Abstract
Applying toxicogenomics to improving the safety profile of drug candidates and crop protection molecules is most useful when it identifies relevant biological and mechanistic information that highlights risks and informs risk mitigation strategies. Pathway-based approaches, such as gene set enrichment analysis, integrate toxicogenomic data with known biological process and pathways. Network methods help define unknown biological processes and offer data reduction advantages. Integrating the 2 approaches would improve interpretation of toxicogenomic information. Barriers to the routine application of these methods in genome-wide transcriptomic studies include a need for “hands-on” computer programming experience, the selection of 1 or more analysis methods (eg pathway analysis methods), the sensitivity of results to algorithm parameters, and challenges in linking differential gene expression to variation in safety outcomes. To facilitate adoption and reproducibility of gene expression analysis in safety studies, we have developed Collaborative Toxicogeomics, an open-access integrated web portal using the Django web framework. The software, developed with the Python programming language, is modular, extensible and implements “best-practice” methods in computational biology. New study results are compared with over 4000 rodent liver experiments from Drug Matrix and open TG-GATEs. A unique feature of the software is the ability to integrate clinical chemistry and histopathology-derived outcomes with results from gene expression studies, leading to relevant mechanistic conclusions. We describe its application by analyzing the effects of several toxicants on liver gene expression and exemplify application to predicting toxicity study outcomes upon chronic treatment from expression changes in acute-duration studies.
Keywords: toxicogenomics, systems biology, gene expression analysis, mechanism inference, toxicity prediction
Despite significant advances, determining the relevance of toxicity findings from animal safety studies to human health effects is challenging. Identifying the mechanistic underpinnings of a toxicity finding is key, and characterization of tissue samples using gene expression is often performed toward this end. Initiatives to study a broad range of toxicants and reference molecules has produced rich contextual information aiding the interpretation of gene expression changes produced by a molecule of interest (Ganter et al., 2005; Igarashi et al., 2015). However, these data are challenging to process, search, and analyze. Furthermore, many analysis algorithms are available, ranging from mature approaches such as overrepresentation analysis (widely used in the DAVID web application) (Huang et al., 2009), the related gene set enrichment analysis (GSEA) method (Subramanian et al., 2005) and Connectivity Map (CMAP) (Lamb et al., 2006; Smalley et al., 2010), to newer methods purporting to offer additional insights (Bell et al., 2016; Lee et al., 2014; Tawa et al., 2014; Te et al., 2016). These methods are generally not integrated and utility is often limited to computational specialists, who themselves may struggle to implement methods or reproduce results from the literature without having access to source code and detailed protocols. Finally, for those approaches that offer user-friendly interfaces, a user must navigate the process of producing “analysis-ready” fold change results for each gene and upload them in multiple applications.
Herein, we sought to improve the integration of standard methods with newer network-based methods to improve usability for toxicologists and mechanistic interpretation relevant to risk assessment. We describe the creation of an open-source, publicly available platform for the analysis of gene expression results from toxicity studies. Although the platform was developed using rat liver studies, it can be used to analyze data from any study comparing the effect of a treatment or other intervention to control samples. We have processed and loaded within the application results from Drug Matrix (DM) (Ganter et al., 2005) and TG-GATEs (TG) (Igarashi et al., 2015), which between them provide results on 4182 liver experiments. This includes both expression results and the histology/clinical chemistry data within those repositories, analyzed using a shared lexicon and standardized between the sources. In addition to analyzing their uploaded data, users can query DM and TG experiments and analyze results at the level of individual genes and gene sets. Connectivity Map (Lamb et al., 2006; Smalley et al., 2010) and related approaches attempt to infer properties of a treatment of interest by identifying other drugs or tool compounds having similar effects on the global transcriptome. Users can identify the experiments from DM/TG having the most similar transcriptional effects.
Previously, we described an approach linking perturbation of co-expression networks (or “modules”) to a variety of toxicity phenotypes (TXG-MAP analysis) (Sutherland et al., 2018). This allows the identification of networks perturbed by a molecule of interest that are also generally associated with its observed toxicity across a broad range of conditions. Here, we extend this approach by identifying a nonredundant collection of gene ontology (GO) and canonical pathways that can be modulated in liver toxicity studies.
MATERIALS AND METHODS
Web application and availability
A web application was created using the Python and R programming languages. The application consists of a PostgreSQL database, Python, and R backend computation scripts, celery queuing system for job management (http://www.celeryproject.org/; accessed August 29, 2018). The user interface was developed using the Django web framework and the HighCharts JavaScript library for visualizations (https://www.highcharts.com/; accessed August 29, 2018). All data files and source code are provided in the GitHub repository hosted at https://github.com/IndianaBiosciences/toxapp; accessed April 26, 2019. References to file names below refer to their location in the repository. The public instance of the application is hosted on Linux servers from Amazon Web Services. The application has been thoroughly tested on Ubuntu 16.04 LTS and Centos 7.4. A deployment script (setup_server.sh) allows private instances to be created with all requirements satisfied.
Gene-level data preparation
Throughout, an “experiment” denotes a comparison of treatment versus control samples; a treatment consists of a combination of drug, dose, duration of dosing, route of administration. More broadly, an experiment (or intervention) can be used to compare animals harboring null alleles of a gene of interest (ie a knockout animal) versus wild-type animals. In DM (Ganter et al., 2005) and TGs (Igarashi et al., 2015), experiments compare the effects of drug treatment in livers from 2 to 5 treated animals versus 2 to 5 vehicle matched controls. The complete list of 3528 TG and 654 DM experiments viewable in the application is available in Supplementary Table 1 of our prior work (Sutherland et al., 2018) and the repository file data/experiments_DM_TG.txt.
Affymetrix CEL files for individual rat liver samples analyzed with RG230-2 microarrays were obtained from DM (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE57815, accessed April 26, 2019) and the TGs repositories (http://toxico.nibiohn.go.jp/english/; accessed August 29, 2018). Normalized log intensities were calculated using the University of Michigan “Brain Array” version 19 assignment of microarray probes to genes (http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/19.0.0/entrezg.asp; accessed August 29, 2018) and the RMA algorithm (Irizarry et al., 2003) as implemented in the Affy package in Bioconductor (scripts/NORM.R). For any given experiment, fold change values for each gene were obtained using the Bioconductor package Limma (scripts/Limma.R). Preprocessed fold-change results, consisting of log2 fold change (log2_fc), number of treated animals (n_trt), number of control animals (n_ctl), average gene intensity in controls (expression_ctl0), unadjusted p value (p), and Benjamini Hochberg-adjusted p value (p_bh) are available in the data repository at location data/groupFC. For example, repeat-dose results for gemfibrozil (from TGs) are in the file data/groupFC/gemfibrozil.Rat.in_vivo.Liver.Repeat-groupFC.txt.
Gene model and orthology
The rat genome was selected as the reference organism, using the Entrez gene nomenclature. The application supports the upload of rat, mouse, and human gene expression data. Mapping of human and mouse genes to rat was performed using orthology information from RGD (Shimoyama et al., 2015) as described in Sutherland et al. (2016). The gene model includes all rat genes according to RGD, whether or not they are tested on a given measurement technology. However, human or mouse genes having no rat ortholog were excluded, and users uploading data for these organisms will not obtain fold-change results for such genes. At the level of pathways or modules where results are aggregated across several genes (see below), the impact is trivial. The file data/gene_info.txt includes 18 004 rat Entrez gene ids, mapped to 17 540 mouse and 17 820 human genes.
TXG-MAP co-expression analysis (modules)
Co-expression network analysis seeks to identify genes that respond similarly to perturbations in a biological system of interest. We used the Bioconductor package WGCNA to identify groups of co-expressed genes (or “modules”) from the DM rat liver data. Algorithm parameters and details on the creation the 415 modules were provided in Sutherland et al. (2016). Because the modules are associated with several toxicity phenotypes, we refer to the method as “Toxicogenomic Module Association with Pathogenesis” (TXG-MAP). A visual representation of the modules on a phylogenetic tree provides a consistent visual frame of reference for analysis. Module scores are presented as Z-scores, such that a score of 2 is achieved by fewer than 4% of drug treatments, a score of 3 in fewer than 1%, etc. This indicates the magnitude of an effect in the context of other drug treatments.
Here, we updated the modules by replacing the original Affymetrix probe sets with Brain Array definitions (via rat Entrez gene IDs to which both were mapped 1:1). The assignment of genes to modules is provided in data/WGCNA_modules.txt, and module scores for a given experiment calculated from the method score_modules in the source package src/computation.py.
Gene set analysis
Gene set analysis seeks to identify canonical pathways, GO and other gene sets that are enriched among the most differentially expressed genes. We used the PAGE algorithm (Kim and Volsky, 2005) as implemented in the Bioconductor package Piano, given similar performance compared with the popular GSEA algorithm and significantly faster calculation (Varemo et al., 2013). Gene sets used in the analysis were taken from GO and Molecular Signatures Database (MSigDB), as described in Sutherland et al. (2016). The assignment of genes to gene sets is provided in data/rgd_vs_GO_expansion.txt and data/MSigDB_and_TF_annotation.txt. The method score_gsa in the source package src/computation.py performs the calculations in R. Gene sets from GO and MSigDB are numerous and overlapping. Limiting to GO and curated pathways from MSigDB, there are 11 581 gene sets for consideration. In prior work, we noted that 1839 of these gene sets were inducible or repressible in 1% or more of 3528 TG experiments (in brief, “inducible gene sets”). The vast majority of gene sets represent molecular processes that are either absent from liver or rarely affected by drug perturbation.
To eliminate redundancy among related gene sets, the pvclust R library was used to hierarchically cluster 1839 gene sets versus gene set analysis (GSA) scores from all 3528 TG liver experiments (command in R: pvclust(data, method.dist=“cor”, method.hclust=“ward.D”, nboot = 1000, parallel = TRUE)). Clustering algorithms may create too many clusters, or divisions of samples that are no longer observed when the dataset is changed (here, the term sample from statistics is a liver experiment comparing treated versus control liver expression, not a liver sample). The algorithm evaluates the significance of edges in the dendrogram while performing bootstrap resampling of the samples. The method reduce_tree in the src/treemap.py package takes as input the dendrogram exported from R, traces up the dendrogram from terminal to root nodes and finds the edge nearest the root that is nonsignificant. All gene sets below the edge are merged into a single cluster. The related method reduce_tree_pca uses principal components analysis on a table of GSA scores having clustered gene sets on columns and 3528 liver experiments on rows. The gene set with highest loading in the first principal component is selected as a representative for the cluster.
A visual representation of the nonredundant gene sets was created from the pruned dendrogram returned by the reduce_tree_pca method. The tree was truncated at a height of 15, a threshold selected to separate large branches near the tree root. Visually, we found this to improve the aesthetics and interpretability of results by making better use of 2D plane (ie web browser screen space). The truncated dendrogram was rendered using the yFiles circular layout algorithm in Cytoscape.
Clinical chemistry and histology results
Clinical chemistry and histology results from DM and TG experiments are available from the respective repositories. A simplified view of these results are provided within our application, by focusing on a limited number of nonredundant endpoints encountered 10 or more times across the combined DM and TG datasets; details on the curation process and lexicon creation are given in Sutherland et al. (2018). The following histology endpoints are included: Apoptosis/SingleCellNecrosis: Hepatocellular; Congestion/Hemorrhage/Edema: Vascular; Degeneration/Necrosis: Hepatocellular; Dilation/Dilatation/Ectasia/Distension: Vascular; Fibrosis; Glycogen_Increased: Hepatocellular; Hematopoiesis; Hyperplasia: Biliary; Hypertrophy: Hepatocellular; Infiltration/Inflammation; Mitosis_Increased: Hepatocellular; Vacuolation: Hepatocellular. The following clinical chemistry endpoints are albumin (ALB), alkaline phosphatase (ALP), alanine aminotransferase (ALT), aspartate aminotransferase (AST), cholesterol (chol), gamma-glutamyl transpeptidase (GGT), glucose (Glu), total bilirubin (T Bili), total protein (TP), triglycerides (Trig). The source file containing these results for each experiment is data/toxicology_results_DM_TG.txt.
Association of gene sets with toxicity phenotypes
Previously, we described the assignment of DM and TG experiments to 13 representative toxicity phenotypes (Supplementary Table 4 in Sutherland et al. [2018]). A toxicity phenotype denotes the occurrence of 1 or more dominant histological or clinical chemistry changes. For example, the phenotype “Necrosis, 1) no other findings” denotes the occurrence of 2 minimal grades, 1 slight grade, or greater severity of hepatocellular necrosis among 3 animals in a dose group, without other significant co-occurring histological changes (“pure” necrosis). In addition, we defined an additional composite phenotype of “adverse at 29 days” which included histological changes such as necrosis, single cell necrosis, bile duct hyperplasia that are deemed adverse by pathologists in studies of 29-day duration. The 14 toxicity phenotypes were “adverse at 29 days,” “bile duct hyperplasia,” “cholesterol decrease,” “cholesterol increase,” “fibrosis,” “hematopoiesis,” “hypertrophy,” “increased glycogen,” “increased mitosis,” “necrosis,” “single cell necrosis,” “trigs (triglyceride) decrease,” “trigs increase,” “vacuolation.” These abbreviated phenotype terms correspond to the detailed curated histology labels above: “necrosis” aligns to “Degeneration/Necrosis: Hepatocellular,” meaning that a lesion denoted as necrosis, degeneration, or degeneration and necrosis of hepatocytes would be assigned to the “necrosis” toxicity phenotype.
DM and TG experiments were labeled as positive or negative for each of the 14 toxicity phenotypes. Repeating our earlier analysis with modules (Sutherland et al., 2018), the statistical association between induction or repression of each gene set (in the 1839 “inducible” set) and the occurrence of the liver phenotype was calculated using logistic regression. The models in the R statistical language were computed as: glm(toxic ∼ avgAbsEG + pathway_GSA_score, family=“binomial”). As described in Sutherland et al. (2018), avgAbsEG is the average absolute module score, a measure which is equivalent to computing the percentage of genes that are differentially expressed. This covariate removes the variation in the odds of observing toxicity that is explained by overall effects of the treatment on gene expression, such that attributing significance to a particular gene set genuinely reflects its role in explaining toxicity, not surrogacy with overall gene expression changes. Finally, we report separately the association between gene sets in toxicity for concurrent injury (expression analysis performed on samples where the lesion is present) versus predictive (expression analysis performed on samples collected at 3, 6, 9, or 24 h of dosing, before the lesion was noted). The source file containing these association results is data/geneset_vs_tox_association.txt.
Expression similarity from microarray versus RNA-seq measurement technologies
To support the validity of comparing experiments analyzed using RNA-seq to experiments from DM and TG using Affymetrix microarrays, datasets were obtained from the Gene Expression Omnibus repository (GEO; https://www.ncbi.nlm.nih.gov/geo/, accessed March 22, 2019) and analyzed with the Collaborative Toxicogenomics (CTox) application.
The first comparison evaluated RNA-seq results from the Sequencing Quality Control (SEQC) consortium (Wang et al., 2014). The file GSE55347_TGxSEQC_GeneExpressionIndex_Magic_20120831_116samplesGSE47875_series_matrix.txt was obtained for GEO series GSE55347. The authors’ mapping of Aceview/Refseq transcripts to NCBI Entrez Gene IDs was used. Because our RNA-seq workflow uses Ensemble identifiers, we mapped Entrez Gene IDs to Ensemble Rat version 6.0 (Rnor_6.0) using the NCBI mapping file (ftp://ftp.ncbi.nih.gov/gene/DATA/gene2ensembl.gz; accessed March 18, 2019). Where multiple Aceview/Refseq transcripts mapped to a single Ensemble gene, only the highest abundance transcript was retained, as assessed by the average abundance across the 116 samples. Finally, because the authors’ pipeline produced continuous abundance estimates with quasi-normal distribution, these were analyzed directly using the R package Limma, producing fold-change and p value of each gene. Computation of pathway, module and experiment similarity were performed in the same manner as all other results processed within the CTox application. Because the SEQC study used DM microarray results, there was no need to process Affymetrix CEL files from GSE47875, as they were already processed and available in the application.
A second comparison evaluated results from a recent study, referred to as “Abbvie study” in results (Rao et al., 2019). As RNA-seq pipelines have become increasingly standardized because the SEQC study, this afforded the opportunity to evaluate results from a representative “industry-standard” approach. The file GSE122315_Raw_counts.txt was obtained from GSE122315. Except for rounding the counts to the nearest integer, this file was processed as-is by the CTox application. Affymetrix RG230-2 CEL files were obtained from the file GSE122184_RAW.tar for series GSE122184, and uploaded using the standard workflow.
The above datasets produced pairs of treatment versus control experiments that differed only in the measurement technology employed. We evaluated the similarity of experiment pairs by calculating the Pearson R correlation coefficient using the 382 nonredundant pathways or the 405 TXG-MAP modules. The experiments in Supplementary Dataset 6 were sorted by the average absolute module score (avgAbsEG), calculated as described in Sutherland et al. (2018).
RESULTS
Web Application
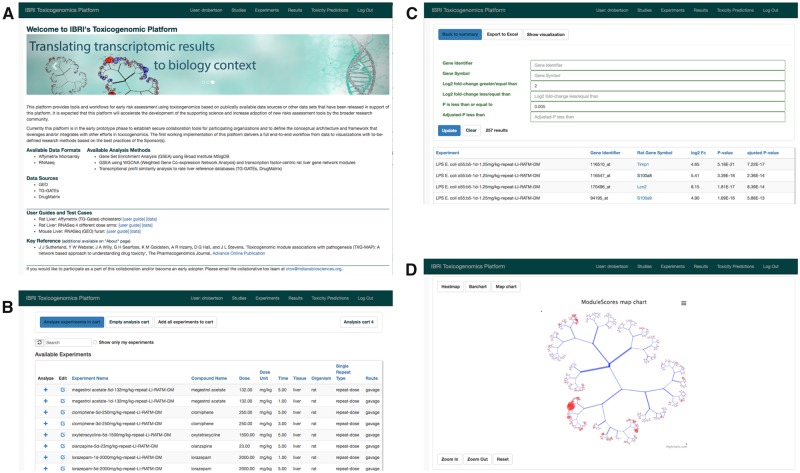
A web application for analysis of gene expression study data was created using the Django web framework. The use of Python, the R statistical language, and PostgreSQL database, technologies with a large user base among scientific developers, will facilitate its extension by others. The CTox application allows users to describe the purpose of a study, treatments/perturbations under investigation and analyze sample results in the form of Affymetrix CEL files or RNA-seq count data. Expression results from rat, mouse, and human are currently supported. Representative application dialogs are shown in Figure 1.
Figure 1.
Illustrating key functionality in the CTox web application. A, Background and user guide links when accessing the application. B, Dialog for searching and adding experiments of interest to an analysis “cart.” C, Viewing gene-level results for experiments in the analysis cart. D, Visualization of co-expression results on the TXG-MAP (see Results section).
Available Gene Expression and Histology/Clinical Chemistry Results
Drug Matrix (Ganter et al., 2005) and the open TGs (Igarashi et al., 2015) repositories are rich resources describing the effects of 308 drugs, toxicants and other perturbations across 4182 liver experiments (an experiment denotes the administration of a drug for at a given dose and duration compared with a control group). We reprocessed all Affymetrix CEL files using updated probe set definitions (Dai et al., 2005) and robust p value estimation using Limma (Irizarry et al., 2003).
Several result types are available when viewing 1 or more experiments (Table 1). Gene-level analysis, whereby the expression change of each gene is reported as a fold change and p value, can be used to identify the most induced or repressed genes. Gene set analysis, implemented using the PAGE algorithm (Kim and Volsky, 2005), is applied to gene sets from GO terms, canonical pathways, and others from the MSigDB (Subramanian et al., 2005). Co-expression networks summarize results across 415 co-expressed gene sets (ie “modules”) obtained by analysis of DM rat liver experiments (Sutherland et al., 2018). When an experiment of interest strongly resembles the global transcriptional profile of a reference compound, they may share a similar mode of action. To that end, comparison of a new experiment to all TG and DM rat liver experiments is easily achieved. Finally, a simplified view of clinical chemistry and histology results across the 2 repositories allows the identification of treatments that lead to a toxicity phenotype (or apical endpoint) of interest and their further characterization in the application.
Table 1.
Analysis Methods Available Within the CTox Web Application
| Method | Summary | Strengths/Weaknesses |
|---|---|---|
| Gene-level analysis | A list of all genes probed by the measurement technology, with fold change and associated p value for treatment versus control | Ease of interpretation, but high risk of false positives due to large number of hypotheses being tested.a High risk of “confirmation bias,” ie focusing on the induction/repression of genes of interest in experiments where many (unrelated) genes are more significantly perturbed by the experimental conditions. |
| GSA | A list of gene sets (GO terms, pathways, and others) for which the constituent genes are disproportionally induced or repressed compared with all measured genes; returns a score and p value for treatment versus control | Simplifies by summarizing effects aggregated across related genes. Ease of interpretation depends on the perturbed gene set; eg “cholesterol biosynthesis” is clear in the context of liver, but “generation of neurons” is not. In theory, lower risk of false positives, however the MSigDB collection now approaches 18 000 gene sets. |
| TXG-MAP module analysis | A list of scores, 1 for each of 415 modules in the TXG-MAP. Modules consist of co-expressed genes. | Simplifies by summarizing across co-expressed genes. Unlike other methods, provides context on magnitude of effect compared with other liver perturbations. Captures co-regulated biology not described by canonical pathways, but interpretation requires study of constituent genes. Results depend on quality/comprehensiveness of training datasets. Low risk of false positives. |
| Similar experiments analysis | A list of most similar experiments, compared with user’s experiment. Global transcriptional similarity, assessed by comparing approximately 400 GSA scores or TXG-MAP scores between experiments. | The fastest way to help understand mechanism, in those cases where a compound of interest is similar to compounds/drugs with well-understood mechanistic effects. Intermediate risk of false positives (ca. 4000 comparisons performed). Requires global similarity (ie expression profile similarity across all pathways/modules); often no high similarity reference compounds and therefore no insights from approach. |
| Clinical chemistry/histology findings | For DM and TGs, standardized clinical chemistry, and histology findings | NA—not expression analysis. When associated with gene expression, results yield meaningful association with standard toxicology interpretations. |
High risk of false positives if not applying a multiple hypothesis testing correction to the p values; when applying such a correction, it becomes a high risk of false negatives—ie a gene is genuinely differentially expressed with a small non-adjusted p value, but after adjusting, the p value (or q value) is no longer small enough to be recognized as significant. The application described herein returns both unadjusted and adjusted p values.
Nonredundant Gene Sets for Liver Analysis
The identification of gene sets for which the constituent genes are disproportionally induced or repressed is a common analysis method for transcriptomic studies. The popular GSEA approach can be applied to gene sets from GO, KEGG, REACTOME, and the MSigDB compilation of curated pathways and other sources. MSigDB currently approaches 18 000 gene sets, resulting in as many hypothesis tests as analysis of all individual genes in the genome (gene-level analysis). As such, large adjustments to p values occur when correcting for multiple hypothesis testing, introducing many false-negative results. Several approaches for reducing the number of gene sets have been proposed (Cantini et al., 2018; Liberzon et al., 2015). Minimal, nonredundant gene sets will vary by tissue, owing to varying expression of the underlying genes and tissue-specific functions. Previously, we described a subset of GO terms (biological process and cellular component [CC]) and curated pathways (KEGG, REACTOME, Biocarta, and others) that are perturbed in 1% or more of TG liver experiments (Sutherland et al., 2016). This reduced the number of gene sets for consideration from 11 581 to 1839 (henceforth, the “inducible/repressible” set, or “inducible pathways” for conciseness).
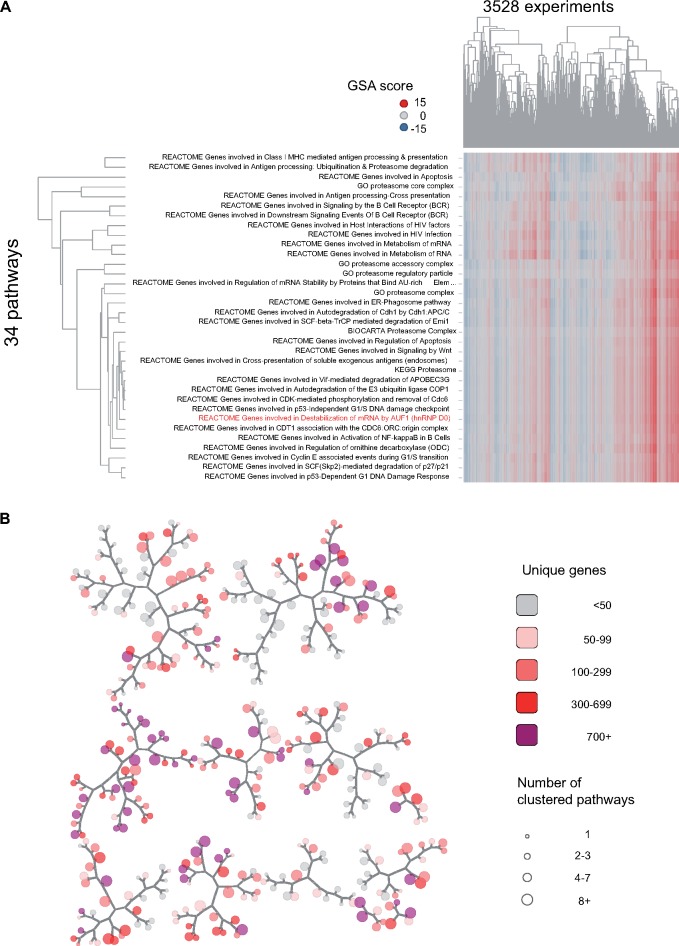
Within the 1839 inducible set, inclusion of gene sets from different sources suggested a significant level of redundancy (eg cholesterol biosynthesis from GO, KEGG, REACTOME). Using scores for each gene set versus 3528 TG liver experiments, hierarchical clustering was used to organize the gene sets into clusters of gene sets that are induced/repressed in the same experiments. By testing the stability of clusters upon repeated sampling of experiments (see Materials and Methods section), we obtained 382 clusters containing 1 or more gene sets (Supplementary Dataset 1). Within each cluster, a single gene set that best captures the variation within the cluster was identified, serving as a surrogate for the others (no consideration of whether the gene set in question best summarizes biological themes represented in the cluster). The 382 nonredundant gene sets were graphically organized to facilitate visual interpretation of GSA results for liver gene expression studies (Figure 2). The example in Figure 2 shows a cluster of 34 terms for which the REACTOME term “Genes involved in Destabilization of mRNA by AUF1” captured the maximum variation for the 34 clustered terms. Inclusion in the cluster of 4 GO-CC terms relating to the proteasome and inspection of other pathway annotations suggests degradation of cell signaling factors by the proteasome, a common method of regulation (Rousseau and Bertolotti, 2018).
Figure 2.
Network map of nonredundant inducible pathways in rat liver. The full set of 1839 inducible/repressible pathways was clustered using gene set analysis scores for 3528 TG liver experiments; (A) showing 1 cluster (cluster 1578), which includes 30 pathways and 4 GO-CC terms (all relating to the proteasome) with highly correlated scores (see Materials and Methods section). One pathway (REACTOME Genes involved in Destabilization of mRNA by AUF1) best captured the variation in scores for all terms and was selected as a representative for the cluster. Other selections, based on maximal perturbation across experiments of interest, or most significant association with a toxicity phenotype are possible. The grouping is based solely on correlation of pathway scores, not gene membership or biological themes among the gene sets. B, The set of 1839 pathways reduced to 382 clusters, with each cluster represented as a node on the map; the cluster in (A) is represented as 1 node on the map. Proximity in the map, as measured by traversal of branches, corresponds to correlation of scores between clusters. Clusters range in size from 1 through 92 gene sets (node size), and when counting all unique genes among their members, include from 5 to 2039 unique genes (shading).
Association of Inducible Gene Sets With Toxicity Phenotypes
Gene expression profiling can be used to explain putative mechanisms responsible for drug-induced morphologic changes in the liver. Because many differentially expressed genes or gene sets will have no relation to the observed morphologic changes, identifying the subset of expression changes linked to injury is difficult. Previously, we described the statistical association between induction or repression of networks of co-expressed gene sets, TXG-MAP modules, and the occurrence of various histology-anchored toxicity phenotypes (Sutherland et al., 2018). Here, we performed similar analyses to establish the relationship between perturbation of inducible gene sets and the occurrence of liver toxicity phenotypes. We distinguish predictive relationships (ie expression profiling from tissue collected 24 h after a single dose, before the appearance of injury) from concurrent relationships (ie expression profiling from the injured tissue). Mirroring our findings with liver modules, we found a larger number of statistically significant relationships for concurrent injury, compared with single dose exposures (Supplementary Dataset 2). For each combination of a timepoint and toxicity phenotype, pathways were ranked from most to least associated with a given phenotype. For any experiment, the odds of observing a toxicity phenotype can be computed from pathway scores (Table 2). This allows for the prediction of toxicity in longer duration studies when performing expression studies of ≤ 24 h treatment duration.
Table 2.
Top Three Nonredundant Pathways for Association With Toxicity Phenotype
| Tox Phenotype a | Time b | Top-Ranked Pathways c |
|---|---|---|
| Adverse at 29 days | 1d | GO: cellular amino acid metabolic process (−0.7), GO: drug metabolic process (−0.6), GO: unsaturated fatty acid metabolic process (0.8) |
| Bile duct hyperplasia | 1d | GO: positive regulation of leukocyte apoptotic process (1.2), GO: cofactor metabolic process (−0.6), KEGG: Cell adhesion molecules (0.6) |
| C | REACTOME: Genes involved in Apoptotic execution phase (1.8), GO: positive regulation of extrinsic apoptotic signaling pathway in absence of ligand (1.7), GO: regulation of triglyceride metabolic process (−1.7) | |
| Cholesterol decrease | 1d | GO: regulation of membrane potential (−0.9), GO: germ cell nucleus (1), GO: regulation of feeding behavior (−1) |
| C | REACTOME: Genes involved in Cell surface interactions at the vascular wall (−0.8), GO: extracellular matrix (−0.5), REACTOME: Genes involved in Association of TriC/CCT with target proteins during biosynthesis (1.2) | |
| Cholesterol increase | C | REACTOME: Genes involved in Ethanol oxidation (0.7), GO: ethanol catabolic process (0.6), GO: canalicular bile acid transport (0.5) |
| Hematopoiesis | 1d | GO: negative regulation of programed cell death (1.3), REACTOME: Genes involved in Sulfur amino acid metabolism (−1.4), GO: dicarboxylic acid metabolic process (−1.1) |
| C | GO: apoptotic cell clearance (2.3), GO: complement activation, classical pathway (2.2), GO: blood coagulation (2.2) | |
| Hypertrophy | 1d | REACTOME: Genes involved in Orc1 removal from chromatin (0.6), REACTOME: Genes involved in SCF(Skp2)-mediated degradation of p27/p21 (0.7), GO: ribosomal large subunit export from nucleus (1.5) |
| C | GO: protein homotetramerization (1.2), GO: sterol esterification (−1.1), REACTOME: Genes involved in Recycling of bile acids and salts (1.1) | |
| Increased glycogen | C | PID: TNF receptor signaling pathway PMID: 18832364 (−1.6), GO: cellular response to lipoprotein particle stimulus (−0.9), GO: intrinsic apoptotic signaling pathway in response to DNA damage (−1.5) |
| Increased mitosis | 1d | KEGG: Pyruvate metabolism (1.4), KEGG: Propanoate metabolism (1), GO: single-organism catabolic process (0.6) |
| Increased mitosis | C | GO: nuclear ubiquitin ligase complex (1.6), PID: Aurora A signaling PMID: 18832364 (1.1), GO: homologous chromosome segregation (1) |
| Necrosis | C | GO: macrophage chemotaxis (0.8), GO: myeloid leukocyte migration (0.5), GO: monocarboxylic acid catabolic process (−0.8) |
| Single cell necrosis | C | REACTOME: Genes involved in Activation of Genes by ATF4 (1), GO: endoplasmic reticulum unfolded protein response (1.1), PID: ATF-2 transcription factor network PMID: 18832364 (0.6) |
| Trigs decrease | C | BIOCARTA: Apoptotic DNA fragmentation and tissue homeostasis (−1.3), GO: pronucleus (−1), GO: protein depolymerization (−0.9) |
| Trigs increase | C | KEGG: Steroid biosynthesis (−0.3), GO: sterol esterification (0.7), GO: response to drug (−0.4) |
| Vacuolation | C | KEGG: Steroid biosynthesis (0.7), GO: isoprenoid biosynthetic process (0.7), GO: extracellular matrix assembly (−0.8) |
Representative toxicity phenotype as defined in methods; “adverse at 29 days” is an aggregate endpoint and included any treatments that produced 1 or more adverse morphologic changes at 29 days of dosing; only relationships at 1 day (1d) were evaluated.
Whether expression analysis is performed from samples collected after 1 day (1d) or concurrent with the observed toxicity phenotype (C).
Showing the top 3-ranked pathways for the toxicity phenotype and time, ranked using p-adj per (Sutherland et al., 2018), selecting only the most highly ranked pathway within each cluster. The value in parentheses is the pathway’s coefficient in the logistic regression model, interpreted as the natural log of odds-ratio for observing toxicity given a 1 unit increase in the pathway score. The sign indicates whether induction (positive) or repression (negative) associates with increased odds of toxicity. A coefficient value of 0.69 corresponds to 2× odds of toxicity, 1.1 corresponds to 3×, etc. The full results are provided in Supplementary Dataset 2.
Omeprazole-induced Liver Hypertrophy
In addition to their use for predictive applications, the relationship between pathway perturbation and toxicity phenotypes can be used to identify putative mechanisms linked to an observed toxicity phenotype. When studying a compound that causes a given toxicity phenotype (eg hepatocellular hypertrophy), this allows a user to observe whether pathways highly ranked by expression are also highly ranked in their association with the toxicity phenotype: “(1) this new compound causes hypertrophy and induces pathway x; (2) pathway x associates with hypertrophy across many other compounds; (3) therefore pathway x may contribute to the occurrence of hypertrophy for this new compound.”
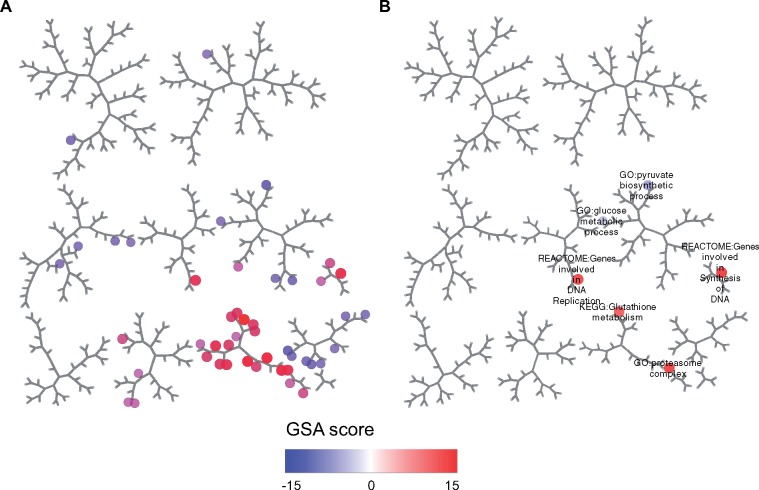
Hepatocellular hypertrophy is frequently observed in toxicity studies and is generally considered an adaptive response to molecule exposure. When hypertrophy is observed, understanding its putative causes may help rationalize its relationship to the compound’s pharmacological effects and/or relevance in other species. Administration of omeprazole at 300 and 1000 mg/kg caused hypertrophy at 4 days or more of exposure (TG histology results). To identify potential mechanisms linked to hypertrophy, we identified 46 nonredundant gene sets having an adjusted p value of < .001 at both doses, 24 h after administration of a single dose (Figure 3; Supplementary Dataset 3). The significantly perturbed gene sets encompass a wide range of biological functions, as underscored by their distribution throughout the pathway map (recall that gene sets that tend to be perturbed by the same experiments are nearby on the map). We subsequently filtered these to the top 50 most strongly associated with hypertrophy across the entire TG database (ie exhibit a statistically significant relationship versus the phenotype across a wide range of conditions; Supplementary Dataset 2, p-adj). This led to the identification of 8 significant gene sets, including glutathione metabolism, proteasome complex, DNA replication, and mitotic cell cycle gene sets. These results are consistent with reports indicating that omeprazole induces Nrf2-mediated response to oxidative stress. Thus, filtering a larger list of differentially expressed gene sets against those associated with the observed morphologic changes facilitates the process of identifying molecular processes causally linked to injury.
Figure 3.
Pathways perturbed by omeprazole treatment in rat liver. Expression results were analyzed for omeprazole 24 h following the administration of single doses of 300 and 1000 mg/kg. A, Only pathways with gene set analysis score p-adj < .001 at both doses were retained. Within each of 382 pathway clusters, the pathway with the highest average absolute score across the 2 doses was selected. B, Only the subset of 8 pathways ranked in the top 50 out of 1839 for association with hypertrophy was considered.
Comparison of Nonredundant Gene Sets Versus TXG-MAP Modules
Gene sets from GO terms and pathways represent biological processes as they are understood and curated from the biomedical literature (henceforth, “pathways”). Notably, whether or not a gene is transcriptionally active in response to stress is not a consideration in associating genes with terms or pathways. Several approaches have been described for identifying putative biological networks from gene expression data, with no reference to pathways. TXG-MAP modules (Sutherland et al., 2018) were obtained by analyzing co-expression patterns of genes across the DM liver database. Biological processes driving co-expression of genes in response to treatments that are not represented in pathways are identified, but those drivers may be difficult to discern from the constituent genes and interpretation is more challenging than pathway analysis.
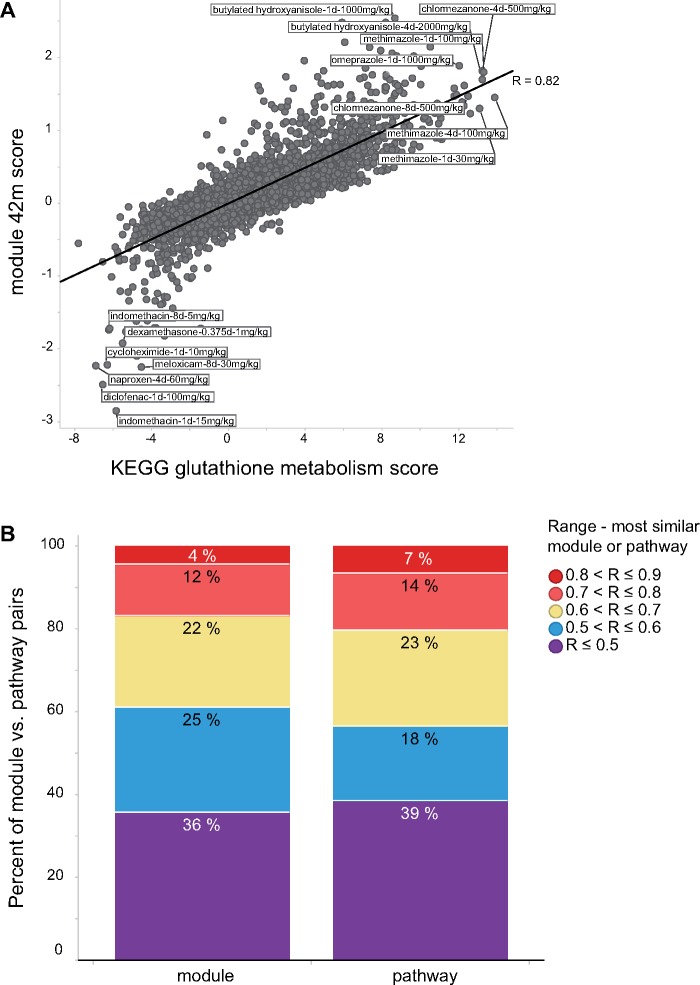
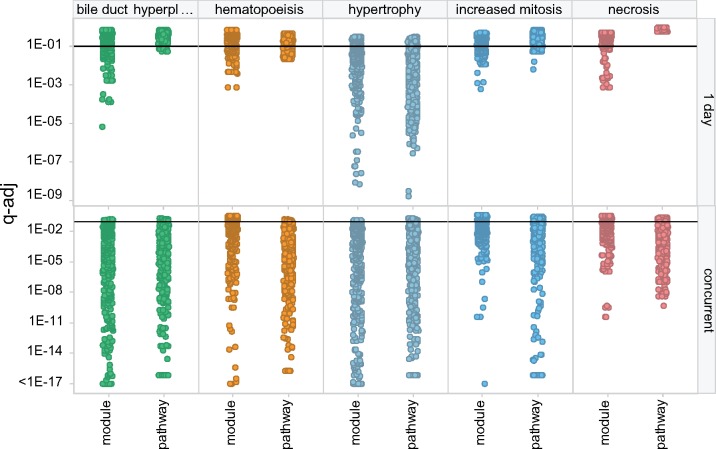
The CTox web application allows users to analyze their experiments using pathways and co-expression networks. To study the extent to which pathways recapitulate analysis using co-expression modules, and vice versa, each TG liver experiment was analyzed using the nonredundant pathways and TXG-MAP modules. When comparing module and pathway scores across 3528 TG experiments, a small proportion of modules are observed to be highly correlated with pathways and therefore summarize similar effects (eg module 42 m and KEGG glutathione metabolism; Figure 4A). Overall, only 16% of TXG-MAP modules capture co-expression behavior well-represented by pathways (defined as having similarity ≥ 0.7; Figure 4B). Conversely, only 20% of nonredundant pathways are well-represented by modules. Modules and pathways are similarly predictive of concurrent toxicity phenotypes, whereas modules outperform for predictive applications (ie analyzing samples at 1 day to predict later toxicity; Figure 5). Hence, orthogonal approaches are more likely to lead to mechanistic insights for a treatment of interest.
Figure 4.
Comparison of TG liver experiment scores via nonredundant pathways and TXG-MAP modules. A, When comparing scores for 3528 TG experiments, the most similar pathway for module 42 m was KEGG glutathione metabolism, and vice versa, with Pearson R = 0.82. B, The analysis in (A) was repeated for each module, identifying the most similar pathway, and assigning the calculated Pearson R values to ranges as shown; stacked bar graphs show the cumulative distribution of similarities across the 415 modules. Likewise, each pathway was compared with modules, the most similar identified and assigned to similarity ranges in the same manner.
Figure 5.
Module and pathway association with toxicity phenotypes. For selected toxicity phenotypes (trellis panel columns), the significance of the relationship (q-adj) between module/pathway scores and occurrence of toxicity is shown for predictive applications (ie expression analysis at 1 day) and concurrent with injury (trellis panel rows). Small q-adj values indicate greater significance. The horizontal line at q-adj = .1 denotes a common cutoff for statistical significance. Points are jittered horizontally for clarity.
Identification of Similar Experiments
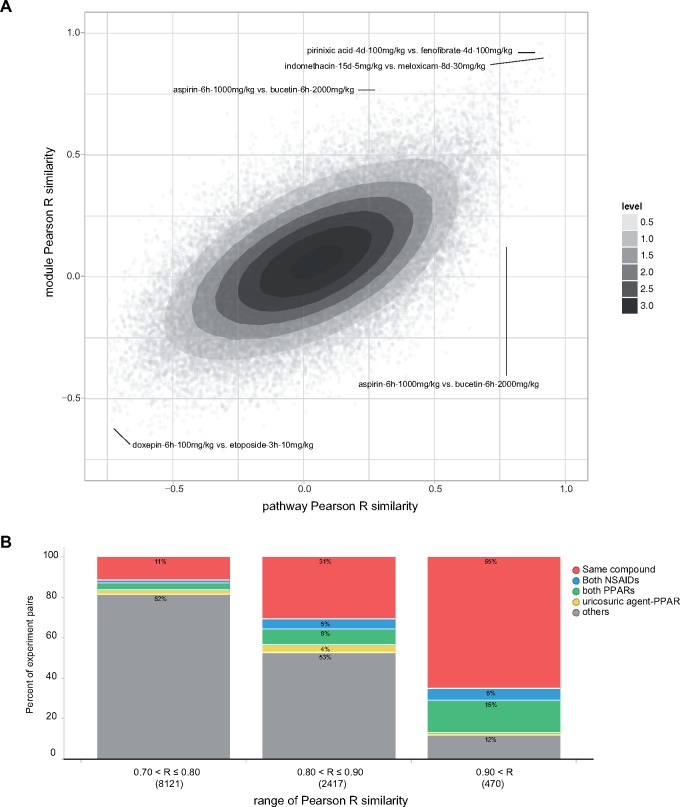
Compounds that exhibit similar effects on the transcriptome may share pharmacological and/or mechanistic effects, an observation that underpins CMAP and related approaches (Lamb et al., 2006; Smalley et al., 2010). Similarity of transcriptional responses can be assessed by describing the effects of each compound as a vector of pathway (GSA) or module scores (eg a vector of 382 pathway scores for the 382 nonredundant pathways). Taking each pair of TG liver experiments, we compared their similarity using the nonredundant pathways versus TXG-MAP modules. Experiment similarities between pathway and modules scores were modestly correlated (Figure 6A; Pearson R = 0.59). Using the average similarity from nonredundant pathways and TXG-MAP modules, most pairs of highly similar experiments involved the same compound at different doses or timepoints, or comparisons of different compounds in the same pharmacological class (eg NSAIDs, PPAR modulator; Figure 6B). High similarity pairs of experiments involving compounds in different pharmacological classes may indicate that they share previously unrecognized pharmacological activity (or similar mechanisms of toxicity).
Figure 6.
Similar experiments analysis using modules and pathways. A, Comparing pairwise experiment similarities for nonredundant pathways (gene set analysis) scores and co-expression modules scores; Pearson R = 0.59 on ca 6.2 M pairs of TG experiments. Darker contours show increasing density of points, and contour values are an arbitrary linear scale. Labeled experiment pairs indicate compound name, treatment duration (h-hours, d-days) and dose. B, Composition of high similarity experiment pairs; similarities were averaged for pathways and modules, and assigned to ranges of 0.7–0.8, 0.8–0.9 and > 0.9. The proportion of experiment pairs involving the same compound (at different doses and/or timepoints), or both compounds being NSAIDs or PPAR modulators accounted for the majority of very high similarity pairs.
Uricosuric Agents Benzbromarone and Benziodarone Are PPAR Modulators
Among TG liver experiment pairs having similarity > 0.9, a high proportion were PPAR modulators (clofibrate, fenofibrate, gemfibrozil, pirinixic acid, rosiglitazone). In addition, we noted 5 pairs of experiments where 1 agent was a PPAR modulator and the other was one of the uricosuric agents benzbromarone or benziodarone, commonly used to treat gout. A further 95 such pairs were found in the next lower similarity range from 0.8 to 0.9. Compared with the lower similarity ranges, this represented a significant enrichment in prevalence of PPAR-uricosuric agents (4.6% or 100 of 2887 pairs with ≥ 0.8 similarity, versus 0.2% of 2.6 M pairs with < 0.8 similarity; p = < 2e-16, χ2 test). The 100 pairs included 12 benzbromarone or benziodarone experiments, with treatment duration ranging from 9 h to 29 days (Supplementary Dataset 4). This suggested that benzbromarone and benziodarone are PPAR modulators.
Clinical chemistry and histology changes for the 12 benzbromarone/benziodarone experiments included 6 causing > 50% decrease in triglyceride levels and 7 causing hepatocellular hypertrophy. The 39 PPAR experiments that comprise the 100 high similarity pairs also lower triglyceride levels, without causing hepatocellular hypertrophy. However, most caused hepatocellular necrosis, not observed with benzbromarone/benziodarone (Supplementary Dataset 4).
To further validate triglyceride-lowering mechanistic effects, we evaluated pathway and module scores for the 6 uricosuric agents and 26 PPAR modulator experiments that caused > 50% decreases in triglyceride levels, ranking them from most to least perturbed across agents of each class (Table 3). As expected from global similarity of gene expression profiles, the most highly induced pathways and modules for uricosuric agent experiments were also highly ranked for PPAR modulators. In addition, most of the gene sets and modules had strong association with triglyceride lowering in general but not hypertrophy. The utility of considering induction/repression in the experiments of interest and the general association with the tox phenotype can be seen for module 285; it is induced across the uricosuric agents and PPAR modulators but ranks poorly in associating with triglyceride lowering in general. Hence, it may not play a role in the observed phenotype.
Table 3.
Most Perturbed Modules and Pathways by the Uricosuric Agents Benzbromarone and Benziodarone, and Relationship to Observed Toxicity Phenotypes
| Gene Set | Urico Agent Expression Rank a | PPAR Mod Expression Rank a | q-Adj Decreased Trigs b | Rank Decreased Trigs c | q-Adj Hypertrophy b | Rank Hypertrophy c |
|---|---|---|---|---|---|---|
| GO: fatty acid catabolic process | 1 | 1 | 2.64E-10 | 20 | 0.0000131 | 117 |
| DM: liver: 26 | 2 | 2 | 0.000041 | 36 | 0.000043 | 195 |
| DM: liver: 17 | 2.5 | 1.5 | 0.0000064 | 21 | 9.2E-08 | 130 |
| GO: negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 3 | 9 | 5.06E-08 | 26 | 2.94E-11 | 25 |
| DM: liver: 285 | 5.5 | 16.5 | 0.025 | 177 | 2.7E-12 | 73 |
| GO: coenzyme metabolic process | 5.5 | 8 | 0.0000429 | 43 | 8.73E-15 | 15 |
| REACTOME: Genes involved in destabilization of mRNA by AUF1 (hnRNP D0) | 5.5 | 7.5 | 0.00026007 | 51 | 1.15E-07 | 72 |
| REACTOME: Genes involved in metabolism of lipids and lipoproteins | 6.5 | 7 | 6.16E-13 | 7 | 0.00000173 | 92 |
Each pathway or module was ranked from most to least perturbed (absolute value of pathway/module score) within each of 6 uricosuric agent and 26 PPAR modulator experiments, and the rank averaged across the 6/26 experiments.
The adjusted q value indicating the pathway or module’s general association with the phenotype across all TGs experiments.
The corresponding rank for the phenotype. These are “concurrent” associations, because expression results were taken from samples where the phenotype is present. Abbreviation: Trigs, triglycerides. Full results are provided in Supplementary Dataset 5.
Our results strongly suggest that benzbromarone and benziodarone are PPAR modulators in rodent liver. Others have investigated the PPAR activity of benzbromarone, confirming our findings in rodents (Kunishima et al., 2007; Lee et al., 2016) and human patients being treated for gout (Inokuchi et al., 2009).
Comparing Results From RNA-seq and Microarray Studies
Users of the CTox application may upload results obtained by microarray or RNA-seq analysis. Several studies support the validity of comparing results from different measurement technologies (Black et al., 2014; Rao et al., 2019; Wang et al., 2014). To further corroborate these conclusions, we loaded into the application results from 2 studies: (1) RNA-seq from the SEQC consortium (Wang et al., 2014), and (2) recently published Affymetrix microarray and RNA-seq results generated concurrently from the same samples (Abbvie study) (Rao et al., 2019) (Supplementary Dataset 6). For the SEQC comparison, the median Pearson R expression similarity was .80 and .86 for nonredundant pathways and TXG-MAP modules, respectively. For the Abbvie study, the median expression similarity was .82 and .86 for pathways and modules, respectively. These results describe concordance across measurement technologies (ie same tissue samples, different technology). By comparison, expression similarity assessed from samples generated by different laboratories using the same measurement technology and treatment conditions are notably lower (Sutherland et al., 2016). Consistent with our prior results (Sutherland et al., 2016), the level of agreement between microarrays and RNA-seq increases as the level of expression perturbation increases (Supplementary Dataset 6). These results support the validity of comparing results obtained using microarray and RNA-seq in the CTox application.
DISCUSSION AND CONCLUSION
Numerous reports and algorithms have been described for predicting and understanding mechanism of liver injury using transcriptomic results from nonclinical safety studies as a means to identify relevant mechanistic information. However, nearly 2 decades after transcript profiling was widely available for use by toxicity researchers in academia and industry, barriers to fuller adoption of toxicogenomic approaches in risk assessment remain. A notable technical barrier is the need for computational skills held by a minority of researchers in the field. Other technical challenges include the difficulty of implementing methods described in scientific reports without having access to computer source code or detailed protocols. In addition, analysis usually relies on 1 method rather than an integrated approach using multiple analysis methods. These (and other) barriers contribute to challenges in assuring the reproducibility of results. Finally, gaining greater acceptance of the significance of particular findings (eg induction of the glutathione metabolism pathway) requires an understanding of results for 1 compound in a broad context and rapid interpretation, preferably by the toxicologist doing the risk assessment analysis.
To facilitate progress toward these goals, we have created a CTox application that allows non-computational specialists to evaluate liver gene expression results for their studies. Users can describe their experiments, upload the corresponding samples, evaluate their results using a variety of established and emerging systems biology analysis methods, compare them to the extensive DM and TG repositories, and share them with other researchers. Users can also search for and analyze data from those repositories using the same analysis methods. Analyses in other public applications are enabled, via exports of suitably formatted data. The CTox software is hosted in the Amazon Web Services cloud for use by the scientific community, or can be downloaded and installed within organizations’ private networks. Finally, the source code is publicly available on the GitHub.com repository, and all software components are available via open source licenses. This enables the extension and improvement of the application by others without licensing barriers.
To illustrate the utility of the CTox application, we described its use for identifying pathways predictive of an adaptive response in liver after treatment with omeprazole. In this case there was good overlap between modules and GO terms and canonical pathways that are predictive of hypertrophy. We also elucidated a putative role of PPAR modulation by the uricosuric agents benzbromarone and benziodarone, in explaining their effects in rat liver. We demonstrated the importance of combining multiple lines of investigation to arrive at a small number of gene sets and/or co-expression modules (“features”) putatively linked to the observed phenotype: (1) identify features consistently affected by the treatment across doses and/or timepoints, (2) identify the subset of those features robustly associated with the phenotype across a variety of treatments, and (3) corroborate if possible in molecules showing overall similar expression effects compared with the treatment of interest. In the absence of known toxicity study outcomes for a compound of interest, the subset of affected features with statistically significant relation to 1 or more toxicity phenotypes can be used to understand the odds of toxicity for the molecule of interest. These analyzes are possible either in the application itself, or by exporting small subsets of the data and further manipulating them in Excel.
We seek to further develop the application in collaboration with the toxicology community. Improvements to the suite of visualization tools will permit more detailed analysis within the application itself, such as exploring the connection between co-expression networks, canonical gene sets and their linkage to gene-level results. Also, we are actively implementing other analysis methods, such as the benchmark dose methodology to identify points of departure for multi-dose expression studies. Finally, whereas the application can be used to analyze data from any system, we are developing additional tissue-specific nonredundant gene set collections and co-expression modules, and annotating their association to tissue injury (heart and kidney).
In summary, we have developed an open-source toxicogenomics analysis application which helps to increase accessibility, transparency, and collaboration between researchers in the field. The application is available to the scientific community at http://ctox.indianabiosciences.org.
DATA AVAILABILITY
Supplementary data are available at https://datadryad.org/resource/doi:10.5061/dryad.159h65k.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and publication of this article.
ACKNOWLEDGMENTS
The authors wish to thank Steve Evans, Bino John, Jon Klinginsmith, Derek Marren, and Terry Wright for helpful discussions during the course of this work, and Meeta Pradhan for contributions toward coding microarray and RNA-seq fold change calculation scripts.
FUNDING
This work was supported by financial contributions of Corteva Agriscience and Eli Lilly and Company to the Indiana Biosciences Research Institute.
REFERENCES
- Bell S. M., Angrish M. M., Wood C. E., Edwards S. W. (2016). Integrating publicly available data to generate computationally predicted adverse outcome pathways for fatty liver. Toxicol. Sci. 150, 510–520. [DOI] [PubMed] [Google Scholar]
- Black M. B., Parks B. B., Pluta L., Chu T.-M., Allen B. C., Wolfinger R. D., Thomas R. S. (2014). Comparison of microarrays and RNA-seq for gene expression analyses of dose-response experiments. Toxicol. Sci. 137, 385–403. [DOI] [PubMed] [Google Scholar]
- Cantini L., Calzone L., Martignetti L., Rydenfelt M., Blüthgen N., Barillot E., Zinovyev A. (2018). Classification of gene signatures for their information value and functional redundancy. NPJ Syst. Biol. Appl. 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M., Wang P., Boyd A. D., Kostov G., Athey B., Jones E. G., Bunney W. E., Myers R. M., Speed T. P., Akil H., et al. (2005). Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 33, e175.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganter B., Tugendreich S., Pearson C. I., Ayanoglu E., Baumhueter S., Bostian K. A., Brady L., Browne L. J., Calvin J. T., Day G. J., et al. (2005). Development of a large-scale chemogenomics database to improve drug candidate selection and to understand mechanisms of chemical toxicity and action. J. Biotechnol. 119, 219–244. [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Igarashi Y., Nakatsu N., Yamashita T., Ono A., Ohno Y., Urushidani T., Yamada H. (2015). Open TG-GATEs: A large-scale toxicogenomics database. Nucleic Acids Res. 43, D921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi T., Tsutsumi Z., Takahashi S., Ka T., Yamamoto A., Moriwaki Y., Masuzaki H., Yamamoto T. (2009). Effects of benzbromarone and allopurinol on adiponectin in vivo and in vitro. Horm. Metab. Res. 41, 327–332. [DOI] [PubMed] [Google Scholar]
- Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Kim S. Y., Volsky D. J. (2005). PAGE: Parametric analysis of gene set enrichment. BMC Bioinform. 6, 144.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunishima C., Inoue I., Oikawa T., Nakajima H., Komoda T., Katayama S. (2007). Activating effect of benzbromarone, a uricosuric drug, on peroxisome proliferator-activated receptors. PPAR Res. 2007, 36092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J., Crawford E. D., Peck D., Modell J. W., Blat I. C., Wrobel M. J., Lerner J., Brunet J. P., Subramanian A., Ross K. N., et al. (2006). The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935. [DOI] [PubMed] [Google Scholar]
- Lee M., Liu Z., Huang R., Tong W. (2016). Application of dynamic topic models to toxicogenomics data. BMC Bioinform. 17, 368.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Liu Z., Kelly R., Tong W. (2014). Of text and gene—using text mining methods to uncover hidden knowledge in toxicogenomics. BMC Syst. Biol. 8, 93.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J. P., Tamayo P. (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M. S., Van Vleet T. R., Ciurlionis R., Buck W. R., Mittelstadt S. W., Blomme E. A. G., Liguori M. J. (2019). Comparison of RNA-seq and microarray gene expression platforms for the toxicogenomic evaluation of liver from short-term rat toxicity studies. Front. Genet. 9, 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau A., Bertolotti A. (2018). Regulation of proteasome assembly and activity in health and disease. Nat. Rev. Mol. Cell Biol. 19, 697–712. [DOI] [PubMed] [Google Scholar]
- Shimoyama M., De Pons J., Hayman G. T., Laulederkind S. J., Liu W., Nigam R., Petri V., Smith J. R., Tutaj M., Wang S. J., et al. (2015). The Rat Genome Database 2015: Genomic, phenotypic and environmental variations and disease. Nucleic Acids Res. 43, D743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley J. L., Gant T. W., Zhang S.-D. (2010). Application of connectivity mapping in predictive toxicology based on gene-expression similarity. Toxicology 268, 143–146. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland J. J., Jolly R. A., Goldstein K. M., Stevens J. L. (2016). Assessing concordance of drug-induced transcriptional response in rodent liver and cultured hepatocytes. PLoS Comput. Biol. 12, e1004847.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland J. J., Webster Y. W., Willy J. A., Searfoss G. H., Goldstein K. M., Irizarry A. R., Hall D. G., Stevens J. L. (2018). Toxicogenomic module associations with pathogenesis: A network-based approach to understanding drug toxicity. Pharmacogenomics J. 18, 377–390. [DOI] [PubMed] [Google Scholar]
- Tawa G. J., AbdulHameed M. D. M., Yu X., Kumar K., Ippolito D. L., Lewis J. A., Stallings J. D., Wallqvist A. (2014). Characterization of chemically induced liver injuries using gene co-expression modules. PLoS One 9, e107230.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te J. A., AbdulHameed M. D. M., Wallqvist A. (2016). Systems toxicology of chemically induced liver and kidney injuries: Histopathology-associated gene co-expression modules. J. Appl. Toxicol. 36, 1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varemo L., Nielsen J., Nookaew I. (2013). Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 41, 4378–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Gong B., Bushel P. R., Thierry-Mieg J., Thierry-Mieg D., Xu J., Fang H., Hong H., Shen J., Su Z., et al. (2014). The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat. Biotechnol. 32, 926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplementary data are available at https://datadryad.org/resource/doi:10.5061/dryad.159h65k.