Many bacteria have been shown to differentiate into genetically identical yet morphologically distinct cell types. Such population heterogeneity is especially prevalent among biofilms, where multicellular communities are primed for unexpected environmental conditions and can efficiently distribute metabolic responsibilities. Bacillus subtilis is a model system for studying population heterogeneity; however, a role for c-di-GMP in these processes has not been thoroughly investigated. Herein, we introduce a fluorescent reporter, based on a c-di-GMP-responsive riboswitch, to visualize the relative abundance of c-di-GMP for single B. subtilis cells. Our analysis shows that c-di-GMP levels are conspicuously different among B. subtilis cellular subtypes, suggesting a role for c-di-GMP during biofilm formation. These data highlight the utility of riboswitches as tools for imaging metabolic changes within individual bacterial cells. Analyses such as these offer new insight into c-di-GMP-regulated phenotypes, especially given that other biofilms also consist of multicellular communities.
KEYWORDS: Bacillus subtilis, riboswitch, biofilm, cyclic di-GMP, microscopy
ABSTRACT
The synthesis of signaling molecules is one strategy bacteria employ to sense alterations in their environment and rapidly adjust to those changes. In Gram-negative bacteria, bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) regulates the transition from a unicellular motile state to a multicellular sessile state. However, c-di-GMP signaling has been less intensively studied in Gram-positive organisms. To that end, we constructed a fluorescent yfp reporter based on a c-di-GMP-responsive riboswitch to visualize the relative abundance of c-di-GMP for single cells of the Gram-positive model organism Bacillus subtilis. Coupled with cell-type-specific fluorescent reporters, this riboswitch reporter revealed that c-di-GMP levels are markedly different among B. subtilis cellular subpopulations. For example, cells that have made the decision to become matrix producers maintain higher intracellular c-di-GMP concentrations than motile cells. Similarly, we find that c-di-GMP levels differ between sporulating and competent cell types. These results suggest that biochemical measurements of c-di-GMP abundance are likely to be inaccurate for a bulk ensemble of B. subtilis cells, as such measurements will average c-di-GMP levels across the population. Moreover, the significant variation in c-di-GMP levels between cell types hints that c-di-GMP might play an important role during B. subtilis biofilm formation. This study therefore emphasizes the importance of using single-cell approaches for analyzing metabolic trends within ensemble bacterial populations.
IMPORTANCE Many bacteria have been shown to differentiate into genetically identical yet morphologically distinct cell types. Such population heterogeneity is especially prevalent among biofilms, where multicellular communities are primed for unexpected environmental conditions and can efficiently distribute metabolic responsibilities. Bacillus subtilis is a model system for studying population heterogeneity; however, a role for c-di-GMP in these processes has not been thoroughly investigated. Herein, we introduce a fluorescent reporter, based on a c-di-GMP-responsive riboswitch, to visualize the relative abundance of c-di-GMP for single B. subtilis cells. Our analysis shows that c-di-GMP levels are conspicuously different among B. subtilis cellular subtypes, suggesting a role for c-di-GMP during biofilm formation. These data highlight the utility of riboswitches as tools for imaging metabolic changes within individual bacterial cells. Analyses such as these offer new insight into c-di-GMP-regulated phenotypes, especially given that other biofilms also consist of multicellular communities.
INTRODUCTION
Bacteria use diverse strategies to sense alterations in their environment and rapidly adjust to those changes. Adaptability to cell stresses can sometimes be manifested as phenotypic variation, where individual cells differentially express subsets of their genes, sometimes occurring within an isogenic population (1–3). For example, in the Gram-positive model microorganism Bacillus subtilis, bistability in gene expression can help guide differentiation of distinct cell types (4, 5). Some cells within a bulk B. subtilis population express motility genes, as induced by the sigma factor SigD (6). Simultaneously, a smaller subset are activated by the ComK transcription factor to differentiate into competent cells, proficient in DNA uptake and primed for homologous recombination (7). Other cells within the population are triggered by regulatory proteins (e.g., SinR/SinI, SlrR, and DegU) to differentiate into sessile chains that overproduce biofilm matrix components or extracellular proteases (8–11). Yet another subpopulation is irreversibly directed into the developmental program of endospore formation, which culminates in the formation of a dormant cell type resistant to many stresses (12, 13). For cells that will be encouraged into endospore formation, nutrient limitation stimulates a phosphorelay that results in the phosphorylation of the master regulator Spo0A, which triggers the onset of sporulation. Furthermore, cells that are further into the sporulation program than others will secrete extracellular killing factors to cannibalize their siblings for nutrients (14). In sum, B. subtilis is capable of switching between cell fates through highly coordinated processes that are governed by master regulators, two-component systems, and phosphorylation cascades.
Signaling molecules also influence B. subtilis differentiation pathways. Several types of signaling molecules have been discovered that coordinate gene expression and regulate bacterial behavior. One class of extracellular signaling molecules, called autoinducers, regulate quorum sensing and allow bacterial populations to behave collectively (15). In addition to autoinducers, a variety of nucleotide-based second messengers are specifically produced for intracellular signaling in bacteria (16–18). One particular ribonucleotide signaling molecule, bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP), has been shown in many bacteria to regulate the transition from a unicellular motile state to a multicellular sessile community (19, 20). c-di-GMP signaling networks are harbored by almost every phylum in the bacterial domain, making this molecule a near-universal second messenger, although it has still been incompletely examined in several model microbes (21).
In response to external stimuli, c-di-GMP is synthesized from 2 GTP molecules by GGDEF domain-containing diguanylate cyclases (DGCs) (22–28). The second messenger can then bind intracellular effectors to direct physiological changes. EAL or HD-GYP domain-containing phosphodiesterases (PDEs) hydrolyze cyclic di-GMP into the linear dinucleotide 5′-phosphoguanylyl-(3′-5′)-guanosine (pGpG) (29–38), which is then recycled into nucleoside monophosphate pools through the action of oligoribonucleases (39–41)). Through bioinformatic analyses, GGDEF, EAL, and HD-GYP domain-containing enzymes have readily been identified in almost all bacterial phyla (35, 42). Conversely, there is greater diversity in the classes of receptors that associate with c-di-GMP. These receptors often have no sequence or structural similarity to one another, making the prediction of c-di-GMP receptors sometimes difficult (43). In addition to the large number of protein receptors, riboswitches also have been shown to bind c-di-GMP to regulate gene expression (44, 45). Riboswitches are located in the untranslated regions of mRNA transcripts and coordinate downstream gene expression in response to highly specific interactions with their cognate ligand (46). The presence of c-di-GMP-responsive riboswitches upstream of a diverse array of genes, such as those encoding GGDEF/EAL/HD-GYP proteins, flagella, pili, other motility factors, transcription factors, and membrane transporters, suggests many different targets of c-di-GMP regulation in bacteria (47, 48).
B. subtilis harbors three DGCs (DgcK, DgcP, and DgcW), one PDE (PdeH), and three putative c-di-GMP receptors (MotI, YdaK, and YkuI) (see Fig. S1 in the supplemental material) (49, 50). MotI is a PilZ domain-containing protein that is thought to inhibit flagellar motility by acting as a molecular clutch on the flagellar stator element MotA, to disengage and sequester it from the flagellar rotor FliG (51). Genetic evidence also supports a direct relationship between c-di-GMP and the regulation of motility through MotI and PdeH (49, 50). While deletion of pdeH results in a swarming motility defect, it does not result in any obvious changes to biofilm colonies or pellicles. Overexpression of the DGCs also did not appear to affect biofilms, initially suggesting that c-di-GMP is not important for the control of biofilm formation in this organism. Recently, however, data suggest that the degenerate DGC YdaK is a c-di-GMP receptor that is likely to participate in biofilms. YdaK is encoded within the ydaJKLMN operon. While the exact role of the yda operon has yet to be determined, the operon may be involved in the synthesis of an unknown exopolysaccharide, which may bolster the stress resistance of the biofilm under specific physiological conditions (52, 53). Therefore, more remains to be understood regarding the role of c-di-GMP in regulating B. subtilis biofilm formation. The third putative receptor, YkuI, is a catalytically inactive PDE that has retained the ability to bind c-di-GMP but not hydrolyze it (54). Interestingly, its deletion was shown to confer resistance to inhibitory concentrations of zinc (55). It is possible that deletion of ykuI somehow restricts access to zinc, although a mechanistic model for this phenotype has yet to be identified. Furthermore, its involvement in the regulation of lifestyle switching in B. subtilis has not yet been ascertained.
The genetic malleability and wealth of knowledge regarding B. subtilis development make it an excellent experimental model in which to interrogate the contribution of c-di-GMP regulation to cell differentiation in Firmicutes. Elevated c-di-GMP levels have been shown to inhibit motility in this organism (49, 50); however, it is currently unknown if sporulation and competence are also regulated by this signaling molecule. Developing evidence suggests that c-di-GMP may also affect the composition of extracellular polysaccharides during biofilm formation in this organism (52, 53). Yet, the methods used to examine whether c-di-GMP affects biofilm formation have thus far incompletely considered that B. subtilis biofilms are heterogeneously composed of several subpopulations (4, 5, 56). In this study, we made use of a fluorescent c-di-GMP-responsive riboswitch reporter and quantified its expression in vivo for single cells of B. subtilis. When combined with fluorescent transcriptional reporters that demarcate the primary classes of cell types, the c-di-GMP riboswitch reporter displayed markedly different levels of fluorescence within the multicellular community. The combination of these data revealed that within a single population, some cell fates correlate with high levels of c-di-GMP while other fates exhibit low levels of c-di-GMP. Therefore, these data demonstrate that for some bacteria, c-di-GMP levels are adjusted heterogeneously across bulk populations. Finally, these data highlight the utility of riboswitches as tools for imaging metabolic changes within individual bacterial cells.
RESULTS
Analysis of a c-di-GMP-responsive riboswitch upstream of Bacillus licheniformis lichenysin synthesis genes.
In this study, we sought to identify a riboswitch that could be used to construct a genetic reporter of c-di-GMP. A bioinformatics-based search for c-di-GMP-responsive riboswitches in Bacillales revealed a particularly interesting candidate in the untranslated leader region of the Bacillus licheniformis lch gene cluster. This nearly 27-kb operon (lchA) encodes the subunits of the nonribosomal peptide synthetase that makes lichenysin. Lichenysin is an antimicrobial cyclic lipopeptide that is virtually identical to surfactin, an important specialized metabolite produced by B. subtilis (57–59). The location of this riboswitch is particularly interesting given that riboswitches have not been previously analyzed as being important for the genetic regulation of secondary metabolites. We chose to further examine this class I riboswitch to determine whether it is a good candidate for the development of a genetic reporter of c-di-GMP abundance.
The interaction between a riboswitch and its cognate ligand induces a conformational change in the transcript that modulates downstream gene expression (60). Many riboswitches couple the detection of their target signal to transcription attenuation. In the unbound state, these riboswitches will oftentimes adopt a conformation in which an “antiterminator” helix is created from the left half of the terminator and an upstream sequence. Then, when bound to its cognate ligand, the riboswitch will adopt an alternate conformation that allows for the formation of an intrinsic terminator (61), causing disassociation of the transcription elongation complex (Fig. 1A). Manual inspection of the lchAA putative riboswitch revealed that its sequence included a candidate terminator site. Therefore, to determine if c-di-GMP modulates downstream gene expression of the putative B. licheniformis riboswitch, we performed a transcription termination assay in vitro (Fig. 1B). A DNA template of the lchAA untranslated element that included 50 nucleotides beyond the putative terminator (“runoff”) was generated by PCR and mixed with RNA polymerase holoenzyme in the presence of various amounts of c-di-GMP. Under the conditions that we used, a concentration of 10 μM c-di-GMP began to promote transcription termination at the terminus of the riboswitch, and ligand-responsive termination continued to increase up to the maximum concentration tested, 1 mM c-di-GMP.
FIG 1.
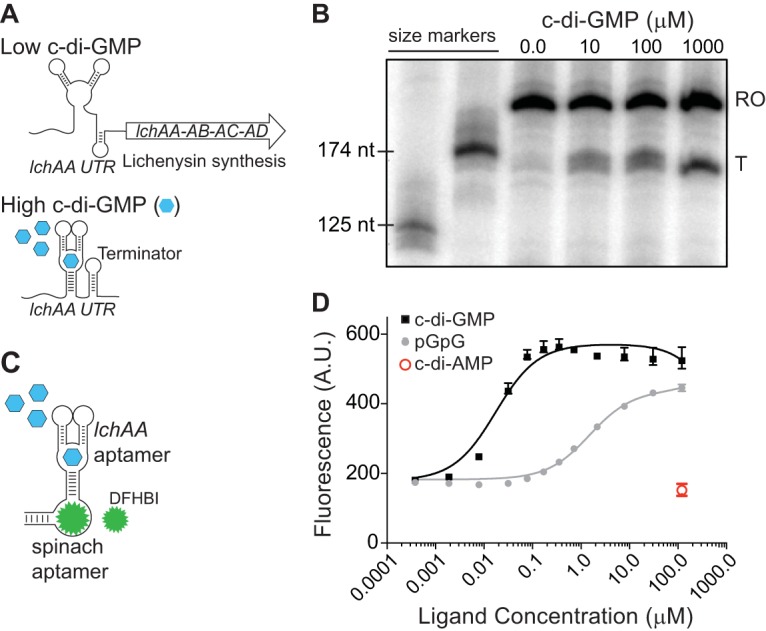
The lchAA riboswitch promotes transcription termination in response to c-di-GMP. (A) Schematic of the mechanism of regulation by the B. licheniformis lchAA riboswitch in response to high and low levels of c-di-GMP. (B) A transcription termination assay of the lchAA untranslated region (UTR) RNA with increasing concentrations of c-di-GMP (0 to 1,000 μM) shows premature termination of runoff (RO) within the leader sequence compared to that of a terminated (T) transcript. (C) Schematic of the Spinach2 RNA allosterically regulated by the lchAA riboswitch aptamer in response to c-di-GMP. (D) In vitro fluorescence assay of the riboswitch-Spinach2 RNA (100 nM) in the presence of saturating DFHBI (10 μM) and increasing concentrations of c-di-GMP or pGpG. Binding affinity measurements are representative of two independent replicates.
To directly measure binding of c-di-GMP to the lchAA riboswitch, we identified the lchAA aptamer domain via manual inspection and fused it to the Spinach2 aptamer. Spinach and Spinach2 RNAs fluoresce upon binding 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI) (62, 63). Spinach variants can be carefully fused to riboswitch aptamer domains such that the Spinach domain fluoresces only in response to binding of the riboswitch ligand (64–66). Therefore, we isolated the lchAA aptamer and fused it to the P2 stem-loop of Spinach2 (see Fig. S2 in the supplemental material). We hypothesized this would create an allosteric version of Spinach2, thereby creating an RNA with two distinct binding sites, for which binding of the lchAA aptamer to its cognate ligand will then trigger binding of DFHBI to Spinach2 (Fig. 1C). We then measured Spinach2 fluorescence in the presence of saturating concentrations of DFHBI and with increasing concentrations of c-di-GMP. This revealed that fluorescence of the lchAA aptamer-Spinach2 RNA was dependent on an appropriate amount of c-di-GMP, confirming that the lchAA aptamer acts as a sensor of c-di-GMP. In contrast, we did not detect Spinach2 fluorescence in the presence of 100 μM c-di-AMP. The lchAA aptamer, as part of a Spinach2 biosensor construct, bound c-di-GMP with an apparent dissociation constant (Kd) of 17 nM. While this apparent Kd is lower than the concentration of c-di-GMP required to promote transcription termination in vitro on purified DNA templates, it has been observed that many riboswitches are not driven by equilibrium ligand interactions. Instead, many riboswitches are driven by coordination of the kinetics of ligand association and RNA polymerization speed (67). In these instances, a higher concentration of c-di-GMP is required to promote a conformational change of the expression platform in vitro. Fluorescence of the lchAA-Spinach2 aptamer was also observed in the presence of pGpG, the linear dinucleotide and c-di-GMP degradation product; however, the apparent Kd for this molecule was 2 orders of magnitude higher than that for c-di-GMP (Fig. 1D). This roughly agrees with previous results in which pGpG binds a c-di-GMP riboswitch with much poorer affinity (68). We conclude from these aggregate data that c-di-GMP specifically interacts with the lchAA riboswitch to promote transcription attenuation of the lchAA operon.
A c-di-GMP riboswitch-yfp reporter is heterogeneously expressed in B. subtilis.
We next wanted to assess c-di-GMP levels in single cells of B. subtilis in vivo. Since intracellular expression of Spinach has not been examined for this organism, we first ectopically integrated a constitutively expressed Spinach2 sequence into the B. subtilis PY79 genome at a nonessential locus. When cells reached late log phase (optical density at 600 nm [OD600] of 1.0), they were incubated with saturating DFHBI for 1 h and examined by fluorescence microscopy. These cells exhibited a roughly 2-fold increase in fluorescence compared to that of untreated cells. However, many cells remained fully inactivated for fluorescence in the presence of DFHBI, indicating broad heterogeneity in fluorescence by a constitutive Spinach2 reporter (see Fig. S3). Incubation with DFHBI for 1 h had no apparent effect on bacterial growth, either for cells in culture or upon examination by microscopy.
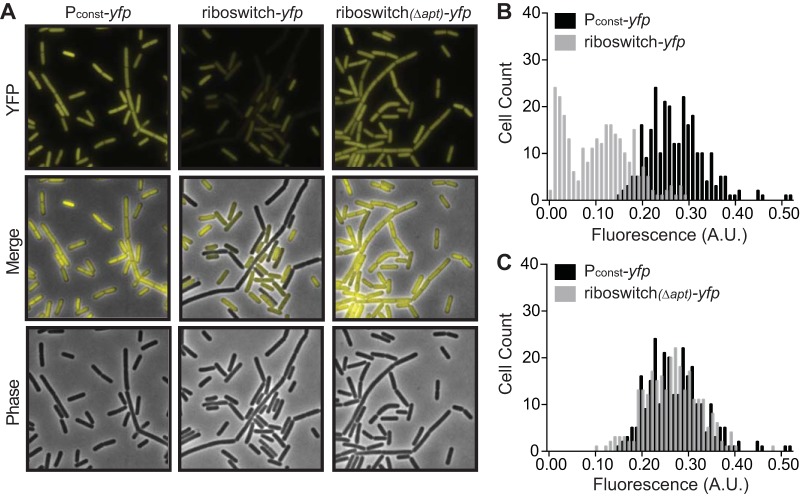
As the Spinach2-based reporter was unacceptably nonuniform in B. subtilis, we constructed a reporter based on expression of yellow fluorescent protein (YFP). We ectopically integrated a constitutively expressed yfp sequence into the B. subtilis genome and analyzed cells at late log phase (OD600 of 1.0) by fluorescence microscopy. Unlike the strain that constitutively expressed Spinach2, the constitutive yfp strain exhibited more-uniform fluorescence. This population exhibited a unimodal distribution of fluorescence per cell (Fig. 2A and B). We then constructed a corresponding reporter based on the c-di-GMP riboswitch. The entire leader sequence of B. licheniformis lchAA was inserted between the constitutively active promoter sequence and yfp. Unlike the constitutive yfp reporter, a strain containing the riboswitch-yfp reporter exhibited two prominently distinct levels of fluorescence within the population. Nearly 70% of the cells were moderately fluorescent compared to the constitutive yfp strain, while the remaining cells exhibited significantly diminished fluorescence (Fig. 2A and B). Therefore, unlike the unimodal distribution of fluorescence exhibited by the constitutive yfp reporter, the riboswitch-yfp reporter strain appeared to show a bimodal distribution of fluorescence. To determine whether this distribution was due to riboswitch regulation of YFP, we mutated the riboswitch element. Specifically, we deleted 65 nucleotides from the 5′ end of the lchAA leader, which includes part of the c-di-GMP-binding aptamer, and reexamined cellular fluorescence. This population of cells uniformly expressed YFP at levels similar to that of the constitutive yfp reporter construct (Fig. 2A and C). Long considered a B. subtilis “legacy” laboratory strain, PY79 has been widely used in a plethora of genetic analyses that have provided profound insight into the molecular pathways that govern cellular differentiation. Indeed, the regulation of cell-specific gene expression in B. subtilis was determined in this genetic background (69–74). Historically, however, undomesticated strains such as 3610 have often been used to study biofilm formation (10, 75–77). We therefore also assessed our riboswitch-yfp reporter in the undomesticated 3610 background. We observed an identical distribution of fluorescence to the PY79 background, suggesting that c-di-GMP dynamics behave similarly in both strains (see Fig. S4). Therefore, we chose to continue our studies with PY79 for interrogating gene expression differences between different cellular subtypes. Taken together, these findings suggest that the c-di-GMP riboswitch-yfp reporter is bimodally expressed in a population of B. subtilis.
FIG 2.
Expression of a lchAA riboswitch-yfp reporter in vivo results in bimodal distribution of fluorescence. (A) Representative microscopy images of wild-type B. subtilis PY79 expressing a constitutive Pconst-yfp reporter, a riboswitch reporter construct Pconst-riboswitch-yfp, or a riboswitch reporter construct comprising a deletion corresponding to the riboswitch aptamer (Pconst-riboswitchΔapt-yfp). (B and C) Histograms of the quantification of fluorescence intensity per cell of wild-type B. subtilis PY79 expressing Pconst-yfp compared to Pconst-riboswitch-yfp (B) or Pconst-yfp compared to Pconst-riboswitchΔapt-yfp (C) (n ≈ 300).
The riboswitch reduces downstream gene expression in response to elevated c-di-GMP.
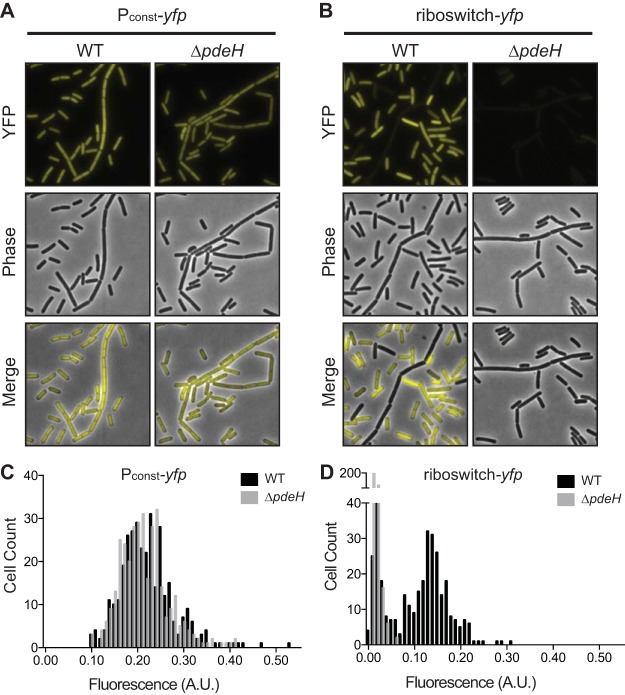
Of the two phosphodiesterases encoded in the B. subtilis genome, previous work has shown only that encoding PdeH to be active (49, 50). With this in mind, we wanted to confirm that the bimodal distribution of fluorescence observed in single cells of B. subtilis was solely due to differences in intracellular c-di-GMP levels. To do this, we integrated the constitutive yfp and riboswitch-yfp reporter fusions into a ΔpdeH knockout strain. We predicted that loss of PdeH should inhibit hydrolysis of c-di-GMP and consequently elevate intracellular levels, which should promote premature transcription termination in the riboswitch-yfp reporter fusion and thereby reduce the YFP signal. Indeed, the riboswitch-yfp reporter resulted in significantly decreased yfp expression in the ΔpdeH background, suggesting intracellular cyclic di-GMP was elevated in all cells at late log phase (OD600 of 1.0) (Fig. 3B and D). Conversely, constitutive yfp expression was unaffected by the ΔpdeH mutation, confirming the specificity of the riboswitch-yfp reporter for c-di-GMP (Fig. 3A and C).
FIG 3.
Deletion of pdeH results in increased c-di-GMP. (A and B) Representative microscopy images of B. subtilis PY79 wild-type (WT) or ΔpdeH cells expressing the constitutive Pconst-yfp reporter (A) or the riboswitch reporter construct Pconst-riboswitch-yfp (B). (C and D) Histograms of the quantification of fluorescence intensity per cell comparing B. subtilis PY79 WT or ΔpdeH expressing Pconst-yfp (C) or Pconst-riboswitch-yfp (D) (n ≈ 300).
To independently assess the contribution of PdeH to c-di-GMP in cells, we employed liquid chromatography-tandem mass spectrometry (LC-MS/MS) to detect c-di-GMP levels in both PY79 wild-type and ΔpdeH strains grown to late log phase (OD600 of 1.0). A roughly 3-fold increase in c-di-GMP was observed by LC-MS/MS for the bulk population of ΔpdeH cells (see Fig. S5). We also attempted to quantify intracellular pGpG levels, but levels of pGpG were below the limit of detection for both wild-type and ΔpdeH cells (data not shown). A previous study quantified intracellular pGpG by LC-MS/MS in a B. subtilis mutant devoid of NrnA and NrnB, which are the two exoribonucleases thought to degrade pGpG in Firmicutes and other organisms that do not encode oligoribonuclease (Orn) (41). Similar to our current study, pGpG was undetectable in wild-type cells and could only be detected in the ΔnrnA ΔnrnB double mutant. Therefore, we conclude that pGpG does not accrue to levels sufficiently high to interfere with the c-di-GMP riboswitch-yfp reporter.
Expression of the riboswitch reporter correlates with specific cell types.
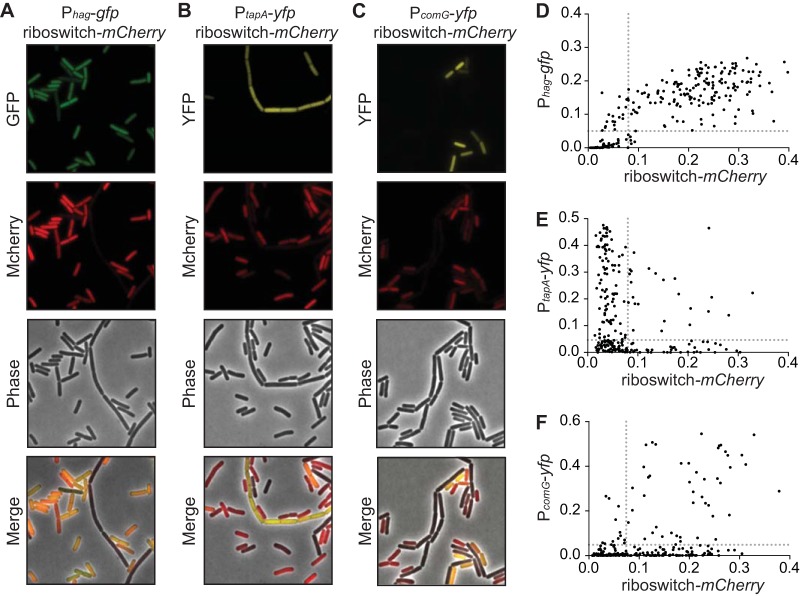
We next sought to correlate c-di-GMP levels with subpopulations of B. subtilis. We replaced yfp with another gene encoding a red fluorescent protein, mCherry, and introduced the riboswitch reporter in strains that already harbored transcriptional gfp or yfp reporters that demarcate the most common B. subtilis cell types, including motile, matrix producing, or competent. When analyzed by fluorescence microscopy at late log phase (OD600 of 1.0), a majority of cells exhibited high expression of a Phag-gfp reporter (denoting motility) and high expression of the riboswitch-mCherry reporter (signifying low c-di-GMP). Of the cells that were transcriptionally active for motility gene expression, 89.8% also exhibited fluorescence from the riboswitch reporter. This suggests that c-di-GMP abundance is uniformly low in motile cells (Fig. 4A and D), and that a positive correlation exists between the genetic reporters. Low c-di-GMP levels were also observed in competent cells. While roughly 20% of cells in the population were competent, as indicated by a PcomG-yfp reporter, 79.7% of these cells exhibited high fluorescence from the c-di-GMP riboswitch-mCherry reporter (Fig. 4C and F). However, when we analyzed the riboswitch-mCherry reporter in the context of matrix-producing cells, as identified by a PtapA-yfp reporter, we observed an anticorrelation between both reporters. Cells activated for producing extracellular matrix (as denoted by tapA gene expression) were observed mainly as long chains. Of these activated cells, 81.0% exhibited diminished fluorescence from the riboswitch-mCherry reporter (Fig. 4B and E). This suggests that c-di-GMP levels are high in matrix-producing cells. Conversely, cells that showed high fluorescence for the riboswitch-mCherry reporter were almost never activated for tapA expression. We did, however, observe a small population of cells that had c-di-GMP levels sufficiently high to shut off the riboswitch reporter but that had not activated the transcriptional reporter for biofilm formation (PtapA-yfp). We speculate that these cells had not been activated yet for extracellular matrix production, which might suggest that c-di-GMP levels change prior to the activation of matrix production. Finally, we introduced the riboswitch-yfp reporter in a strain that also harbored a Pconst-mCherry reporter and assessed c-di-GMP levels in cells that were activated for sporulation. Cells that progressed through spore development maintained expression of mCherry but were uniformly low in riboswitch-yfp expression, suggesting that c-di-GMP levels are high during endospore formation (Fig. 5).
FIG 4.
Expression of the riboswitch reporter varies nonrandomly among B. subtilis cell types. (A) Representative microscopy images of wild-type B. subtilis PY79 expressing the riboswitch reporter construct Pconst-riboswitch-mCherry and the motility reporter Phag-gfp. Statistical analyses show these reporters are significantly correlated (P < 0.0001). (B) Expression of Pconst-riboswitch-mCherry and the biofilm reporter PtapA-yfp are anticorrelated (P = 0.0039). (C) Expression of Pconst-riboswitch-mCherry and the competence reporter PcomG-yfp are correlated (P < 0.0001). (D to F) Quantification of the fluorescence intensity per cell of Pconst-riboswitch-mCherry compared to each cell type reporter in each construct (n ≈ 300). Dotted lines represent the cutoff that divides fluorescent cells from nonfluorescent cells. Statistical significance was determined by chi-square analysis.
FIG 5.
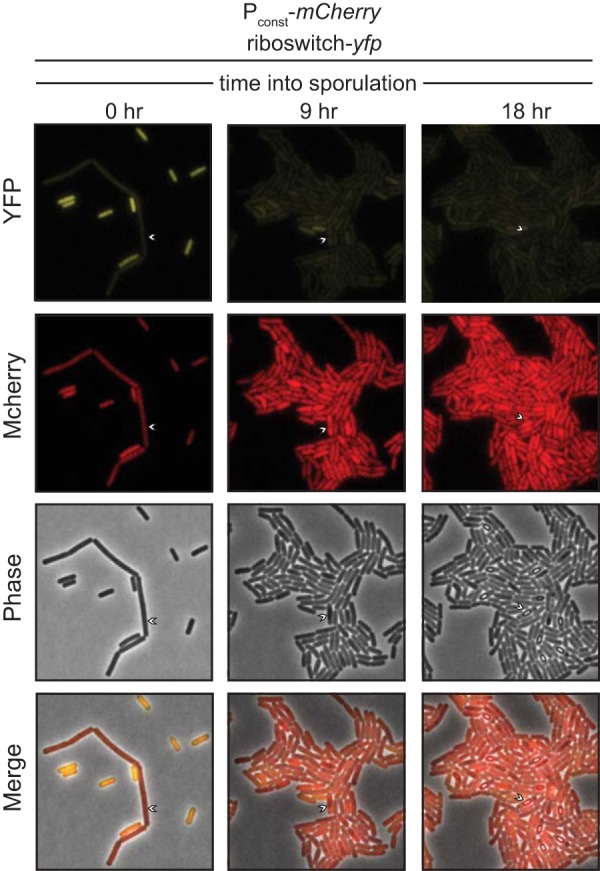
High c-di-GMP levels correlate with sporulation. Representative microscopy images of a time course through sporulation of the wild-type B. subtilis PY79 expressing the riboswitch reporter construct Pconst-riboswitch-yfp and the constitutive reporter Pconst-mCherry.
An increase in c-di-GMP does not appear to change cell type identity.
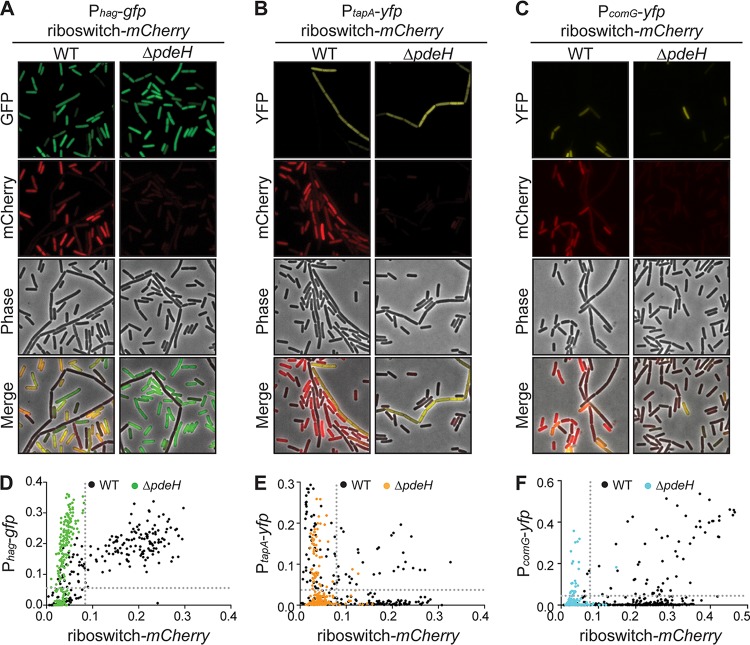
To see if the manipulation of intracellular c-di-GMP levels could influence the proportion of cells that became matrix producers (as well as motile or competent), we used the “high c-di-GMP” strain, ΔpdeH cells, and integrated both transcriptional reporters for c-di-GMP levels and cell type in this background. When cells were analyzed by fluorescence microscopy, we observed uniformly low levels of mCherry fluorescence, thereby confirming the elevated levels of c-di-GMP in the ΔpdeH strain (Fig. 6). Additionally, we observed no obvious consequence on cell fate at the transcriptional level. In many instances, however, c-di-GMP regulates phenotypic outputs at the posttranslational level (78–80). This is exemplified in B. subtilis motile cells, where the binding of MotI to c-di-GMP directs MotI to directly disengage MotA from the rotor of the flagellar apparatus (51). It is possible that c-di-GMP posttranslationally regulates YdaK as well, and this interaction activates production of the unknown exopolysaccharide (EPS). So, while the proportions of cells in each subtype were consistent between the wild type and the ΔpdeH mutant, and the levels of activation for motility and matrix gene expression as measured by the reporters were the same (Fig. 6A, B, D, and E), it remains possible that c-di-GMP is a regulator of B. subtilis’ differentiation at the posttranslational level. Interestingly, mild repression of the competence transcriptional reporter was observed in the ΔpdeH strain compared to that in the wild type (Fig. 6C and F).
FIG 6.
Deletion of pdeH does not change expression of each cell type reporter. (A to C) Representative microscopy images of B. subtilis PY79 wild-type (WT) or ΔpdeH expressing the riboswitch reporter construct Pconst-riboswitch-mCherry and the motility reporter Phag-gfp (A), Pconst-riboswitch-mCherry and the biofilm reporter PtapA-yfp (B), or Pconst-riboswitch-mCherry and the competence reporter PcomG-yfp (C). (D to F) Quantification of the fluorescence intensity per cell of Pconst-riboswitch-mCherry compared to each cell type reporter, expressed in B. subtilis PY79 WT or ΔpdeH cells (n ≈ 300).
pdeH expression is regulated transcriptionally.
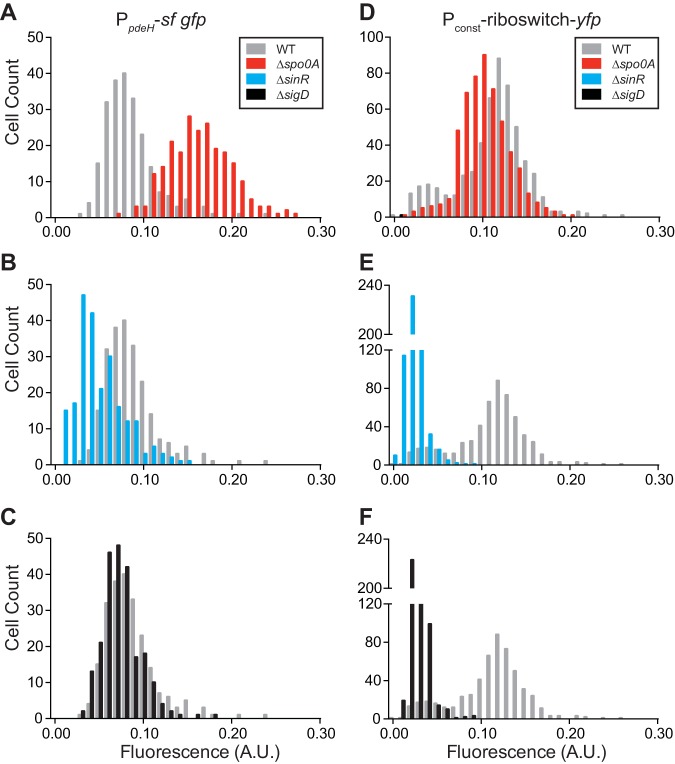
In addition to the catalytic EAL domain that provides phosphodiesterase activity, some PDEs also contain sensory domains (e.g., PAS) that allow for posttranslational regulation of phosphodiesterase activity through activation by an extracellular signal. No such domain has been identified in B. subtilis PdeH. However, pdeH transcription was previously suggested to be repressed by Spo0A (49, 81) indicating that pdeH might be regulated transcriptionally. We therefore sought to ascertain if heterogeneous expression of the riboswitch reporter was due to heterogeneous expression of pdeH in each cellular subtype. We created a transcriptional fusion of the pdeH promoter to a reporter gene encoding Superfolder GFP (PpdeH-sf gfp) and ectopically integrated it into wild-type and Δspo0A background strains. Mean fluorescence for the Δspo0A background (0.162; 95% confidence interval [CI], 0.158 to 0.167) increased by roughly 2-fold compared to that of the wild type (0.086; 95% CI, 0.082 to 0.090) (Fig. 7A). This agrees with prior data showing Spo0A inhibition of pdeH expression (49). Furthermore, quantification of the PpdeH-sf gfp reporter revealed a single peak, implying a normal distribution of pdeH expression across the population. While a ΔsigD mutation had little effect on mean fluorescence (0.075; 95% CI, 0.071 to 0.080) (Fig. 7C), a ΔsinR mutation (0.050; 95% CI, 0.046 to 0.054) led to a roughly 2-fold decrease in mean fluorescence (Fig. 7B). From this, we speculate that removal of SinR might lead to physiological changes that enhance the activation of Spo0A, thereby maximizing the Spo0A-mediated inhibition of pdeH transcription. Alternatively, SinR might activate expression of pdeH, directly or indirectly, through an unknown mechanism. We also quantified fluorescence from the riboswitch reporter in these genetic backgrounds to correlate c-di-GMP levels with activity from pdeH. Our data (Fig. 7A to C) predict that derepression of pdeH due to Δspo0A should lead to lower intracellular c-di-GMP levels, exhibited by uniformly high fluorescence from the riboswitch reporter. Conversely, repression of pdeH due to ΔsinR should result in higher intracellular c-di-GMP levels and result in uniformly lower fluorescence from the riboswitch reporter. As predicted, our riboswitch reporter followed the hypothesized trends. A Δspo0A mutation led to loss of a bimodal distribution of fluorescence, and the population exhibited similar fluorescence to the “low c-di-GMP” population of wild-type cells (Fig. 7D). Mean fluorescence in a ΔsinR mutant was uniformly low (0.023; 95% CI, 0.022 to 0.023), indicating high intracellular c-di-GMP levels (Fig. 7E). Unexpectedly, however, we observed uniformly low fluorescence from the riboswitch reporter in a ΔsigD background (0.029; 95% CI, 0.028 to 0.030). This suggests ΔsigD cells have an increase in intracellular c-di-GMP levels that cannot be explained by pdeH activity alone (Fig. 7C and F).
FIG 7.
Deletion of global regulators affects pdeH expression and c-di-GMP levels. (A to C) Quantification of the fluorescence intensity per cell of PpdeH-sf-gfp in B. subtilis PY79 wild-type (WT) and Δspo0A (A), ΔsinR (B), and ΔsigD (C) genetic backgrounds (n ≈ 300). (D to F) Quantification and comparison of the fluorescence intensity per cell of Pconst-riboswitch-mCherry in B. subtilis PY79 wild-type (WT) and Δspo0A (D), ΔsinR (E), and ΔsigD (F) genetic backgrounds (n ≈ 500).
DISCUSSION
To study c-di-GMP abundance for single cells of B. subtilis, we employed a constitutively expressed c-di-GMP riboswitch fused to yfp as our biosensor. This riboswitch reporter revealed apparent differences in intracellular c-di-GMP levels among B. subtilis subpopulations. From these results, we suggest that c-di-GMP riboswitch-yfp reporters could be used in other bacteria—Bacillales in particular—for single-cell analyses of c-di-GMP abundance. These riboswitch-reporters may prove to be useful tools in elucidating the increasingly diverse c-di-GMP regulatory mechanisms used by Gram-positive bacteria. Studies on Bacillus cereus group organisms, Clostridioides difficile, and Streptomyces coelicolor have suggested a link between c-di-GMP metabolism and spore formation, which is a developmental program almost exclusive to the three genera (20). Interestingly, there is an ortholog of B. subtilis YkuI in the B. cereus bacterial group, which is named CdgJ. Overexpression of Bacillus thuringiensis cdgJ resulted in increased biofilm formation and earlier entry into sporulation (82). Conversely, no sporulation was observed in a B. thuringiensis cdgJ mutant. Given the high similarity in protein sequences between YkuI and CdgJ, it is therefore possible that YkuI also exhibits a similar role in B. subtilis, although it has yet to be explored. Sporulation is also affected by c-di-GMP in S. coelicolor. Overexpression of DGCs cdgA or cdgB or deletion of PDEs rmdA or rmdB led to an increase in intracellular c-di-GMP and inhibition of sporulation (83–85). Studies on S. coelicolor have also revealed the only c-di-GMP-sensing transcriptional regulator to date among Gram-positive organisms (BldD). Deletion of bldD accelerates sporulation (86), which is inhibited through overexpression of cdgA and cdgB, which are also direct targets of BldD. c-di-GMP-bound BldD also represses expression of antibiotic synthesis genes and sporulation genes during vegetative growth (83, 84). Together, these studies generally suggest a broadening role for c-di-GMP signaling in Gram-positive bacteria. Our study herein extends this even further by showing that a c-di-GMP riboswitch is likely to control expression of a B. licheniformis lichenysin biosynthesis gene cluster. This observation suggests that in some bacteria, antibiotic synthesis is under the purview of c-di-GMP signaling. By extension, these data suggest that surfactin biosynthesis could also be subject to c-di-GMP regulation in other Bacillus species. Interestingly, a peptide essential for competence (ComS) is encoded within the surfactin operon srfAA-AD in B. subtilis (87). srfAA-AD is also regulated by the transcription factor ComA, which is ultimately activated by the quorum sensing molecule ComX (88–90). This regulatory arrangement allows B. subtilis to integrate a single signaling pathway into multiple adaptive processes. Prior studies have shown that surfactin-producing cells coexist with, but are phenotypically distinct from, cells that produce the extracellular matrix components (91). Therefore, our observation that c-di-GMP is low in competent cells implies that c-di-GMP should also be low for surfactin-producing cells. Indeed, the presence of the c-di-GMP riboswitch upstream of B. licheniformis lchAA operon indicates that lichenysin is produced only when c-di-GMP levels are low. ComA has been shown to also recognize the B. licheniformis lchAA promoter, further supporting a relationship between lichenysin and competence gene expression (92, 93). While a c-di-GMP-responsive riboswitch does not appear to be located in the leader of B. subtilis srfAA, it is possible that c-di-GMP could regulate this gene cluster by an as yet undiscovered mechanism. In summary, c-di-GMP influences a variety of cellular functions in Gram-positive bacteria, including sporulation and secondary metabolite production, and these discoveries may also foreshadow roles for c-di-GMP signaling during competence development in B. subtilis.
Other types of highly selective fluorescent c-di-GMP biosensors have also been developed in recent years. For example, a protein biosensor for c-di-GMP was previously developed from the Salmonella enterica serovar Typhimurium c-di-GMP binding protein YcgR (94, 95), which coupled binding of c-di-GMP to Förster resonance energy transfer (FRET). This genetically encoded biosensor protein was previously used for FRET-based microscopy of Gram-negative organisms such as Caulobacter crescentus and Pseudomonas aeruginosa (94, 96). However, FRET-based biosensors exhibit lower fluorescence intensities than individual fluorescent proteins, which can result in an overall narrowing of their dynamic range. Furthermore, many c-di-GMP protein receptors remain to be discovered in Gram-positive organisms for use in FRET microscopy. Riboswitch aptamers have also been used in prior studies as allosteric regulators of the conditionally fluorescent Spinach RNA for measuring c-di-GMP dynamics (64, 97). Yet, while they exhibit ideal performance characteristics in vitro, they require further optimization for routine usage in bacterial cells. For example, Spinach-based biosensors exhibit lower fluorescence than common fluorescent proteins. Also, significant amounts of DFHBI have to be added to cells for sufficient quantities of the chromophore to pass through the cell membrane. Some researchers have circumvented this limitation by tagging targeted RNAs with up to 64 spinach aptamers in tandem, thereby increasing the brightness of tagged RNA molecules (98); however, this approach is not ideal for biosensor purposes. In our experiments, we found that the constitutive YFP control in B. subtilis displayed more uniform fluorescence in a population of B. subtilis than the constitutive Spinach aptamer (see Fig. S3 in the supplemental material). Based on all of these considerations, we chose to pursue development of a riboswitch-yfp reporter fusion that could provide useful information on B. subtilis c-di-GMP abundance. The fluorescent proteins used in this study have also been used in multiple studies regarding B. subtilis cell differentiation (99, 100). They have been shown to exhibit a shortened half-life and therefore can be expected to report relative differences in gene expression; however, it remains to be determined whether these reporters are sufficient to measure rapid c-di-GMP dynamics.
Given the importance of B. subtilis as a model system for the study of bacterial development, it is essential that its c-di-GMP regulatory mechanisms be further elucidated. One of the most common assays to measure the impact of mutations, such as those affecting c-di-GMP homeostasis, on biofilm formation is to visually assess the complexity of colony topology. While this can be qualitatively useful, it may not be representative of changes that occur at the cellular level. We take this to be due to the inherently heterogeneous composition of cell types within biofilm communities. Instead, the recent development of cell-type-specific fluorescent reporters allows investigators to examine features of B. subtilis subpopulations and their relationship to c-di-GMP effector proteins, DGCs, and PDEs. (51–53).
Herein, we coupled cell-type-specific transcriptional reporters with a c-di-GMP riboswitch-yfp reporter to ascertain whether c-di-GMP varies between B. subtilis subpopulations. Indeed, our single-cell microscopy shows definitively that intracellular c-di-GMP differs among B. subtilis cell types within a single population. Cells that have made the decision to become matrix producers maintain higher intracellular c-di-GMP levels than motile cells. This study also shows that the transition into endospore formation correlates with high c-di-GMP levels, while competent cells correlate with lower c-di-GMP levels. This trend was not previously examined in other studies. However, while our data demonstrate that c-di-GMP levels vary significantly between different cell types, it is unclear whether these changes in c-di-GMP abundance occur as a result of cellular decision making or whether they participate in initiating the choice of cell fate. Instead, we provide evidence that a general increase in c-di-GMP abundance is not likely to change the proportion of each cell type at the transcriptional level. While our data suggest that c-di-GMP metabolic enzymes act downstream of the master regulators that drive genetic regulation of cellular differentiation such as Spo0A and SinR (Fig. 7A and B), the molecular details of how c-di-GMP levels affect cellular differentiation require further exploration. Indeed, the general importance of c-di-GMP has yet to be fully ascertained for this organism. It is possible that the primary purpose of c-di-GMP signaling in this organism is to influence the motility apparatus. Alternatively, it is possible that effector targets for c-di-GMP-binding proteins have yet to be identified, as might be suggested by the recently discovered link to the yda exopolysaccharide pathway (52, 53). Overall, the abundance of c-di-GMP in a bulk ensemble of B. subtilis is likely averaged across the population, and we propose that single-cell analysis of c-di-GMP levels offers unique insight into how the abundance of this signaling molecule varies among cellular subpopulations. Analyses such as these are likely to offer new insight into c-di-GMP-regulated phenotypes, especially given that other biofilms also consist of multicellular communities.
MATERIALS AND METHODS
Transcription termination assay.
PCR amplification was performed on the B. licheniformis lchAA leader sequence using primers that place it downstream of a constitutive promoter Pconst and that ended 216 nucleotides (nt) downstream of the transcription start site (see Fig. S2 in the supplemental material). Transcription reactions were performed on the PCR-generated DNA template, resulting in a terminated (T) transcript of approximately 174 nt, or a 217-nt runoff (RO) transcript. These reaction mixtures comprised 5 μM template, 250 μM nucleoside triphosphates (NTPs), 1× Escherichia coli RNA polymerase reaction buffer (40 mM Tris-HCl [pH 7.5], 150 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol [DTT], 0.01% Triton X-100; NEB), 20 μCi [α-32P]UTP, and 0.5 unit of E. coli RNA polymerase, holoenzyme (NEB). c-di-GMP was added to the runoff transcription reactions to final concentrations of 10 μM, 100 μM, and 1 mM. A 125-nt size marker was also transcribed. All reaction mixtures were incubated at 37°C for 2 h. Reaction results were resolved by 6% urea-denaturing polyacrylamide gel electrophoresis (PAGE).
Spinach activation assay.
PCR was performed to amplify two aptamers: a tRNALys-lchAA-Spinach2 template and a tRNALys-Spinach 2 control template (Fig. S2). A forward annealing primer was used to introduce the T7 promoter sequence, resulting in 230-bp and 168-bp DNA templates, respectively. Transcription reactions were performed, and the products were purified and quantified as previously described (101). The in vitro Spinach2 fluorescence activation assay was modified from methods described previously (64). Briefly, the two RNA aptamers were each diluted to 2 μM, added to an equal volume of 2× renaturation buffer (80 mM HEPES [pH 7.5], 250 mM KCl, 6 mM MgCl2), heated to 70°C for 3 min, and cooled at room temperature for 5 min. To test the binding affinity of c-di-GMP for the lchAA aptamer, binding reactions were prepared for each RNA aptamer (40 mM HEPES [pH 7.5], 125 mM KCl, 3 mM MgCl2, 100 nM RNA, 10 μM DFHBI) and incubated in the dark at room temperature for 30 min. Stocks of c-di-GMP or pGpG were prepared and added to the binding reaction mixture every 30 min to achieve the final concentrations that are graphed in the figures. Ligands were added until saturation of fluorescence was reached. A Quantus fluorometer (Promega) was used to excite the reaction mixture at 495 nm every 30 min after incubation with increasing amounts of c-di-GMP and pGpG. Binding to c-di-AMP was also assessed. The experiment was replicated twice, and the background fluorescence of DFHBI alone was subtracted from all data points.
Culture conditions and construction of B. subtilis strains.
All B. subtilis strains in this study were derived from PY79 (unless otherwise noted) and are listed in Table S1. Assessment of B. subtilis 3610 was performed with the use of DS7187, a competent 3610 ΔcomI mutant (provided by D. Kearns, Indiana University) (102). A table of the primers and plasmids used is available upon request. Strains were grown at 37°C on lysogeny broth (LB) plates supplemented with 1.5% Bacto agar and, when appropriate, with final concentrations of the following antibiotics: 5 μg/ml chloramphenicol, and 1 μg/ml erythromycin added alongside 25 μg/ml lincomycin. Integration at the amyE locus was performed with plasmids derived from pJG019 (GenBank accession no. KX499653.1) or pVMZ006, both derivatives of pDG1662 (BGSC). To construct pRSL_F4, the lchAA leader sequence was synthesized (GenScript) and subcloned into the HindIII restriction site of pJG019. For construction of the PpdeH-sf gfp reporter, the constitutive promoter of pVMZ006 was replaced with the pdeH promoter sequence that was described previously (49). The sequence of sf gfp was amplified from pJ204:102624 (DNA2.0, Inc.) and was used to replace the yfp sequence of pVMZ006 via Gibson assembly (103). The fluorescent transcriptional cell type reporters Phag-gfp, PtapA-yfp, and PcomG-yfp that were used in this study were described previously (4, 5, 104, 105). To construct the plasmids harboring these reporters, promoter sequences of tapA and comG were amplified from B. subtilis and used to replace the constitutive promoter upstream of yfp in pVMZ006 by Gibson assembly (103). The amyE::Phag-gfp fusion from DS4432 (provided by D. Kearns, Indiana University) was introduced into the PY79 chromosome through double homologous recombination of competent cells. Plasmids derived from pDG1664 (BGSC) were used for integration at the thrC locus. To make markerless deletions of ΔpdeH, Δspo0A, ΔsinR, and ΔsigD, strains harboring an erythromycin resistance cassette inserted in loci BSU31740 (pdeH), BSU24220 (spo0A), BSU24610 (sinR), and BSU16470 (sigD) were obtained from the BKE collection (BGSC). Markerless deletions were created through transformation with pDR244 (BGSC), as previously described (106). Removal of the erythromycin-resistance cassette was verified by Sanger sequencing. Transformation of PY79 was performed using a previously described protocol (107).
Fluorescence microscopy and quantification.
Single colonies were used to inoculate liquid minimal salts glycerol glutamate (MSgg) medium (75) and grown at 37°C with shaking overnight. The following morning, cultures of each strain were inoculated 1:50 in fresh medium and grown at 37°C with shaking until reaching an optical density at 600 nm (OD600) of 1.0. Aliquots of these cultures were placed on 1.5% low-melting-point agarose MSgg pads and allowed to dry for 10 min. Agarose pads were inverted on a glass bottom dish (Willco Wells). Cells were imaged at room temperature using a Zeiss Axio-Observer Z1 inverted fluorescence microscope equipped with a Rolera EM-C2 electron-multiplying charge-coupled (EMCC) camera, enclosed within a temperature-controlled environmental chamber. Fluorescence intensity per cell was quantified using Oufti analysis software (108). Images were analyzed and adjusted with FIJI software (109).
Supplementary Material
ACKNOWLEDGMENTS
Research for this project was supported by NSF MCB1051440 and NIH AI110432. C.A.W. was supported by NIH T32 GM080201.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00247-19.
REFERENCES
- 1.Dubnau D, Losick R. 2006. Bistability in bacteria. Mol Microbiol 61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 2.Smits WK, Kuipers OP, Veening J-W. 2006. Phenotypic variation in bacteria: the role of feedback regulation. Nat Rev Microbiol 4:259–271. doi: 10.1038/nrmicro1381. [DOI] [PubMed] [Google Scholar]
- 3.Veening J-W, Smits WK, Kuipers OP. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol 62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- 4.Lopez D, Vlamakis H, Kolter R. 2009. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev 33:152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- 5.Vlamakis H, Aguilar C, Losick R, Kolter R. 2008. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev 22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kearns DB, Losick R. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev 19:3083–3094. doi: 10.1101/gad.1373905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leisner M, Stingl K, Frey E, Maier B. 2008. Stochastic switching to competence. Curr Opin Microbiol 11:553–559. doi: 10.1016/j.mib.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 8.Chai Y, Chu F, Kolter R, Losick R. 2008. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol 67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson FA, Seon-Yi C, Stanley-Wall NR. 2012. Selective heterogeneity in exoprotease production by Bacillus subtilis. PLoS One 7:e38574. doi: 10.1371/journal.pone.0038574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol 55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 11.Marlow VL, Porter M, Hobley L, Kiley TB, Swedlow JR, Davidson FA, Stanley-Wall NR. 2014. Phosphorylated DegU manipulates cell fate differentiation in the Bacillus subtilis biofilm. J Bacteriol 196:16–27. doi: 10.1128/JB.00930-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins D, Dworkin J. 2012. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev 36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piggot PJ, Hilbert DW. 2004. Sporulation of Bacillus subtilis. Curr Opin Microbiol 7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 14.González-Pastor JE. 2011. Cannibalism: a social behavior in sporulating Bacillus subtilis. FEMS Microbiol Rev 35:415–424. doi: 10.1111/j.1574-6976.2010.00253.x. [DOI] [PubMed] [Google Scholar]
- 15.Papenfort K, Bassler BL. 2016. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol 14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenal U, Reinders A, Lori C. 2017. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol 15:271–284. doi: 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- 17.Gomelsky M. 2011. cAMP, c-di-GMP, c-di-AMP and now cGMP: bacteria use them all! Mol Microbiol 79:562–565. doi: 10.1111/j.1365-2958.2010.07514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hengge R, Gründling A, Jenal U, Ryan R, Yildiz F. 2016. Bacterial signal transduction by cyclic di-GMP and other nucleotide second messengers. J Bacteriol 198:15–26. doi: 10.1128/JB.00331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purcell EB, Tamayo R. 2016. Cyclic diguanylate signaling in Gram-positive bacteria. FEMS Microbiol Rev 40:753–773. doi: 10.1093/femsre/fuw013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Römling U, Gomelsky M, Galperin MY. 2005. c-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol 57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 22.Ausmees N, Mayer R, Weinhouse H, Volman G, Amikam D, Benziman M, Lindberg M. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol Lett 204:163–167. doi: 10.1111/j.1574-6968.2001.tb10880.x. [DOI] [PubMed] [Google Scholar]
- 23.Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci U S A 101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol 187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 26.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, Meuwly M, Jenal U. 2006. Allosteric control of cyclic di-GMP signaling. J Biol Chem 281:32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- 29.Simm R, Morr M, Kader A, Nimtz M, Römling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility: cyclic di-GMP turnover by GGDEF and EAL domains. Mol Microbiol 53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 30.Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, Benziman M. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol 180:4416–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol 53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL Domains. J Bacteriol 187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem 280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 34.Tamayo R, Tischler AD, Camilli A. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J Biol Chem 280:33324–33330. doi: 10.1074/jbc.M506500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galperin MY, Nikolskaya AN, Koonin EV. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett 203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 36.Galperin MY, Natale DA, Aravind L, Koonin EV. 1999. A specialized version of the HD hydrolase domain implicated in signal transduction. J Mol Microbiol Biotechnol 1:303–305. [PMC free article] [PubMed] [Google Scholar]
- 37.Hammer BK, Bassler BL. 2009. Distinct sensory pathways in Vibrio cholerae El Tor and classical biotypes modulate cyclic dimeric GMP levels to control biofilm formation. J Bacteriol 191:169–177. doi: 10.1128/JB.01307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miner KD, Kurtz DM. 2016. Active site metal occupancy and cyclic di-GMP phosphodiesterase activity of Thermotoga maritima HD-GYP. Biochemistry 55:970–979. doi: 10.1021/acs.biochem.5b01227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen D, Mechold U, Nevenzal H, Yarmiyhu Y, Randall TE, Bay DC, Rich JD, Parsek MR, Kaever V, Harrison JJ, Banin E. 2015. Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:11359–11364. doi: 10.1073/pnas.1421450112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orr MW, Donaldson GP, Severin GB, Wang J, Sintim HO, Waters CM, Lee VT. 2015. Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover. Proc Natl Acad Sci U S A 112:E5048–E5057. doi: 10.1073/pnas.1507245112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orr MW, Weiss CA, Severin GB, Turdiev H, Kim S-K, Turdiev A, Liu K, Tu BP, Waters CM, Winkler WC, Lee VT. 2018. A subset of exoribonucleases serve as degradative enzymes for pGpG in c-di-GMP signaling. J Bacteriology 200:e00300-18. doi: 10.1128/JB.00300-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chou S-H, Galperin MY. 2016. Diversity of Cyclic Di-GMP-Binding Proteins and Mechanisms. J Bacteriol 198:32–46. doi: 10.1128/JB.00333-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson JW, Breaker RR. 2017. The lost language of the RNA World. Sci Signal 10:eaam8812. doi: 10.1126/scisignal.aam8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henkin TM. 2008. Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev 22:3383–3390. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramesh A. 2015. Second messenger-sensing riboswitches in bacteria. Semin Cell Dev Biol 47–48:3–8. doi: 10.1016/j.semcdb.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 48.Lee ER, Sudarsan N, Breaker RR. 2010. Riboswitches That Sense Cyclic Di-GMP, p 215–229. In Wolfe AJ, Visick KL (ed), The second messenger cyclic di-GMP. ASM Press, Washington, DC. [Google Scholar]
- 49.Chen Y, Chai Y, Guo J.-h, Losick R. 2012. Evidence for Cyclic Di-GMP-mediated signaling in Bacillus subtilis. J Bacteriol 194:5080–5090. doi: 10.1128/JB.01092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao X, Mukherjee S, Matthews PM, Hammad LA, Kearns DB, Dann CE. 2013. Functional characterization of core components of the Bacillus subtilis cyclic-di-GMP signaling pathway. J Bacteriol 195:4782–4792. doi: 10.1128/JB.00373-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subramanian S, Gao X, Dann CE, Kearns DB. 2017. MotI (DgrA) acts as a molecular clutch on the flagellar stator protein MotA in Bacillus subtilis. Proc Natl Acad Sci U S A 114:13537–13542. doi: 10.1073/pnas.1716231114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bedrunka P, Graumann PL. 2017. New functions and subcellular localization patterns of c-di-GMP components (GGDEF domain proteins) in B. subtilis. Front Microbiol 8:794. doi: 10.3389/fmicb.2017.00794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bedrunka P, Graumann PL. 2017. Subcellular clustering of a putative c-di-GMP-dependent exopolysaccharide machinery affecting macro colony architecture in Bacillus subtilis. Environ Microbiol Rep 9:211–222. doi: 10.1111/1758-2229.12496. [DOI] [PubMed] [Google Scholar]
- 54.Minasov G, Padavattan S, Shuvalova L, Brunzelle JS, Miller DJ, Baslé A, Massa C, Collart FR, Schirmer T, Anderson WF. 2009. Crystal structures of YkuI and its complex with second messenger cyclic di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains. J Biol Chem 284:13174–13184. doi: 10.1074/jbc.M808221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chandrangsu P, Helmann JD. 2016. Intracellular Zn(II) intoxication leads to dysregulation of the PerR regulon resulting in heme toxicity in Bacillus subtilis. PLoS Genet 12:e1006515. doi: 10.1371/journal.pgen.1006515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aguilar C, Vlamakis H, Losick R, Kolter R. 2007. Thinking about Bacillus subtilis as a multicellular organism. Curr Opin Microbiol 10:638–643. doi: 10.1016/j.mib.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grangemard I, Wallach J, Maget-Dana R, Peypoux F. 2001. Lichenysin. Appl Biochem Biotechnol 90:199–210. doi: 10.1385/ABAB:90:3:199. [DOI] [PubMed] [Google Scholar]
- 58.Sen R. 2010. Surfactin: biosynthesis, genetics and potential applications. Adv Exp Med Biol 672:316–323. doi: 10.1007/978-1-4419-5979-9_24. [DOI] [PubMed] [Google Scholar]
- 59.Veith B, Herzberg C, Steckel S, Feesche J, Maurer KH, Ehrenreich P, Bäumer S, Henne A, Liesegang H, Merkl R, Ehrenreich A, Gottschalk G. 2004. The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. J Mol Microbiol Biotechnol 7:204–211. doi: 10.1159/000079829. [DOI] [PubMed] [Google Scholar]
- 60.Serganov A, Nudler E. 2013. A decade of riboswitches. Cell 152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winkler WC, Breaker RR. 2005. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol 59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 62.Paige JS, Wu KY, Jaffrey SR. 2011. RNA mimics of green fluorescent protein. Science 333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.You M, Jaffrey SR. 2015. Structure and mechanism of RNA mimics of green fluorescent protein. Annu Rev Biophys 44:187–206. doi: 10.1146/annurev-biophys-060414-033954. [DOI] [PubMed] [Google Scholar]
- 64.Kellenberger CA, Hammond MC. 2015. In vitro analysis of riboswitch-Spinach aptamer fusions as metabolite-sensing fluorescent biosensors. Methods Enzymol 550:147–172. doi: 10.1016/bs.mie.2014.10.045. [DOI] [PubMed] [Google Scholar]
- 65.Ponchon L, Dardel F. 2007. Recombinant RNA technology: the tRNA scaffold. Nat Methods 4:571–576. doi: 10.1038/nmeth1058. [DOI] [PubMed] [Google Scholar]
- 66.You M, Litke JL, Jaffrey SR. 2015. Imaging metabolite dynamics in living cells using a Spinach-based riboswitch. Proc Natl Acad Sci U S A 112:E2756–2765. doi: 10.1073/pnas.1504354112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. 2005. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell 18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 68.Smith KD, Lipchock SV, Strobel SA. 2012. Structural and biochemical characterization of linear dinucleotide analogues bound to the c-di-GMP-I aptamer. Biochemistry 51:425–432. doi: 10.1021/bi2016662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eichenberger P, Jensen ST, Conlon EM, van Ooij C, Silvaggi J, González-Pastor J-E, Fujita M, Ben-Yehuda S, Stragier P, Liu JS, Losick R. 2003. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J Mol Biol 327:945–972. doi: 10.1016/S0022-2836(03)00205-5. [DOI] [PubMed] [Google Scholar]
- 70.Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. 1991. Forespore-specific transcription of a gene in the signal transduction pathway that governs pro-sigma K processing in Bacillus subtilis. Genes Dev 5:456–466. doi: 10.1101/gad.5.3.456. [DOI] [PubMed] [Google Scholar]
- 71.Camp AH, Losick R. 2009. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev 23:1014–1024. doi: 10.1101/gad.1781709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roels S, Driks A, Losick R. 1992. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J Bacteriol 174:575–585. doi: 10.1128/jb.174.2.575-585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng L, Halberg R, Roels S, Ichikawa H, Kroos L, Losick R. 1992. Sporulation regulatory protein gerE from Bacillus subtilis binds to and can activate or repress transcription from promoters for mother-cell-specific genes. J Mol Biol 226:1037–1050. doi: 10.1016/0022-2836(92)91051-P. [DOI] [PubMed] [Google Scholar]
- 74.Cutting S, Panzer S, Losick R. 1989. Regulatory studies on the promoter for a gene governing synthesis and assembly of the spore coat in Bacillus subtilis. J Mol Biol 207:393–404. doi: 10.1016/0022-2836(89)90262-3. [DOI] [PubMed] [Google Scholar]
- 75.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A 98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chai Y, Kolter R, Losick R. 2010. Reversal of an epigenetic switch governing cell chaining in Bacillus subtilis by protein instability. Mol Microbiol 78:218–229. doi: 10.1111/j.1365-2958.2010.07335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chai Y, Kolter R, Losick R. 2009. Paralogous antirepressors acting on the master regulator for biofilm formation in Bacillus subtilis. Mol Microbiol 74:876–887. doi: 10.1111/j.1365-2958.2009.06900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fang X, Gomelsky M. 2010. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol Microbiol 76:1295–1305. doi: 10.1111/j.1365-2958.2010.07179.x. [DOI] [PubMed] [Google Scholar]
- 79.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell 38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 81.Molle V, Fujita M, Jensen ST, Eichenberger P, González‐Pastor JE, Liu JS, Losick R. 2003. The Spo0A regulon of Bacillus subtilis. Mol Microbiol 50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- 82.Fagerlund A, Smith V, Røhr ÅK, Lindbäck T, Parmer MP, Andersson KK, Reubsaet L, Økstad OA. 2016. Cyclic diguanylate regulation of Bacillus cereus group biofilm formation. Mol Microbiol 101:471–494. doi: 10.1111/mmi.13405. [DOI] [PubMed] [Google Scholar]
- 83.Tran NT, Hengst CDD, Gomez-Escribano JP, Buttner MJ. 2011. Identification and characterization of CdgB, a diguanylate cyclase involved in developmental processes in Streptomyces coelicolor. J Bacteriol 193:3100–3108. doi: 10.1128/JB.01460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Den Hengst CD, Tran NT, Bibb MJ, Chandra G, Leskiw BK, Buttner MJ. 2010. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol Microbiol 78:361–379. doi: 10.1111/j.1365-2958.2010.07338.x. [DOI] [PubMed] [Google Scholar]
- 85.Hull TD, Ryu M-H, Sullivan MJ, Johnson RC, Klena NT, Geiger RM, Gomelsky M, Bennett JA. 2012. Cyclic di-GMP phosphodiesterases RmdA and RmdB are involved in regulating colony morphology and development in Streptomyces coelicolor. J Bacteriol 194:4642–4651. doi: 10.1128/JB.00157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tschowri N, Schumacher MA, Schlimpert S, Chinnam NB, Findlay KC, Brennan RG, Buttner MJ. 2014. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158:1136–1147. doi: 10.1016/j.cell.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.D'Souza C, Nakano MM, Zuber P. 1994. Identification of comS, a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis. Proc Natl Acad Sci U S A 91:9397–9401. doi: 10.1073/pnas.91.20.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nakano MM, Zuber P. 1989. Cloning and characterization of srfB, a regulatory gene involved in surfactin production and competence in Bacillus subtilis. J Bacteriol 171:5347–5353. doi: 10.1128/jb.171.10.5347-5353.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hamoen LW, Venema G, Kuipers OP. 2003. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 149:9–17. doi: 10.1099/mic.0.26003-0. [DOI] [PubMed] [Google Scholar]
- 90.Dubnau D. 1991. Genetic competence in Bacillus subtilis. Microbiol Mol Biol Rev 55:395–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lopez D, Vlamakis H, Losick R, Kolter R. 2009. Paracrine signaling in a bacterium. Genes Dev 23:1631–1638. doi: 10.1101/gad.1813709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yakimov MM, Golyshin PN. 1997. ComA-dependent transcriptional activation of lichenysin A synthetase promoter in Bacillus subtilis cells. Biotechnol Prog 13:757–761. doi: 10.1021/bp9700622. [DOI] [PubMed] [Google Scholar]
- 93.Jakobs M, Hoffmann K, Liesegang H, Volland S, Meinhardt F. 2015. The two putative comS homologs of the biotechnologically important Bacillus licheniformis do not contribute to competence development. Appl Microbiol Biotechnol 99:2255–2266. doi: 10.1007/s00253-014-6291-5. [DOI] [PubMed] [Google Scholar]
- 94.Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI. 2010. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328:1295–1297. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kulasekara BR, Kamischke C, Kulasekara HD, Christen M, Wiggins PA, Miller SI. 2013. c-di-GMP heterogeneity is generated by the chemotaxis machinery to regulate flagellar motility. Elife 2:e01402. doi: 10.7554/eLife.01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Abel S, Bucher T, Nicollier M, Hug I, Kaever V, Abel Zur Wiesch P, Jenal U. 2013. Bi-modal distribution of the second messenger c-di-GMP controls cell fate and asymmetry during the Caulobacter cell cycle. PLoS Genet 9:e1003744. doi: 10.1371/journal.pgen.1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kellenberger CA, Sales-Lee J, Pan Y, Gassaway MM, Herr AE, Hammond MC. 2015. A minimalist biosensor: quantitation of cyclic di-GMP using the conformational change of a riboswitch aptamer. RNA Biol 12:1189–1197. doi: 10.1080/15476286.2015.1062970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang J, Fei J, Leslie BJ, Han KY, Kuhlman TE, Ha T. 2015. Tandem Spinach array for mRNA imaging in living bacterial cells. Sci Rep 5:17295. doi: 10.1038/srep17295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Süel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. 2007. Tunability and noise dependence in differentiation dynamics. Science 315:1716–1719. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- 100.Campo N, Rudner DZ. 2007. SpoIVB and CtpB are both forespore signals in the activation of the sporulation transcription factor K in Bacillus subtilis. J Bacteriol 189:6021–6027. doi: 10.1128/JB.00399-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dann CE, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. 2007. Structure and mechanism of a metal-sensing regulatory RNA. Cell 130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 102.Konkol MA, Blair KM, Kearns DB. 2013. Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. J Bacteriol 195:4085–4093. doi: 10.1128/JB.00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 104.Veening J-W, Smits WK, Hamoen LW, Kuipers OP. 2006. Single cell analysis of gene expression patterns of competence development and initiation of sporulation in Bacillus subtilis grown on chemically defined media. J Appl Microbiol 101:531–541. doi: 10.1111/j.1365-2672.2006.02911.x. [DOI] [PubMed] [Google Scholar]
- 105.Smits WK, Eschevins CC, Susanna KA, Bron S, Kuipers OP, Hamoen LW. 2005. Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol Microbiol 56:604–614. doi: 10.1111/j.1365-2958.2005.04488.x. [DOI] [PubMed] [Google Scholar]
- 106.Koo B-M, Kritikos G, Farelli JD, Todor H, Tong K, Kimsey H, Wapinski I, Galardini M, Cabal A, Peters JM, Hachmann A-B, Rudner DZ, Allen KN, Typas A, Gross CA. 2017. Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst 4:291.e7–305.e7. doi: 10.1016/j.cels.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jarmer H, Berka R, Knudsen S, Saxild HH. 2002. Transcriptome analysis documents induced competence of Bacillus subtilis during nitrogen limiting conditions. FEMS Microbiol Lett 206:197–200. doi: 10.1111/j.1574-6968.2002.tb11009.x. [DOI] [PubMed] [Google Scholar]
- 108.Paintdakhi A, Parry B, Campos M, Irnov I, Elf J, Surovtsev I, Jacobs-Wagner C. 2016. Oufti: an integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol Microbiol 99:767–777. doi: 10.1111/mmi.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open source platform for biological image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.