Abstract
Exposure to endocrine disrupting chemicals is an established risk factor for obesity. The most commonly used pesticide active ingredients have never been tested in an adipogenesis assay. We tested for the first time the potential of glyphosate, 2, 4-dichlorophenoxyacetic acid, dicamba, mesotrione, isoxaflutole, and quizalofop-p-ethyl (QpE) to induce lipid accumulation in murine 3T3-L1 adipocytes. Only QpE caused a dose-dependent statistically significant triglyceride accumulation from a concentration of 5 up to 100 µM. The QpE commercial formulation Targa Super was 100 times more cytotoxic than QpE alone. Neither the estrogen receptor antagonist ICI 182, 780 nor the glucocorticoid receptor antagonist RU486 was able to block the QpE-induced lipid accumulation. RNAseq analysis of 3T3-L1 adipocytes exposed to QpE suggests that this compound exerts its lipid accumulation effects via a peroxisome proliferator-activated receptor gamma (PPARγ)-mediated pathway, a nuclear receptor whose modulation influences lipid metabolism. QpE was further shown to be active in a PPARγ reporter gene assay at 100 µM, reaching 4% of the maximal response produced by rosiglitazone, which acts as a positive control. This indicates that lipid accumulation induced by QpE is only in part caused by PPARγ activation. The lipid accumulation capability of QpE we observe suggest that this pesticide, whose use is likely to increase in coming years may have a hitherto unsuspected obesogenic property.
Keywords: quizalofop, pesticide, adipogenesis, obesogen, glyphosate, 3T3-L1
Obesity has become a global health problem over the past 30 years (Collaboration, 2017). The worldwide prevalence of obesity has tripled since 1975 and it is estimated that 39% of the global adult population is overweight (BMI ≥25) and 13% obese (BMI ≥ 30). The most dramatic increase has been reported in children and adolescents of 5–19 years of age, where the incidence of obesity has risen from 4% to 18% between 1975 and 2016 (Collaboration, 2017). Obesity is a complex and multifactorial condition with both genetic and behavioral causative components (Ghosh and Bouchard, 2017). Exposure to environmental pollutants and toxicants is now recognized to be a risk factor for obesity (Newbold, 2010). Among the environmental factors predisposing to obesity, there has been increasing interest in the role of endocrine disruptive chemicals (EDCs) such as pesticides (Thayer et al., 2012).
EDCs promote adipogenesis and induce obesity by altering lipid metabolic pathways, deregulating adipogenesis and energy balance, or promoting lipid accumulation in adipocytes (Janesick and Blumberg, 2016). A large number of in vitro studies have shown that some xenobiotics, such as tributyltin chloride and bisphenol A, can promote adipocyte differentiation (Chamorro-Garcia et al., 2012; Li et al., 2011). Furthermore, in vivo animal studies and human epidemiological investigations have linked exposure to certain chemicals to an increased fat deposition and consequently an increase in body weight (Grun et al., 2006; Janesick and Blumberg, 2011). Of even greater concern is the fact that prenatal or early life exposure to environmental obesogens can predispose individuals to an increased probability to develop obesity in adult life (Janesick and Blumberg, 2011). A predisposition to obesity and some associated diseases such as nonalcoholic fatty liver disease can also be transmitted transgenerationally via epigenetic mechanisms (Chamorro-Garcia et al., 2013).
An increasing number of studies report the association between the use of pesticides and the onset of obesity (Rosenbaum et al., 2017; Vassilopoulou et al., 2017). The extensive use of pesticides in an agricultural setting leads to the consumption of foodstuffs contaminated by pesticide residues (Damalas and Eleftherohorinos, 2011). An emblematic obesogen is the pesticide active ingredient dichlorodiphenyltrichloroethane (DDT), which is widely used to control mosquito populations. Although DDT was subsequently banned in the 1970s in most countries due to its carcinogenic and reproductive toxicity in humans, DDT is very persistent and its residues continue to this day to be associated to increased adiposity in humans and rodents (Cano-Sancho et al., 2017).
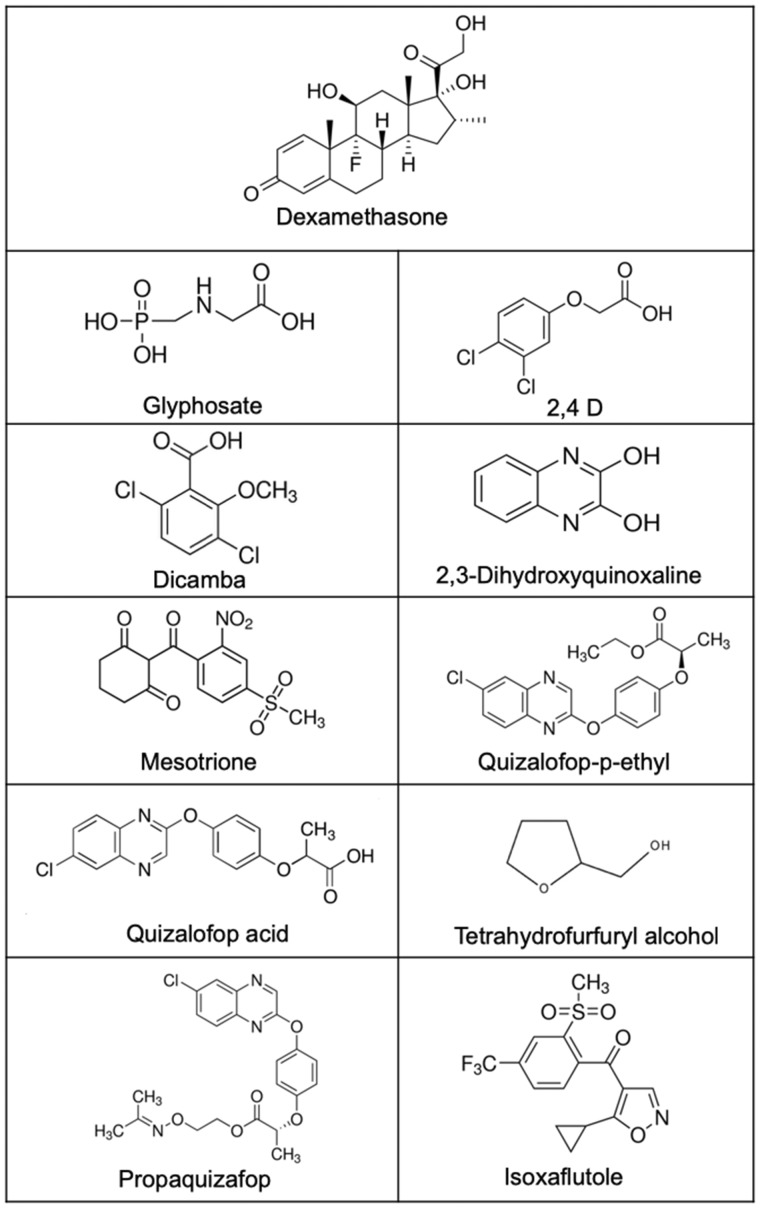
The most widely used pesticides around the world are glyphosate-based herbicides (Benbrook, 2016). Many studies have investigated toxic effects of glyphosate-based herbicides including the possible endocrine disrupting properties of their ingredients (Mesnage et al., 2015, 2017). The reliance on a single herbicide in glyphosate-tolerant genetically modified (GM) cropping systems has increased the spread of glyphosate-resistant weeds (Heap and Duke, 2018), which has forced farmers to resort to using mixtures of glyphosate and additional pesticide active ingredients (Myers et al., 2016). They include 2, 4-dichlorophenoxyacetic acid (2, 4-D), dicamba, mesotrione, isoxaflutole and quizalofop-p-ethyl (QpE) (Figure 1). Moreover, the agricultural biotechnology industry has developed GM crops tolerant to multiple herbicides, including glyphosate, to control resistant weeds (Myers et al., 2016). It could thus be expected that this change in agricultural practices will lead to an increased human exposure to these compounds. Given the alarming global rate of obesity and the increasing use of pesticides in agriculture, there is an urgent need to evaluate their adipogenic potential.
Figure 1.
Molecular structures of the different pesticide active ingredients tested in this study in comparison to the positive control dexamethasone.
We describe here the evaluation of the adipogenic potential of glyphosate, 2, 4-D, dicamba, mesotrione, isoxaflutole, and QpE in the murine 3T3-L1 preadipocyte lipid accumulation assay system. The use of 3T3-L1 cells is a well-established in vitro system for the investigation of adipogenesis and lipid metabolism with many studies conducted to validate its use as an adipocyte model system (Kassotis et al., 2017; Morrison and McGee, 2015). We found that QpE is a potent inducer of adipogenesis in this 3T3-L1 cell assay. Isoxaflutole and dicamba also caused lipid accumulation but to a lesser degree. Further evaluation of the adipogenic effects of QpE by testing its metabolites and a commercial formulation, as well as its mechanisms of action in a variety of bioassays including an RNA-seq analysis, suggested that this compound exerts its effects in part via a peroxisome proliferator-activated receptor gamma (PPARγ)-mediated pathway.
MATERIALS AND METHODS
Reagents
All reagents and chemicals, unless otherwise specified, were of analytical grade and were purchased from Sigma-Aldrich (Gillingham, Dorset, UK). The pesticide active principles were N-(phosphonomethyl)glycine (glyphosate; CAS: 1071-83-6, XLogP3-AA = −4.6, purity ≥ 96%, catalog No.: 337757), 2, 4-D (CAS: 94-95-7, XLogP3-AA = 2.8, purity ≥ 97%, catalog No.: D70724), 3, 6-dichloro-2-methoxybenzoic acid (dicamba; CAS: 1918-00-9, XLogP3-AA = 2.2, purity ≥ 98%, Santa Cruz Biotechnology catalog No.: sc-239692), QpE (CAS: 100646-51-3, XLogP3-AA = 4.3, purity ≥ 98%, Santa Cruz Biotechnology, catalog No.: sc-224246), isoxaflutole (CAS: 141112-29-0, XLogP3-AA = 2.4, purity ≥ 98%, Santa Cruz Biotechnology, catalog No. : sc-228380), mesotrione (CAS: 104206-82-8, XLogP3-AA = 0.7, purity > 99%, Santa Cruz Biotechnology, catalog No.: sc-224051), quizalofop acid (CAS: 76578-12-6, XLogP3-AA = 3.6, purity ≥ 99.5%, ChemService Inc, Milford, NH, catalog No.: MET-13174A-50MG), propaquizafop (CAS: 111479-05-1, XLogP3-AA = 4.6, catalog No.: 31572), tetrahydrofurfuryl alcohol (CAS: 97-99-4, XLogP3-AA = −0.1, purity ≥ 99%, catalog No.: 185396), 2, 3-dihydroxyquinoxaline (CAS: 15804-19-0, XLogP3-AA = 0.2, purity ≥ 98%, catalog No.: 144789), dexamethasone (CAS: 50-02-2, XLogP3-AA = 1.9, purity ≥ 98%, catalog No.: sc-29059). The QpE commercial formulation tested was Targa Super (Nissan Chemical Europe S.A.R.L, France, which contained 50 g/l QpE, below 5% of calcium dodecylbenzene sulfonate, below 25% of ethoxylated lauryl alcohol C12, below 75% solvent naphtha (petroleum) and super heavy aromatic (<1% naphthalene); homologation F11-5-004), fulvestrant (ICI 182, 780; CAS: 129453-61-8, purity >98%), mifepristone (RU-486; CAS: 84371-65-3, purity ≥98%). Stock solutions of glyphosate was prepared in serum-free medium and adjusted to pH 7.2. Stock solutions of all other compounds were prepared in dimethyl sulfoxide (DMSO). Dilutions of these stock solutions to be administered to cell cultures always resulted in a DMSO concentration below 0.5%.
Cell culture
The murine fibroblast 3T3-L1 cell line was purchased from ZenBio (Cambridge Bioscience, Cambridge, UK) and was not used past passage 10. Undifferentiated 3T3-L1 cells were grown at 37°C (5% CO2) in 75 cm2 flasks (Corning, Tewksbury) in a maintenance medium composed of phenol red free Dulbecco's Modified Eagle Medium (DMEM) (Life Technologies, Warrington, UK), 10% newborn calf serum (New Zealand origin, Thermo Fisher Scientific [Life Technologies]), 2 mM glutamine (GE Healthcare Life Sciences, Fisher Scientific, Loughborough, UK) and 10 µg/ml penicillin/streptomycin (Life Technologies). Cells were released from the flask substratum using 0.05% trypsin-EDTA (Life Technologies) and counted using a hemocytometer prior to seeding.
Adipocyte differentiation
For differentiation of murine 3T3-L1 cultures to adipocytes, cells were seeded into 96-well plates (Dutscher Scientific, Brentwood, UK) at a density of 20 000 cells per well in 100 µl maintenance medium. Following a 2-day stabilization period, they were switched to differentiation medium consisting of DMEM, 2 mM glutamine, 10 µg/ml penicillin/streptomycin, 10% fetal bovine calf serum (GE Healthcare Life Sciences, Buckinghamshire, UK), 500 μM 3-isobutyl-1-methylxanthine (Sigma-Aldrich Ltd) and 100 nM insulin from bovine pancreas (Sigma-Aldrich Ltd). After a further 2 days of culture, the medium was refreshed to start the incubation with various concentrations of the test substances. Dexamethasone was used as a positive control. Media were replenished every 2 days for a further 6 days. Lipid accumulation was visualized on day 8 using fluorescent Nile Red staining in accordance with the manufacturer’s instructions (AdipoRed Assay Reagent, Lonza Walkersville Inc, USA) and quantified using a microplate reader (GloMax Multi Microplate Multimode Reader, Promega, Madison). The fluorescence was measured with a filter giving an excitation at 490 nm and emission at 510–570 nm. The adipogenic effect was expressed as a fold change in emission signal intensity between untreated differentiated and treated differentiated 3T3-L1 cells.
Cell viability assay
Cell viability was assessed using a colorimetric 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay, which indirectly measures cell number by testing for activity of mitochondrial succinate dehydrogenase. The MTT assay has also been proven to be a reliable measure of toxicity (Mosmann, 1983). The 3T3-L1 cells were seeded into 96-well plates (Dutscher Scientific) and differentiated as described above for 8 days with the indicated treatments. The MTT assay was performed as follows. Cells were incubated with 100 µl of MTT solution (1 mg/ml) for 2 h and the test terminated by lysing the cells by adding 100 µl DMSO. As a measure of cell number, the optical density of the cell lysate was determined at 570 nm using a GloMax Multi Microplate Multimode Reader (Promega). The number of cells was directly proportional to the intensity of the signal. Cell viability was expressed as a percentage of the control, untreated samples.
PPARγ transactivation assay
The PPARγ assay kit was obtained from INDIGO Biosciences (Bertin Bioreagent, Montigny le Bretonneux, France). QpE was tested in this assay in comparison to the positive control compound rosiglitazone in a 96-well plate assay following the manufacturer’s instructions. This assay utilizes a nonhuman cell line engineered to constitutively express human PPARγ and also harbor a luciferase reporter gene under control of a PPARγ-responsive promoter. Compounds that bind to and activate the human PPARγ lead to a proportional increase in luciferase reporter gene expression. This provides a quantitative and functional measurement of PPARγ activity when cells are treated with either agonists or antagonists of human PPARγ.
Intracellular lipid staining
The 3T3-L1 cells were seeded into 96-well clear bottom black tissue culture treated plates (Corning) and differentiated as described above for 8 days with the indicated treatments. Medium was then removed and cells fixed by addition of 100 μl 4% paraformaldehyde (Thermo Fisher Scientific). Cells were then stained for intracellular lipid accumulation by adding 50 μl of 1 µg/ml Nile Red (Yorlab, York, UK) and 1 µg/ml 4',6-diamidino-2-phenylindole (DAPI) (Santa Cruz Biotechnology) in 0.2% Triton X-100-phosphate-buffered saline for 15 min in the dark (Boucher et al., 2015). Nile Red staining for lipid droplets and DAPI staining for cell nuclei were imaged at 530 and 405 nm, respectively, using fluorescence imaging on a Nikon Eclipse Ts2 microscope (40× objective).
RNA extraction and library preparation
Independent cultures of 3T3-L1 cells, either treated or untreated with 50 μM QpE, were harvested and total RNA extracted at 6, 24, and 288 h (12 days) posttreatment. A total of 5 biological replicates, each in triplicate, were performed (n = 5). RNA extraction was performed using the Qiagen RNeasy kit (Qiagen, Manchester, UK) according to the manufacturer's instructions. RNA samples were assessed for quantity and integrity using the NanoDrop 8000 spectrophotometer V2.0 (ThermoScientific, USA) and Agilent 2100 Bioanalyser (Agilent Technologies, Waldbronn, Germany), respectively. All samples displayed a RIN score of 10 and were taken forward for RNA library preparation.
Library generation and RNA sequencing
A 100 ng aliquot of total RNA from each sample was used to prepare mRNA libraries using the NEBNext mRNA isolation kit in conjunction with the NEBNext Ultra II Directional RNA Library preparation kit (New England Biolabs, Massachusetts), and samples randomized before preparation. Fragmentation of isolated mRNA prior to first strand cDNA synthesis was conducted using incubation conditions recommended by the manufacturer for a fragment size of 300 bp (94°C for 10 min). A total of 13 cycles of PCR were performed for final library amplification. Resulting libraries were quantified using the Qubit 2.0 spectrophotometer (Life Technologies, California) and average fragment size assessed using the Agilent 2200 Tapestation (Agilent Technologies). A final sequencing pool was created using equimolar quantities of each sample with compatible indexes. Paired-end reads of 75 bp were generated for each library using the Illumina NextSeq500 in conjunction with the NextSeq500 v2 High-output 150-cycle kit (Illumina Inc., Cambridge, UK).
Statistical analysis
The statistical analysis of the dose-response results from the adipogenesis assay was performed by ANOVA. Pair-wise comparisons were made using a Mann-Whitney test. Nonlinear regression analysis was performed using 5-parameter logistic dose-response curve models. These statistical analyses were performed using GraphPad Prism version 7.00 for MAC OS X (GraphPad Software, La Jolla, California, www.graphpad.com). The analysis of the RNA-seq data was performed using the new version of the Tuxedo protocol with HISAT2, StringTie and Ballgown as previously described (Mesnage et al., 2018a). Briefly, sequences were aligned to the mouse genome 91, GRCm38, using the program HISAT2. Then, transcripts were assembled and quantified using the program StringTie. Differential gene analysis was conducted using Ballgown, in R environment (R Core Team, 2017). We identified statistically significant increases or decreases in gene expression with a minimum fold change of 1.2 and a p-value < .05. The gene set enrichment analysis was performed using Enrichr web servers (Kuleshov et al., 2016). These RNA-seq data have been submitted to Gene Omnibus and are accessible through accession number GSE121419.
RESULTS
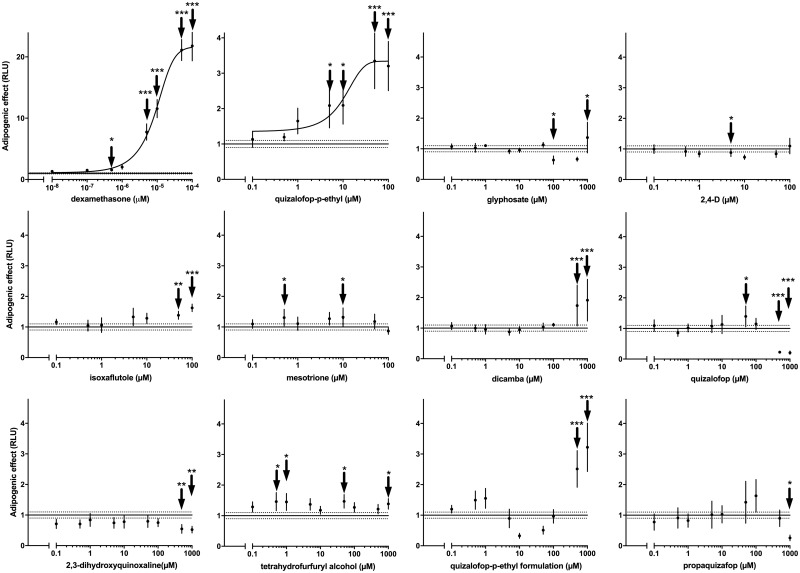
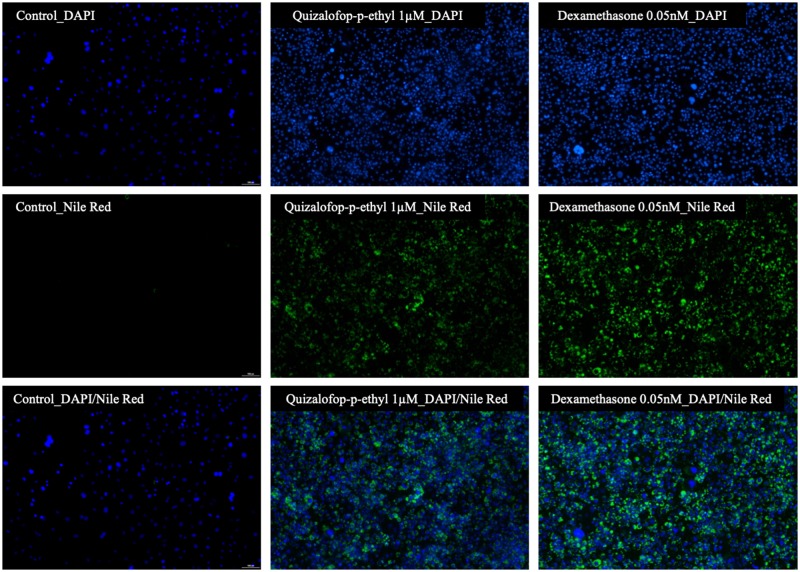
The aim of this study was to investigate the adipogenic potential of major chemicals declared as active principles in commercial herbicide formulations. The adipogenic assay employed involves looking for lipid accumulation in response to test substances in murine 3T3-L1 cells, which had been induced to undergo differentiation to adipocytes. Using dexamethasone as a positive control shows this to be a sensitive assay system causing a maximum 21-fold increase in lipid accumulation by comparison to untreated differentiated cells (Figure 2). The dose response of dexamethasone was used to determine the concentration that caused a half-maximal response (EC50), using nonlinear dose-response relationships. The EC50 for dexamethasone was 9.4 pM. Some of the tested compounds showed an effect on lipid accumulation with different dose-response patterns. Treatment with glyphosate, mesotrione and 2, 4-D scored negative. Some of the tested concentrations for these compounds were statistically significant, but there was no clear dose-response relationship, which indicates that these effects are probably random outcomes. The active ingredient dicamba caused an approximately 2.5-fold increase (p < .001) in lipid accumulation from a concentration of 500 µM. Isoxaflutole caused adipogenesis at 50 (p < .01) and 100 µM (p < .001) resulting in a 2-fold increase in lipid accumulation (Figure 2). We found that the active ingredient with the greatest adipogenic potency was QpE, which gave increases in lipid accumulation starting at 1 µM, becoming statistically significant at 5 µM (p < .05) and reaching a 3-fold maximal increase at 50 and 100 µM (p < .001) (Figure 2). The EC50 determined by nonlinear regression analysis was 8.8 µM for QpE. The adipogenic effect of QpE was further confirmed by Nile Red lipid staining (Figure 3). The 3T3-L1 cells treated with 1 µM QpE for 8 days showed a clear increase in lipid accumulation as was observed with cells treated with dexamethasone (0.05 nM). We thus decided to focus further investigations on the mode of action of QpE.
Figure 2.
QpE induces lipid accumulation in 3T3-L1 adipocytes. Cells of the 3T3-L1 line, which had been induced to undergo differentiation to adipocytes, were treated with several pesticide active ingredients and 0.05 µM dexamethasone (Dex), which acted as a positive control. QpE metabolites, propaquizafop and a commercial QpE herbicide formulation (Targa Super) were also tested. Note that the effects of the QpE formulation are an artifact due to the pesticide co-formulants since we observed an increase in fluorescence intensity in the absence of cells (Supplementary Figure 1). After 8 days of treatment with the indicated compounds, lipid accumulation was evaluated by measuring fluorescence intensity following Nile Red staining. Results are expressed as fluorescence intensity relative to the treatment-free control cells. Data are the mean ± SE of at least 5 independent experiments, with each performed in triplicate (*p < .05; **p < .01; ***p < .001). Nonlinear regression analysis was performed for dexamethasone and QpE using the Hill equation, as they were the only compounds to show a dose response of sufficient quality to obtain convergence of a nonlinear model.
Figure 3.
The adipogenic effect of QpE confirmed using fluorescence imaging. The 3T3-L1 adipocyte cells were cultured with standard treatment-free medium (control), or in the presence of 1 µM QpE or 0.05 nM dexamethasone (positive control). After 8 days of differentiation with the indicated treatments, lipid accumulation was visualized by fluorescent Nile Red staining and the cells visualized by fluorescence microscopy. Nile Red staining for lipid droplets and DAPI staining for cell nuclei were imaged at 530 and 405 nm, respectively, using fluorescence imaging on a Nikon Eclipse Ts2 microscope (40× objective).
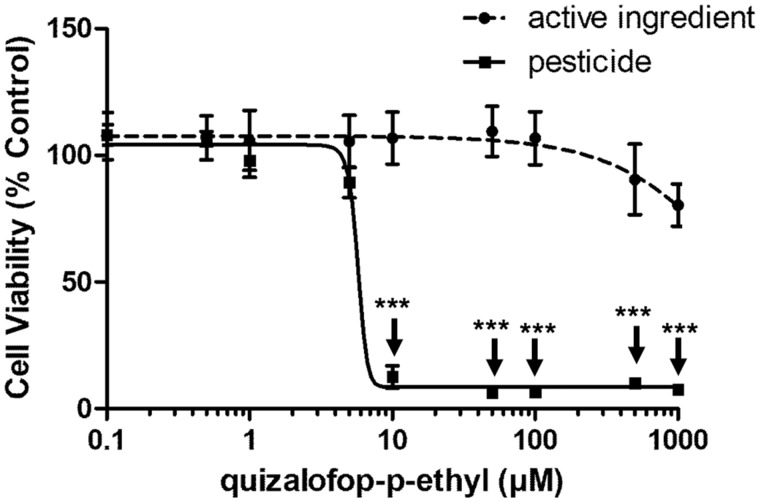
It is important to note that QpE undergoes rapid metabolism upon ingestion (European Food Safety Authority, 2009). Therefore, 3 metabolites of QpE (quizalofop acid, tetrahydrofurfuryl alcohol, and 2, 3-dihydroxyquinoxaline) and another active principle from the same class of pesticides (propaquizafop), were tested in the 3T3-L1 cell lipid accumulation assay. Neither the QpE metabolites nor propaquizafop were able to induce lipid accumulation in 3T3-L1 preadipocytes (Figure 2). A commercial formulation of QpE (Targa Super) also appeared to cause an increase in lipid accumulation albeit at the highest concentrations tested of 500 and 1000 µM. However, this appears to be an artifact due to the pesticide coformulants since we observed an increase in fluorescence intensity in the absence of cells (Supplementary Figure 1). We then compared the toxicity of these chemicals using an MTT viability assay. The results showed that the QpE commercial formulation was approximately 1000 times more toxic than its active principle (Figure 4). In addition, we found that the metabolite quizalofop acid was approximately 10-times more toxic than its parent compound QpE (Supplementary Figure 2).
Figure 4.
Viability of 3T3-L1 adipocytes exposed to QpE and its commercial formulation Targa Super. The 3T3-L1 adipocyte differentiated cells were treated with QpE and Targa Super (0.1–1000 µM). The quantity of formulated Targa Super is expressed as QpE equivalent concentrations. After 8 days of differentiation with the indicated treatments, cell viability was determined by an MTT colorimetric assay. Results are expressed as cell viability percentage relative to the viability cell count under treatment-free conditions. Data are the mean ± SE of 3 independent experiments, with each performed in triplicate (***p < .001).
Pesticides are often applied as a mixture in a simultaneous or sequential manner during crop cultivation, or found as mixtures in the environment, making the combined effects of active principles an important question. We therefore next tested QpE in combination with 10 µM of glyphosate, 2, 4 D, dicamba, mesotrione, and isoxaflutole to determine if they were able to potentiate the adipogenic effect of QpE. No additive effects of these active ingredients on QpE action were observed under any of the mixture conditions (Supplementary Figure 3).
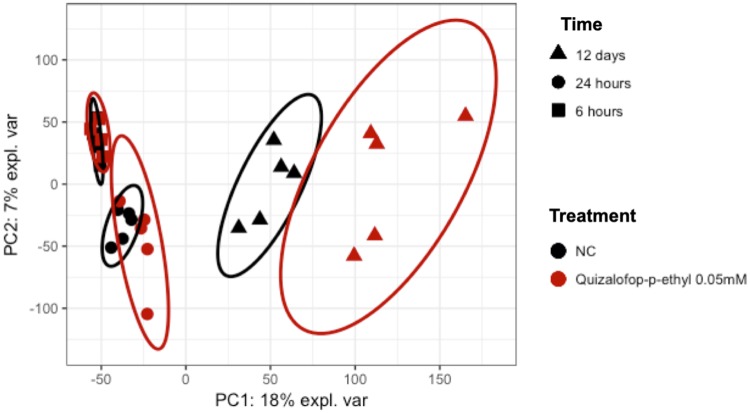
We next conducted a transcriptome analysis of QpE-exposed 3T3-L1 cells in an effort to obtain mechanic insight into the adipogenic capability of this compound through differential gene expression analysis. RNA-seq was conducted on the Illumina sequencing platform with samples from 3T3-L1 cells either treated or untreated with 50 μM QpE. Cells were harvested and RNA extracted at 6, 24, and 288 h (12 days) following treatment. Initially, a principal component analysis (PCA) was conducted on the FPKM (Fragments Per Kilobase Million) values (Figure 5). The first 2 components accounted for 25% of the total variability in our data, which was mainly driven by the separation between the different time point groups. However, there was also a clear separation between the treatment groups showing that QpE had a significant effect on the gene expression profile of the 3T3-L1 cells (Figure 5). We used the Enrichr gene set enrichment analysis web server to investigate pathway over-representation in the gene expression alterations caused by QpE (Table 1) in conjunction with the pathway databases Kyoto Encyclopedia of Genes and Genomes (KEGG), Wikipathways, and Reactome. No pathways were found to be significantly disturbed after 6 h of treatment as indicated. Contrastingly, there was a statistically significant enrichment of adipogenesis pathways after 24 h treatment. In particular the enrichment of the pathway “Nuclear receptors in lipid metabolism and toxicity” was a clear confirmation that QpE had an adipogenic effect. Although this doesn’t directly provide information on the modulators of gene expression involved in the observed adipogenesis process, this result suggests that the effects of QpE could be mediated by a nuclear receptor whose modulation influences lipid metabolism. The pathway enrichment analysis of the transcriptome profile alterations caused by a 12-day exposure to QpE mostly reflected the consequence of the lipid accumulation. The most altered metabolic pathways reflected mitochondrial functional disturbances. The most significantly altered pathway was that for nonalcoholic fatty liver disease. Then, we attempted to identify the receptor mediating QpE adipogenic effects by analyzing the overrepresentation of transcription factor binding sites in the promoters of the genes having their expression altered by QpE (Supplementary Figure 4). The transcription factor, which was likely to bind to the genes whose expression was altered after 6 h QpE exposure was the upstream stimulatory factor-2, which is a known modulator of lipid metabolism, but which does not provide a mechanism of action for QpE. The genes with altered expression highlighted by the transcriptome profiles after 24 h and 12 days exposure were enriched in PPARγ binding sites. However, key adipogenic genes such as Fabp4, Lpl, and Adipoq did not have their levels altered by QpE.
Figure 5.
RNA-seq reveals alterations of adipocyte transcriptome profiles by QpE. PCA of the 3T3-L1 cell transcriptome following exposure to 50 μM QpE for 6, 24, and 288 h (12 days) compared with untreated control cultures (NC).
Table 1.
Pathway Enrichment Analysis of the Transcriptome of 3T3-L1 Cells Exposed to QpE Shows Lipid Metabolism Dysregulation Associated With Markers of Nuclear Receptor Activation
| Source | pathway_name | p | Adj-p |
|---|---|---|---|
| 6 h | |||
| WP | Exercise-induced Circadian Regulation_Mus musculus_WP544 | 2.8E-03 | 2.3E-01 |
| Diurnally Regulated Genes with Circadian Orthologs_Mus musculus_WP1268 | 3.2E-02 | 5.6E-01 | |
| G Protein Signaling Pathways_Mus musculus_WP232 | 9.4E-02 | 5.6E-01 | |
| Fatty Acid Biosynthesis_Mus musculus_WP336 | 1.2E-01 | 5.6E-01 | |
| Signal Transduction of S1P Receptor_Mus musculus_WP57 | 1.2E-01 | 5.6E-01 | |
| KEGG | Fanconi anemia pathway_hsa03460 | 3.7E-03 | 3.4E-01 |
| Circadian rhythm_hsa04710 | 1.3E-02 | 6.2E-01 | |
| Toxoplasmosis_hsa05145 | 3.2E-02 | 6.2E-01 | |
| Rap1 signaling pathway_hsa04015 | 3.6E-02 | 6.2E-01 | |
| Spliceosome_hsa03040 | 4.4E-02 | 6.2E-01 | |
| 24 h | |||
| WP | Adipogenesis genes_Mus musculus_WP447 | 1.8E-05 | 2.6E-03 |
| Nuclear receptors in lipid metabolism and toxicity_Mus musculus_WP431 | 1.8E-03 | 9.3E-02 | |
| Focal Adhesion-PI3K-Akt-mTOR-signaling pathway_Mus musculus_WP2841 | 8.1E-03 | 1.6E-01 | |
| miRNA regulation of DNA Damage Response_Mus musculus_WP2087 | 5.5E-03 | 1.5E-01 | |
| p53 signaling_Mus musculus_WP2902 | 5.5E-03 | 1.5E-01 | |
| KEGG | Malaria_Homo sapiens_hsa05144 | 2.2E-04 | 4.6E-02 |
| Proteoglycans in cancer_hsa05205 | 1.0E-03 | 1.1E-01 | |
| MAPK signaling pathway_hsa04010 | 2.1E-03 | 1.4E-01 | |
| Cytokine-cytokine receptor interaction_hsa04060 | 7.9E-03 | 3.0E-01 | |
| Melanoma_hsa05218 | 8.5E-03 | 3.0E-01 | |
| 228 h | |||
| WP | Electron Transport Chain_Mus musculus_WP295 | 2.3E-10 | 4.0E-08 |
| Proteasome Degradation_Mus musculus_WP519 | 6.7E-09 | 7.6E-07 | |
| PodNet: protein–protein interactions in the podocyte_Mus musculus_WP2310 | 2.5E-05 | 1.1E-03 | |
| Adipogenesis genes_Mus musculus_WP447 | 1.0E-05 | 5.7E-04 | |
| Oxidative phosphorylation_Mus musculus_WP1248 | 3.1E-05 | 1.2E-03 | |
| KEGG | Nonalcoholic fatty liver disease (NAFLD_hsa04932 | 7.0E-11 | 1.8E-08 |
| Parkinson’s disease _hsa05012 | 3.0E-10 | 3.8E-08 | |
| Alzheimer’s disease _hsa05010 | 1.2E-09 | 8.1E-08 | |
| Oxidative phosphorylation_hsa00190 | 1.3E-09 | 8.1E-08 | |
| Huntington’s disease_hsa05016 | 1.2E-08 | 6.0E-07 |
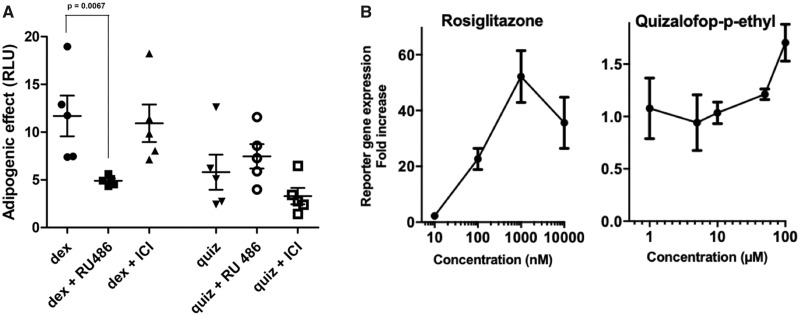
As PPARγ activation is known to be a master regulator of adipogenesis (Kershaw et al., 2007; Shao et al., 2016), we then conducted a series of experiments to further investigate the mode of action of QpE. Neither the estrogen receptor antagonist ICI 182, 780 nor the glucocorticoid receptor antagonist RU486 was able to block QpE-mediated lipid accumulation (Figure 6A). We also tested the ability of QpE to bind and activate PPARγ in a luciferase reporter gene assay. The basis of this commercially available system is a double-transgenic cell line, which constitutively expresses human PPARγ and harbors a luciferase reporter gene driven by a PPARγ-responsive promoter. Thus, in the presence of PPARγ agonists an increase in luciferase gene expression will be observed. Treatment with the positive control substance rosiglitazone gave a maximum stimulation of approximately 50-fold at 1 μM concentration (Figure 6B). QpE also resulted in a modest but consistent increase in luciferase expression of approximately 2-fold at 100 μM, which is albeit only 4% of the maximal response caused by rosiglitazone. This suggests that the QpE activation of PPARγ is only in part a contributory mechanism to the lipid accumulation capability displayed by this compound (Figure 6B).
Figure 6.
QpE effects on nuclear receptors. A, The 3T3-L1 adipocyte differentiated cells were treated with Dex (10 nM) and QpE (quiz, 100 µM) in the presence or absence of either 1 µM ICI 182, 780 or RU486. B, PPARγ assay using a luciferase reporter gene system shows a consistent but modest increase in expression by QpE in comparison to rosiglitazone. (Note: different scales in the y-axis). Note: cells treated with 17β-estradiol alone did not cause any lipid accumulation, the results of which in the interests of clarity of presentation are not shown.
DISCUSSION
The global epidemic of obesity is multifactorial in origin and includes exposure to chemicals acting as endocrine disruptors. We have tested for the first time commonly used herbicide active principles in an adipogenesis assay to evaluate their obesogenic potential. Using the 3T3-L1 cell assay system, which scores for lipid accumulation following differentiation to adipocytes, isoxaflutole and dicamba were found to be adipogenic at high concentrations only, whilst glyphosate and mesotrione scored negative at all concentrations tested. Contrastingly, we found that QpE was a potent and efficient inducer of increased lipid accumulation in 3T3-L1 adipocytes (Figs. 2 and 3). Transcriptomics analysis revealed that QpE possesses the profile of a PPAR-γ agonist (Table 1; Supplementary Figure 4). The potential for QpE to activate PPAR-γ was suggested in a luciferase reporter gene assay (Figure 6B). However, PPAR-γ activation occurred at QpE concentrations 100-times higher than those which induced lipid accumulation, and reached only 4% of the maximal response caused by the potent agonist rosiglitazone. This suggests that the adipogenic effect of QpE in 3T3-L1 cells is only partially accountable by PPAR-γ activation.
Further research is required to understand the molecular mechanisms by which QpE causes lipid accumulation. It is likely that QpE acts on the PPAR-γ pathway independently of the activation of the receptor. In support of this possibility is the recently reported observation that the fungicide active ingredient pyraclostrobin caused lipid accumulation by inducing mitochondrial dysfunction independent of PPARγ activation (Luz et al., 2018).
The liver has been proposed to be the main target of toxicity for QpE in studies conducted for regulatory purposes (European Food Safety Authority, 2009). The administration of 3.7 mg/kg bw/day QpE to laboratory rats increased the number of liver masses and centrilobular enlargement of this organ in both sexes (EPA, 2016). Few peer-reviewed studies are available, but a case report has confirmed that commercial formulations of pesticides containing QpE can also be a probable inducer of cholestatic/hepatocellular liver injury in human (Elefsiniotis et al., 2007). Hepatic cholestasis can be accompanied by fatty changes (Jüngst et al., 2013), and it is likely that the mechanisms of action of QpE-induced lipid accumulation in our study can be related to a role of QpE as a probable inducer of occupational liver injury. HepaRG human liver cells have been described as a reliable model to understand the mechanism of toxicant-induced steatosis such as for lipid uptake, lipid efflux, fatty acid oxidation, and lipid accumulation (Angrish et al., 2017). We recently showed that QpE caused alterations in gene expression that are associated with changes in fatty acid degradation and response to alcoholism in HepaRG cells (Mesnage et al., 2018b).
Our study indicated that QpE but not his metabolites induced lipid accumulation in 3T3-L1 adipocytes. Quizalofop herbicides show a relatively rapid absorption and distribution, and rather slow elimination in urine and feces with only 56%–70% of QpE being eliminated after administration to rats (European Food Safety Authority, 2009). Upon ingestion, it is rapidly metabolized in quizalofop acid, followed by hydroxylation. Only quizalofop acid but not QpE was detected after the administration of 10 mg/kg QpE in rats (Liang et al., 2014). It is interesting to note that QpE can be found as 2 different enantiomers potentially having different toxicological properties. For instance, R(+)-forms of aryloxypropanoates are more potent acetyl coenzyme A carboxylase inhibitors and thus have higher herbicidal activities (Kurihara et al., 1997). In a toxicokinetic study in rats, the concentrations of R(+)-quizalofop-acid exceeded those of R(−)-quizalofop-acid (Liang et al., 2014). Only the R(+) enantiomers of quizalofop were present in large excess in rat body fluids. The differential toxicity of the different forms of quizalofop is unclear. However, a recent study investigating the toxicity of QpE and its metabolite quizalofop acid on the worm Eisenia foetida found that the metabolite was far more toxic than the parent compounds (Ma et al., 2016). The same study also showed QpE and quizalofop acid enantioselective toxicity to these earthworms, with the racemic mixtures being more toxic than the R(+) forms alone (Ma et al., 2016).
We have also compared the toxicity of glyphosate, QpE, mesotrione, and isoxaflutole on HepaRG human liver cells (Mesnage et al., 2018b). The differences in toxicity between these compounds were similar to what we found in this study. QpE was the most toxic and affected the expression of genes involved in lipid metabolism (Mesnage et al., 2018b). Isoxaflutole was less toxic to HepaRG cells but caused alterations in gene expression profiles correlated with activation of nuclear receptors such as the pregnane X receptor and PPAR-γ. By comparison, glyphosate and mesotrione caused only few and subtle changes, as observed here.
Our study uses the well-established murine 3T3-L1 cell line model system for obesogenic screening. However, this cell line can only address a limited number of possible modes of action. The 3T3-L1 cells consist of unipotent preadipocytes, which can only differentiate into mature adipocytes. Mesenchymal stem cells may constitute a suitable alternative model system due to their multipotent nature and which can also be used to study earlier stages of the adipocyte differentiation pathway (Kirchner et al. 2010). Another important limitation of 3T3-L1 cells that needs to be born in mind is that they are of murine origin, and may not fully be representative of human metabolism. In addition, stocks of 3T3-L1 cells from different sources can have different metabolic capabilities (Kassotis et al., 2017). A recent comparison of different protocols and cell lines used in adipogenesis assays revealed that cell source (ATCC vs Zenbio 3T3-L1) and culture protocols have a significant impact on the results of this assay (Kassotis et al., 2017).
The commercial formulated product Targa Super, containing QpE as an active ingredient, failed to induce adipogenicity (Figure 2). This appears to be due to the fact that we found Targa Super to be approximately 1000 times more toxic than QpE itself. This does not come as a surprise since this pesticide formulation contained naphtha (petroleum) solvent and super heavy aromatic compounds (<1% naphthalene). However, it is still surprising that the authors of scientific studies keep confounding the declared active principles of a pesticide with the pesticide itself (Mesnage and Antoniou, 2017). For example, the report describing liver injury induced by the spraying of a QpE-based formulation refers to QpE as a herbicide, and does not discuss the composition of the pesticide sprayed by the subject of this case study (Elefsiniotis et al., 2007). Thus, it is not possible to be certain that the effects described in this report are due to QpE itself or to a coformulant in the commercial preparation, or combination of the 2, making the conclusion of the authors unsubstantiated. Other commercial formulations of QpE, or of other pesticide ingredients, may have different effects because they contain different mixtures of coformulants, some of which have been showed to cause adipogenic effects (Kassotis et al., 2018).
In conclusion, we reveal alterations of adipocyte metabolism by major pesticide active ingredients. In particular, we found that QpE was a potent stimulator of lipid accumulation and thus potentially a hitherto unsuspected obsogen. QpE is promoted as a new selective postemergence herbicide to control weeds resistant to glyphosate, and thus its agricultural use is expected to increase in coming years. Although our results cannot be readily translated to policy recommendations, it is clear from the study presented here and work conducted by others that a proper comparative risk-benefit analysis should be performed when new pesticide active ingredients are promoted as substitutes.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Sustainable Food Alliance (USA) whose support is gratefully acknowledged.
DECLARATION OF CONFLICTING INTERESTS
RM has served as a consultant on glyphosate risk assessment issues as part of litigation in the US over glyphosate health effects. All other authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
AUTHOR CONTRIBUTIONS
M.B. performed the cell culture experiment, the RNA extraction, and drafted the article. R.M. performed the transcriptome data analysis and drafted the article. E.W., T.X., and C.A.M. performed the RNA-seq. R.F. assisted with the transcriptome data analysis and performed the PPARγ reporter gene activation assays. MNA coordinated the investigation and assisted in the drafting of the article.
REFERENCES
- Angrish M. M., McQueen C. A., Cohen-Hubal E., Bruno M., Ge Y., Chorley B. N. (2017). Editor’s highlight: Mechanistic toxicity tests based on an adverse outcome pathway network for hepatic steatosis. Toxicol. Sci. 159, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbrook C. M. (2016). Trends in glyphosate herbicide use in the United States and globally. Environ. Sci. Eur. 28, 3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher J. G., Boudreau A., Ahmed S., Atlas E. (2015). In vitro effects of bisphenol A beta-D-glucuronide (BPA-G) on adipogenesis in human and murine preadipocytes. Environ. Health Perspect. 123, 1287–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Sancho G., Salmon A. G., La Merrill M. A. (2017). Association between exposure to p, p'-DDT and its metabolite p, p'-DDE with obesity: Integrated systematic review and meta-analysis. Environ. Health Perspect. 125, 096002.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro-Garcia R., Kirchner S., Li X., Janesick A., Casey S. C., Chow C., Blumberg B. (2012). Bisphenol A diglycidyl ether induces adipogenic differentiation of multipotent stromal stem cells through a peroxisome proliferator-activated receptor gamma-independent mechanism. Environ. Health Perspect. 120, 984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro-Garcia R., Sahu M., Abbey R. J., Laude J., Pham N., Blumberg B. (2013). Transgenerational inheritance of increased fat depot size, stem cell reprogramming, and hepatic steatosis elicited by prenatal exposure to the obesogen tributyltin in mice. Environ. Health Perspect. 121, 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaboration N. C. D. R. F. (2017). Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: A pooled analysis of 2416 population-based measurement studies in 128.9 million children, adolescents, and adults. Lancet 390, 2627–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damalas C. A., Eleftherohorinos I. G. (2011). Pesticide exposure, safety issues, and risk assessment indicators. Int. J. Environ. Res. Public Health 8, 1402–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elefsiniotis I. S., Liatsos G. D., Stamelakis D., Moulakakis A. (2007). Case report: Mixed cholestatic/hepatocellular liver injury induced by the herbicide quizalofop-p-ethyl. Environ. Health Perspect. 115, 1479–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPA. (2016). Quizalofop Ethyl; Pesticide Tolerances. Federal Register/Vol. 81, No. 231/Thursday, December 1, 2016/Rules and Regulations. [EPA–HQ–OPP–2015–0412; FRL–9950–89].
- European Food Safety Authority. (2009). Conclusion regarding the peer review of the pesticide risk assessment of the active substance quizalofop-P. EFSA J. 205, 1–216. [Google Scholar]
- Ghosh S., Bouchard C. (2017). Convergence between biological, behavioural and genetic determinants of obesity. Nat. Rev. Genet. 18, 731–748. [DOI] [PubMed] [Google Scholar]
- Grun F., Watanabe H., Zamanian Z., Maeda L., Arima K., Cubacha R., Gardiner D. M., Kanno J., Iguchi T., Blumberg B. (2006). Endocrine-disrupting organotin compounds are potent inducers of adipogenesis in vertebrates. Mol. Endocrinol. 20, 2141–2155. [DOI] [PubMed] [Google Scholar]
- Heap I., Duke S. O. (2018). Overview of glyphosate-resistant weeds worldwide. Pest Manag. Sci. 74, 1040–1049. [DOI] [PubMed] [Google Scholar]
- Janesick A., Blumberg B. (2011). Endocrine disrupting chemicals and the developmental programming of adipogenesis and obesity. Birth Defects Res. C Embryo Today 93, 34–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janesick A. S., Blumberg B. (2016). Obesogens: An emerging threat to public health. Am. J. Obstet. Gynecol. 214, 559–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jüngst C., Berg T., Cheng J., Green R. M., Jia J., Mason A. L., Lammert F. (2013). Intrahepatic cholestasis in common chronic liver diseases. Eur. J. Clin. Invest. 43, 1069–1083. [DOI] [PubMed] [Google Scholar]
- Kassotis C. D., Masse L., Kim S., Schlezinger J. J., Webster T. F., Stapleton H. M. (2017). Characterization of adipogenic chemicals in three different cell culture systems: Implications for reproducibility based on cell source and handling. Sci. Rep. 7, 42104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassotis C. D., Kollitz E. M., Ferguson P. L., Stapleton H. M. (2018). Nonionic ethoxylated surfactants induce adipogenesis in 3T3-L1 cells. Toxicol. Sci. 162, 124–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw E. E., Schupp M., Guan H. P., Gardner N. P., Lazar M. A., Flier J. S. (2007). PPARgamma regulates adipose triglyceride lipase in adipocytes in vitro and in vivo. Am. J. Physiol. Endocrinol. Metab. 293, E1736–E1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner S., Kieu T., Chow C., Casey S., Blumberg B. (2010). Prenatal exposure to the environmental obesogen tributyltin predisposes multipotent stem cells to become adipocytes. Mol. Endocrinol. 24, 526–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandez N. F., Duan Q., Wang Z., Koplev S., Jenkins S. L., Jagodnik K. M., Lachmann A., et al. (2016). Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara N., Miyamoto J., Paulson G. D., Zeeh B., Skidmore M. W., Hollingworth R. M., Kuiper H. A. (1997). Pesticides report 37: Chirality in synthetic agrochemicals: Bioactivity and safety consideration (Technical Report). Pure Appl. Chem., 69, 2007. [Google Scholar]
- Li X., Ycaza J., Blumberg B. (2011). The environmental obesogen tributyltin chloride acts via peroxisome proliferator activated receptor gamma to induce adipogenesis in murine 3T3-L1 preadipocytes. J. Steroid Biochem. Mol. Biol. 127, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz A. L., Kassotis C. D., Stapleton H. M., Meyer J. N. (2018). The high-production volume fungicide pyraclostrobin induces triglyceride accumulation associated with mitochondrial dysfunction, and promotes adipocyte differentiation independent of PPARγ activation, in 3T3-L1 cells. Toxicology 393, 150–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Wang P., Liu D., Shen Z., Liu H., Jia Z., Zhou Z. (2014). Enantioselective metabolism of quizalofop-ethyl in rat. PLoS One 9, e101052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Liu H., Qu H., Xu Y., Wang P., Sun M., Zhou Z., Liu D. (2016). Chiral quizalofop-ethyl and its metabolite quizalofop-acid in soils: Enantioselective degradation, enzymes interaction and toxicity to Eisenia foetida. Chemosphere 152, 173–180. [DOI] [PubMed] [Google Scholar]
- Mesnage R., Antoniou M. N. (2017). Ignoring adjuvant toxicity falsifies the safety profile of commercial pesticides. Front. Public Health 5, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage R., Biserni M., Balu S., Frainay C., Poupin N., Jourdan F., Wozniak E., Xenakis T., Mein C. A., Antoniou M. A. (2018a). Integrated transcriptomics and metabolomics reveal signatures of lipid metabolism dysregulation in HepaRG liver cells exposed to PCB 126. Arch. Toxicol. 92, 2533–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage R., Biserni M., Wozniak E., Xenakis T., Mein C. A., Antoniou M. N. (2018b). Comparison of transcriptome responses to glyphosate, isoxaflutole, quizalofop-p-ethyl and mesotrione in the HepaRG cell line. Toxicol. Rep. 5, 819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage R., Defarge N., Spiroux de Vendomois J., Seralini G. E. (2015). Potential toxic effects of glyphosate and its commercial formulations below regulatory limits. Food Chem. Toxicol. 84, 133–153. [DOI] [PubMed] [Google Scholar]
- Mesnage R., Phedonos A., Biserni M., Arno M., Balu S., Corton J. C., Ugarte R., Antoniou M. N. (2017). Evaluation of estrogen receptor alpha activation by glyphosate-based herbicide constituents. Food Chem. Toxicol. 108, 30–42. [DOI] [PubMed] [Google Scholar]
- Morrison S., McGee S. L. (2015). 3T3-L1 adipocytes display phenotypic characteristics of multiple adipocyte lineages. Adipocyte 4, 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. (1983). Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63. [DOI] [PubMed] [Google Scholar]
- Myers J. P., Antoniou M. N., Blumberg B., Carroll L., Colborn T., Everett L. G., Hansen M., Landrigan P. J., Lanphear B. P., Mesnage R., et al. (2016). Concerns over use of glyphosate-based herbicides and risks associated with exposures: A consensus statement. Environ. Health 15, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbold R. R. (2010). Impact of environmental endocrine disrupting chemicals on the development of obesity. Hormones (Athens) 9, 206–217. [DOI] [PubMed] [Google Scholar]
- R Core Team (2017). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria: Available at: https://www.R-project.org. Accessed April 24, 2019. [Google Scholar]
- Rosenbaum P. F., Weinstock R. S., Silverstone A. E., Sjodin A., Pavuk M. (2017). Metabolic syndrome is associated with exposure to organochlorine pesticides in Anniston, AL, United States. Environ. Int. 108, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X., Wang M., Wei X., Deng S., Fu N., Peng Q., Jiang Y., Ye L., Xie J., Lin Y. (2016). Peroxisome proliferator-activated receptor-gamma: Master regulator of adipogenesis and obesity. Curr. Stem Cell Res. Ther. 11, 282–289. [DOI] [PubMed] [Google Scholar]
- Thayer K. A., Heindel J. J., Bucher J. R., Gallo M. A. (2012). Role of environmental chemicals in diabetes and obesity: A National Toxicology Program workshop review. Environ. Health Perspect. 120, 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilopoulou L., Psycharakis C., Petrakis D., Tsiaoussis J., Tsatsakis A. M. (2017). Obesity, persistent organic pollutants and related health problems. Adv. Exp. Med. Biol. 960, 81–110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.