Picolinic acid is a common metabolite of l-tryptophan and some aromatic compounds and is an important intermediate in organic chemical synthesis. Although the microbial degradation/detoxification of picolinic acid has been studied for over 50 years, the underlying molecular mechanisms are still unknown. Here, we show that the pic gene cluster is responsible for the complete degradation of picolinic acid. The pic gene cluster was found to be widespread in other Alpha-, Beta-, and Gammaproteobacteria. These findings provide a new perspective for understanding the catabolic mechanisms of picolinic acid in bacteria.
KEYWORDS: Alcaligenes faecalis, bacterial degradation, pathway, pic gene cluster, picolinic acid
ABSTRACT
Picolinic acid (PA) is a natural toxic pyridine derivative. Microorganisms can degrade and utilize PA for growth. However, the full catabolic pathway of PA and its physiological and genetic foundation remain unknown. In this study, we identified a gene cluster, designated picRCEDFB4B3B2B1A1A2A3, responsible for the degradation of PA from Alcaligenes faecalis JQ135. Our results suggest that PA degradation pathway occurs as follows: PA was initially 6-hydroxylated to 6-hydroxypicolinic acid (6HPA) by PicA (a PA dehydrogenase). 6HPA was then 3-hydroxylated by PicB, a four-component 6HPA monooxygenase, to form 3,6-dihydroxypicolinic acid (3,6DHPA), which was then converted into 2,5-dihydroxypyridine (2,5DHP) by the decarboxylase PicC. 2,5DHP was further degraded to fumaric acid through PicD (2,5DHP 5,6-dioxygenase), PicE (N-formylmaleamic acid deformylase), PicF (maleamic acid amidohydrolase), and PicG (maleic acid isomerase). Homologous pic gene clusters with diverse organizations were found to be widely distributed in Alpha-, Beta-, and Gammaproteobacteria. Our findings provide new insights into the microbial catabolism of environmental toxic pyridine derivatives.
IMPORTANCE Picolinic acid is a common metabolite of l-tryptophan and some aromatic compounds and is an important intermediate in organic chemical synthesis. Although the microbial degradation/detoxification of picolinic acid has been studied for over 50 years, the underlying molecular mechanisms are still unknown. Here, we show that the pic gene cluster is responsible for the complete degradation of picolinic acid. The pic gene cluster was found to be widespread in other Alpha-, Beta-, and Gammaproteobacteria. These findings provide a new perspective for understanding the catabolic mechanisms of picolinic acid in bacteria.
INTRODUCTION
Picolinic acid (PA) is a dead-end metabolite of l-tryptophan via the kynurenine pathway in humans and other mammals (1, 2). It is often detected in various biological media, such as cell culture supernatants, serum, and human milk (3). PA can also be produced through other biological processes, such as the microbial degradation of 2-aminophenol, nitrobenzene, catechol, anthranilic acid, and 3-hydroxyanthranilic acid (4–7) (see Fig. S1 in the supplemental material). Furthermore, PA is an important intermediate in the organic synthesis of pharmaceuticals (e.g., carbocaine), herbicides (e.g., picloram and diquat), and fungicides (e.g., 3-trifluoromethyl picolinic acid) (8, 9). Due to its chelating properties, PA has also been added to chromium and iron preparations to treat diabetes and anemia (10, 11). Nevertheless, many studies have shown that PA is highly toxic to organisms. PA inhibits the growth of normal rat kidney cells and the proliferation of T cells, enhances seizure activity in mice, and induces cell death via apoptosis (12–14). In particular, PA showed high antimicrobial activity at concentrations as low as 8 μg/liter (15–17).
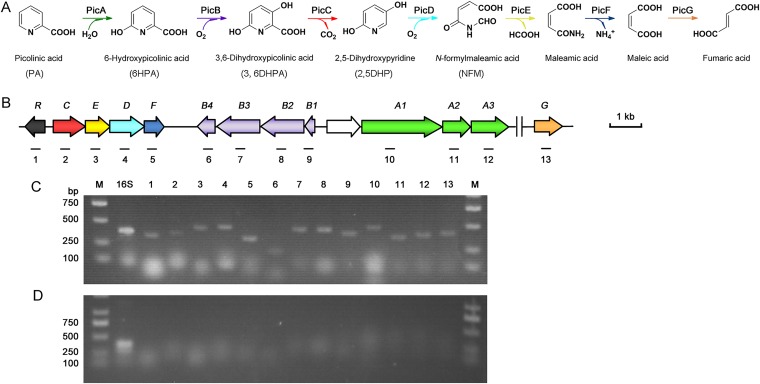
PA cannot be metabolized in the human body and is excreted through urine or sweat (18). Nevertheless, many microorganisms, such as Alcaligenes (19), Arthrobacter (20), Burkholderia (21), and Streptomyces (22), and an unidentified Gram-negative bacterium (designated the UGN strain) were shown to degrade PA (23). Through identification of metabolites, a bacterial degradation pathway for PA was proposed (Fig. 1): PA is 6-hydroxylated to 6-hydroxypicolinic acid (6HPA), which is 3-hydroxylated to 3,6-dihydroxypicolinic acid (3,6DHPA), which is further decarboxylated to 2,5-dihydroxypyridine (2,5DHP), a central degradation intermediate of many pyridine derivatives (24, 25). However, little is known about this catabolic pathway at genetic level.
FIG 1.
PA degradation in A. faecalis JQ135. (A) The proposed PA catabolic pathway and corresponding enzymes. (B) Genetic organization of the pic gene cluster. Genes are annotated according to the color scheme in panel A. (C and D) Agarose gel electrophoresis of RT-PCR products generated using RNA of A. faecalis JQ135 cells grown with PA (C) or citrate (D). Lane M, DNA marker; lane 16S, positive control. Lanes 1 to 13 correspond to the regions marked in panel B.
In our previous studies, the bacterial strain Alcaligenes faecalis JQ135 was isolated from municipal wastewater and was shown to efficiently degrade and utilize PA (26, 27). A mutant of strain JQ135 that was unable to grow on PA was screened through random transposon mutagenesis (28). Bioinformatic analysis indicated that the transposon was inserted into a gene designated picC. PicC was found to be a decarboxylase that catalyzes the decarboxylation of 3,6DHPA to 2,5DHP, which is the third step in PA degradation (28). However, the complete catabolic pathway of PA and its underlying genetic foundation are still unknown. In this study, we found that picC is located within the pic gene cluster (Fig. 1), and the function of each gene in this cluster was identified via in vitro enzymatic assays and gene disruption. In addition, the distribution and organization of pic clusters in other bacteria were also investigated.
RESULTS AND DISCUSSION
Identification of a gene cluster involved in PA degradation.
Previous studies have shown that the 3,6DHPA decarboxylase PicC was involved in PA metabolism (28). In this study, we investigated the genes located upstream and downstream of picC through bioinformatic analysis. The results showed that picC was located in a gene cluster (designated pic) (Fig. 1), consisting of 12 genes, including a putative regulatory gene picR and 11 putative catabolic genes (picA1, picA2, picA3, picB1, picB2, picB3, picB4, picC, picE, picD, and picF). The predicted functions of these genes are summarized in Table 1. Reverse transcription-PCR (RT-PCR) analysis showed that all of the pic genes were induced by PA but not by citrate (Fig. 1). The above results strongly suggested that the pic cluster was involved in PA catabolism in A. faecalis JQ135.
TABLE 1.
The pic genes and assigned functions in Alcaligenes faecalis JQ135
| Gene (locus tag) | Product (aa)a | Assigned function | Sequence identity (%) | Homologous protein in UniProtKB/SwissProt (accession no.) |
|---|---|---|---|---|
| picR (AFA_15150) | 186 | MarR-type negative regulator PicR | 33 | MexR (P52003) |
| picC (AFA_15145) | 323 | 3,6DHPA decarboxylase | 37 | 2,3-Dihydroxybenzoate decarboxylase (P80402) |
| picE (AFA_15140) | 274 | N-Formylmaleamic acid deformylase | 60 | NFM deformylase (Q88FY3) |
| picD (AFA_15135) | 350 | 2,5DHP 5,6-dioxygenase | 55 | 2,5-DHP dioxygenase (Q88FY1) |
| picF (AFA_15130) | 216 | Maleamic acid amidohydrolase | 43 | Maleamic acid amidohydrolase (Q88FY5) |
| picB4 (AFA_15125) | 172 | 6HPA monooxygenase subunit IV | 35 | Anthranilate 1,2-dioxygenase small subunit (Q84BZ2) |
| picB3 (AFA_15120) | 429 | 6HPA monooxygenase subunit III | 45 | Salicylate 5-hydroxylase, oxygenase large subunit (O52379) |
| picB2 (AFA_15115) | 425 | 6HPA monooxygenase subunit II | 36 | Benzene 1,2-dioxygenase ferredoxin reductase (B1LNJ8) |
| picB1 (AFA_15110) | 104 | 6HPA monooxygenase subunit I | 46 | Naphthalene 1,2-dioxygenase ferredoxin (Q51493) |
| picA1 (AFA_15100) | 800 | PA dehydrogenase large subunit | 39 | Caffeine dehydrogenase large subunit (D7REY3) |
| picA2 (AFA_15095) | 278 | PA dehydrogenase small subunit | 33 | Carbon monoxide dehydrogenase medium chain (P19914) |
| picA3 (AFA_15090) | 395 | PA dehydrogenase medium subunit | 46 | Carbon monoxide dehydrogenase small chain (P19915) |
| picG (AFA_16520) | 253 | Maleic acid cis-trans isomerase | 99 | Maleic acid isomerase (O24766) |
aa, amino acid.
The picA1A2A3 genes encode the PA dehydrogenase (PicA).
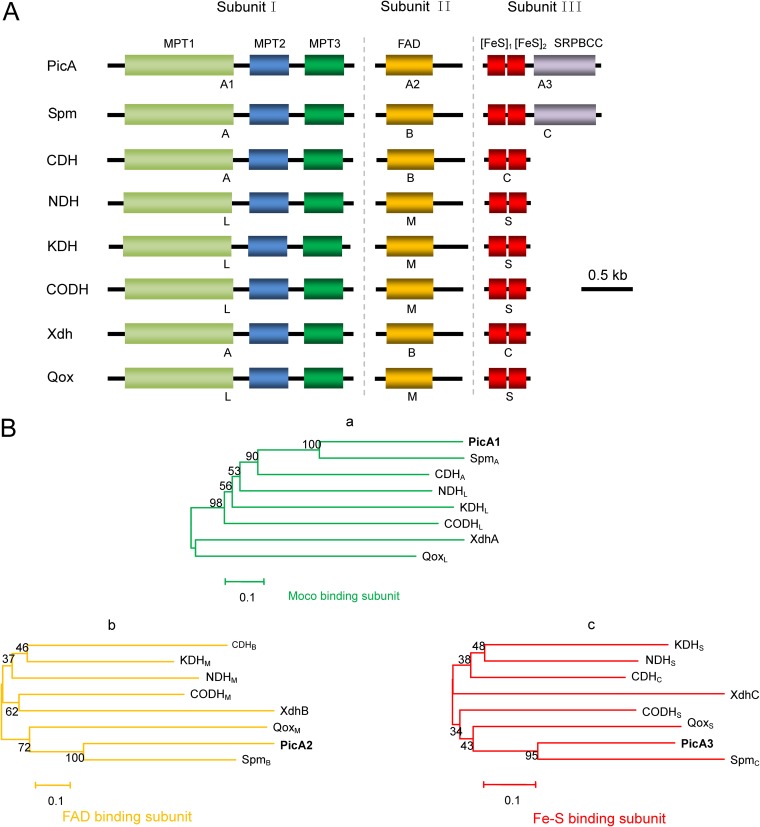
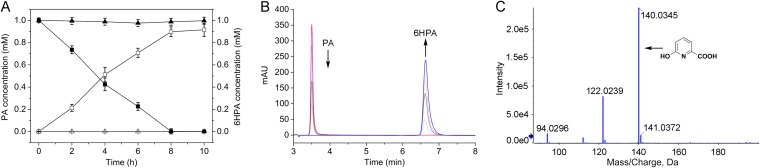
Previous studies have reported that PA was initially 6-hydroxylated to 6HPA (19, 23, 27, 29). The hydroxylation of N-heterocyclic aromatic compounds is usually catalyzed by multicomponent molybdopterin-containing dehydrogenases (30, 31). In the pic gene cluster, the picA1, picA2, and picA3 gene products showed the highest similarities (∼40%) to the respective subunits of three-component molybdopterin-containing dehydrogenases, such as the carbon monoxide dehydrogenase and caffeine dehydrogenase (Table 1). PicA1 and PicA2 were predicted to contain binding domains for the molybdopterin cytosine dinucleotide (MCD) and flavin adenine dinucleotide (FAD), respectively, and PicA3 contained two predicted [Fe-S] clusters (Fig. 2). Thus, picA1, picA2, and picA3 were predicted to encode a three-component molybdopterin-containing dehydrogenase that catalyzes the initial step in the PA catabolic pathway. When picA1A2A3 were deleted, the resulting mutant JQ135 ΔpicA1A2A3 lost the ability to grow on PA but could still grow on 6HPA (see Fig. S2 in the supplemental material). The complemented strain JQ135 ΔpicA1A2A3/pBBR-picA1A2A3 regained the ability to grow on PA. Moreover, the picA1A2A3 genes were then cloned into pBBR1MCS-5 (pBB-picA1A2A3) and transferred into Pseudomonas putida KT2440, which does not degrade PA, 6HPA, and 3,6DHPA (30). High-performance liquid chromatography (HPLC) results showed that the recombinant KT/pBBR-picA1A2A3 strain acquired the ability to convert 1 mM PA into approximately equivalent amounts of 6HPA (Fig. 3). Liquid chromatography-time of flight mass spectrometry (LC/TOF-MS) analysis also showed that the product was 6HPA {the molecular ion peak ([M+H]+) 140.0345}. When lacking one component, the recombinants KT/pBBR-picA2A3 and KT/pBBR-picA1A2 did not show the conversion ability (data not shown), indicating that all of the three components were essential.
FIG 2.
Bioinformatic analysis of PicA. (A) Molecular architecture of several multicomponent molybdenum-containing hydroxylases. Subunits I, II, and III are molybdopterin cytosine dinucleotide (MCD)-, FAD-, and two [Fe-S] cluster-containing components, respectively. SpmABC (GenBank accession numbers AEJ14617 and AEJ14616), 3-succinoylpyridine dehydrogenase from P. putida; CDHABC (D7REY3, D7REY4, and D7REY5), caffeine dehydrogenase from Pseudomonas sp. strain CBB1; NDHLMS (CAA53088, CAA53087, and CAA53086), nicotine dehydrogenase from Arthrobacter nicotinovorans; KDHLMS (WP_016359451, WP_016359456, and WP_016359457), ketone dehydrogenase from A. nicotinovorans; CODHLMS (P19913, P19914, and P19915), carbon monoxide dehydrogenase from Hydrogenophaga pseudoflava; XDHABC (Q46799, Q46800, and Q46801), xanthine dehydrogenase from E. coli; QoxLMS (CAD61045, CAD61046, and CAD61047), quinaldine 4-oxidase from Arthrobacter ilicis. The letters depicted below the proteins indicate the subunit names of the corresponding proteins. The conserved domains are as follows: MPT, domain for binding to the molybdopterin cytosine dinucleotide cofactor (MoCo); FAD, FAD-binding domain; [FeS], ferredoxin-like [2Fe-2S]-binding domain; SRPBCC, SRPBCC ligand-binding domain. (B) Phylogenetic analysis of PicA and related molybdenum-containing hydroxylases: PicA1 and the Moco binding subunit of other enzymes (a); PicA2 and the FAD binding subunit of other enzymes (b); PicA3 and the [2Fe-2S] binding subunit of other enzymes (c). The phylogenetic trees were constructed by using the neighbor-joining method (with a bootstrap of 1,000) with software MEGA, version 6.0. The bar represents the number of amino acid substitutions per site.
FIG 3.
Conversion of PA into 6HPA by recombinant strain KT/pBBR-picA1A2A3. (A) The time course of PA degradation and 6HPA accumulation. Filled symbols, PA degradation by KT2440 (▲) and KT/pBBR-picA1A2A3 (■); open symbols, product 6HPA by KT2440 (△) and KT/pBBR-picA1A2A3 (□). (B) HPLC profiles of PA degradation and 6HPA accumulation by the KT/pBBR-picA1A2A3 strain. The detection wavelength was set at 280 nm. (C) LC/TOF-MS profile of the transformation product 6HPA. AU, arbitrary units.
To investigate the PA dehydrogenase activity of KT/pBBR-picA1A2A3, the recombinant strain was cultured in mineral salts medium (MSM) containing 1.0 mM citrate and 1.0 mM PA for 12 h, and the cell lysate of KT/pBBR-picA1A2A3 was tested for PA dehydrogenase activity with phenazine methosulfate (PMS) as an electron acceptor. The Km value for PA at pH 7.0 and 25°C was 0.65 ± 0.14 μM, and the Vmax was 44.89 ± 2.45 mU/mg (Fig. S3). No PA dehydrogenase activity was detected under anaerobic conditions, which was similar to results for the nicotinate hydroxylase (NicAB) in P. putida KT2440 (30). All of these results demonstrated that the picA1A2A3 genes are responsible for the hydroxylation of PA to 6HPA in A. faecalis JQ135.
The picB1B2B3B4 genes encode a four-component 6HPA monooxygenase (PicB).
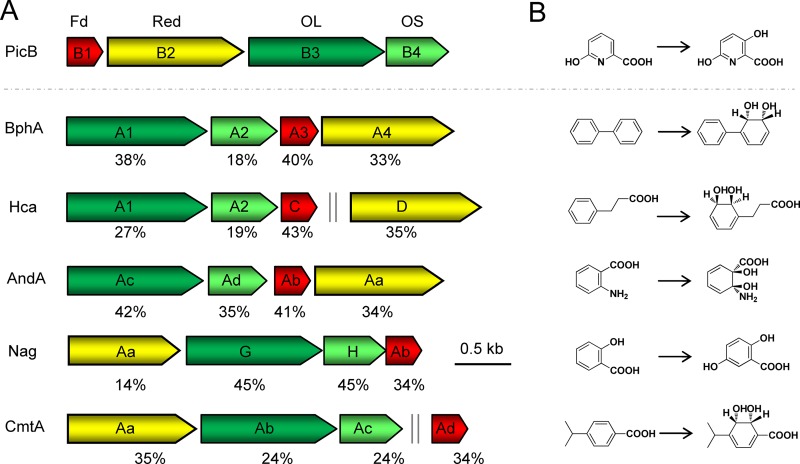
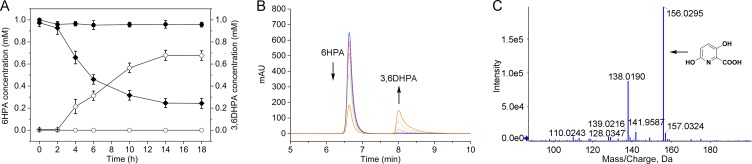
The second step in PA catabolism is predicted to be the 3-hydroxylation of 6HPA to 3,6DHPA (23). BLAST homology searches against the NCBI database showed that picB1, picB2, picB3, and picB4 had high levels of identity (35% to 45%) at the amino acid level with the respective components of four-component Rieske non-heme iron aromatic ring-hydroxylating oxygenases (RHOs) (Table 1; Fig. 4). RHOs are usually involved in the hydroxylation of aromatic compounds (32–34). Therefore, the genes picB1B2B3B4 were predicted to encode a four-component RHO catalyzing the 3-hydroxylation of 6HPA to 3,6DHPA. The picB1B2B3B4 genes were deleted from the genome of A. faecalis JQ135, and the resulting mutant JQ135 ΔpicB1B2B3B4 lost the ability to grow on PA or 6HPA but could still grow on 3,6DHPA (Fig. S4). The complemented strain JQ135 ΔpicB1B2B3B4/pBBR-picB1B2B3B4 regained the ability to grown on PA and 6HPA. The picB1B2B3B4 genes were cloned into pBBR1MCS-5 and transferred into P. putida KT2440. HPLC analysis showed that the recombinant KT/pBBR-picB1B2B3B4 strain could degrade 6HPA with the appearance of one new peak in the HPLC chromatogram (Fig. 5). LC/TOF-MS analysis showed that this peak corresponded to 3,6DHPA [with a molecular ion peak ([M+H]+) at 156.0295]. After degradation for 24 h, 0.75 mM 6HPA was depleted, and an approximately equal molar concentration of 3,6DHPA accumulated. Moreover, the recombinants KT/pBBR-picB2B3B4 and KT/pBBR-picB1B2B3 could not convert 6HPA (data not shown), suggesting that all four of these components were essential. These results indicated that the picB1B2B3B4 genes encode a four-component RHO-type monooxygenase responsible for the conversion of 6HPA into 3,6DHPA in A. faecalis JQ135.
FIG 4.
Genetic organizations of the PicB-encoding genes compared with similar Rieske non-heme iron aromatic ring-hydroxylating oxygenase genes (A) and the corresponding enzyme reactions (B). The arrows in panel A indicate the size and direction of each gene. Homologous genes are shown in the same color. Fd, ferredoxin; Red, reductase; OL, oxygenase large component; OS, oxygenase small component. Double vertical gray lines indicate discontinuous genes. Numbers below the arrows indicate the percent amino acid sequence identity with the ortholog picB gene product. BphA1A2A3A4 (GenBank accession numbers Q53122, Q53123, Q53124, and Q0S032), biphenyl 2,3-dioxygenase from Rhodococcus jostii; HcaA1A2CD (P0ABR5, Q47140, P0ABW0, and P77650), 3-phenylpropionate dioxygenase from E. coli; AndAaAbAcAd (Q84BZ0, Q84BZ1, Q84BZ3, and Q84BZ2), anthranilate 1,2-dioxygenase from Burkholderia cepacia; NagAaGHAb (O52378, O52379, O52380, and O52381), salicylate 5-hydroxylase from Ralstonia sp.; CmtAaAbAcAd (Q51973, Q51974, Q51975, and Q51978), p-cumate 2,3-dioxygenase from Pseudomonas putida.
FIG 5.
Conversion of 6HPA into 3,6DHPA by recombinant strain KT/pBBR-picB1B2B3B4. (A) The time course of 6HPA degradation and 3,6DHPA accumulation. Filled symbols, 6HPA degradation by KT2440 (●) and KT/pBBR-picB1B2B3B4 (◆); open symbols, transformation product 3,6DHPA by KT2440 (○) and KT/pBBR-picB1B2B3B4 (◇). (B) HPLC profiles of 6HPA degradation and 3,6DHPA accumulation by the KT/pBBR-picB1B2B3B4 strain. The detection wavelength was set at 310 nm. (C) LC/TOF-MS profile of the transformation product 3,6DHPA.
Interestingly, cell lysates of KT/pBBR-picB1B2B3B4 grown in the presence of 6HPA were unable to convert 6HPA even though several cofactors (e.g., FAD, flavin mononucleotide [FMN], NADH, or NADPH) were added. Previous studies have attempted to purify the 6HPA monooxygenase from cell lysates of Arthrobacter picolinophilus DSM 20665 and the UGN strain; however, these studies were also unable to detect the PicB activity (23, 35). These results suggested that the PicB was unstable and readily lost activity.
PicDEF and PicG (MaiA) convert 2,5DHP into fumaric acid.
Our previous study demonstrated that 3,6DHPA was converted to 2,5DHP by PicC in A. faecalis JQ135 (28). 2,5DHP is a central intermediate of many pyridine derivatives, including nicotinate (30), nicotine (36), 2-hydroxy-pyridine (24), and 5-hydroxypicolinic acid (26). Bacterial catabolism of 2,5DHP has been previously studied, and two 2,5DHP catabolic pathways have been reported: the hydroxylation and maleamate pathways. In the hydroxylation pathway, 2,5DHP is hydroxylated to 2,3,6-trihydroxypyridine, which spontaneously converts to a blue pigment (25). In the maleamate pathway, however, 2,5DHP is cleaved to N-formylmaleamic acid (NFM) by a dioxygenase, and NFM is then converted by a deformylase to maleamic acid, which is further deaminated by an amidohydrolase to maleic acid (24, 25, 30, 36, 37). Previous studies have shown that the four enzymes involved in this pathway are highly conserved (Fig. S5). In the pic gene cluster, three genes, picD, picE, and picF, are located downstream of picC (Fig. 1). PicD, PicE, and PicF show high similarities (40 to 60%) to 2,5DHP 5,6-dioxygenase, NFM deformylase, and maleamic acid amidohydrolase, respectively (Table 1; Fig. S5). This strongly implies that 2,5DHP is degraded via the maleamate pathway in A. faecalis JQ135. We overexpressed PicD in Escherichia coli BL21(DE3), and SDS-PAGE analysis showed an intense band at the predicted size of 6×His-tagged PicD (38 kDa) (Fig. S6). Purified PicD was able to convert 2,5DHP (Fig. S6), as measured spectrophotometrically by a decrease in absorbance at 320 nm (36, 37). No blue pigment accumulated in the enzymatic reaction, further suggesting that 2,5DHP was degraded via the maleamate pathway and not the hydroxylation pathway. Unfortunately, the expected product (NFM) was not detected, which might be due to the instability of NFM. PicD had a Km value of 65.72 ± 6.27 μM and showed no activity without the addition of Fe2+ (Fig. S6), which resembles previous reports of other 2,5DHP dioxygenases (30, 36, 37). To further investigate the function of the picEDF genes, a DNA fragment containing picEDF was cloned into Sphingomonas wittichii DC-6, which cannot degrade 2,5DHP (Fig. S7) (38). Recombinant DC-6/pBBR-picEDF acquired the ability to convert 2,5DHP to maleic acid (Fig. S8); however, the recombinant strain could not utilize 2,5DHP as a carbon source for growth (Fig. S7), which might be due to its inability to further degrade maleic acid to fumaric acid. These results further confirmed that the picEDF genes were responsible for conversion of 2,5DHP to maleic acid.
Maleic acid can be isomerized into fumaric acid, an intermediate of the Krebs cycle. However, the pic cluster lacks an isomerase-encoding gene. Bioinformatic analysis revealed that the gene AFA_16520 (maiA, also named picG in this study) in the genome of A. faecalis JQ135, which is physically separated from the pic cluster, shares 99% similarity at the amino acid level with a maleic acid cis-trans isomerase MaiA from A. faecalis IFO13111 (39). Our previous study showed that PicG was essential for the utilization of PA, nicotinic acid, and 5-hydroxypicolinic acid (26). To further investigate the function of picG, this gene was deleted from A. faecalis JQ135, resulting in the mutant JQ135 ΔpicG. The mutant strain could still degrade 2,5DHP but lost the ability to utilize the substrate for growth and accumulated maleic acid in the culture (Fig. S8). This result indicated that PicG was responsible for the isomerization of maleic acid to fumaric acid. Furthermore, a DNA fragment containing the picEDFG genes was cloned into Sphingomonas wittichii DC-6. Recombinant DC-6/pBBR-picEDFG acquired the ability to utilize 2,5DHP for growth (Fig. S7). These results confirmed that PicDEF and PicG (MaiA) are responsible for the conversion of 2,5DHP to fumaric acid.
Diversity of pic genes in other bacteria.
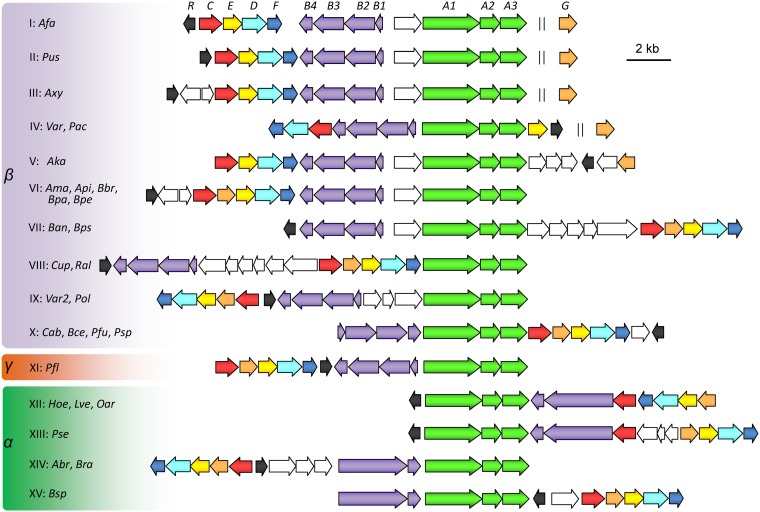
Six complete genome sequences of A. faecalis strains (ZD02, P156, DSM 30030, J481, FDAARGOS_491, and BDB4) are available in NCBI, and pic gene clusters were identified in all of these genomes (Fig. 6; Table S1). Orthologous pic gene clusters were also found in other Alpha-, Beta-, and Gammaproteobacteria (21 genera and over 160 strains) (Table S1). Most of these strains belong to the order Burkholderiales of the class Betaproteobacteria, including the following genera: Achromobacter, Alcaligenes, Advenella, Bordetella, Burkholderia, Caballeronia, Cupriavidus, Pandoraea, Paraburkholderia, Polaromonas, Pseudacidovorax, Pusillimonas, Ralstonia, and Variovorax. Interestingly, some of these strains are toxic-compound degraders (e.g., Achromobacter xylosoxidans A8 and Variovorax sp. strain JS1663 (40, 41), human pathogens (e.g., Bordetella pertussis Tohama I and Bordetella bronchiseptica RB50) (42), and nematicidal bacteria (e.g., A. faecalis ZD02) (43), whereas other strains represent typical soil bacteria (e.g., Pseudomonas), plant rhizosphere-associated bacteria (e.g., Bradyrhizobium), arctic bacteria (Octadecabacter arcticus 238) (44), and Antarctic bacteria (Hoeflea sp. strain IMCC20628) (45).) Three of predicted degraders listed in Table S1 (A. faecalis ATCC 8750, Cupriavidus pinatubonensis JMP134 [46], and Pusillimonas sp. strain T2) were tested for their abilities to utilize PA. The results confirmed that these threes strains could utilize PA as a sole carbon source (Fig. S9). Previous reports also showed that bacteria in the Aerococcus, Arthrobacter, and Streptomyces genera could degrade PA (20, 22, 29). However, no orthologues of the A. faecalis pic genes were found in these bacteria, suggesting that other new and unknown catabolic genes may exist.
FIG 6.
Predicted PA catabolism gene clusters in bacterial genomes. α, β, and γ indicate the Alpha-, Beta-, and Gammaproteobacteria. I to XV, the 15 different types of PA catabolism loci. Abbreviations and representative strains are as follows: Afa, Alcaligenes faecalis JQ135; Pus, Pusillimonas sp. strain T2; Axy, Achromobacter xylosoxidans A8; Var, Variovorax paradoxus S110; Pac, Pseudacidovorax sp. strain RU35E; Aka, Advenella kashmirensis W13003; Ama, Achromobacter marplatensis B2; Api, Achromobacter piechaudii ATCC 43553; Bbr, Bordetella bronchiseptica RB50; Bpa, Bordetella parapertussis ATCC BAA-587; Bpe, Bordetella pertussis Tohama I; Ban, Bordetella ansorpii NCTC13364; Bps, Bordetella pseudohinzii HI4681; Cup, Cupriavidus necator N-1; Ral, Ralstonia pickettii DTP0602; Var2, Variovorax sp. JS1663; Pol, Polaromonas sp. strain OV174; Cab, Caballeronia arvi LMG 29317; Bce, Burkholderia cepacia JBK9; Pfu, Paraburkholderia fungorum NBRC 102489; Psp., Pandoraea sputorum DSM 21091; Pfl, Pseudomonas fluorescens C8; Hoe, Hoeflea sp. IMCC20628; Lve, Loktanella vestfoldensis SMR4r; Oar, Octadecabacter arcticus 238; Pse, Paracoccus seriniphilus DSM 14827; Abr, Afipia broomeae GAS525; Bra, Bradyrhizobium canariense GAS369; Bsp., Bradyrhizobium sp. strain C9. The detailed genomic accession numbers and the gene locus tags are listed in Table S1 in the supplemental material. Identities (percent) of amino acid sequences between Pic proteins of strain A. faecalis JQ135 and representative homologs are listed in Table S2.
The genetic organization of the pic gene clusters in these bacteria was highly diverse (Fig. 6). The picA genes usually adjoin picB genes, except in Cupriavidus strains. In most genera, picG was located between picC and picE. However, in Alcaligenes, Pusillimonas, and some Achromobacter strains, picG was located at a site distant from the pic gene cluster. In βeta- and Gammaproteobacteria, PicB consists of four components. Interestingly, in Alphaproteobacteria, including the Bradyrhizobium, Hoeflea, and Octadecabacter genera, PicB consists of two components in which the large subunit PicB123 appears to be a fusion of PicB1, PicB2, and PicB3 (Fig. S10).
Conclusions.
PA is generated through cell metabolism or synthesized artificially in industry and is thus ubiquitous in the environment. The degradation and detoxification of PA by microorganisms have been studied for more than 50 years (29). This study revealed that the pic gene cluster is responsible for the complete degradation of PA. PicA initially 6-hydroxylates PA to 6HPA, and then PicB 3-hydroxylates 6HPA to 3,6DHPA, which is then decarboxylated to 2,5DHP by PicC. 2,5DHP is further converted to fumaric acid via the maleamate pathway by the catalysis of PicD, PicE, PicF, and PicG. The genetic functions of picA and picB genes are reported here for the first time. In addition, the findings of homologous pic gene clusters in other Alpha-, Beta-, and Gammaproteobacteria help us understand the growth, competition, and environmental adaptability of bacteria in nature.
MATERIALS AND METHODS
Chemicals.
PA, 6HPA, and 2,5DHP were purchased from J&K Scientific, Ltd. (Shanghai, China). 3,6DHPA was chemically synthesized (28), and other reagents were purchased from Sangon Biotech Co., Ltd. (Shanghai, China).
Strains, plasmids, and culture conditions.
All bacterial strains and plasmids used in this study are listed in Table 2. Mineral salts medium (MSM) and Luria-Bertani (LB) medium have been previously described (26). Alcaligenes faecalis JQ135 is a wild-type PA-degrading strain (27). Alcaligenes faecalis ATCC 8750 was obtained from the China General Microbiological Culture Collection Center. Cupriavidus pinatubonensis JMP134 was a gift from Luying Xun (Shandong University) (46). Pusillimonas sp. strain T2 is a nicotinic acid-degrading bacterium (47). P. putida KT2440 is a model Gram-negative organism that cannot degrade or convert PA (30). Sphingomonas wittichii DC-6 is a chloroacetanilide herbicide degrader that is unable to degrade PA or 2,5DHP (38). E. coli DH5α acted as the host for the construction of plasmids. E. coli BL21(DE3) was used for protein overexpression. E. coli strains were grown in LB medium at 37°C. Antibiotics were added as required at the following concentrations: chloramphenicol (Cm), 34 mg/liter; gentamicin (Gm), 50 mg/liter; kanamycin (Km), 50 mg/liter; and streptomycin (Str), 50 mg/liter.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or sourcea |
|---|---|---|
| Alcaligenes faecalis strains | ||
| ATCC 8750 | Strr; type strain | CGMCC 1.924 |
| JQ135 | Strr; PA-degrading bacterium, Gram negative, wild type | CCTCC M 2015812 |
| JQ135 ΔpicA1A2A3 | Strr; picA1A2A3 deletion mutant of JQ135 | This study |
| JQ135 ΔpicA1A2A3/pBBR-picA1A2A3 | Strr Gmr; JQ135 ΔpicA1A2A3 complementation with pBBR-picA1A2A3 | This study |
| JQ135 ΔpicB1B2B3B4 | Strr; picB1B2B3B4 deletion mutant of JQ135 | This study |
| JQ135 ΔpicB1B2B3B4/pBBR-picB1B2B3B4 | Strr Gmr; JQ135 ΔpicB1B2B3B4 complementation with pBBR-picB1B2B3B4 | This study |
| JQ135 ΔpicG | Strr; maiA(AFA_16520) deletion mutant of JQ135 | 26 |
| Pseudomonas putida strains | ||
| KT2440 | Cmr; metabolically versatile saprophytic soil strain, PA-nondegrading bacterium | ATCC 47054 |
| KT/pBBR-picA1A2A3 | Cmr Gmr; KT2440 containing plasmid pBBR-picA1A2A3 | This study |
| KT/pBBR-picA1A2 | Cmr Gmr; KT2440 containing plasmid pBBR-picA1A2 | This study |
| KT/pBBR-picA2A3 | Cmr Gmr; KT2440 containing plasmid pBBR-picA2A3 | This study |
| KT/pBBR-picB1B2B3B4 | Cmr Gmr; KT2440 containing plasmid pBBR-picB1B2B3B4 | This study |
| KT/pBBR-picB1B2B3 | Cmr Gmr; KT2440 containing plasmid pBBR-picB1B2B3 | This study |
| KT/pBBR-picB2B3B4 | Cmr Gmr; KT2440 containing plasmid pBBR-picB2B3B4 | This study |
| Sphingomonas wittichii DC-6 | Strr; chloroacetanilide herbicide degrader, PA-nondegrading bacterium | KACC 16600 |
| DC-6/pBBR-picEDFG | Strr Gmr; S. wittichii DC-6 containing pBBR-picEDFG | This study |
| DC-6/pBBR-picEDF | Strr Gmr; S. wittichii DC-6 containing pBBR-picEDF | This study |
| Pusillimonas sp. strain T2 | Nicotinic acid-degrading bacterium | CCTCC M 2014272 |
| Cupriavidus pinatubonensis JMP134 | Hydrogen sulfide-oxidizing bacterium | 46 |
| E. coli strains | ||
| DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | TaKaRa |
| BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm (DE3) | TaKaRa |
| SM10 λpir | Donor strain for biparental mating | Lab stock |
| Plasmids | ||
| pET29a(+) | Kmr; expression plasmid | Novagen |
| pJQ200SK | Gmr Mob+ orip15A lacZα+ sacB; suicide plasmid | Lab stock |
| pBBR1MCS-5 | Gmr; broad-host-range cloning plasmid | Lab stock |
| pJQ-ΔpicA1A2A3 | Gmr; picA1A2A3 gene deletion plasmid; the upstream region of picA1 gene and downstream region of picA3 gene fused into SacI/PstI-digested pJQ200SK | This study |
| pJQ-ΔpicB1B2B3B4 | Gmr; picB1B2B3B4 gene deletion plasmid; the upstream region of picB1 gene and downstream region of picB4 gene fused into SacI/PstI-digested pJQ200SK | This study |
| pBBR-picA1A2A3 | Gmr; the fragment containing the picA1A2A3 gene inserted into XhoI/HindIII-digested pBBR1MCS-5 | This study |
| pBBR-picA1A2 | Gmr; the fragment containing the picA1A2 gene inserted into XhoI/HindIII-digested pBBR1MCS-5 | This study |
| pBBR-picA2A3 | Gmr; the fragment containing the picA2A3 gene inserted into XhoI/HindIII-digested pBBR1MCS-5 | This study |
| pBBR-picB1B2B3B4 | Gmr; the fragment containing the picB1B2B3B4 gene inserted into XhoI/HindIII-digested pBBR1MCS-5 | This study |
| pBBR-picB1B2B3 | Gmr; the fragment containing the picB1B2B3 gene inserted into XhoI/HindIII-digested pBBR1MCS-5 | This study |
| pBBR-picB2B3B4 | Gmr; the fragment containing the picB2B3B4 gene inserted into XhoI/HindIII-digested pBBR1MCS-5 | This study |
| pET-PicD | Kmr; NdeI-XhoI fragment containing picD gene inserted into pET29a(+) | This study |
CGMCC, China General Microbiological Culture Collection; CCTCC, China Center for Type Culture Collection; KACC, Korean Agricultural Culture Collection.
General DNA techniques.
Routine isolation of genomic DNA, extraction of plasmids, restriction digestion, transformations, PCR, and electrophoresis were performed according to standard procedures described by Sambrook et al. (48). Primer synthesis and the sequencing of PCR products or plasmids were performed by Genscript Biotech (Nanjing, China). The primers used in this study are listed in Table 3.
TABLE 3.
Primers used in this study
| Primer | Sequence (5→3) | Description |
|---|---|---|
| kopicA1A2A3-UF | AGCTTGATATCGAATTCCTGCAGCTGATTTTGCCAAGATCTATC | To construct plasmid pJQ-ΔpicA1A2A3 |
| kopicA1A2A3-UR | TCTAGAACTAGTGGATCCTTCCATCGGCAACATGCACTGC | |
| kopicA1A2A3-DF | GGATCCACTAGTTCTAGACCGTCGTGGCCGAGTTCAATCC | |
| kopicA1A2A3-DR | AGGGAACAAAAGCTGGAGCTCGGCAGGCAACATGAACAGCAC | |
| kopicB1B2B3B4-UF | AGCTTGATATCGAATTCCTGCAGACTCGGAGCGCGACTGCTCAC | To construct plasmid pJQ-ΔpicB1B2B3B4 |
| kopicB1B2B3B4-UR | TCTAGAACTAGTGGATCCGTATAAGTCCGTGGTTTGCATC | |
| kopicB1B2B3B4-DF | GGATCCACTAGTTCTAGAGTTTCGGAAATTTTTATCCAGT | |
| kopicB1B2B3B4-DR | AGGGAACAAAAGCTGGAGCTCTCTGCGCGTAAGGCACCAGTTC | |
| picA1A2A3-F | CAGGAATTCGATATCAAGCTTTACGACAAGTCTCAGGAGCTGTGG | To construct plasmid pBBR-picA1A2A3 |
| picA1A2A3-R | GGTACCGGGCCCCCCCTCGAGGTACCATTGCACTTCCAGCCAGG | |
| picA1A2-F | CAGGAATTCGATATCAAGCTTTACGACAAGTCTCAGGAGCTGTGG | To construct plasmid pBBR-picA1A2 |
| picA1A2-R | GGTACCGGGCCCCCCCTCGAGGTCGCGGCTGGTGGCCAACATGC | |
| picA2A3-F | CAGGAATTCGATATCAAGCTTATCGCCCAATACGGAGTTTGG | To construct plasmid pBBR-picA2A3 |
| picA2A3-R | GGTACCGGGCCCCCCCTCGAGGTACCATTGCACTTCCAGCCAGG | |
| picB1B2B3B4-F | GGTACCGGGCCCCCCCTCGAGGTTGCGGCCGTTGCCGATTTCAG | To construct plasmid pBBR-picB1B2B3B4 |
| picB1B2B3B4-R | CAGGAATTCGATATCAAGCTTGGAATCAGAAGACCTTCTGACTC | |
| picB1B2B3-F | GGTACCGGGCCCCCCCTCGAGGTTGCGGCCGTTGCCGATTTCAG | To construct plasmid pBBR-picB1B2B3 |
| picB1B2B3-R | CAGGAATTCGATATCAAGCTTACGATCCAACGTGGCACAGTAC | |
| picB2B3B4-F | GGTACCGGGCCCCCCCTCGAGGTAGAAACTGGCCCTGCACAAACTGG | To construct plasmid pBBR-picB2B3B4 |
| picB2B3B4-R | CAGGAATTCGATATCAAGCTTGGAATCAGAAGACCTTCTGACTC | |
| picEDF-F | CAGGAATTCGATATCAAGCTTCATCGGCAGCAATATTCTGATCACC | To construct plasmid pBBR-picEDF |
| picEDF-R | GGTACCGGGCCCCCCCTCGAGGTCACATCGTTCTTCTCAAATAC | |
| picEDFG-F1 | CAGGAATTCGATATCAAGCTTCATCGGCAGCAATATTCTGATCACC | To construct plasmid pBBR-picEDFG |
| picEDFG-R1 | CCAAGCATCGTCGCACATTTCCCACATCGTTCTTCTCAAATAC | |
| picEDFG-F2 | GGAAATGTGCGACGATGCTTGG | |
| picEDFG-R2 | GGTACCGGGCCCCCCCTCGAGGTGGCAGGGCTGTGTACTGCTAAG | |
| expPicD-F | CTTTAAGAAGGAGATATACATATGAGCACATTTTTGTATGGC | To construct plasmid pET-PicD |
| expPicD-R | GTGGTGGTGGTGGTGGTGCTCGAGCACCAAGCGCTGGCCCAG | |
| RT-16S F | CGCGGTAATACGTAGGGTG | To amplify fragment 16S shown in Fig. 1 |
| RT-16S R | AACTTCACGCGTTAGCTGCG | |
| RT-1 F | TCGTCGGGGTTGATACGACG | To amplify fragment 1 shown in Fig. 1 |
| RT-1 R | GGCCGACATCCTGGGTCCAGTG | |
| RT-2 F | GTACGTCGTGCTGTCGCAGACT | To amplify fragment 2 shown in Fig. 1 |
| RT-2 R | ATAGAACGGCACATCCAGCTT | |
| RT-3 F | GGCAGTAACTTGGGGTTTTGTGG | To amplify fragment 3 shown in Fig. 1 |
| RT-3 R | AGAAGGCGCGCATATCCTCAG | |
| RT-4 F | AGATTGCCGCTCAGTCCATGG | To amplify fragment 4 shown in Fig. 1 |
| RT-4 R | TCACGAATCGCAGGGAATTC | |
| RT-5 F | GCAACGGCACGTGAGCAG | To amplify fragment 5 shown in Fig. 1 |
| RT-5 R | CAACCGCTGGTAACCGCAC | |
| RT-6 F | ATACCGGGAACACCAGATCGTTCG | To amplify fragment 6 in Fig. 1 |
| RT-6 R | ATTGAGGCCGAGGGTAATTTCATGC | |
| RT-7 F | GCATACTTGTCCTCGGACTTGG | To amplify fragment 7 in Fig. 1 |
| RT-7 R | ACCAGTGGTCTTACAGCCTGAAGG | |
| RT-8 F | ATCCTGCCCGACAATGGCG | To amplify fragment 8 in Fig. 1 |
| RT-8 R | CACTGTCATTGAAGCGAATG | |
| RT-9 F | AGGCCTGGCTAAAGTCAGCC | To amplify fragment 9 in Fig. 1 |
| RT-9 R | AAATTTCCGAAACCGATGC | |
| RT-10 F | CGCCGCCCGTGCTGACCGC | To amplify fragment 10 in Fig. 1 |
| RT-10 R | GCAGGCGGCCTTCATTGC | |
| RT-11 F | CCCACGCTCAGATTGAAGA | To amplify fragment 11 in Fig. 1 |
| RT-11 R | ACATCCACAGACTCAATAAAC | |
| RT-12 F | GAACGGCTTTACGACGTTTG | To amplify fragment 12 in Fig. 1 |
| RT-12 R | TCCAACGTGGCATTGCCCTTGAAGG | |
| RT-13 F | CCGCATGAGCGTGATGGC | To amplify fragment 13 in Fig. 1 |
| RT-13 R | TCGTGGTTCAGGCGCTTGAC |
Construction of recombinant plasmids and heterologous expression.
Genes from A. faecalis JQ135 were PCR amplified using the corresponding primers (Table 3). Amplified DNA fragments were cloned into digested plasmids using a ClonExpress MultiS One Step Cloning kit (Vazyme Biotech Co., Ltd., Nanjing, China).
The plasmid pBBR1MCS-5 (49) was used for heterologous expression in P. putida KT2440. PCR products and XhoI/HindIII-digested plasmids were ligated, and the resulting recombinant plasmids were transferred into P. putida KT2440 by biparental mating using SM10 λpir.
To test the components of the PA dehydrogenase, pBBR1MCS-5-based plasmid clones containing different gene combinations (picA1A2A3, picA1A2, and picA2A3) were transferred into P. putida KT2440 to yield the recombinants KT/pBBR-picA1A2A3, KT/pBBR-picA1A2, and KT/pBBR-picA2A3, respectively. Similarly, to test the components of the 6HPA monooxygenase, the strains KT/pBBR-picB1B2B3B4, KT/pBBR-picB1B2B3, and KT/pBBR-picB2B3B4 were constructed.
For overexpression of the picD gene in E. coli BL21(DE3), the complete open reading frame (ORF) without its corresponding stop codon was amplified and inserted into NdeI/XhoI-digested plasmid pET29a(+), resulting in the plasmid pET-PicD. The induction and purification of 6×His-tagged PicD from E. coli BL21(DE3) (containing pET-PicD) were performed as previously described (26). Purified 6×His-tagged PicD was then analyzed by 12.5% SDS-PAGE. Protein concentration was determined using the Bradford method (50).
Gene knockout and genetic complementation of A. faecalis JQ135.
Deletion mutants of the picA1A2A3 and picB1B2B3B4 genes in A. faecalis JQ135 were constructed using a two-step homologous recombination method with the suicide plasmid pJQ200SK (51). Using the deletion of picA1A2A3 as an example, two homologous recombination-directing sequences (500 to 1,000 bp) were amplified using primers kopicA1A2A3-UF/-UR and kopicA1A2A3-DF/-DR, respectively. The two PCR fragments were subsequently ligated into SacI/PstI-digested pJQ200SK yielding pJQ-ΔpicA1A2A3. The plasmid pJQ-ΔpicA1A2A3 was then introduced into A. faecalis JQ135 cells. Single-crossover mutants were screened on LB plates containing Str and Gm. Gentamicin-resistant colonies were then subjected to repeated cultivation in LB medium containing 10% sucrose and no gentamicin. Double-crossover mutants, which had lost their vector backbones and were sensitive to gentamicin, were selected on LB-Str plates. Deletion of the picA1A2A3 genes was confirmed by PCR. This procedure resulted in construction of the deletion mutant strain JQ135 ΔpicA1A2A3.
Knockout mutants were complemented with the corresponding gene(s). For example, the complete picA1A2A3 genes were amplified with primers picA1A2A3-F/-R and then ligated with XhoI/HindIII-digested pBBR1-MCS5, generating pBBR-picA1A2A3. The pBBR-picA1A2A3 vector was transferred into the mutant strain JQ135 ΔpicA1A2A3 to generate the complemented strain JQ135 ΔpicA1A2A3/pBBR-picA1A2A3.
Preparation of resting cells and biodegradation assays.
Resting cells were prepared as follows. Alcaligenes, Pseudomonas, and Sphingomonas strains and their derivates were grown in 250-ml Erlenmeyer flasks with 100 ml of LB medium at 30°C and 180 rpm. When late exponential phase was reached, the cells were harvested by centrifugation at 4°C and 6,000 rpm for 10 min. The cells were washed twice with MSM and finally resuspended in MSM. The optical density at 600 nm (OD600) was adjusted to 2.0. These cells were named resting cells.
For the growth experiment, resting cells (such as JQ135 ΔpicA1A2A3) were added to MSM with 1 mM substrate (PA, 6HPA, or 3,6DHPA) for preincubation. After 24 h, the precultured cells were recollected, washed with MSM, and resuspended in MSM at a final OD600 of 2.0. Precultured cells were used for inoculation of MSM with 1 mM substrate. The initial OD600 was set at 0.15 to 0.2.
For biotransformation experiments, resting cells (such as KT/pBBR-picA1A2A3) were added to MSM with 1 mM substrate (PA or 6HPA) and 1 mM citrate for preincubation. After 12 h, the precultured cells were recollected, washed with MSM, and resuspended in MSM. The initial OD600 was set as 2.0, and then 1 mM substrate was added.
The growth and biotransformation experiments were all conducted in triplicate in 50-ml Erlenmeyer flasks with 20 ml of MSM at 30°C and 180 rpm. Samples (0.5 ml per time point) were collected periodically from the cultures. The growth of cells was monitored by measuring the OD600. The concentrations of substrate or products were measured by HPLC.
RT-PCR.
A. faecalis JQ135 resting cells were inoculated into 50-ml flasks with 20 ml of MSM containing 1 mM PA or citrate and cultured at 30°C and 180 rpm. The initial OD600 of the cultures was adjusted to 0.5. After 8 h, 5 ml of the culture was harvested by centrifugation at 4°C and 12,000 rpm for 10 min. Total RNA was isolated using an RNA isolation kit (TaKaRa), and reverse transcription-PCR (RT-PCR) was carried out with a PrimeScript RT reagent kit (TaKaRa) according to the manufacturer’s protocol. The cDNA was then used as a template in the following PCR amplification reaction mixtures: 10 μl of 2×SYBRPremix Ex Taq (TaKaRa), 0.8 μl of primers (10 μM), 2 μl of cDNA, and 7.2 μl of H2O. The primers used for RT-PCR are listed in Table 3. All samples were run in triplicate.
Enzymatic assays.
PA dehydrogenase (PicA) assays were performed using cell extracts of KT/pBBR-picA1A2A3. Resting KT/pBBR-picA1A2A3 cells were added into MSM containing 1 mM citrate and 1 mM PA. Following incubation for 12 h, cells were harvested by centrifugation at 6,000 rpm for 10 min at 4°C. Cell pellets were then resuspended in 50 mM phosphate buffered saline (PBS) (pH 7.0) and disrupted by sonication in an ice water bath. Cell-free extracts were obtained by centrifugation of cell lysates at 16,000 × g for 30 min at 4°C. The supernatant was used for PA dehydrogenase assays, while PicA activity was analyzed spectrophotometrically at 25°C by the formation of 6HPA at 310 nm (ε = 4.45 cm−1 mM−1) using a UV2450 spectrophotometer (Shimadzu) in 1-cm-path-length quartz cuvettes (23). The 1-ml reaction mixture contained 50 mM PBS (pH 7.0), 1 to 10 μg of total protein from the cell extract, 1 mM PA, and 1 mM electron acceptor PMS, and the reaction was started by adding PA. One unit of activity was defined as the amount of enzyme that catalyzed the formation of 1 μmol of 6HPA in 1 min.
For the 6HPA monooxygenase (PicB) assays, cell extracts of KT/pBBR-picB1B2B3B4 were used. The preparation of cell extracts was the same as that for KT/pBBR-picA1A2A3. PicB activity was analyzed spectrophotometrically by observing the formation of 3,6DHPA at 360 nm (ε = 4.4 cm−1 mM−1) (28). The 1-ml reaction mixture contained 50 mM PBS (pH 7.0), 1 to 10 μg of total protein from cell extract, 1 mM 6HPA, and 1 mM electron donor (NADH, NADPH, FAD, or FMN), and the reaction was started by the addition of 6HPA. One unit of activity was defined as the amount of enzyme that catalyzed the formation of 1 μmol of 3,6DHPA in 1 min.
The 2,5DHP dioxygenase (PicD) assays were performed similarly to those previously performed for NicX from P. putida KT2440 (30) and VppE in Ochrobactrum sp. strain SJY1 (36). A 1-ml reaction mixture containing 50 mM PBS (pH 7.0), 0.2 mM 2,5DHP, 0.1 μg of purified PicD, and 1 mM Fe2+ was incubated at 25°C. Activity was assayed spectrophotometrically by measuring the reduction in absorbance at 320 nm (ε = 5.2 cm−1 mM−1), which corresponded to the disappearance of 2,5DHP (30, 36, 37). One unit of activity was defined as the amount of enzyme that catalyzed the consumption of 1 μmol of 2,5DHP in 1 min.
Analytical methods.
The determination of PA and 6HPA, 3,6DHPA, 2,5DHP, and maleic acid was performed by UV-visible spectroscopy, HPLC, and LC/TOF-MS analysis as described previously (28).
Data availability.
The pic gene cluster sequence and the complete genome sequence of Alcaligenes faecalis JQ135 have been deposited in the GenBank/DDBJ/EMBL database under the accession numbers KY264362 and CP021641, respectively.
Comparisons of the Pic proteins were performed against the nonredundant protein (nr) sequence database using BLASTP (protein-protein BLAST) on the NCBI website, employing an expect (E) value inclusion threshold of 10. Conserved protein domains were analyzed using the Conserved Domain Database (CDD [https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi]). The genome sequence accession numbers of other strains and the corresponding locus tags of the pic gene clusters are listed in Table S1 in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Luying Xun (Shandong University) for his kind provision of strain Cupriavidus pinatubonensis JMP134.
This work was supported by the National Science and Technology Major Project (2018ZX0800907B-002), the National Natural Science Foundation of China (grants 41630637, 31870092, 31770117, and 31600080), and the Key R&D Program Project in Jiangsu Province (BE2016374).
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00077-19.
REFERENCES
- 1.Heyes MP, Chen CY, Major EO, Saito K. 1997. Different kynurenine pathway enzymes limit quinolinic acid formation by various human cell types. Biochem J 326:351–356. doi: 10.1042/bj3260351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryleva EY, Brundin L. 2017. Kynurenine pathway metabolites and suicidality. Neuropharmacology 112:324–330. doi: 10.1016/j.neuropharm.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esquivel DG, Ramirez-Ortega D, Pineda B, Castro N, Rios C, de la Cruz VP. 2017. Kynurenine pathway metabolites and enzymes involved in redox reactions. Neuropharmacology 112:331–345. doi: 10.1016/j.neuropharm.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 4.Nishino SF, Spain JC. 1993. Degradation of nitrobenzene by a Pseudomonas pseudoalcaligenes. Appl Environ Microbiol 59:2520–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehler AH. 1956. Formation of picolinic and quinolinic acids following enzymatic oxidation of 3-hydroxyanthranilic acid. J Biol Chem 218:241–254. [PubMed] [Google Scholar]
- 6.Asano Y, Yamamoto Y, Yamada H. 1994. Catechol 2,3-dioxygenase-catalyzed synthesis of picolinic acids from catechols. Biosci Biotechnol Biochem 58:2054–2056. doi: 10.1271/bbb.58.2054. [DOI] [Google Scholar]
- 7.Chirino B, Strahsburger E, Agullo L, Gonzalez M, Seeger M. 2013. Genomic and functional analyses of the 2-aminophenol catabolic pathway and partial conversion of its substrate into picolinic acid in Burkholderia xenovorans LB400. PLoS One 8:e75746. doi: 10.1371/journal.pone.0075746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdullaev M, Klyuev M, Abdullaeva ZS, Kurbanov B, Idrisova A. 2008. Preparation of lidocaine, bipuvacaine, mepivacaine, trimecaine, and pyromecaine by reductive acylation on palladium catalysts. Pharm Chem J 42:357–359. doi: 10.1007/s11094-008-0126-6. [DOI] [Google Scholar]
- 9.McCall PJ, Agin GL. 1985. Desorption kinetics of picloram as affected by residence time in the soil. Environ Toxicol Chem 4:37–44. doi: 10.1002/etc.5620040106. [DOI] [Google Scholar]
- 10.Broadhurst CL, Domenico P. 2006. Clinical studies on chromium picolinate supplementation in diabetes mellitus—a review. Diabetes Technol Ther 8:677–687. doi: 10.1089/dia.2006.8.677. [DOI] [PubMed] [Google Scholar]
- 11.Martin J, Wang ZQ, Zhang XH, Wachtel D, Volaufova J, Matthews DE, Cefalu WT. 2006. Chromium picolinate supplementation attenuates body weight gain and increases insulin sensitivity in subjects with type 2 diabetes. Diabetes Care 29:1826–1832. doi: 10.2337/dc06-0254. [DOI] [PubMed] [Google Scholar]
- 12.Ogata S, Inoue K, Iwata K, Okumura K, Taguchi H. 2001. Apoptosis induced by picolinic acid-related compounds in HL-60 cells. Biosci Biotechnol Biochem 65:2337–2339. doi: 10.1271/bbb.65.2337. [DOI] [PubMed] [Google Scholar]
- 13.Cioczek-Czuczwar A, Czuczwar P, Turski WA, Parada-Turska J. 2017. Influence of picolinic acid on seizure susceptibility in mice. Pharmacol Rep 69:77–80. doi: 10.1016/j.pharep.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Prodinger J, Loacker LJ, Schmidt RL, Ratzinger F, Greiner G, Witzeneder N, Hoermann G, Jutz S, Pickl WF, Steinberger P. 2015. The tryptophan metabolite picolinic acid suppresses proliferation and metabolic activity of CD4+ T cells and inhibits c-Myc activation. J Leukoc Biol 99:583–594. doi: 10.1189/jlb.3A0315-135R. [DOI] [PubMed] [Google Scholar]
- 15.Nakata HM, Halvorson HO. 1960. Biochemical changes occurring during growth and sporulation of Bacillus cereus. J Bacteriol 80:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamer Ö, Tamer SA, İdil Ö, Avcı D, Vural H, Atalay Y. 2018. Antimicrobial activities, DNA interactions, spectroscopic (FT-IR and UV-Vis) characterizations, and DFT calculations for pyridine-2-carboxylic acid and its derivates. J Mol Struct 1152:399–408. doi: 10.1016/j.molstruc.2017.09.100. [DOI] [Google Scholar]
- 17.Suksrichavalit T, Prachayasittikul S, Nantasenamat C, Isarankura-Na-Ayudhya C, Prachayasittikul V. 2009. Copper complexes of pyridine derivatives with superoxide scavenging and antimicrobial activities. Eur J Med Chem 44:3259–3265. doi: 10.1016/j.ejmech.2009.03.033. [DOI] [PubMed] [Google Scholar]
- 18.Smythe GA, Poljak A, Bustamante S, Braga O, Maxwell A, Grant R, Sachdev P. 2003. ECNI GC-MS analysis of picolinic and quinolinic acids and their amides in human plasma, CSF, and brain tissue, p 705–712. In Allegri G, Cost CVL, Ragazzi E, Steinhart H, Varesio L (ed), Developments in tryptophan and serotonin metabolism. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 19.Kiener A, Glockler R, Heinzmann K. 1993. Preparation of 6-oxo-1,6-dihydropyridine-2-carboxylic acid by microbial hydroxylation of pyridine-2-carboxylic acid. J Chem Soc Perkin 1:1201–1202. doi: 10.1039/P19930001201. [DOI] [Google Scholar]
- 20.Siegmund I, Koenig K, Andreesen JR. 1990. Molybdenum involvement in aerobic degradation of picolinic acid by Arthrobacter picolinophilus. FEMS Microbiol Lett 67:281–284. doi: 10.1016/0378-1097(90)90009-F. [DOI] [Google Scholar]
- 21.Zheng C, Wang Q, Ning Y, Fan Y, Feng S, He C, Zhang TC, Shen Z. 2017. Isolation of a 2-picolinic acid-assimilating bacterium and its proposed degradation pathway. Bioresource Technol 245:681–688. doi: 10.1016/j.biortech.2017.09.031. [DOI] [PubMed] [Google Scholar]
- 22.Zheng C, Zhou J, Wang J, Qu B, Wang J, Lu H, Zhao H. 2009. Aerobic degradation of 2-picolinic acid by a nitrobenzene-assimilating strain: Streptomyces sp. Z2. Bioresource Technol 100:2082–2084. doi: 10.1016/j.biortech.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 23.Orpin CG, Knight M, Evans WC. 1972. The bacterial oxidation of picolinamide, a photolytic product of Diquat. Biochem J 127:819–831. doi: 10.1042/bj1270819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petkevičius V, Vaitekūnas J, Stankevičiūtė J, Gasparavičiūtė R, Meškys R. 2018. Catabolism of 2-hydroxypyridine by Burkholderia sp. strain MAK1: a 2-hydroxypyridine 5-monooxygenase encoded by hpdABCDE catalyzes the first step of biodegradation. Appl Environ Microbiol 84:e00387-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaiser JP, Feng YC, Bollag JM. 1996. Microbial metabolism of pyridine, quinoline, acridine, and their derivatives under aerobic and anaerobic conditions. Microbiol Rev 60:483–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu J, Liu B, Zhao L, Zhang Y, Cheng D, Yan X, Jiang J, Hong Q, He J. 2018. A novel degradation mechanism for pyridine derivatives in Alcaligenes faecalis JQ135. Appl Environ Microbiol 84:e00910-18. doi: 10.1128/AEM.00910-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu J, Zhang J, Zhang Y, Wang Y, Tong L, Hong Q, He J. 2017. Biodegradation of picolinic acid by a newly isolated bacterium Alcaligenes faecalis strain JQ135. Curr Microbiol 74:508–514. doi: 10.1007/s00284-017-1205-2. [DOI] [PubMed] [Google Scholar]
- 28.Qiu J, Zhang Y, Yao S, Ren H, Qian M, Hong Q, Lu Z, He J. 2019. Novel 3, 6-dihydroxypicolinic acid decarboxylase mediated picolinic acid catabolism in Alcaligenes faecalis JQ135. J Bacteriol 201:e00665-18. doi: 10.1128/JB.00665-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dagley S, Johnson PA. 1963. Microbial oxidation of kynurenic, xanthurenic and picolinic acids. Biochim Biophys Acta 78:577–587. doi: 10.1016/0006-3002(63)91023-0. [DOI] [PubMed] [Google Scholar]
- 30.Jiménez JI, Canales A, Jiménez-Barbero J, Ginalski K, Rychlewski L, García JL, Díaz E. 2008. Deciphering the genetic determinants for aerobic nicotinic acid degradation: the nic cluster from Pseudomonas putida KT2440. Proc Natl Acad Sci U S A 105:11329–11334. doi: 10.1073/pnas.0802273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grether-Beck S, Igloi GL, Pust S, Schilz E, Decker K, Brandsch R. 1994. Structural analysis and molybdenum-dependent expression of the pAO1-encoded nicotine dehydrogenase genes of Arthrobacter nicotinovorans. Mol Microbiol 13:929–936. doi: 10.1111/j.1365-2958.1994.tb00484.x. [DOI] [PubMed] [Google Scholar]
- 32.Chakraborty J, Ghosal D, Dutta A, Dutta TK. 2012. An insight into the origin and functional evolution of bacterial aromatic ring-hydroxylating oxygenases. J Biomol Struct Dyn 30:419–436. doi: 10.1080/07391102.2012.682208. [DOI] [PubMed] [Google Scholar]
- 33.Vaitekūnas J, Gasparavičiūtė R, Rutkienė R, Tauraitė D, Meškys R. 2016. A 2-hydroxypyridine catabolism pathway in Rhodococcus rhodochrous strain PY11. Appl Environ Microbiol 82:1264. doi: 10.1128/AEM.02975-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li A, Qu Y, Zhou J, Ma F. 2009. Enzyme-substrate interaction and characterization of a 2,3-dihydroxybiphenyl 1, 2-dioxygenase from Dyella ginsengisoli LA-4. FEMS Microbiol Lett 292:231–239. doi: 10.1111/j.1574-6968.2009.01487.x. [DOI] [PubMed] [Google Scholar]
- 35.Tate R, Ensign J. 1974. Picolinic acid hydroxylase of Arthrobacter picolinophilus. Can J Microbiol 20:695–702. doi: 10.1139/m74-106. [DOI] [PubMed] [Google Scholar]
- 36.Yu H, Tang H, Zhu X, Li Y, Xu P. 2015. Molecular mechanism of nicotine degradation by a newly isolated strain, Ochrobactrum sp. strain SJY1. Appl Environ Microbiol 81:272–281. doi: 10.1128/AEM.02265-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang H, Yao Y, Wang L, Yu H, Ren Y, Wu G, Xu P. 2012. Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation. Sci Rep 2:377. doi: 10.1038/srep00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Q, Wang C-H, Deng S-K, Wu Y-D, Li Y, Yao L, Jiang J-D, Yan X, He J, Li S-P. 2014. Novel three-component Rieske non-heme iron oxygenase system catalyzing the N-dealkylation of chloroacetanilide herbicides in sphingomonads DC-6 and DC-2. Appl Environ Microbiol 80:5078–5085. doi: 10.1128/AEM.00659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hatakeyama K, Asai Y, Uchida Y, Kobayashi M, Terasawa M, Yukawa H. 1997. Gene cloning and characterization of maleate cis-trans isomerase from Alcaligenes faecalis. Biochem Biophys Res Commun 239:74–79. doi: 10.1006/bbrc.1997.7430. [DOI] [PubMed] [Google Scholar]
- 40.Strnad H, Ridl J, Paces J, Kolar M, Vlcek C, Paces V. 2011. Complete genome sequence of the haloaromatic acid-degrading bacterium Achromobacter xylosoxidans A8. J Bacteriol 193:791–792. doi: 10.1128/JB.01299-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahan KM, Zheng H, Fida TT, Parry RJ, Graham DE, Spain JC. 2017. Iron-dependent enzyme catalyzes the initial step in biodegradation of N-nitroglycine by Variovorax sp. strain JS1663. Appl Environ Microbiol 83:e00457-17. doi: 10.1128/AEM.00457-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Holden MTG, Churcher CM, Bentley SD, Mungall KL, Cerdeño-Tárraga AM, Temple L, James K, Harris B, Quail MA, Achtman M, Atkin R, Baker S, Basham D, Bason N, Cherevach I, Chillingworth T, Collins M, Cronin A, Davis P, Doggett J, Feltwell T, Goble A, Hamlin N, Hauser H, Holroyd S, Jagels K, Leather S, Moule S, Norberczak H, O'Neil S, Ormond D, Price C, Rabbinowitsch E, Rutter S, Sanders M, Saunders D, Seeger K, Sharp S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Unwin L, Whitehead S, Barrell BG, Maskell DJ. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet 35:32. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- 43.Ju S, Lin J, Zheng J, Wang S, Zhou H, Sun M. 2016. Alcaligenes faecalis ZD02, a novel nematicidal bacterium with an extracellular serine protease virulence factor. Appl Environ Microbiol 82:2112–2120. doi: 10.1128/AEM.03444-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vollmers J, Voget S, Dietrich S, Gollnow K, Smits M, Meyer K, Brinkhoff T, Simon M, Daniel R. 2013. Poles apart: Arctic and Antarctic Octadecabacter strains share high genome plasticity and a new type of xanthorhodopsin. PLoS One 8:e63422. doi: 10.1371/journal.pone.0063422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cubillas C, Miranda-Sánchez F, González-Sánchez A, Elizalde JP, Vinuesa P, Brom S, García-De L. 2017. A comprehensive phylogenetic analysis of copper transporting P1B ATPases from bacteria of the Rhizobiales order uncovers multiplicity, diversity and novel taxonomic subtypes. Microbiologyopen 6:e452. doi: 10.1002/mbo3.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Li J, Lü C, Xia Y, Xin Y, Liu H, Xun L, Liu H. 2017. FisR activates σ54-dependent transcription of sulfide‐oxidizing genes in Cupriavidus pinatubonensis JMP 134. Mol Microbiol 105:373–384. doi: 10.1111/mmi.13725. [DOI] [PubMed] [Google Scholar]
- 47.Yuan M, Zhang Y, Zhao L, Ma Y, He Q, He J, Qiu J. 2018. Identification and characterization of a new three-component nicotinic acid hydroxylase NahAB1B2 from Pusillimonas sp. strain T2. Lett Appl Microbiol 66:321–328. doi: 10.1111/lam.12850. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 49.Kovach M, Elzer P, Hill D, Robertson G, Farris M, Roop R, Peterson K. 1995. Four new derivatives of the broad-hostrange cloning vector pBBR1MCS, carrying different antibiotic resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 50.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 51.Quandt J, Hynes M. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The pic gene cluster sequence and the complete genome sequence of Alcaligenes faecalis JQ135 have been deposited in the GenBank/DDBJ/EMBL database under the accession numbers KY264362 and CP021641, respectively.
Comparisons of the Pic proteins were performed against the nonredundant protein (nr) sequence database using BLASTP (protein-protein BLAST) on the NCBI website, employing an expect (E) value inclusion threshold of 10. Conserved protein domains were analyzed using the Conserved Domain Database (CDD [https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi]). The genome sequence accession numbers of other strains and the corresponding locus tags of the pic gene clusters are listed in Table S1 in the supplemental material.