Abstract
Most idiosyncratic drug-induced liver injury appears to result from an adaptive immune attack on the liver. Recent evidence suggests that the T-cell response may be facilitated by the loss of immune tolerance. In this study, we explored the hypothesis that constitutively released hepatocyte-derived exosomes (HDE) are important for maintaining normal liver immune tolerance. Exosomes were isolated from the conditioned medium of primary human hepatocytes via polymer precipitation. Mock controls were prepared by processing fresh medium that was not hepatocyte exposed with precipitation reagent. THP-1 monocytes were then treated with HDE or an equivalent volume of mock control for 24 h, followed by a 6-h stimulation with LPS. HDE exposure resulted in a significant decrease in the LPS-induced media levels of interleukin-1β and interleukin-8. Gene expression profiling performed in THP-1 cells just prior to LPS-induced stimulation identified a significant decrease among genes associated with innate immune response. MicroRNA (miRNA) profiling was performed on the HDE to identify exosome contents that may drive immune suppression. Many of the predicted mRNA target genes for the most abundant microRNAs in HDE were among the differentially expressed genes in THP-1 cells. Taken together, our data suggest that HDE play a role in maintaining normal liver immune tolerance. Future experiments will explore the possibility that drugs causing idiosyncratic liver injury promote the loss of homeostatic HDE signaling.
Keywords: exosomes, idiosyncratic drug-induced liver injury, immune response, hepatocytes, monocytes
It is now generally believed that most idiosyncratic drug-induced liver injury (IDILI) with prolonged latency results from an adaptive immune attack on the liver (Usui and Naisbitt, 2017). This is supported, in part, by the identification of strong genetic associations between specific human leukocyte antigen (HLA) alleles and the risk of liver injury for an increasing number of IDILI drugs (Urban et al., 2014). The autoimmune response in IDILI is directed at neoantigens (eg, drug-modified self-proteins and altered self-peptide repertoires) presented on HLA molecules in response to drug exposure. However, significant evidence suggests that the cascade of T-cell mediated events culminating in IDILI begins with drug-induced hepatocyte stress, the release of danger signals, and activation of the innate immune system—sequential steps necessary to promote an adaptive immune attack (Mosedale and Watkins, 2017). Recent work from our group has demonstrated that hepatocyte-derived exosomes (HDE) are altered in response to hepatotoxic drugs prior to or in the absence of overt injury and therefore may play a role in these early IDILI events (Holman et al., 2016; Mosedale et al., 2018).
By virtue of their small size (<150 nm), HDE can readily cross through fenestrations in the sinusoidal endothelium and enter the bloodstream carrying hepatocyte-specific contents (Conde-Vancells et al., 2008; Wetmore et al., 2010). HDE can also stimulate a variety of liver nonparenchymal cell (NPC) types with immune signaling capacity, including monocytes and their derivatives (Dreux et al., 2012; Momen-Heravi et al., 2015), lymphocytes (Cobb et al., 2018; Seo et al., 2016), stellate cells (Chen et al., 2015; Seo et al., 2016), and liver sinusoidal endothelial cells (Cobb et al., 2018; Giugliano et al., 2015). The potential for HDE to mediate immune signaling in IDILI is supported by several studies demonstrating alterations in the immune phenotype of NPCs exposed to conditioned medium from hepatocytes treated with subtoxic concentrations of IDILI drugs (Kato and Uetrecht, 2017; Kegel et al., 2015; Oda et al., 2016; Ogese et al., 2017). It has been proposed that IDILI-induced changes in HDE number and/or content are responsible for promoting innate immune activation, a critical early event for a T-cell response. However, recent evidence suggests that the T-cell response in IDILI may in fact be facilitated by the loss of immune tolerance.
Perhaps the strongest evidence demonstrating a role for the loss of immune tolerance in IDILI comes from the development of a mouse model that recapitulates some of the clinical characteristics of idiosyncratic reactions, including a delay in onset and immune infiltrate in the liver (Mak et al., 2018; Mak and Uetrecht, 2015; Metushi et al., 2015). This phenotype is achieved by inhibiting functions of programmed cell death 1 (PD-1) and cytotoxic T-lymphocyte-associated protein 4 (CTLA4), negative regulators of T-cell activation that are important for the induction of immune tolerance. Cancer drugs that target CTLA4 and PD-1, as well as other molecules involved in immune tolerance, have also been associated with IDILI and other immune-mediated adverse reactions in the clinic (Suzman et al., 2018). Finally, drugs associated with IDILI typically cause asymptomatic elevations in serum aminotransferases that resolve despite continuation of treatment with the offending drug (Mosedale and Watkins, 2017). In some cases, these transient liver injuries have been associated with the same HLA risk alleles as the clinically important liver injuries (Singer et al., 2010). This suggests that the initiation of a drug-induced adaptive immune attack on the liver is usually reversible, presumably through immune tolerance mechanisms.
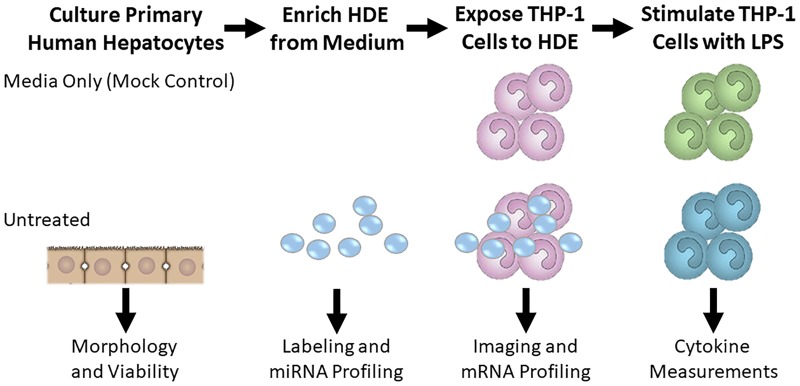
IDILI-induced changes in HDE number and/or content may therefore play a more important role in the loss of immune tolerance than in immune activation. To our knowledge, the impact of constitutively released HDE on immune cells has not been previously explored. In the present study, we tested the hypothesis that HDE may be essential for maintaining immune homeostasis by treating a human monocyte cell line with exosomes collected from the culture medium of untreated primary human hepatocytes and evaluating the cytokine response after LPS stimulation (Figure 1). We also profiled the microRNA (miRNA) content of the HDE and compared it with gene expression changes observed in the HDE-exposed monocytes prior to LPS stimulation. Findings from our research suggest that HDE may be essential for maintaining immune homeostasis and support the assertion that IDILI drugs promote loss of homeostatic HDE signaling.
Figure 1.
Hepatocyte-derived exosomes (HDE, small circles) were collected from the conditioned medium of primary human hepatocytes or fresh medium that was not hepatocyte exposed (mock control). These exosomes were used for miRNA profiling, fluorescent labeling, and/or treatment of THP-1 monocytes. THP-1 monocytes were then treated with HDE or an equivalent volume of mock control for 24 h. THP-1 monocytes were subjected to fluorescent microscopy, global mRNA profiling, or an additional 6 h stimulation with LPS. Medium from LPS-stimulated THP-1 cells was then collected for cytokine analysis.
MATERIALS AND METHODS
Hepatocyte culture
Primary human hepatocytes from N = 5 donors (Supplementary Table 1) were obtained from commercial sources (QPS Hepatic Biosciences and Triangle Research Labs). Freshly isolated hepatocytes were used when available. Individual batches of hepatocytes were seeded into 10 cm dishes coated with collagen type I (Corning) at a density of 10 × 106 cells per dish in hepatocyte plating medium (William’s E medium, penicillin-streptomycin, GlutaMax, HEPES, sodium pyruvate, fetal bovine serum [Thermo Fisher Scientific], insulin, and dexamethasone [Sigma-Aldrich]). Culture plates were incubated at 37°C with 5% CO2. After attachment, nonadherent cells were removed, and fresh plating medium was added. Cultures were then transitioned to hepatocyte maintenance medium without fetal bovine serum within 6–12 h of plating. Following a 24-h acclimation period, hepatocyte cultures were exposed to fresh medium for an additional 24 h to allow for exosome accumulation.
Keratinocyte culture
Primary human keratinocytes (PCS-200-011, ATCC) were subcultured and then seeded onto 10 cm dishes at a density of 0.4 × 106 cells per dish in complete growth medium (Dermal Cell Basal Medium, PCS-200-030, ATCC; Keratinocyte Growth Kit, PCS-200-040, ATCC). At approximately 80% confluence, cells were given fresh medium for an additional 24 h to allow for exosome accumulation.
Exosome enrichment
Donor-specific preparations of HDE and keratinocyte exosomes were obtained via modification of an established protocol (Holman et al., 2016). Briefly, conditioned medium was collected after the 24 h exposure and centrifuged at 3000 × g for 15 min to remove cellular debris. Supernatants were filtered at 0.45 µm and subsequently concentrated using Centricon-70 centrifugal filter units with a 10 000-Da cut-off (Millipore) per manufacturer’s instructions. The resultant concentrates were subjected to a second filtration (0.22 µm) and processed under sterile conditions for the remainder of the preparation. Exosomes were precipitated from these concentrates with ExoQuick-TC (EQ; System Biosciences) following the protocol for tissue culture supernatants.
Preparation of mock controls
To control for effects of the EQ solution and possible residual cell culture medium that may have carried over during exosome isolation, portions of unconditioned media were reserved from each primary human hepatocyte and the keratinocyte experiment. A volume of unconditioned medium equal to that of conditioned (hepatocyte- or keratinocyte-exposed) medium from each treatment was processed in parallel for mock “exosome enrichment” as described above. Following the centrifugation step in our precipitation protocol, the majority of supernatant was removed and a volume equivalent to the volume of the corresponding true exosome pellet was saved as the mock control. This solution was diluted in monocyte culture medium exactly as the matching exosomes were diluted for monocyte treatment.
Monocyte exosome exposures
THP-1 human monocytes (TIB-202, ATCC) and primary human peripheral blood monocytes from a single healthy donor (HPBMs, 6906-50A, Sigma) were cultured in suspension in RPMI-1640 (ATCC) supplemented with 10% (v/v) exosome-depleted FBS (Gibco), penicillin-streptomycin (Gibco), and 0.05 mM β-mercaptoethanol (THP-1 only, Sigma). Exposures were conducted in 96-well round-bottom plates (Corning) at a density of 1 × 105 cells per well. Monocytes were exposed to 200 µg/ml exosomes or the matched volume of mock control in separate experiments for each hepatocyte donor and for the keratinocytes. In each well, monocytes were seeded at the desired density in 100 µl of medium and a 2× solution of exosomes in 100 µl medium was added for a total of 200 µl final volume. For the keratinocyte experiment, these volumes were halved due to the concentration of keratinocyte-exosomes obtained. Exosome protein concentrations were determined using a BCA Protein Assay Kit (Thermo). Protein concentrations were also measured in mock controls which were nominal for HDE but similar to exosome preparations for keratinocytes. Normalizing exosome doses for in vitro exposure based on total protein concentration is a common practice in mechanistic experiments (Chiba et al., 2012; Giugliano et al., 2015; Li et al., 2013). The final concentration of 200 µg/ml exosome protein was modeled after total exosomal protein concentrations in human plasma (Momen-Heravi et al., 2015; Muller et al., 2014). In a dose-response experiment conducted using Donor 1 HDE, this concentration of HDE also showed a greater effect on THP-1 cells compared with 100 µg/ml (Supplementary Figure 1). Following 24 h of co-culture, a portion of the THP-1 monocytes were lysed in QIAzol (Qiagen) for gene expression profiling as described below. LPS (Escherichia coli 0111: B4; Sigma) at 5 ng/ml was spiked into the remaining monocytes and medium was harvested 6 h later for the quantification of pro-inflammatory cytokine release.
Cytokine and chemokine analysis
Release of the chemokine monocyte chemoattractant-1 (MCP-1) was quantified using an MSD Single Spot assay according to the manufacturer’s instructions. Pro-inflammatory cytokines interleukin-1β (IL-1β), interleukin-8 (IL-8), and tumor necrosis factor alpha (TNF-α) were measured in clarified culture supernatant using a multiplex MSD V-plex assay (Meso Scale Discovery) according to the manufacturer’s protocol for cell culture supernatant samples. MSD plates were read on a Meso QuickPlex SQ 120 instrument and analyzed using Discovery Workbench 4.0 software (Meso-Scale Discovery). For each analyte from donor-pooled and single-donor analyses, values from mock- and exosome-treated monocytes were divided by the mean of the mock-treated monocytes and compared using a t test. A Sidak’s multiple comparison test was used to compare exosome-treated and mock-treated analyte values for individual donors in multidonor comparisons.
Monocyte gene expression profiling
Total RNA from THP-1 monocytes was isolated with the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. RNA integrity was assessed using the Agilent 2200 Tape Station System (Agilent Technologies). Optimal purity and minimum degradation for subsequent microarray analysis were defined by A260/280 > 1.8, A230/280 > 1.8, and RNA integrity number (RIN) > 8.0. Whole genome microarray profiling was performed by the UNC Functional Genomic Core facility. For microarrays, sense-strand cDNA (ss-cDNA) was synthesized from 100 ng of total RNA using the GeneChip WT PLUS Reagent Kit (Affymetrix). Following purification, 5.5 µg of ss-cDNA were fragmented and end labeled with biotin before hybridizing to the array plate. The Affymetrix Clariom S HT Human 24-peg array was used with the Affymetrix Gene Titan Multi-Channel system. GeneTitan robotic instrumentation was used to perform the hybridization, washing, and scanning of the peg array using the Affymetrix GeneTitan Hybridization, Wash, and Stain Kit for WT Arrays.
Data summarization and QC were performed using the Transcriptome Analysis Console (TAC) software version 4.0 (ThermoFisher). Affymetrix CEL files were normalized using the Robust Multi-array Average method with a log base 2 (log2) transformation (Irizarry et al., 2003). All subsequent analyses were performed using Partek Genomics Suite version 6.6. Principal component analysis (PCA) was used to evaluate the overall performance of the arrays and identify outliers. Samples >3 SD away from the mean were omitted from analysis. This procedure resulted in removal of one sample. Data summarization, QC, normalization, and PCA were then repeated with this sample removed. A filtering step was performed on the final data set to remove low expression probe sets. Differential expression was determined using an ANOVA model with linear contrasts. Hepatocyte donor was identified as a significant factor contributing to the variability in probe intensity levels and was therefore used to normalize analyses where appropriate. An unadjusted p-value of .05 and absolute value fold change (FC) cut-off of >2.25 was used as noted below. Pathways enriched and predicted miRNA regulators among statistically significant, differentially expressed genes in the data were identified using the Tox Analysis module in Ingenuity Pathway Analysis (Ingenuity Systems; Build version 486068M; Content version: 46901286). Gene expression data generated for this manuscript can be downloaded in its entirety from the Gene Expression Omnibus repository under the accession number GSE125690. All data are MIAME compliant.
Exosome microRNA profiling
Targeted miRNA profiling using next-generation sequencing techniques was carried out using the HTG EdgeSeq miRNA Whole Transcriptome assay (HTG Molecular). Precipitated, intact HDE were washed 1× with PBS and centrifuged at 10 000 × g for 60 min. The supernatants were removed and 30 µl of HTG Lysis Buffer was added to each HDE sample. Lysates (25 µl) were loaded onto an EdgeSeq plate and processed according to the manufacturer’s instructions for cells. Briefly, libraries were prepared in two steps: nuclease protection on an HTG Edge processor followed by PCR labeling and amplification. Library concentration and quality were determined using a High Sensitivity DNA Assay Kit on an Agilent 2100 Bioanalyzer. Libraries were pooled and sequenced in a single lane on an Ilumina MiSeq system for 50 bp single-end reads. Data were parsed using HTG EdgeSeq software; one sample did not pass quality control metrics and was removed from further analysis. Median-normalized counts for all 2083 miRNAs were used for data analysis (Anders and Huber, 2010), and signal intensities averaged across samples from N = 4 donors passing quality control were used to identify the most abundant miRNAs. Disease categories and predicted mRNA targets among the most abundant miRNA in the data were identified using the Tox Analysis module in Ingenuity Pathway Analysis Ingenuity Systems; Build version 486068M; Content version: 46901286).
Exosome uptake imaging
HDE for uptake imaging were stained using a PKH67 dye kit (Sigma). Exosomes were diluted to 200 µg/ml (the same concentration used for activity analysis) with diluent C and were combined with an equal volume of 1:250 dye solution and incubated according to the manufacturer’s protocol. Following incubation, 3 ml of complete exosome-free medium was added to terminate the labeling reaction. Labeled HDE were harvested and washed once with 1× PBS by centrifugation (100 000 × g for 1 h at 4°C). HDE were suspended in warm monocyte culture medium and were incubated with THP-1 cells for 8 h at 37°C. Monocytes were then harvested and labeled for visualization of exosome uptake. Monocytes were incubated with Alexa Fluor 647 antihuman CD32 antibody (BioLegend) to define cell boundaries and permeabilized with Cytofix/Cytoperm Fixation and Permeabilization Kit (BD). DAPI nucleic acid stain (BD Pharmingen) was added to delineate nuclei post-fixation. Image acquisition was performed on an Amnis ImageStreamX Mark II Imaging Flow Cytometer with INSPIRE software at 60× magnification. Focused, single, and DAPI-positive (+) cells were first gated and an internalization score was generated to quantify uptake of PKH67+ HDE by CD32+ cells with the IDEAS software package (v6.2) as described previously (Ackerman et al., 2011). Briefly, a mask representing the cell membrane was defined by the brightfield image, and an internal mask was defined by eroding the whole cell mask by 6 pixels, resulting in an area significantly smaller than the cell membrane. An internalization score was calculated as follows: (number of fluorescent pixels in internal mask) – (number of fluorescent pixels in the entire field of view). Scores ≥1 were interpreted as internalization (Ackerman et al., 2011).
RESULTS
Exosomes From Primary Human Hepatocytes Are Internalized by Monocytes
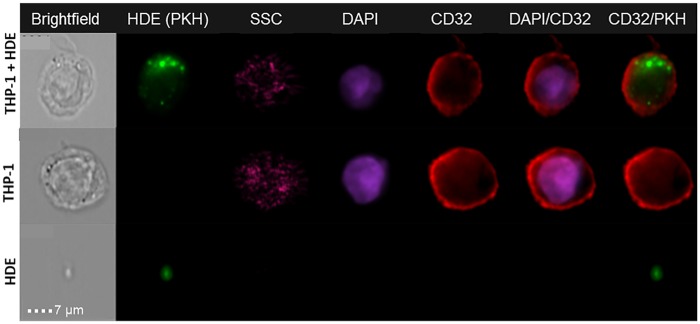
HDE released over 24-h culture period were enriched from the conditioned medium of primary human hepatocytes. A confluent monolayer of viable hepatocytes was observed in all plates from each of N = 5 hepatocyte donors (data not shown). To determine if HDE would be internalized by THP-1 monocytes and to examine HDE subcellular localization, we performed uptake imaging using PKH dye and an ImageStream X flow cytometry imager, an approach that has been validated for resolution of exosome internalization (Franzen et al., 2014; Headland et al., 2014). THP-1 monocytes were incubated with PKH-labeled HDE, washed, and stained for visualization. Uptake of HDE was visually confirmed (Figure 2) and quantified via internalization scores generated from approximately 10 000 monocytes from each treatment. Mean internalization scores were 2.1 ± 0.9 for HDE.
Figure 2.
Representative ImageStream acquisitions of PKH-stained HDE and internalization by CD32+ monocytes. Individual preparations of monocytes and HDE are also shown. SSC, side scatter; DAPI, nuclear counterstain.
Exosomes Dampen Immune Response of Monocytes to Endotoxin
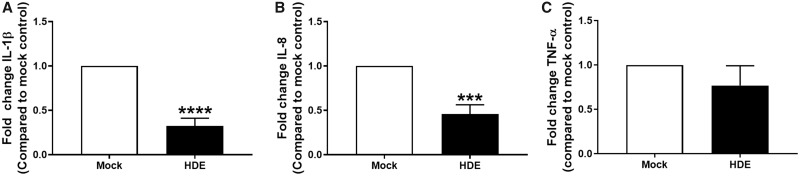
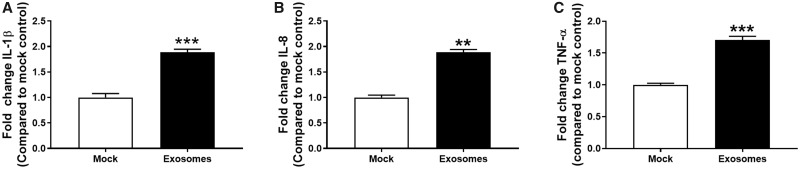
Next, we sought to assess whether the uptake of HDE by THP-1 cells would have a functional effect on monocyte immune response. HDE activity was analyzed on a per-donor basis such that HDE preparations from individual donors (N = 5) and donor-specific mock controls were tested in separate THP-1 experiments. THP-1 monocytes were exposed to HDE or mock controls for 24 h. Monocytes were then stimulated with LPS or vehicle for another 6 h. HDE induced a significant decrease in the LPS-induced media levels of IL-1β and IL-8 (Figs. 3A and 3B). A trend toward a decrease in TNF-α was also observed (Figure 3C). Donor-specific data are provided in Supplementary Figure 2.
Figure 3.
Fold change in protein levels of (A) IL-1β, (B) IL-8, and (C) TNF-α in supernatant collected from HDE- or mock-exposed THP-1 cells following a 6 h stimulation with LPS. Data represent the mean + SEM from N = 5 donors and significance is given as p < ***.001 or ****.0001.
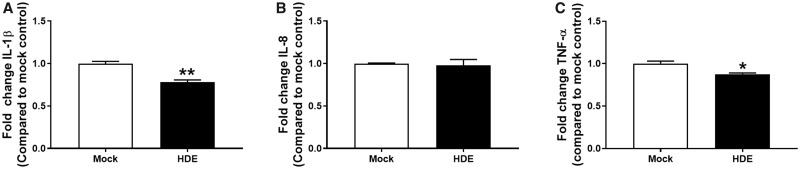
Because THP-1 cells are a model for human peripheral blood monocytes, we sought to confirm these responses in their primary HPBM counterparts. In a separate experiment, additional HDE were collected from Donor 1 and applied to primary HPBMs using the same experimental design. HDE induced a significant decrease in the LPS-induced media levels of IL-1β and TNF-α (Figs. 4A and 4C), but no effect on IL-8 (Figure 4B). To evaluate the specificity of this response to HDE, exosomes collected from primary human keratinocytes were also applied to THP-1 cells under the same experimental conditions. Keratinocyte-derived exosomes induced significant increases in the LPS-induced media levels of IL-1β, IL-8, and TNF-α (Figure 5).
Figure 4.
Fold change in protein levels of (A) IL-1β, (B) IL-8, (C) TNF-α in supernatant collected from HDE- or mock-exposed primary HPBM following a 6 h stimulation with LPS. Data represent the mean + SEM from N = 3 technical replicates and significance is given as p < *.05 or **.01.
Figure 5.
Fold change in protein levels of (A) IL-1β, (B) IL-8, and (C) TNF-α in supernatant collected from keratinocyte exosome- or mock-exposed THP-1 cells following a 6 h stimulation with LPS. Data represent the mean + SEM from N = 2–3 technical replicates and significance is given as p < **.01 or ***.001.
Exosomes Alter the Monocyte Transcriptome Prior to Endotoxin Stimulation
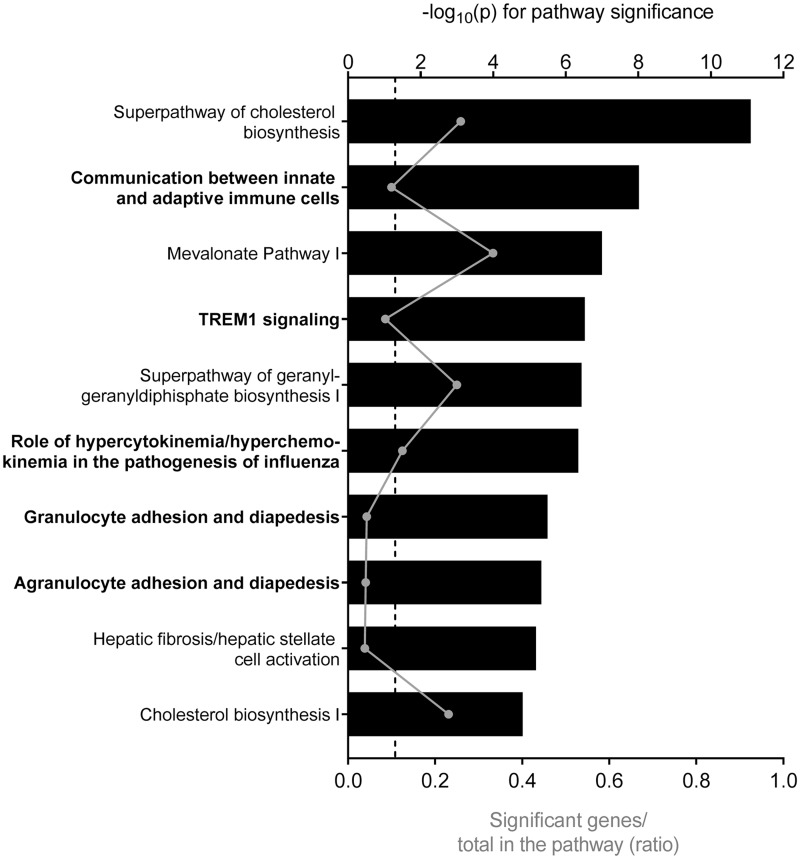
To identify HDE-induced changes that might underlie the altered immune response, gene expression profiling was performed on RNA isolated from HDE- and mock-exposed THP-1 monocytes prior to LPS stimulation. A total of 72 THP-1 mRNAs were significantly differentially expressed in the HDE-exposed THP-1 monocytes compared with mock controls (p < .05 and |FC|> 2.25). All 72 transcripts were decreased and reflect suppression of immune response as 7 of the top 10 most altered mRNAs (Table 1) and 5 of the 10 the most affected pathways are related to immune function (Figure 6). A targeted interrogation was also performed to examine the effect of HDE on the mRNA levels of IL-1β, IL-8, and TNF-α in the monocytes. All 3 cytokine mRNAs were significantly decreased in HDE-exposed THP-1 cells compared with mock controls (Supplementary Figure 3).
Table 1.
Top 10 Most Altered mRNA in Exosome-Exposed THP-1 Cells
| Symbol | Gene Name | Fold Change |
|---|---|---|
| CCL3 | C-C motif chemokine ligand 3 | −6.53 |
| KCNE1 | Potassium voltage-gated channel subfamily E regulatory subunit 1 | −5.25 |
| FLT1 | Fms-related tyrosine kinase 1 | −5.04 |
| CCL3L3 | C-C motif chemokine ligand 3 like 3 | −5.00 |
| CXCL8 a | C-X-C motif chemokine ligand 8 | −4.87 |
| C3AR1 | complement C3a receptor 1 | −4.76 |
| IFI44L | Interferon-induced protein 44 like | −4.53 |
| CCL4L1/CCL4L2 | C-C motif chemokine ligand 4 like 1 | −4.41 |
| IGFBP3 | Insulin-like growth factor binding protein 3 | −4.22 |
| MX1 | MX dynamin-like GTPase1 | −4.21 |
CXCL8 encodes IL-8. Bold text represents immune-related mRNA. Fold change represents gene alterations in THP-1 cells exposed to HDE compared with mock control (p < .05 and |FC|>2.25).
Figure 6.
Top 10 most significantly enriched pathways among genes differentially expressed in THP-1 cells exposed to HDE compared with mock control (p < .05 and |FC|> 2.25). Pathway significance is plotted on the upper x-axis and represented by the black bars on the graphs. The ratio of significant genes to total genes in the pathway is plotted on the lower x-axis and represented by the gray dots (connected by a gray line) on the graphs. The dashed line represents a threshold for significance set at −log10(p) > 1.3. Pathways in bold are related to immune function.
miRNA Species Enriched in Exosomes May Be Responsible for Monocyte Immune Suppression
Exosomes are well known for carrying regulatory miRNAs which can be transferred to and stimulate immune cells (Yu et al., 2016). To explore the hypothesis that the immunomodulatory activity of HDE is mediated in part by the delivery of functional miRNAs, we performed global miRNA profiling on the HDE from all 5 hepatocyte donors and investigated correlations between specific miRNAs enriched in HDE and the exosome-induced transcriptional changes observed in monocytes. Data generated from N = 1 donor failed to pass quality control and were removed from the analysis. Initially, we compared the mean miRNA levels quantified in this study to miRNA profiling data collected from a separate study where exosomes were enriched from N = 5 different primary human hepatocyte donors using ultracentrifugation, which has been recently shown to yield a purer population of HDE (Thacker et al., 2018). A strong correlation was observed between HDE miRNA levels in both studies (Supplementary Figure 4).
Next, we identified the top 50 most enriched miRNA in primary human HDE for further analysis (Supplementary Table 2). Three of the 10 most significantly enriched disease categories associated with the top 50 most abundant HDE miRNAs were immune related (Table 2). We then investigated the mRNA targets of the top 10 most abundant miRNAs in relation to the transcripts differentially expressed in exosome-exposed THP-1 cells. Many of the predicted mRNA target genes for the most abundant miRNAs in HDE were among the differentially expressed genes observed in the THP-1 cells (Table 3). We also compared predicted miRNA regulators of the top 10 most altered mRNAs in THP-1 cells to the top miRNAs in the HDE and a similar concordance was observed (Supplementary Table 3).
Table 2.
Most Significant Disease Categories Among the Top 50 Exosomal miRNAs
| Categories | p-Value | No. of miRNAs |
|---|---|---|
| Organismal injury and abnormalities, reproductive system disease | 5.05E-11 | 8 |
| Cancer, organismal injury, and abnormalities | 7.34E-11 | 6 |
| Cancer, gastrointestinal disease, hepatic system disease, organismal injury, and abnormalities | 1.31E-10 | 12 |
| Cancer, organismal injury and abnormalities, reproductive system disease | 1.47E-10 | 6 |
| Cancer, organismal injury, and abnormalities | 2.54E-08 | 8 |
| Endocrine system disorders, gastrointestinal disease, metabolic disease, organismal injury, and abnormalities | 3.88E-08 | 8 |
| Inflammatory response, organismal injury, and abnormalities | 4.89E-08 | 12 |
| Inflammatory disease, inflammatory response, organismal injury and abnormalities, renal, and urological disease | 8.38E-08 | 5 |
| Cancer, hematological disease, hereditary disorder, organismal injury, and abnormalities | 2E-07 | 4 |
| Cancer, hematological disease, immunological disease, organismal injury, and abnormalities | 2.6E-07 | 5 |
Disease categories in bold are immune-related.
Table 3.
Regulation of Significant mRNAs in Monocytes by Top 10 miRNAs in Exosomes
| miRNA | Predicted No. of Regulated Genes | Genes Predicted to Be Regulated |
|---|---|---|
| miR-6126 | 1 | DHCR7 |
| miR-6780b-5p | 3 | KCNE1, LSS, MSC |
| miR-3197 | 1 | NUPR1 |
| miR-22-3p | 4 | CCL4L1, INSIG1, KLF6, RGS2 |
| miR-122-5p | 3 | CD83, KCNJ2, SLC7A11 |
| miR-1273h-5p | 5 | CXCL10, RHOB, SLC12A8, SLCO4C1, TNFRSF9 |
| miR-149-3p | 12 | ACSS2, BTG2, CCL3, CSF1, CXCL10, IFI44L, IFIT3, IL32, MVK, RHOB, SLCO4C1, TNF |
| miR-192-5p | 6 | ATF3, CSF1, FABP3, FLT1, IFI44L, IL21R |
| miR-148a-3p | 4 | CSF1, FLT1, KLF6, LIPG |
| miR-6875-5p | 3 | CHAC1, CXCL10, SH2D3C |
Genes in bold function in an immune response.
Abbreviations: DHCR7, 7-dehydrocholesterol reductase; KCNE1, potassium voltage-gated channel subfamily E regulatory subunit 1; LSS, lanosterol synthase; MSC, musculin; NUPR1, nuclear protein 1; CCL4L1, C-C motif chemokine ligand 4 like 1; INSIG1, insulin-induced gene 1; KLF6, Kruppel-like factor 6; RGS2, regulator of G protein signaling 2; CD83, cluster of differentiation 83; KCNJ2, potassium voltage-gated channel subfamily J member 2; SLC7A11, solute carrier family 7 member 11; CXCL10, C-X-C motif chemokine ligand 10; RHOB, Ras homolog family member B; SLC12A8, solute carrier family 12 member 8; SLCO4C1, solute carrier organic anion transporter family member 4C1; TNFRSF9, tumor necrosis factor (TNF) receptor superfamily member 9; ACSS2, acyl-CoA synthetase short-chain family member 2; BTG2, BTG antiproliferative factor 2; CCL3, C-C motif chemokine ligand 3; CSF1, colony stimulating factor 1; IFI44L, interferon-induced protein 44 like; IFIT3, interferon-induced protein with tetratricopeptide repeats 3; IL32, interleukin (IL) 32; MVK, mevalonate kinase; ATF3, activating transcription factor 3; FABP3, fatty acid-binding protein 3; FLT1, fms-related tyrosine kinase 1; IL21R, IL-21 receptor; LIPG, lipase G, endothelial type; CHAC1, ChaC glutathione-specific gamma-glutamylcyclotransferase 1; SH2D3C, SH2 domain containing 3C.
DISCUSSION
The role of exosomes in immunologically relevant liver diseases, such as viral hepatitis, alcoholic liver injury, and NASH have been substantiated by a handful of studies over the last 5 years (Dreux et al., 2012; Hirsova et al., 2016; Kakazu et al., 2016; Momen-Heravi et al., 2015; Verma et al., 2016). However, the mechanistic importance of exosomes derived from healthy primary human hepatocytes in immune homeostasis had yet to be investigated. Given the potential role for exosomes in the early events of IDILI (Holman et al., 2016; Mosedale et al., 2018) and recent evidence suggesting that these steps include the loss of immune tolerance (Mak et al., 2018; Mak and Uetrecht, 2015; Metushi et al., 2015), we sought to understand the functional impact of constitutively released HDE on innate immune cells. In the present study, HDE from 5 individual human hepatocyte donors were applied to human monocytes to evaluate the immunomodulatory capacity of HDE released under standard culture conditions.
Exosome-induced alterations in endotoxin-mediated cytokine production suggested that HDE from primary human hepatocytes had pre-disposed monocytes to respond differently to LPS stimulation, with HDE having a predominantly immune-dampening effect. These results suggest that under normal conditions, HDE from healthy hepatocytes promote a tolerogenic immune state. In contrast, exosomes from primary human keratinocytes increased the endotoxin-mediated secretion of proinflammatory cytokines from monocytes. This is in keeping with the understanding that the liver is a uniquely immune tolerant organ and expands on the known mechanisms by which hepatocytes contribute to this phenotype (Tiegs and Lohse, 2010).
The use of the THP-1 cell line and selection of proinflammatory cytokines measured in our study was, in part, motivated by previous reports of IL-1 β, IL-8, and TNF-α elevations in THP-1 and HL-60 cells exposed to conditioned medium from drug-treated hepatocytes (Kato and Uetrecht, 2017; Mak et al., 2018; Momen-Heravi et al., 2015; Oda et al., 2016). Oda et al. (2016) also reported an effect on MCP-1 mRNA levels. Unlike the other cytokines measured in our study, we did not observe a statistically significant HDE-induced decrease in MCP-1 mRNA. We also measured MCP-1 levels in the medium from exosome-treated monocytes, but the responses to HDE were highly variable across hepatocyte donors (data not shown). Furthermore, we found that HDE treatment of primary HPBMs increased production of MCP-1, whereas keratinocyte-derived exosome treatment of THP-1 cells decreased production of MCP-1 (data not shown). This suggests that HDE do not influence all cytokines equally and that MCP-1 may not play a role in liver immune tolerance.
In our study, we specifically chose to use undifferentiated monocytes stimulated with LPS, as this model was previously used in the only similar study reporting an effect of exosomes on the THP-1 response (Momen-Heravi et al., 2015). Furthermore, stimulation with LPS can simulate the continuous exposure of the liver to bacterial products from the gut (Tiegs and Lohse, 2010). However, it would be of interest to explore the effect of constitutively released HDE on unstimulated, differentiated THP-1 cells (macrophages), Kupffer cells (liver-specific macrophages), and/or other liver NPCs as HDE collected under treated conditions are known to stimulate a variety of cell types with immune signaling capacity (Chen et al., 2015; Cobb et al., 2018; Dreux et al., 2012; Giugliano et al., 2015; Momen-Heravi et al., 2015; Seo et al., 2016).
In an effort to understand the mechanism by which HDE influence the THP-1 response, differential gene expression was assessed in HDE-exposed monocytes relative to mock controls. HDE exposure resulted in the decrease of 72 transcripts, many of which are involved in immune function. Interestingly the second most enriched pathway among differentially expressed genes was “Communication between Innate and Adaptive Immune Cells” (Figure 3) indicating the potential for changes within innate immune cells like monocytes to influence an adaptive immune response. The suppression of monocyte gene expression suggests that functional changes observed in cytokine release may be driven by the exosome-induced suppression of genes controlling cytokine response and/or the cytokines themselves, as indicated by the significant decreases in the mRNAs encoding IL-1β, IL-8, and TNF-α.
After confirming that gene expression changes induced by HDE can potentially explain the altered LPS response in monocytes, we sought to establish a relationship between these transcriptomic effects and the regulatory content within HDE. Exosomes have been shown to carry miRNA which can be transferred to recipient cells via internalization and release of the exosomal contents (Yu et al., 2016). Here we demonstrated that the THP-1 cells in our system internalize the HDE suggesting that HDE are capable of transferring cargo to monocytes via internalization by the recipient cells. Interestingly, we noted areas of diffuse PKH67 (green) cytoplasmic staining in HDE-treated monocytes (Figure 2), which may be due to the distribution of exosomal material within the recipient cells.
Exosomal miRNA profiling revealed that many of the miRNA enriched in HDE target the significantly down-regulated genes in monocytes. Using experimentally validated miRNA-mRNA relationships, we also demonstrated concordance between the predicted miRNA regulators of differentially expressed mRNA and those observed in HDE. These results suggest that HDE regulation of monocyte gene expression may be caused by the horizontal transfer of functional miRNAs, a mechanism that has been substantiated for miR-122 delivery to monocytes via Huh7.5 exosomes (Momen-Heravi et al., 2015). To confirm the immunoregulatory capacity of miRNA transferred by HDE, future work will require the examination of the functional effect of each individual miRNA species on monocyte immune response. It is also important to note that miRNAs regulate post-transcriptional gene expression via multiple mechanisms and using transcriptomics as an endpoint only captures miRNAs that regulate expression via mRNA degradation and destabilization mechanisms. A comprehensive analysis of HDE-based miRNA activity in monocytes would require measuring the protein levels of specific miRNA target genes, ideally by proteomic profiling.
The findings presented here have implications for the future study of DILI mechanisms, as they designate HDE as potentially critical regulators of immune function in early events. A natural progression of this work would utilize the insights and approaches described here to investigate the role of HDE in early responses to IDILI drugs. In vitro experiments may be used to study the response of multiple liver NPC types to exosomes harvested from drug-treated hepatocytes. Translational work may evaluate changes in exosome number and/or content in clinical samples collected from IDILI patients. In all of these experiments, it will be important to consider the potential for patient-specific factors to influence response. It should be noted that significant donor variability was observed among the responses induced by HDE collected from hepatocyte donors in this study (Supplementary Figure 1). However, it has been proposed that patient-specific factors contributing to IDILI susceptibility can be overcome at the hepatocyte level by using higher concentrations of drug (Mosedale and Watkins, 2017).
Finally, the present work may also implicate HDE as mediators of inflammatory responses in other organs and highlights the importance of contextualizing the systemic effects of HDE release. For example, an as-yet undefined relationship exists between immune responses in the liver and lung (Hilliard et al., 2015). This axis, and others like it, may be mediated in part by the immunomodulatory functions of HDE. It is also important to consider other factors that may contribute to immune tolerance within the liver. For example, NPCs produce a variety of anti-inflammatory cytokines such as interleukin-10 and transforming growth factor beta 1 and express the immune checkpoint molecules PD-1 and CTLA-4 which promote immune suppression. These factors may work independently or in concert with HDE to promote immune tolerance in the liver.
Taken together, our data suggest that constitutively released HDE are taken up by monocytes and deliver functional miRNAs. These regulatory miRNAs and possibly other exosomal contents target a variety of immune-mediated transcripts and result in a suppression of LPS-stimulated cytokine release. To our knowledge, this research presents the first evidence of direct, potentially tolerogenic immunomodulation by primary human HDE. Future experiments will explore the possibility that IDILI drugs promote the loss of homeostatic HDE signaling.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
CONFLICTS OF INTEREST
Monicah A. Otieno is an employee of Janssen Research and Development, LLC.
Supplementary Material
ACKNOWLEDGMENTS
We thank multiple core facilities at University of North Carolina Chapel Hill for their assistance. We extend our gratitude to Michael Vernon of the UNC Functional Genomics Core for running the gene expression microarrays, the staff of the UNC High-Throughput Genomics Core for performing the exosomal miRNA profiling, and Sebastien Coquery of the UNC Flow Cytometry Core for his contributions to the exosome internalization analysis and imaging.
FUNDING
This work was supported, in part, by the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health [5UL1TR001111], and an Eshelman Institute for Innovation Award to P.BW. N.S.H. was supported, in part, by the National Institutes of Health, National Institute of Environmental Health Sciences (NIEHS) Toxicology Training Grant [T32-ES007126]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Funding to produce “Data Set 2” in Supplementary Figure 4 and support for S.E.T. was provided by Janssen Research and Development, LLC.
REFERENCES
- Ackerman M. E., Moldt B., Wyatt R. T., Dugast A. S., McAndrew E., Tsoukas S., Jost S., Berger C. T., Sciaranghella G., Liu Q., et al. (2011). A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J. Immunol. Methods 366, 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chen R., Kemper S., Charrier A., Brigstock D. R. (2015). Suppression of fibrogenic signaling in hepatic stellate cells by Twist1-dependent microRNA-214 expression: Role of exosomes in horizontal transfer of Twist1. Am. J. Physiol. Gastroint. Liver Physiol. 309, G491–G499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba M., Kimura M., Asari S. (2012). Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol. Rep. 28, 1551–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb D. A., Kim O. K., Golden-Mason L., Rosen H. R., Hahn Y. S. (2018). Hepatocyte-derived exosomes promote T follicular regulatory cell expansion during hepatitis C virus infection. Hepatology 67, 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde-Vancells J., Rodriguez-Suarez E., Embade N., Gil D., Matthiesen R., Valle M., Elortza F., Lu S. C., Mato J. M., Falcon-Perez J. M. (2008). Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J. Proteome Res. 7, 5157–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreux M., Garaigorta U., Boyd B., Décembre E., Chung J., Whitten-Bauer C., Wieland S., Chisari F. V. (2012). Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe 12, 558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzen C. A., Simms P. E., Van Huis A. F., Foreman K. E., Kuo P. C., Gupta G. N. (2014). Characterization of uptake and internalization of exosomes by bladder cancer cells. BioMed Res. Int. 2014, 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giugliano S., Kriss M., Golden-Mason L., Dobrinskikh E., Stone A.E., Soto-Gutierrez A., Mitchell A., Khetani S.R., Yamane D., Stoddard M., et al. (2015). Hepatitis C virus infection induces autocrine interferon signaling by human liver endothelial cells and release of exosomes, which inhibits viral replication. Gastroenterology 148, 392–402.e313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Headland S. E., Jones H. R., D'Sa A. S., Perretti M., Norling L. V. (2014). Cutting-edge analysis of extracellular microparticles using ImageStream(X) imaging flow cytometry. Sci. Rep. 4, 5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard K. L., Allen E., Traber K. E., Yamamoto K., Stauffer N. M., Wasserman G. A., Jones M. R., Mizgerd J. P., Quinton L. J. (2015). The lung-liver axis: A requirement for maximal innate immunity and hepatoprotection during pneumonia. Am. J. Resp. Cell Mol. Biol. 53, 378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsova P., Ibrahim S. H., Krishnan A., Verma V. K., Bronk S. F., Werneburg N. W., Charlton M. R., Shah V. H., Malhi H., Gores G. J. (2016). Lipid-induced signaling causes release of inflammatory extracellular vesicles from hepatocytes. Gastroenterology 150, 956–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman N. S., Mosedale M., Wolf K. K., LeCluyse E. L., Watkins P. B. (2016). Subtoxic alterations in hepatocyte-derived exosomes: An early step in drug-induced liver injury? Toxicol. Sci. 151, 365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R. A., Hobbs B., Collin F., Beazer‐Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Kakazu E., Mauer A. S., Yin M., Malhi H. (2016). Hepatocytes release ceramide-enriched pro-inflammatory extracellular vesicles in an IRE1alpha-dependent manner. J. Lipid Res. 57, 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato R., Uetrecht J. (2017). Supernatant from hepatocyte cultures with drugs that cause idiosyncratic liver injury activates macrophage inflammasomes. Chem. Res. Toxicol. 30, 1327–1332. [DOI] [PubMed] [Google Scholar]
- Kegel V., Pfeiffer E., Burkhardt B., Liu J. L., Zeilinger K., Nüssler A. K., Seehofer D., Damm G. (2015). Subtoxic concentrations of hepatotoxic drugs lead to Kupffer cell activation in a human in vitro liver model: An approach to study DILI. Mediat. Inflamm. 2015, 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Liu K., Liu Y., Xu Y., Zhang F., Yang H., Liu J., Pan T., Chen J., Wu M., et al. (2013). Exosomes mediate the cell-to-cell transmission of IFN-alpha-induced antiviral activity. Nat. Immunol. 14, 793–803. [DOI] [PubMed] [Google Scholar]
- Mak A., Kato R., Weston K., Hayes A., Uetrecht J. (2018). Editor's highlight: An impaired immune tolerance animal model distinguishes the potential of troglitazone/pioglitazone and tolcapone/entacapone to cause IDILI. Toxicol. Sci. 161, 412–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak A., Uetrecht J. (2015). The combination of anti-CTLA-4 and PD1–/– mice unmasks the potential of isoniazid and nevirapine to cause liver injury. Chem. Res. Toxicol. 28, 2287–2291. [DOI] [PubMed] [Google Scholar]
- Metushi I. G., Hayes M. A., Uetrecht J. (2015). Treatment of PD-1-/- mice with amodiaquine and anti-CTLA4 leads to liver injury similar to idiosyncratic liver injury in patients. Hepatology 61, 1332–1442. [DOI] [PubMed] [Google Scholar]
- Momen-Heravi F., Bala S., Kodys K., Szabo G. (2015). Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Sci. Rep. 5, 9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosedale M., Eaddy J. S., Trask J. O. J., Holman N. S., Wolf K. K., LeCluyse E., Ware B. R., Khetani S. R., Lu J., Brock W. J., et al. (2018). miR-122 release in exosomes precedes overt tolvaptan-induced necrosis in a primary human hepatocyte micropatterned coculture model. Toxicol. Sci. 161, 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosedale M., Watkins P. B. (2017). Drug-induced liver injury: Advances in mechanistic understanding that will inform risk management. Invited “state-of-the-art” review. Clin. Pharmacol. Ther. 101, 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller L., Hong C. S., Stolz D. B., Watkins S. C., Whiteside T. L. (2014). Isolation of biologically-active exosomes from human plasma. J. Immunol. Methods 411, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda S., Matsuo K., Nakajima A., Yokoi T. (2016). A novel cell-based assay for the evaluation of immune- and inflammatory-related gene expression as biomarkers for the risk assessment of drug-induced liver injury. Toxicol. Lett. 241, 60–70. [DOI] [PubMed] [Google Scholar]
- Ogese M. O., Faulkner L., Jenkins R. E., French N. S., Copple I. M., Antoine D. J., Elmasry M., Malik H., Goldring C. E., Park B. K., et al. (2017). Characterization of drug-specific signaling between primary human hepatocytes and immune cells. Toxicol. Sci. 158, 76–89. [DOI] [PubMed] [Google Scholar]
- Seo W., Eun H. S., Kim S. Y., Yi H. S., Lee Y. S., Park S. H., Jang M. J., Jo E., Kim S. C., Han Y. M., et al. (2016). Exosome-mediated activation of toll-like receptor 3 in stellate cells stimulates interleukin-17 production by gammadelta T cells in liver fibrosis. Hepatology 64, 616–631. [DOI] [PubMed] [Google Scholar]
- Singer J. B., Lewitzky S., Leroy E., Yang F., Zhao X., Klickstein L., Wright T. M., Meyer J., Paulding C. A. (2010). A genome-wide study identifies HLA alleles associated with lumiracoxib-related liver injury. Nat. Genet. 42, 711.. [DOI] [PubMed] [Google Scholar]
- Suzman D. L., Pelosof L., Rosenberg A., Avigan M. I. (2018). Hepatotoxicity of immune checkpoint inhibitors: An evolving picture of risk associated with a vital class of immunotherapy agents. Liver Int. 38, 976–987. [DOI] [PubMed] [Google Scholar]
- Thacker S. E., Nautiyal M., Otieno M. A., Watkins P. B., Mosedale M. (2018). Optimized methods to explore the mechanistic and biomarker potential of hepatocyte-derived exosomes in drug-induced liver injury. Toxicol. Sci. 163, 92–100. [DOI] [PubMed] [Google Scholar]
- Tiegs G., Lohse A. W. (2010). Immune tolerance: What is unique about the liver. J. Autoimmun. 34, 1–6. [DOI] [PubMed] [Google Scholar]
- Urban T. J., Daly A. K., Aithal G. P. (2014). Genetic basis of drug-induced liver injury: Present and future. Semin. Liver Dis. 34, 123–133. [DOI] [PubMed] [Google Scholar]
- Usui T., Naisbitt D. J. (2017). Human leukocyte antigen and idiosyncratic adverse drug reactions. Drug Metab. Pharmacokinet. 32, 21–30. [DOI] [PubMed] [Google Scholar]
- Verma V. K., Li H., Wang R., Hirsova P., Mushref M., Liu Y., Cao S., Contreras P. C., Malhi H., Kamath P. S., et al. (2016). Alcohol stimulates macrophage activation through caspase-dependent hepatocyte derived release of CD40L containing extracellular vesicles. J. Hepatol. 64, 651–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore B. A., Brees D. J., Singh R., Watkins P. B., Andersen M. E., Loy J., Thomas R. S. (2010). Quantitative analyses and transcriptomic profiling of circulating messenger RNAs as biomarkers of rat liver injury. Hepatology 51, 2127–2139. [DOI] [PubMed] [Google Scholar]
- Yu X., Odenthal M., Fries J. W. (2016). Exosomes as miRNA carriers: Formation-function-future. Int. J. Mol. Sci. 17, pii: E2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.