predictive performance was assessed by comparing the prediction results with the classification based on in vivo reference data. Therefore, 2 × 2 contingency tables (S vs NS) were constructed and sensitivity (probability of predicting S given the reference classification is S), specificity (probability of predicting NS given the reference classification is NS), and accuracy were calculated. In addition, the performance was evaluated based on the positive predictive value (PPV, proportion of positive classifications that are truly positive) and the negative predictive value (NPV, proportion of negative classifications that are truly negative). It was agreed by the VMG, prior to the commencement of the validation phase, to only consider data from valid experiments when assessing WLR, BLR, and predictive performance, while documenting failed runs to report their occurrence and their underlying cause. All statistical analysis and strategic decisions relating to statistical analysis were done by the VMG, without the knowledge or influence of participating laboratories.
RESULTS
Training and Transfer
The method transfer was initiated by sharing of the SOP, web-based dissemination and discussions and material transfers. During the training phase, naïve laboratories were given the opportunity of on-site hands on training for a duration of 4 days, after which continued training pursued with reduced guidance. Naïve laboratories were given a chance to provide feedback on the SOP, and revisions were introduced into a finalized document (Supplementary Material S1). Using the updated SOP, naïve laboratories demonstrated their proficiency in the method by assaying and accurately classifying the transfer phase chemical set (Table 1, data not shown), estimating the transferability of GARDskin to 100%.
GARD Predictions of Validation Phase Test Chemicals
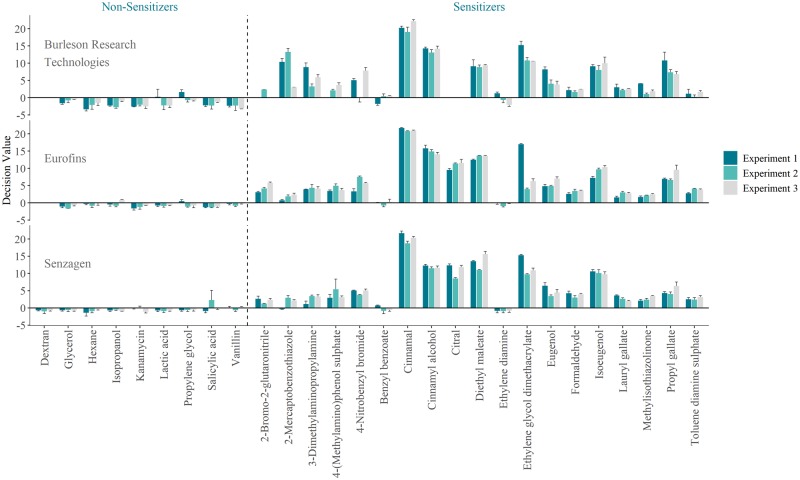
The results from the blind validation phase are summarized in Figure 1, presented as generated GARD™ DVs for each tested compound, across 3 independent experiments in the 3 laboratories. A complete classification table, as defined by the results presented in Figure 1, is attached to this publication as Supplementary Material S2, together with all calculations performed by the VMG of WLR, BLR, and predictive capacity, as summarized below.
Figure 1.
Summarized results generated in the ring trial, presented as mean decision values for each test chemical across 3 independent experiments and 3 independent laboratories. Error bars represent standard deviation in biological triplicate samples.
Each compound within each experiment across the different laboratories generated a classification (sensitizer [S]/nonsensitizer [NS]) or a missing value (N/A). From a total of 252 unique assessments, 12 missing values were generated. Missing values originated from solubility issues (dextran, all 3 experiments at Eurofins and BRT), incompatibility with flow cytometry-based viability assessments due to autofluorescence (citral, all 3 experiments at BRT), or noncompliance with cell viability quality control criteria defined by the SOP (2-bromo-2-gluataronitrile and 4-(methylamino)phenol sulfate, in 2 and 1 experiments at BRT, respectively). Based on this data, the failure rate was estimated to 4.7%. Missing values were excluded when estimating the prediction model performance, as agreed by the VMG in advance of the validation study.
Within-Laboratory Reproducibility
WLR calculations were based on concordance between experiments. The result of a test chemical was considered concordant if 3 experiments generate the same outcome, irrespective of the true class of the test chemical. Based on the available data, WLR was estimated to 82.1%, 88.9%, and 83.3%, for SenzaGen, Eurofins, and BRT, respectively.
Between-Laboratory Reproducibility
BLR calculations were based on concordance between each laboratory’s majority outcome from 3 experiments. The result of a test chemical was considered concordant if all laboratories generate the same outcome, irrespective of the true class of a test chemical. Based on available data, BLR was estimated to 92.0%.
Discordant prediction results between the laboratories were observed for benzyl benzoate (false negative, SenzaGen) and vanillin (false positive, SenzaGen). Although binary classifications indicate dissimilarities, it is noticed that generated signals (GARD™ DV) are highly reproducible (Figure 1). As such, no apparent technical variation could be attributed to observed discrepancies.
Predictive Capacity
Predictive capacity calculations were based on concordance between each laboratory’s majority outcome from 3 experiments and the true class of each test chemical. Contingency tables are presented in Table 3. The predictive accuracy was estimated to 89.3%, 96.3%, and 96.0%, for SenzaGen, Eurofins, and BRT, respectively. Taken together, the cumulative accuracy was estimated to 93.8%. Similarly, the method’s cumulative sensitivity and specificity was estimated to 92.7% and 96%, respectively. The PPV and NPV for each laboratory was 94.4% and 80.0% (SenzaGen), 100.0% and 88.9% (Eurofins), and 100.0% and 88.9% (BRT).
Table 3.
Contingency Tables
| True Group | SenzaGen | Eurofins | BRT | Cumulative | ||||
|---|---|---|---|---|---|---|---|---|
| (19 + 9) |
(19 + 8) |
(17 + 8) |
(55 + 25) |
|||||
| S | NS | S | NS | S | NS | S | NS | |
| S | 17 | 2 | 18 | 1 | 16 | 1 | 51 | 4 |
| NS | 1 | 8 | 0 | 8 | 0 | 8 | 1 | 24 |
| Accuracy (%) | 89.3 | 96.3 | 96.0 | 93.8 | ||||
| Sensitivity (%) | 89.5 | 94.7 | 94.1 | 92.7 | ||||
| Specificity (%) | 88.9 | 100.0 | 100.0 | 96.0 | ||||
Predictive Capacity (Historical Data)
In order to increase the confidence in reported data and to support estimated figures of predictive capacity, the GARD™skin validation report was complemented with historical data, generated by the lead laboratory and analyzed using the GARD™ assay SOP v.05.01. The results are attached to this publication as Supplementary Material S3. In short, the predictive performance was summarized as follows: accuracy, 95.8%; sensitivity, 100.0%; specificity, 92.3%; PPV, 91.7%; NPV, 100.0% (1 false positive, n = 24). Of important note, the obtained results were identical to those previously published (Forreryd et al., 2016).
DISCUSSION
Predictive toxicology is a field that seeks to identify hazards associated with chemical exposure. Traditionally, predictive toxicology has been conducted primarily using animal experimentation. However, due to legislation, public opinion, concern for human environmental health, and economic interests, there is not only a desire, but also a demand that toxicologists move away from animal tests and replace these with novel assays based on state-of-the-art technologies.
Decades of intensive research have resulted in a good understanding of the key biological mechanisms associated with and required for skin sensitization induction and progression. This knowledge has been used for the development of the OECD AOP for skin sensitization.
A variety of nonanimal test methods that seek to reflect the various KEs of skin sensitization are available as OECD TGs. Furthermore, several testing strategies have emerged that combine the data from 2 or more of these test methods in the context of integrated approaches to testing and assessment (IATA) for skin sensitization (OECD, 2016), which in their practical implementation are referred to as integrated testing strategies (ITS) or DA. Despite these developments, there is more to be done in the development of novel nonanimal tests for hazard identification. Furthermore, a key remaining challenge is the accurate assessment of the skin sensitizing potency of contact allergens using in vitro test methods.
GARD™skin is an assay for assessment of skin sensitizers, based on in vitro chemical exposure of DC-like cells. The assay interrogates high informational content gene expression data and provides machine-learning-assisted classifications based on pattern recognition, by utilizing a training data set and a predictive genomic biomarker signature, both interconnectedly defined during assay development. The aim has been to provide a more holistic examination of the biological changes that are associated with exposure to contact allergens than can be expected from methods that are based upon the exclusive use of a single endpoint.
In the present study, we report the results from an inter-laboratory ring trial. The study was conducted by strict adherence to OECD GD and aims to provide the basis for a formal regulatory validation of the GARDskin assay.
In summary, GARD™skin was found to be transferrable from the lead laboratory to 2 naïve laboratories, both of which were able to reproduce the expected outcomes from a limited set of chemicals used for training and demonstration of proficiency. Following the closure of a subsequent blind validation phase, comprising a larger set of chemicals across 3 independent experiments, GARD™skin was found to be reproducible, with a WLR ranging between 82.1% and 88.9%, and an inter-laboratory concordance of 92.0%. Lastly, GARD™skin was found to exhibit predictive accuracies ranging between 89.3% and 96.0%, with a cumulative accuracy of 93.8%.
Examining the discrepancies between the laboratories, valid predictions could not be consistently obtained for 3 of the substances. Dextran were deemed inapplicable by 2 of the laboratories due to solubility issues. Because ocular inspection is used to determine if compounds are dissolvable in vehicle solutions, results will be affected by the laboratory personals subjective judgement, which is likely the reason for this obtained difference. Citral was another substance that was not consistently analyzed by all laboratories. In one of the laboratories, the substance was deemed inapplicable in all 3 experiments due to difficulties in determining suitable exposure concentration caused by autofluorescence during the flow-cytometry analysis. Autofluorescence was also reported by the other 2 laboratories but no definitive explanation could be identified for the conflicting outcomes. Finally, though all laboratories reported challenges when analyzing 2-bromo-2-glutaronitrile due to steep response curves during the viability assessment, only 1 laboratory failed to identify a suitable exposure concentration within the acceptable time frame. It is possible that improved strategies for approaching difficult chemicals could further increase the consistency between laboratories.
The prediction performance attained by the GARD™skin assay in this study should be regarded as comparable and competitive to the results produced by currently validated methods. Considering an excerpt of historical predictions on the set of herein studied substances, currently validated assays reached accuracies of 81% (n = 26), 96% (n = 27), and 89% (n = 27) for DPRA, Keratinosens, and h-CLAT, respectively (Hoffmann et al., 2018). Notable discrepancies compared with expected outcomes include 3-dimethylaminopropylamine (false negative, DPRA), ethylene diamine (false negative, GARD™skin), isoeugenol (false negative, h-CLAT), benzyl benzoate (false negative, DPRA, and h-CLAT), eugenol (false negative, Keratinosens), kanamycin (false positive, DPRA), and salicylic acid (false positive, DPRA and h-CLAT). Although a review of historical data is preferably performed in a large chemical space in conjunction with a mechanistic discussion of reasonable explanations, such meta-analysis of historical data is outside of the scope of this article. However, ambitious efforts toward this end have recently been made (Roberts, 2018).
Results obtained in this study closely mirror previous estimations of its predictive accuracy (Forreryd et al., 2016; Johansson et al., 2014, 2017), which, in comparison with the current state of the art, must be considered consistently high and previously unsurpassed. Recently, a third-party evaluation conducted by the Cosmetics Europe explored the predictive capacity of several methods in a coherent chemical test set (n = 128) (Hoffmann et al., 2018). Predictive accuracies of included in vitro methods, including the 3 adopted TGs, ranged between 73.4% and 78.6% when predicting human hazard.
Originally, the loss of the holistic approach provided by animal models was believed to result in alternative methods with lower predictive performances. Therefore, testing strategies incorporating multiple alternative methods and other physicochemical properties of test substances, representative of the known mechanisms required for the induction of sensitization, were proposed (Jowsey et al., 2006). The initially suggested methods were also designed to provide frameworks for ranking substances by their relative potency, which is still lacking in nonanimal alternatives. Since the early proposals of ITSs, the concept has been widely developed and multiple strategies employing different alternative test methods have been presented both for hazard identification and for hazard characterization (Ezendam et al., 2016).
Nonetheless, though the development of nonanimal alternatives has resulted in test methods achieving predictive performances in level with or above those attained by animal models, there is still a lack of confidence in their performance. This is particularly due to the belief that most alternative methods still only monitor a single mechanism or KE required for the induction of skin sensitization. Therefore, none of the currently validated test methods have been recommended for stand-alone use, and integration of test results is still advocated. In this context it is important to note that the study conducted by Cosmetics Europe also included an evaluation of several proposed DAs (Kleinstreuer et al., 2018). In the same chemical test set as used to evaluate individual assays (n = 128), proposed DA accuracies ranged between 75.6% and 85.0%.
These recent estimations of both individual predictive capacities and those generated by DAs are consistent with historical data from previous compilations (Natsch et al., 2013; Urbisch et al., 2015). Furthermore, a meta-analysis of historical data, studying coherent chemical subsets, shows that ITSs primarily based on a majority vote prediction model fail to increase the predictive capacity to and beyond the levels of the otherwise best performing assay (Roberts, 2018). We have previously argued that the benefits of ITSs and DAs appear to be consistently overestimated in the literature, among regulatory bodies and in the scientific community (Johansson and Gradin, 2017). Evidence indicate that this is true both in terms of added value with respect to predictive capacity, as readily available data show, but also in terms of the notion that accurate safety assessment must rely on a complete mechanistic monitoring of all KEs of the AOP. Such information is in no way forwarded to the prediction model, which in the most advocated cases are solely constituted by a majority vote classification. Taken together, we encourage the field to further question the current paradigm in predictive toxicology. Naturally, a final recommendation on the regulatory adoption of GARD™skin will be subject to a thorough peer review of the submitted validation report by regulatory bodies within the frames of the current paradigm. However, a critical review of the possibility of relying on stand-alone tests for hazard assessment in the future, if a sufficiently predictive assay is readily available, is welcomed and encouraged.
It is appropriate to emphasize the unique technology features, which support the high observed predictive accuracy of GARD™skin. The data-driven assay development, the high informational content readout, and the machine-learning assisted prediction model.
First, the genomic biomarkers utilized as predictors were identified by a data-driven approach, as opposed to the hypothesis-driven designs utilized in previously validated methods. In this context, the biomarker selection and prediction model design were not driven by a priori knowledge, eg, an assumption that expression levels of certain disease-associated genes or proteins would change following chemical exposure. Rather, genome-wide data generated by a reference panel of chemicals were interrogated to learn actual measurable effects, in the context of the specified cellular system. Although certain molecules previously described as relevant, eg, CD86 and signaling entities controlled by the NRF2-pathway, were indeed identified, a data-driven and high-dimensional analysis allowed for a holistic view of induced transcriptional effects. As a direct consequence, this also relates to the second unique feature of GARD™skin, which is the high informational content. Generating high-dimensional data allows for models that generalize well. In context, this is an acknowledgment to the fact that sensitizers are indeed quite heterogenous and may induce unique responses depending on a variety of factors (Albrekt et al., 2014). Thus, although not all genes may be affected in the same way by all chemicals, when studied collectively, the GARD™skin prediction signature allows for a distinct separation between sensitizers and nonsensitizers, whereas assays relying on few biomarkers would fail to accurately classify certain test chemicals whenever the measured biomarker(s) fail to respond as hypothesized. The third unique feature of GARD™skin relates to the prediction model, which is based on state-of-the-art machine learning technology and pattern recognition. Although these techniques are today ubiquitously utilized within almost every aspect of modern technology and innovation, including pure in silico applications for chemical risk assessment, to the best of our knowledge, it is still only utilized by GARD™skin in the context of in vitro hazard assessments. These types of techniques allow for a streamlined, scientifically robust, and statistically correct methodology for interpretation of high-dimensional data, which would otherwise be prone to overfitting.
In conclusion, investigations reported here summarizes the results generated in an inter-laboratory ring trial of GARD™skin. It was established that GARD™skin is transferable, reproducible, and highly accurate.
DATA AVAILABILITY
Supplementary data are available at https://doi.org/10.5061/dryad.sc65030.
DECLARATION OF CONFLICTING INTERESTS
The author/authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Astrid Reus for coding and distribution of blinded chemicals and Annika Eriksson and Matthew Thomas for their technical assistance with nanostring sample preparation and data generation. The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
This work was funded in its entirety by SenzaGen AB.
REFERENCES
- Adler S., Basketter D., Creton S., Pelkonen O., van Benthem J., Zuang V., Andersen K. E., Angers-Loustau A., Aptula A., Bal-Price A., et al. (2011). Alternative (non-animal) methods for cosmetics testing: Current status and future prospects-2010. Arch. Toxicol. 85, 367–485. [DOI] [PubMed] [Google Scholar]
- Ainscough J. S., Frank Gerberick G., Dearman R. J., Kimber I. (2013). Danger, intracellular signaling, and the orchestration of dendritic cell function in skin sensitization. J. Immunotoxicol. 10, 223–234. [DOI] [PubMed] [Google Scholar]
- Albrekt A. S., Johansson H., Borje A., Borrebaeck C., Lindstedt M. (2014). Skin sensitizers differentially regulate signaling pathways in MUTZ-3 cells in relation to their individual potency. BMC Pharmacol. Toxicol. 15. doi: 10.1186/2050-6511-15-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikaga T., Yoshida Y., Hirota M., Yoneyama K., Itagaki H., Sakaguchi H., Miyazawa M., Ito Y., Suzuki H., Toyoda H. (2006). Development of an in vitro skin sensitization test using human cell lines: The human cell line activation test (h-CLAT). I. Optimization of the h-CLAT protocol. Toxicol. In Vitro 20, 767–773. [DOI] [PubMed] [Google Scholar]
- Casati S., Aschberger K., Barroso J., Casey W., Delgado I., Kim T. S., Kleinstreuer N., Kojima H., Lee J. K., Lowit A., et al. (2018). Standardisation of defined approaches for skin sensitisation testing to support regulatory use and international adoption: Position of the International Cooperation on Alternative Test Methods. Arch. Toxicol. 92, 611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C., Vapnik V. (1995). Support-vector networks. Mach. Learn. 20, 273–297. [Google Scholar]
- Cottrez F., Boitel E., Auriault C., Aeby P., Groux H. (2015). Genes specifically modulated in sensitized skins allow the detection of sensitizers in a reconstructed human skin model. Development of the SENS-IS assay. Toxicol. In Vitro 29, 787–802. [DOI] [PubMed] [Google Scholar]
- EC (2006). Regulation (EC) No 1907/2006 of the European Parliament and of the Council of 18 December 2006 concerning the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH), establishing a European Chemicals Agency.
- EU (1976). Cosmetics Directive 76/768/EEC. https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri = CONSLEG:1976L0768:20080424:en:PDF. Last accessed May 16, 2019.
- Ezendam J., Braakhuis H. M., Vandebriel R. J. (2016). State of the art in non-animal approaches for skin sensitization testing: From individual test methods towards testing strategies. Arch. Toxicol. 90, 2861–2883. [DOI] [PubMed] [Google Scholar]
- Forreryd A., Johansson H., Albrekt A. S., Lindstedt M. (2014). Evaluation of high throughput gene expression platforms using a genomic biomarker signature for prediction of skin sensitization. BMC Genomics 15, 379.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forreryd A., Zeller K. S., Lindberg T., Johansson H., Lindstedt M. (2016). From genome-wide arrays to tailor-made biomarker readout - Progress towards routine analysis of skin sensitizing chemicals with GARD™. Toxicol. In Vitro 37, 178–188. [DOI] [PubMed] [Google Scholar]
- Geiss G. K., Bumgarner R. E., Birditt B., Dahl T., Dowidar N., Dunaway D. L., Fell H. P., Ferree S., George R. D., Grogan T., et al. (2008). Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 26, 317–325. [DOI] [PubMed] [Google Scholar]
- Gerberick G. F., Vassallo J. D., Bailey R. E., Chaney J. G., Morrall S. W., Lepoittevin J. P. (2004). Development of a peptide reactivity assay for screening contact allergens. Toxicol. Sci. 81, 332–343. [DOI] [PubMed] [Google Scholar]
- Hartung T., Bremer S., Casati S., Coecke S., Corvi R., Fortaner S., Gribaldo L., Halder M., Hoffmann S., Roi A. J., et al. (2004). A modular approach to the ECVAM principles on test validity. Altern. Lab. Anim. 32, 467–472. [DOI] [PubMed] [Google Scholar]
- Hartung T., Luechtefeld T., Maertens A., Kleensang A. (2013). Integrated testing strategies for safety assessments. Altex 30, 3–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann S., Kleinstreuer N., Alepee N., Allen D., Api A. M., Ashikaga T., Clouet E., Cluzel M., Desprez B., Gellatly N., et al. (2018). Non-animal methods to predict skin sensitization (I): The Cosmetics Europe database. Crit. Rev. Toxicol. 48, 344–358. [DOI] [PubMed] [Google Scholar]
- Johansson H., Albrekt A. S., Borrebaeck C. A., Lindstedt M. (2013). The GARD™ assay for assessment of chemical skin sensitizers. Toxicol. In Vitro 27, 1163–1169. [DOI] [PubMed] [Google Scholar]
- Johansson H., Gradin R. (2017). Skin sensitization: Challenging the conventional thinking - A case against 2 out of 3 as integrated testing strategy. Toxicol. Sci. 159, 3–5. [DOI] [PubMed] [Google Scholar]
- Johansson H., Gradin R., Forreryd A., Agemark M., Zeller K., Johansson A., Larne O., van Vliet E., Borrebaeck C., Lindstedt M. (2017). Evaluation of the GARD™ assay in a blind Cosmetics Europe study. Altex 34, 515–523. [DOI] [PubMed] [Google Scholar]
- Johansson H., Lindstedt, A. S. Albrekt M., Borrebaeck C. A. (2011). A genomic biomarker signature can predict skin sensitizers using a cell-based in vitro alternative to animal tests. BMC Genomics 12. doi: 10.1186/1471-2164-12-399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson H., Rydnert F., Kuhnl J., Schepky A., Borrebaeck C., Lindstedt M. (2014). Genomic allergen rapid detection in-house validation – A proof of concept. Toxicol. Sci. 139, 362–370. [DOI] [PubMed] [Google Scholar]
- Jowsey I. R., Basketter D. A., Westmoreland C., Kimber I. (2006). A future approach to measuring relative skin sensitising potency: A proposal. J. Appl. Toxicol. 26, 341–350. [DOI] [PubMed] [Google Scholar]
- Kimber I., Basketter D. A., Gerberick G. F., Ryan C. A., Dearman R. J. (2011). Chemical allergy: Translating biology into hazard characterization. Toxicol. Sci. 120(Suppl. 1), S238–S268. [DOI] [PubMed] [Google Scholar]
- Kleinstreuer N. C., Hoffmann S., Alepee N., Allen D., Ashikaga T., Casey W., Clouet E., Cluzel M., Desprez B., Gellatly N., et al. (2018). Non-animal methods to predict skin sensitization (II): An assessment of defined approaches (*). Crit. Rev. Toxicol. 48, 359–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson B., Kligman A. M. (1969). The identification of contact allergens by animal assay. The guinea pig maximization test. J. Invest Dermatol. 52, 268–276. [DOI] [PubMed] [Google Scholar]
- Martin S. F. (2015). New concepts in cutaneous allergy. Contact Dermatitis 72, 2–10. [DOI] [PubMed] [Google Scholar]
- Martin S. F., Esser P. R., Weber F. C., Jakob T., Freudenberg M. A., Schmidt M., Goebeler M. (2011). Mechanisms of chemical-induced innate immunity in allergic contact dermatitis. Allergy 66, 1152–1163. [DOI] [PubMed] [Google Scholar]
- Natsch A. (2010). The Nrf2-Keap1-ARE toxicity pathway as a cellular sensor for skin sensitizers–functional relevance and a hypothesis on innate reactions to skin sensitizers. Toxicol. Sci. 113, 284–292. [DOI] [PubMed] [Google Scholar]
- Natsch A., Ryan C. A., Foertsch L., Emter R., Jaworska J., Gerberick F., Kern P. (2013). A dataset on 145 chemicals tested in alternative assays for skin sensitization undergoing prevalidation. J. Appl. Toxicol. 33, 1337–1352. [DOI] [PubMed] [Google Scholar]
- OECD (1992). Test No. 406: Skin Sensitization, OECD Guidelines for the Testing of Chemicals, Section 4. OECD Publishing, Paris. [Google Scholar]
- OECD (2005). Series on Testing and Assessment, No 34: Guidance Document on the Validation and International Acceptance of New or Updated Test Methods for Hazard Assessment. OECD Publishing, Paris. [Google Scholar]
- OECD (2009). Series on Testing and Assessment, No 1: Guidance Document for the Development of OECD Guidelines for Testing of Chemicals (as Revised in 2009). OECD Publishing, Paris. [Google Scholar]
- OECD (2010). Test No. 429: Skin Sensitization: Local Lymph Node Assay, OECD Guidelines for the Testing of Chemicals, Section 4. OECD Publishing, Paris. [Google Scholar]
- OECD (2014). The Adverse Outcome Pathway for Skin Sensitization Initiated by Covalent Binding to Proteins OECD Series on Testing and Assessment, No. 168. OECD Publishing, Paris. [Google Scholar]
- OECD (2015). Test No. 442C: In Chemico Skin Sensitisation: Direct Peptide Reactivity Assay (DPRA), OECD Guidelines of the Testing of Chemicals, Section 4. OECD Publishing, Paris. [Google Scholar]
- OECD (2016). Series on Testing and Assessment, No 256: Guidance Document on the Reporting of Defined Approaches and Individual Information Sources to Be Used within Integrated Approaches to Testing and Assessment (IATA) for Skin Sensitisation, Annex 1, Annex 2. OECD Publishing, Paris. [Google Scholar]
- OECD (2018a). Test No. 442D: In Vitro Skin Sensitisation: ARE-Nrf2 Luciferase Test Method, OECD Guidelines of the Testing of Chemicals, Section 4. OECD Publishing, Paris. [Google Scholar]
- OECD (2018b). Test No. 442E: In Vitro Skin Sensitisation: In Vitro Skin Sensitisation Assays Addressing the Key Event on Activation of Dendritic Cells on the Adverse Outcome Pathway for Skin Sensitisation, OECD Guidelines for the Testing of Chemicals, Section 4. OECD Publishing, Paris. [Google Scholar]
- Roberts D. W. (2018). Is a combination of assays really needed for non-animal prediction of skin sensitization potential? Performance of the GARD™ (Genomic Allergen Rapid Detection) assay in comparison with OECD guideline assays alone and in combination. Regul. Toxicol. Pharmacol. 98, 155–160. [DOI] [PubMed] [Google Scholar]
- UN (2009). Globally Harmonized System of Classification and Labelling of Chemicals (GHS). Part 3: Health hazards. https://www.unece.org/fileadmin/DAM/trans/danger/publi/ghs/ghs_rev04/English/ST-SG-AC10-30-Rev4e.pdf. Last accessed May 16, 2019.
- Urbisch D., Mehling A., Guth K., Ramirez T., Honarvar N., Kolle S., Landsiedel R., Jaworska J., Kern P. S., Gerberick F., et al. (2015). Assessing skin sensitization hazard in mice and men using non-animal test methods. Regul. Toxicol. Pharmacol. 71, 337–351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary data are available at https://doi.org/10.5061/dryad.sc65030.