Abstract
Cytoglobin is a heme protein evolutionarily related to hemoglobin and myoglobin. Cytoglobin is expressed ubiquitously in mammalian tissues; however its physiological functions are yet unclear. Phylogenetic analyses indicate that the cytoglobin gene is highly conserved in vertebrate clades, from fish to reptiles, amphibians, birds, and mammals. Most proposed roles for cytoglobin require the maintenance of a pool of reduced cytoglobin (FeII). We have shown previously that the human cytochrome b5 / cytochrome b5 reductase system, considered a quintessential hemoglobin/myoglobin reductant, can reduce human and zebrafish cytoglobins up to 250-fold faster than human hemoglobin or myoglobin. It was unclear whether this reduction of zebrafish cytoglobins by mammalian proteins indicates a conserved pathway through vertebrate evolution. Here we report the reduction of zebrafish cytoglobins 1 and 2 by the zebrafish cytochrome b5 reductase and the two zebrafish cytochrome b5 isoforms. In addition, the reducing system also supports reduction of Globin X, a conserved globin in fish and amphibians. Indeed, the zebrafish reducing system can maintain a fully reduced pool for both cytoglobins, and both cytochrome b5 isoforms can support this process. We determined the P50 for oxygen being 0.5 torr for cytoglobin-1 and 4.4 torr for cytoglobin-2 at 25 °C. Thus, even at low oxygen tension, the reduced cytoglobins may exist in a predominant oxygen-bound form. In these conditions, the cytochrome b5/cytochrome b5 reductase system can support a conserved role for cytoglobins through evolution, providing electrons for redox signaling reactions such as nitric oxide dioxygenation, nitrite reduction or phospholipid oxidation.
Keywords: Cytoglobin, Cytochrome b5, Cytochrome b5 reductase, nitric oxide, nitric oxide dioxygenation
Graphical Abstract
Introduction
Vertebrate organisms have retained at least eight globin genes in their genome,1 with five globin genes identified in mammals to date.2 These proteins are usually associated with oxygen transport functions, following the roles of the most conspicuous hemoglobin (Hb) and myoglobin (Mb). However, newly discovered globins have been associated with functions that defy this paradigm. Neuroglobin, androglobin and cytoglobin (Cygb) show cellular levels nowhere near the high concentrations of hemoglobin and myoglobin and appear to be involved in other processes not necessarily related to oxygen transport and storage. Proposed functions include the detoxification of reactive oxygen species, regulation of nitric oxide (NO) levels, signaling reactions and lipid peroxidation.3–8
Cygb is a heme protein ubiquitously expressed in mammalian tissues.9–11 Unlike hemoglobin and myoglobin, but similar to neuroglobin and androglobin, the heme in Cygb is coordinated by two histidine side chains, resulting in a six-coordinate conformation. The presence of a distal ligand to the heme iron causes important changes in heme properties.4–6, 11–14 Mammals carry one copy of the Cygb gene, however two copies of the gene have been identified in teleost fish (Cygb1 and Cygb2), probably arising from an ancient whole genome duplication event.15, 16 Studies in zebrafish indicate that both genes are expressed.17 The in vitro characterization of the two proteins suggests that Cygb1 may be involved in oxygen transport, whereas Cygb2 has biophysical properties more alike the mammalian Cygb protein.17, 18
As for other globins, the Cygb reactivity is largely dependent on the oxidation state of its heme group. As most biological reactions will oxidize the heme iron to the ferric (FeIII) form, any catalytic cycle requires a source of electrons to reduce Cygb (equations 1–3).
| (Equation 1) |
| (Equation 2) |
| (Equation 3) |
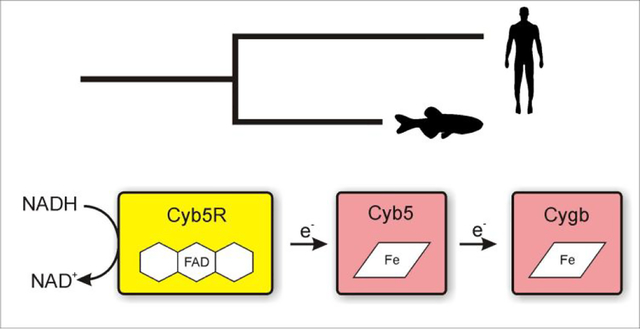
To that purpose, it appears that ascorbic acid can fulfill this role in certain conditions.19 New data suggests that the cytochrome b5 (CYB5) and cytochrome b5 reductase 3 (CYB5R) system is a feasible candidate for the physiological reduction of Cygb.20, 21 To our surprise, we observed that the human CYB5/CYB5R system catalyzes human and zebrafish Cygb reduction at rates up to 250-fold higher than those for their bona fide substrates, Hb and Mb.20
Globin X (GbX) is an ancestral globin protein recently discovered in fish and amphibians.22 The function of this globin is yet unknown. It has been noted that it can be lapidated and associated with membranes23 and it is also present in fish thrombocites (the equivalent to mammalian erythrocites), where it may be involved in oxygen or nitric oxide related functions.24 In our previous work we have observed that human CYB5b/CYB5R has some ability to reduce GbX heme, whereas 5mM Ascorbate shows almost no effect.20
One caveat in our previous study is that we used human CYB5/CYB5R proteins to monitor the reduction of the zebrafish Cygbs and GbX.20 In order to establish unequivocally whether this reaction is functional in zebrafish, a complete system of zebrafish proteins should be used. Moreover, we only tested the ability of human CYB5b to support Cygb and GbX reduction. As the properties of the CYB5a and CYB5b isoforms may differ notably,25, 26 and two CYB5 isoforms are present in zebrafish, as in mammalian genomes, the ability of both proteins to reduce Cygb and GbX may differ and hint towards tissue- or cell-specific capabilities.
Homologous genes for the 4 CYB5R isoforms found in mammals have been identified in the zebrafish genome. It has been reported that a membrane-bound CYB5R protein can reduce fish Hb and maintain proper Hb reduction levels.27, 28 The NADH diaphorase activity of fish red blood cell (RBC) extracts, a surrogate for CYB5R activity, correlates with the resistance of fish to methemoglobinemia-inducing conditions.27, 28 Although the presence of a CYB5R3 homolog – the isoform found in mammalian RBCs and mitochondria – in fish has been described,29, 30 and the protein shows ≈ 65% identity with the characterized mammalian enzymes, the properties of the fish CYB5Rs are unknown. Thus we also characterized the properties of the zebrafish CYB5R3 protein.
Here we study the ability of the reconstituted zebrafish CYB5/CYB5R system to reduce zebrafish Cygbs and GbX. Our results indicate that these zebrafish proteins provide a viable system for the reduction of the Cygbs, with both CYB5 proteins supporting the fast reduction of Cygb1 and Cygb2 at physiological temperatures. The complete zebrafish electron transfer system also catalyzes the reduction of GbX faster than reported for the human CYB5/CYB5R system or ascorbate, and may also provide a physiological source of electrons for GbX in vivo. These observations suggest a conserved role of CYB5/CYB5R supporting Cygb function that possibly predates its role as hemoglobin and myoglobin reductases.
Materials and Methods
Reagents –
All reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified.
Expression and Purification of Recombinant Globins –
Zebrafish GbX, Cygb1 and Cygb2 were cloned into the pET-28 plasmid (Novagen) and expressed in E. coli cells and purified as previously described.20, 24 The cDNA clones for zebrafish CYB5a, CYB5b and CYB5R (cytochrome b5 reductase 3) were obtained from the IMAGE consortium through Thermo Fisher/Open Biosystems. The accession numbers and IMAGE identifiers are as follows: CYB5R, BC_066624 (IMAGE 6960174); CYB5a, BC_154824 (IMAGE 9002123); CYB5b, BC_066748 (IMAGE 6525311). CYB5R was cloned into the pET28a plasmid (Novagen) using the NdeI/HindIII restriction sites. The residues 1–20 necessary for membrane insertion were removed, and the 21–298 sequence, coding the soluble protein, was cloned (Figure 1). The protein sequence includes an N-terminal 6xHis tag. CYB5a and CYB5b were cloned into pET11a plasmid using the NdeI and NcoI restriction sites. The C-terminal residues 101–137 of CYB5a that form a membrane insertion helix were removed. For CYB5b, the N-terminal residues 1–17 and the C-terminal residues 115–153 were removed to produce the soluble protein as reported for mammalian proteins (Figure 1).25, 26 Purification of zebrafish CYB5 proteins and CYB5R was carried out as described for the human proteins.20, 31 Spectral data were recorded using either a Cary 50 spectrophotometer (Agilent) or an Agilent HP8453 diode array spectrophotometer. Proteins were studied in 100 mM sodium phosphate buffer, pH 7.4.
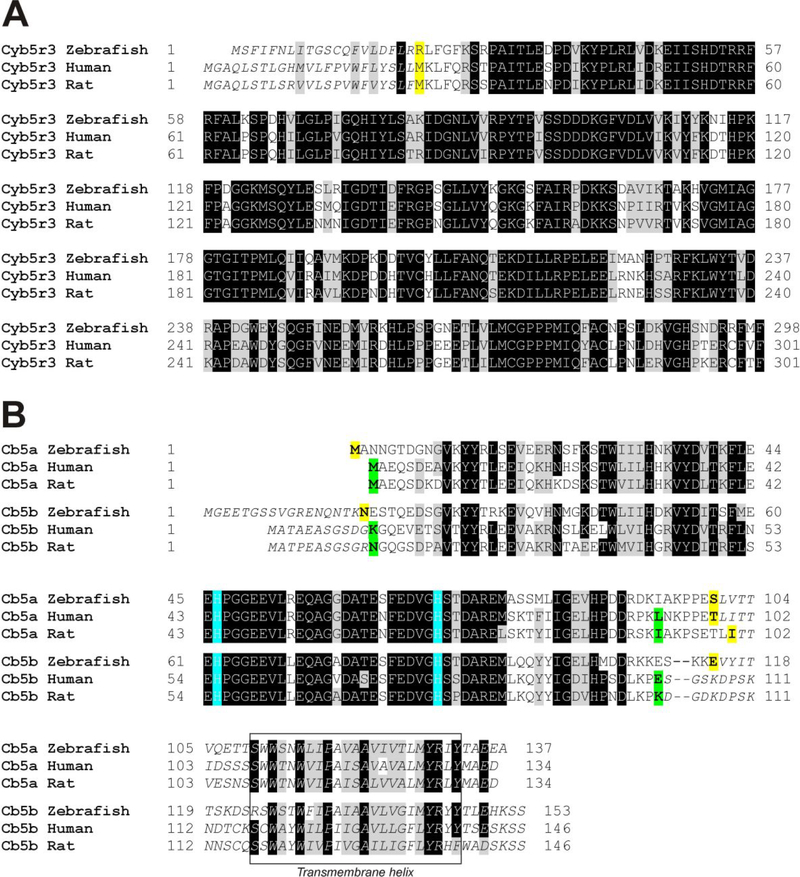
Figure 1. Alignment of cytochrome b5 reductase and cytochromes b5a and b5b protein sequences for zebrafish, human, and rat.
A; alignment of CYB5R3 sequences (Uniprot accession codes: zebrafish, Q6NYE6; human P00387; rat, P20070). Initial portions in italics indicate the missing residues in the soluble isoform (CYB5R3–2) of the mammalian proteins and the homologous residues in zebrafish protein; the first amino acid in the recombinant protein used in this work and the initial amino acid in the mammalian isoform 2 are marked in bold and yellow background. B; alignment of CYB5a (microsomal CYB5) and CYB5b (outer membrane CYB5) sequences (Uniprot accession codes: zebrafish b5a, Q7T341; human b5a, P00167; rat b5a, P00173; zebrafish b5b, Q6NY41; human b5b, O43169; rat b5b, P04166). Conserved heme-binding histidines are indicated in blue background. Amino acids in italics indicate sequences removed in the N-termini of CYB5b and the membrane binding C-terminal regions of CYB5a and CYB5b. The putative membrane intercalating helix region is shown in a rectangle. The first and last amino acids in the recombinant proteins used in this work, along with the splicing sites for the soluble isoform of mammalian cytochrome b5a proteins are marked in bold and yellow background; the first and last amino acids in the recombinant proteins used in other works25, 26 are indicated in bold with green background.
Sequence analysis –
CYB5 sequences were analyzed for possible posttranslational modifications using publicly available software. Acetylation motifs were scanned using Terminus32 and NetAcet33 servers. Palmitoylation sequences were screened using the CSS-Palm 4.0 software.34
Steady-state kinetics of CYB5R with CYB5a and CYB5b –
Steady state parameters for the reaction of CYB5R with NADH and CYB5a/CYB5b were determined as follows: NADH diaphorase activity was assayed using dichlorophenol-indophenol (DCPIP) or potassium ferricyanide as electron acceptors. In the experiments with DCPIP, zebrafish CYB5R (0.028 μM) was incubated with 95 μM DCPIP and the reaction was initiated by adding variable amounts of a 1 mM NADH stock solution for final NADH concentrations between 0 and 22 μM. The reduction of DCPIP was monitored at 600 nm. In the experiments with potassium ferricyanide, zebrafish CYB5R (0.84 nM) was incubated with 1 mM potassium ferricyanide and the reaction was initiated by adding variable amounts of a 1 mM NADH stock solution for final NADH concentrations between 0 and 50 μM. The reduction of potassium ferricyanide was monitored via the decay of NADH absorbance at 340 nm. The initial reduction rates were plotted versus the concentration of NADH and the data was fitted to the Michaelis-Menten equation to determine the kcat and KM parameters. To determine the apparent KM for CYB5a and CYB5b, the reaction samples included CYB5R (7.3 nM), NADH (60 μM) and variable amounts of CYB5a or CYB5b (0 to 140 μM). Reduction of CYB5a or CYB5b was monitored at 555 nm. The initial rates were fitted as a function of the initial CYB5 concentrations and the data was fitted to the Michaelis-Menten equation to determine the kinetic parameters as above. Experiments were carried out in 50 mM Bis-Tris propane buffer, pH 7.4, at 25 °C.
Reduction of Cytoglobins and Globin X by CYB5/CYB5R –
Steady-state reduction of zebrafish Cygbs and GbX by the CYB5/CYB5R was studied as previously reported.20 The reduction was performed in anaerobic conditions and monitored in an Agilent HP8453 spectrophotometer housed in an anaerobic glovebox (Coy Laboratories, Grass Lake, MI). Oxidized globins were prepared by treatment with excess potassium ferricyanide and then the excess ferricyanide was removed by passing the sample through a Sephadex G25 column (PD10, GE healthcare). The globin solution was diluted to a 20 μM final concentration and zebrafish CYB5R (0.2 μM) and either CYB5a or CYB5b (2 μM) was added. The reaction was initiated by addition of 100 μM NADH. The fraction reduced was determined by monitoring the absorbance changes in the characteristic peak for the ferrous form of the globins (≈ 560 nm) as reported.20
Redox potentials –
Oxidation-reduction potentials for the zebrafish CYB5R and CYB5 proteins were determined in anaerobic conditions at 25 °C in 100 mM sodium phosphate buffer, pH 7.0. Spectral data were recorded using an Agilent HP8453 spectrophotometer and redox potential was collected via an Accumet 15 connected to a MI-480 electrode (Microelectrodes, Inc). Proteins (≈10 μM) were titrated with either sodium dithionite (reductive titrations) or potassium ferricyanide (oxidative titrations) in the presence of redox mediators. The redox mediators (1–5 μM) used were phenosafranine (Em = −252 mV) for CYB5R. In the case of CYB5a and CYB5b a modified version of the method described by Efimov et al35, using sodium dithionite as reductant instead of the xanthine/xanthine oxidase system and an anaerobic setup that circumvents the use of the glucose/glucose oxidase system to remove oxygen. Indigo tetrasulfonate (Em = −46 mV) was used as mediator. Spectral and redox potential readings were analyzed using the Nernst equation to calculate the midpoint potentials as described.13, 17, 35
Determination of melting temperatures –
Thermal denaturation of CYB5a and CYB5b proteins in the ferric heme state was monitored by UV-Vis spectroscopy using a Cary50 spectrophotometer. Experiments were carried out in 10 mM phosphate buffered saline, pH 7.4. The temperature was increased from 20 °C to 100 °C; for each step the temperature was maintained for ~5 minutes and the spectral changes were then recorded. Changes in the Soret peak were fitted to the Santoro-Bolen equation36 to determine the melting temperature (Tm) of each protein.
Determination of oxygen binding affinities –
In order to determine the oxygen partial pressures at half-saturation of the ferrous-oxygen complex (P50) of Cygb1 and Cygb2, we measured oxygen equilibrium curves using a thin-layer modified diffusion chamber described elsewhere37–39 with some modifications.14 Samples (5 μl) contained 200 μM heme Cygb1 or Cygb2. As a reducing system, 40 μM human CYB5b, 4 μM human CYB5R and 600 μM NADH were added. 20 mM sodium formate and 10 mU of formate dehydrogenase were added to regenerate the consumed NADH. Samples were equilibrated for 5 minutes at room temperature under N2 atmosphere to allow heme reduction before transfer to the modified diffusion chamber. Experiments were conducted at either 25°C or 37°C in 100 mM potassium phosphate buffer at pH 6.8 or 7.4. Fit of the oxygen saturation data to the Hill equation also provided the Hill coefficient measuring the degree of cooperativity in oxygen binding.
Results
Protein expression and spectral properties –
Zebrafish CYB5R was expressed in soluble form by removing the N-terminus sequence (amino acids 1–20, Figure 1A) to prevent membrane binding. In mammalian CYB5R, Gly2 has been found to be myristoylated to direct the protein to mitochondria and other membrane fractions; the initial fragment (amino acids 1–23 in mammalian proteins) is not present in the soluble isoform expressed in mammalian erythrocytes.40 The sequence of the zebrafish CYB5R lacks the glycine in position 2. Nevertheless, the Protein in fish erythrocytes appears to be membrane-bound28. Although the post translational modification responsible has not been identified, sequence analysis software predicts a possible acetylation site in Ser2 (Terminus32, NetAcet33) and palmitoylation of Cys12 (CSS-Palm34). Zebrafish CYB5a (microsomal isoform) was expressed in its soluble form without its C-terminal membrane binding-domain (amino acids 101–137, Figure 1B). Zebrafish CYB5b (mitochondrial outer membrane isoform) was also expressed in a soluble form, omitting its N-terminal amino acids (amino acids 1–17, Figure 1B) and C-terminal transmembrane domain (amino acids 115–153, Figure 1B).
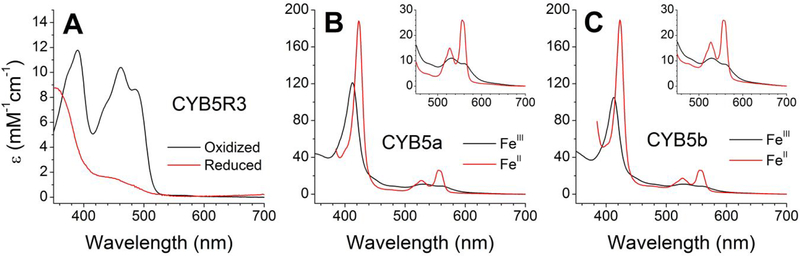
The zebrafish CYB5R and cytochromes CYB5a and CYB5b were expressed in E. coli and purified following similar protocols to those used previously for the mammalian proteins.20, 31 We did not observe notable differences in the expression and purification as compared to the human proteins. The three proteins show spectral properties similar to their mammalian counterparts.25, 41 Spectra for the fully oxidized and fully reduced species are shown in Figure 2. Oxidized CYB5R shows the characteristic peaks of the FAD cofactor, with maxima at 388 nm and 460 nm (Figure 2A). CYB5a and CYB5b showed similar wavelengths for their Soret peaks in the oxidized state (413 nm) and an additional peak at 531 nm for CYB5a or 528mn for CYB5b, consistent with the spectra of other six-coordinated heme proteins in the ferric form. CYB5a and CYB5b in the reduced state show spectra characteristic of a bis-His, six-coordinate heme, with two peaks around 520 nm and 560 nm. The observed maxima were 423 nm, 527 nm and 555 nm (CYB5a) and 423 nm, 527 nm and 556 nm (CYB5b) (Figures 2B, 2C).
Figure 2. Spectral properties of zebrafish cytochrome b5 reductase and cytochromes b5a and b5b.
Panel A, CYB5R; Panel B, CYB5a; Panel C, CYB5b. The red traces represent the reduced state (FADH2 for CYB5R or ferrous (FeII) heme for CYB5a and CYB5b) and the black traces represent the oxidized proteins (FADH2 for CYB5R or ferric (FeIII) heme for CYB5a and CYB5b). Insets in Panels B and C show the 450–700nm range
Steady-state kinetics –
In order to characterize the ability of CYB5R to oxidize NADH and catalyze electron transfer reactions to its electron transfer partner proteins CYB5a and CYB5b, we studied the kinetics of CYB5R with different electron acceptors in steady-state conditions.
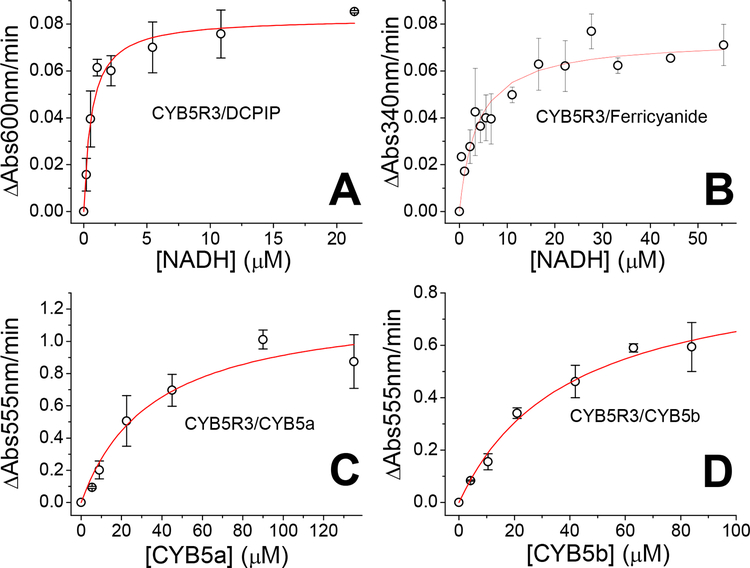
We determined the KM of zebrafish CYB5R for NADH in the presence of DCPIP and ferricyanide as electron acceptors. The observed rates for the reaction of CYB5R with DCPIP are shown in Figure 3A. The values were fitted to a Michaelis-Menten equation yielding a kcat of 26 s−1 and a KM of 0.6 μM (Figure 3A, Table 1). This activity is comparable to that reported for the other CYB5R proteins (Table 1). When potassium ferricyanide was used as electron acceptor, we observed rates also consistent with a Michaelis-Menten fit. Calculated values were kcat of 235 s−1 and a KM of 3.7 μM (Figure 3B, Table 1).
Figure 3. Steady state kinetics of zebrafish Cytochrome b5 reductase.
Panels A and B, determination of the Vmax and KM towards NADH for the NADH-diaphorase activity of CYB5R with DCPIP (Panel A) or ferricyanide (Panel B) as electron acceptor. Panels C and D, determination of the Vmax and KM towards zebrafish CYB5a (Panel C) or CYB5b (Panel D) in the presence of saturating concentrations of NADH (60 μM). Experiments conducted in 50 mM Bis-Tris propane, pH 7.4, 25 °C.
Table 1.
Kinetic parameters for the reaction of zebrafish cytochrome b5 reductase and related proteins with DCPIP, ferricyanide, cytochrome b5a, and cytochrome b5b
| Diaphorase activity - DCPIP | Diaphorase activity - Ferricyanide | Cytochrome b5 reductase activity | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CYB5R | kM NADH (μM) | kM NADH (μM) | Km CYB5a (μM) | Km CYB5a (μM) | Reference | ||||
| Zebrafish | 0.65 ± 0.12 | 3.7 ± 0.9 | 36 ± 12 | 40 ± 8 | This work | ||||
| Fly | nd | 44 | 13 | nd | 42 | ||||
| Rat | nd | 6.0 | 13 | nd | 43 | ||||
| Pig | nd | 2.5 | 8.8 | nd | 44 | ||||
| Human | 0.48 | 0.6 | 35 | nd | 45,46 | ||||
nd, not determined.
The kcat values are comparable to those of the fly CYB5R (Table 1); although 3–4-fold lower than those of mammalian enzymes (~600–800 s−1, Table 1). The KM value is comparable to the reported KM values for other CYB5R enzymes (~0.6–6.0 μM, Table 1) indicating a high affinity towards NADH. We then determined the activity of CYB5R with its partner proteins CYB5a and CYB5b. These experiments were conducted in saturating concentrations of NADH (60 μM).
Experiments on the absence of CYB5R showed that NADH can also reduce directly CYB5a and CYB5b at a significant rate; background activity in the absence of CYB5R was subtracted from the rates in the presence of CYB5R to determine reduction rate due to CYB5R (Figure 3B,C). The fit of the initial rates yielded values of kcat = 165 s−1 and KM = 36 μM for CYB5a and kcat = 121 s−1 KM = 40 μM for CYB5b (Table 1). Thus, CYB5R reduces both substrates with very similar kinetic parameters, though at slightly faster rates with CYB5a. The KM values are higher than the expected physiological CYB5 concentrations, suggesting a linear dependence of the reduction rates with CYB5 concentrations in vivo.
Redox potentials –
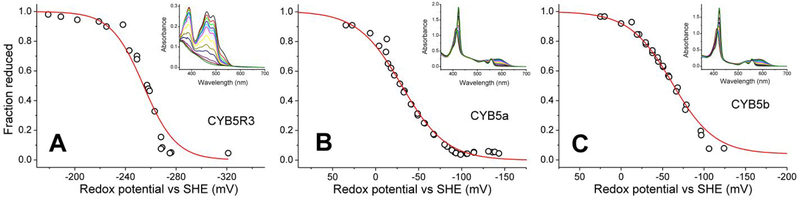
In order to further characterize the properties of the zebrafish CYB5R/CYB5 system, we determined the redox potential of the three zebrafish proteins (Figure 4). The titration of CYB5R shows a single transition, fitted to a slope of ~29.5 mV, consistent with a two-electron step (Figure 1A). Similar observations have been reported for other CYB5R proteins, where the one-electron reduced, semiquinone state is not stable and does not accumulate during the reduction step.43, 47–49 Thus, the two one-electron transitions show a very similar midpoint potential and the two steps are merged in a single apparent Em value. We observe a midpoint potential of 264 mV, in the range of the values reported for mammalian proteins (Table 2).
Figure 4. Redox potentials of zebrafish Cytochrome b5 reductase and cytochromes b5a and b5b.
Panel A, CYB5R; Panel B, CYB5a; Panel C, CYB5b. The plots show the fit of the fraction reduced (as determined from absorbance spectra, open circles) to the Nernst equation (solid lines). The insets show the absorbance changes during the reductive titrations. Peaks at 510nm (CYB5R) or 590nm (CYB5a/b) are due to the mediators.
Table 2.
Redox potential of zebrafish Cytochrome b5 reductase, cytochrome b5a, cytochrome and b5b and related mammalian proteins
Previous work has indicated notable differences in the redox potential of CYB5a and CYB5b in mammalian systems. Reported CYB5a midpoint potential values are around 0 mV, whereas CYB5b proteins show more negative values. In particular, human and rat proteins show values of −40 and −102 mV, respectively (Table 2).25, 26 We observe values in a similar range for the zebrafish proteins (Table 1). The potential for CYB5a is −28 mV, slightly more negative than the value observed for mammalian proteins. In the case of CYB5b, the observed value of −62 mV is between the observed values for rat and human CYB5b proteins. Notably, the observed values for the zebrafish Cygb1 and Cygb2, −58 mV and −26 mV respectively17, are in a similar range to those of the zebrafish CYB5 proteins (Table 2).
Thermal denaturation of zebrafish cytochromes b5a and b5b –
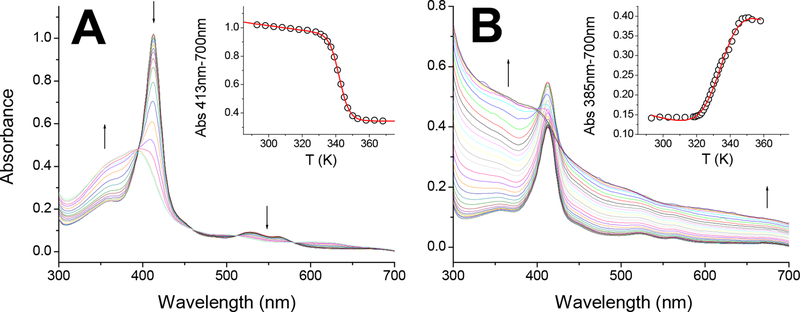
As noted above we observe a 35 mV difference in redox midpotential values between zebrafish CYB5a and CYB5b. This is not unlike the situation for mammalian CYB5 proteins, where differences of up to 95 mV have been reported52–54. Given the high identity and similarity of the CYB5 sequences, the source of these differences is not evident (Figure 1). The changes within CYB5 sequences are limited to structural elements away from the heme binding regions, with nearly 100% identity in the regions close to the heme within CYB5a or CYB5b sequences, and limited changes between the two CYB5s. Given the high variability between mammalian CYB5a and CYB5b redox potentials (Table 3), it has been speculated that the redox potential is in part modulated by the strength of the bond between the FeIII-heme and the proximal histidine.25 Although the heme exposure to the solvent is one of the main contributors to the regulation on the heme redox potential in proteins,55 the differences in heme binding in the ferrous and ferric states can also cause large variations in the redox potential, even in conditions where the heme solvent accessibility is unchanged.56 As a proxy for the FeIII-His bond strength we studied the thermal denaturation of the zebrafish CYB5a and CYB5b proteins. Our experimental determinations of the melting temperatures (Tm) are shown in Figure 5 and Table 3. Our results indicate that the two zebrafish proteins have similar Tm values (Table 3). The value for CYB5a is in line with mammalian CYB5a proteins, however the Tm for zebrafish CYB5b appears much lower than the reported values for mammalian CYB5b (Table 3). The spectral changes also indicate clear differences between the thermal denaturation processes for CYB5a and CYB5b. The denaturation of CYB5a shows a shift from the spectra of the bis-His hexacoordinated globin towards a spectrum similar to that of free heme, with a Soret peak around 385 nm (Figure 5A). A similar increase in the 350–400nm area, indicating heme dissociation, is observed for CYB5b, but in this case the heme release correlates with visible light scattering as noted by the increase of the signal at 700nm and throughout the spectral range (Figure 5B). Without further structural data, the nature of the changes cannot be unequivocally assigned, but we can speculate that the denaturation of the zebrafish CYB5a leads to the formation of a stable apoprotein, whereas the zebrafish CYB5b aggregates when the heme is dissociated form the protein, suggesting a less stable apoprotein in our experimental conditions.
Table 3.
Melting temperatures of zebrafish cytochrome b5a, cytochrome b5b and related mammalian proteins
Figure 5. Thermal denaturation of zebrafish Cytochrome b5a and cytochrome b5b.
Panel A, CYB5a; Panel B, CYB5b. The spectra obtained at increasing temperatures (20–95 °C) are shown, the direction of the absorbance changes is indicated by arrows. The insets show the changes in the Soret peak absorbance with the temperature and the fit of the absorbance changes to the Santoro-Bolen equation (red lines).
Oxygen affinity of zebrafish cytoglobins –
We have shown that zebrafish Cygb1 and Cygb2 can be reduced efficiently by the mammalian CYB5R/CYB5/NADH system.20 This allows for a catalytic cycle of NO dioxygenation with continuous regeneration of the deoxy (FeII) species, and in the presence of oxygen, the subsequent fast formation of the oxygen-bound species (FeII-O2), as shown in the following equations:
| (Equation 2) |
| (Equation 4) |
| (Equation 5) |
| (Equation 6) |
Where equation 4 indicates the regeneration of the Cygb deoxy species by reaction with the pool of reduced CYB5 and the net sum of the three reactions yields the catalytic deoxygenation of NO (Equation 6).
In order for zebrafish Cygbs to support NO dioxygenation reactions, the reduced form of the protein should be able to bind oxygen in physiological conditions, and therefore it is important to determine the oxygen affinity of these Cygbs. We used a met-reducing enzymatic system57 to determine P50 values for Cygb1 and Cygb2. We have used a similar system in the determination of the P50 for other heme proteins, including several mutant Cygbs and GbX.14,23 The system uses ferredoxin NADP+-reductase, ferredoxin and NADPH to regenerate the pool of globin oxidized due to endogenous autoxidation. However this system was unable to maintain zebrafish Cygb2 in the ferrous reduced state due to its very fast autoxidation rate.17 As we previously observed the efficient reduction by the human CYB5R/CYB5 system,20 we used these proteins to keep the zebrafish Cygbs in the reduced state. Formate and formate dehydrogenase were included to regenerate the NADH consumed.
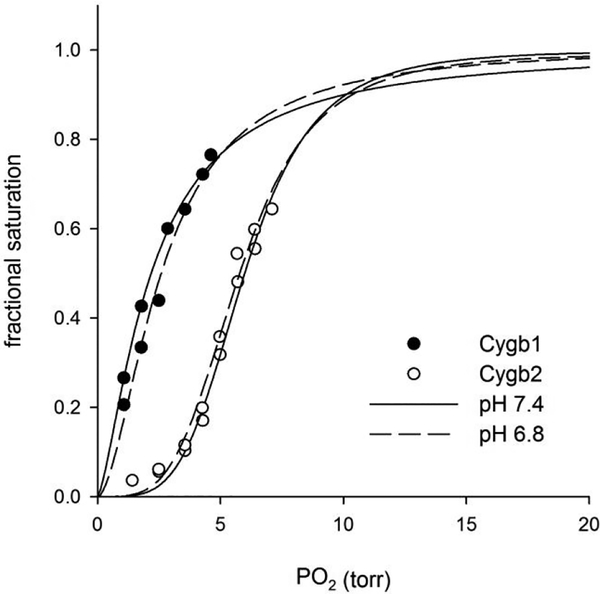
Our experimental results for the determination of the P50 values at pH 6.8 and 7.4 at either 25 °C or 37 °C are shown in Figure 6 and Table 4. We observe that Cygb1 has a higher oxygen affinity than Cygb2, with values between 0.4–2.5 torr for Cygb1 compared to 3.6–6.1 torr for Cygb 2. The P50 values for Cygb2 appear higher than those reported for mammalian Cygbs at pH 7.0 (0.2–2.8 torr)11, 14, 58, 59 which is unexpected given that most properties of Cygb2 are otherwise more similar to the mammalian protein than Cygb1.17 On the other hand, Cygb1 shows very high affinity towards oxygen with similar or lower P50 values than those of the mammalian Cygbs. Comparing the data at different pH values, neither Cygb shows a significant Bohr effect, in agreement with previous data on mammalian Cygbs.14
Figure 6. Oxygen equilibrium curves of zebrafish cytoglobins 1 and 2 at 37 °C.
The points for Cygb1 (solid circles) and Cygb2 (open circles) are indicated. The lines show the fit to the Hill sigmoidal equation.
Table 4.
Oxygen binding parameters for zebrafish Cytoglobins
| Protein | P50 (torr) | Hill coefficient (n) |
|---|---|---|
| Cygb1 | ||
| 25°C, pH 6.8 | 0.39 ± 0.12 | 1.05 ± 0.09 |
| 25°C, pH 7.4 | 0.53 ± 0.06 | 1.43 ± 0.20 |
| 37°C, pH 6.8 | 2.54 ± 0.11 | 1.77 ± 0.01 |
| 37°C, pH 7.4 | 2.20 ± 0.01 | 1.43 ± 0.01 |
| Cygb2 | ||
| 25°C, pH 6.8 | 3.61 ± 0.21 | 3.45 ± 0.08 |
| 25°C, pH 7.4 | 4.44 ± 0.49 | 2.99 ± 0.16 |
| 37°C, pH 6.8 | 5.72 ± 0.07 | 3.78 ± 0.13 |
| 37°C, pH 7.4 | 6.09 ± 0.15 | 4.28 ± 0.11 |
Analysis of the Hill coefficients (n values in Table 4) indicates that both zebrafish proteins bind oxygen cooperatively. In particular, Cygb2 has very high n values (3.0–4.3), as also evident from the highly sigmoidal oxygen equilibrium curves (Fig. 6), suggesting formation of complexes larger than tetramers. It is difficult to speculate further from these data on the nature of these complexes. It has been shown that oligomerization of mammalian Cygbs, with formation of tetramers and octamers, may occur at high concentrations, probably at supraphysiological concentrations.59 It is thus unclear if the oligomers observed in our study are physiologically relevant or are due to the high protein concentrations used and do not necessarily reflect the cellular conditions. Zebrafish GbX binds O2 cooperatively with a overall lower affinity (P50 1.3–12.5 torr at 20 °C)23 compared to zebrafish Cygbs, and thus may not be significantly oxygenated in vivo.
Reduction of zebrafish cytoglobins by the native zebrafish Cytochrome b5 reductase/Cytochrome b5 systems–
In previous work we have shown that human cytochrome b5 reductase, in combination with human CYB5b, can efficiently reduce zebrafish Cygb1 and Cygb2. We were thus interested in evaluating if the native zebrafish systems, using CYB5R in combination with either zebrafish CYB5 isoform, could also reduce the Cygbs, and if so how they perform as compared to the human reducing system.
In order to test the reduction of the Cygbs by CYB5/CYB5R/NADH we monitored the reduction of zebrafish Cygbs 1 or 2 (20 μM) in the presence of zebrafish CYB5a or CYB5b (2 μM, consistent with reported physiological levels,60, 61 either zebrafish or human CYB5R (0.2 μM), and 100 μM NADH to ensure CYB5R saturating conditions.
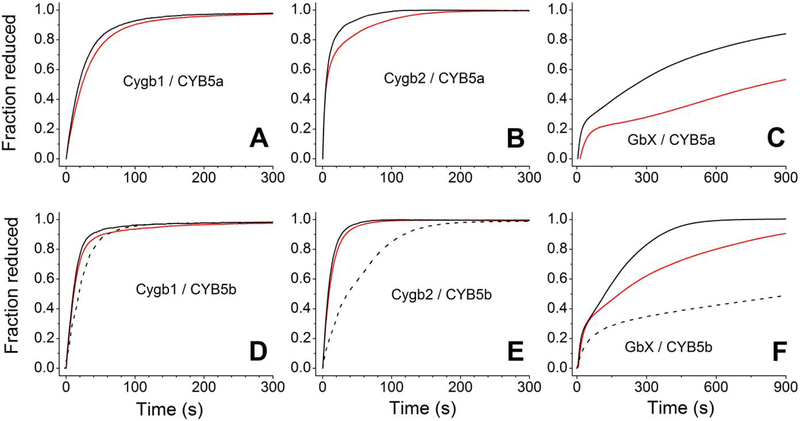
Our results are shown in Figure 7. As in the case of the human Cygb in the presence of human CYB5/CYB5R,20 the zebrafish CYB5/CYB5R system is able to completely reduce Cygb1 or Cygb2 in around 100 s. Both CYB5 isoforms can support fast Cygb1 or Cygb2 reduction, with the CYB5b performing slightly faster than CYB5a (Figure 7). Increasing the temperature from 25 °C to 37 °C shows a modest effect on the reduction rates. In order to compare the reduction of zebrafish Cygbs by the zebrafish CYB5/CYB5R system, traces for previous experiments using the human CYB5b/CYB5R are included (Figure 7 D–E).20 In the case of Cygb1, the human proteins can reduce the zebrafish Cygb1 at a comparable rate (Figure 7D). However in the case of Cygb2, the zebrafish proteins clearly outperform the human CYB5/CYB5R system, and can support full Cygb2 reduction in ≈50 s as compared to ≈200 s for complete reduction with the human proteins (Figure 7E).
Figure 7. Reduction of zebrafish cytoglobins 1 and 2 and GlobinX by cytochrome b5 reductase and cytochrome b5.
Panel A, Reduction of Cygb1 by zebrafish CYB5a and CYB5R. Panel B, Reduction of Cygb2 by zebrafish CYB5a and CYB5R. Panel C, Reduction of GlobinX by zebrafish CYB5a and CYB5R, Panel D, Reduction of Cygb1 by zebrafish CYB5b and CYB5R (solid lines) or the human CYB5b/CYB5R system (dotted lines). Panel E, Reduction of Cygb2 by zebrafish CYB5b and CYB5R (solid lines) or the human CYB5b/CYB5R system (dotted lines). Panel F, Reduction of GlobinX by zebrafish CYB5b and CYB5R (solid lines) or the human CYB5b/CYB5R system (dotted lines). Red lines, reactions monitored at 25 °C, Black lines, reactions monitored at 37 °C.
We have previously hypothesized that GbX could be reduced by the CYB5/CYB5R system in zebrafish.20 The traces for the reaction of the zebrafish CYB5/CYB5R system with GbX are shown in Figure 7 (Panels 7C and 7F). Our results indicate that this reduction system, although not as efficient as for Cygb, can reduce GbX at an appreciable rate. In particular CYB5b is efficient in this reduction even at 25 °C-consistent with zebrafish physiological conditions-catalyzing GbX reduction at a faster rate than the human system at 37 °C (Figure 7F). Thus, we conclude that CYB5/CYB5R can provide a pathway for GbX reduction in zebrafish.
Discussion
Our previous work strongly supported the role of the CYB5/CYB5R/NADH system as physiological reductant of Cygb in human cells.20 It also noted the ability of the human proteins to reduced fish Cygb1 and Cygb2 as well; this observation suggested that this could be a pathway not limited to mammals but conserved in vertebrates.
To test this hypothesis we expressed and purified the components of the CYB5 reducing system from zebrafish in E. coli. This is to our knowledge the first characterization of recombinant CYB5R and CYB5 proteins from fish. Of note, the enzymatic activity of CYB5R in fish is particularly relevant to aquaculture, where the accumulation of nitrites in water can cause methemoglobinemia–accumulation of oxidized hemoglobin in the erythrocytes–63, 64. Fish can recycle methemoglobin to its reduced form through several pathways, including reaction with glutathione, ascorbic acid, or NADPH-dependent methemoglobin reductase; however the main contributor is CYB5R. Indeed, the differences in CYB5R activity can partly explain fish species sensitivity to methemoglobinemia.27
The CYB5R/CYB5R proteins in fish showed comparable properties to the mammalian proteins. The main differences were the more negative midpoint potential for zebrafish CYB5a and the lower thermal stability of CYB5b as compared with the more stable mammalian CYB5b paralogues. However there are differences may be partly related to the lower body temperature of the fish and do not appear to indicate large differences in their physiological properties. As the FeIII-His interaction seems to be less strong in CYB5b, contrary to what is expected given its more negative redox potential,56 it suggests that other factors such as heme exposure53 may be more relevant to zebrafish CYB5b redox potential. The cause for the reduced stability is yet unknown but we notice that important residues from the hydrophobic core (Ala18, Ile32, Leu36, Leu47; rat CYB5b numbering) identified in rat CYB5s25, 26 are not conserved in zebrafish CYB5b; notably, Ala18 and Leu36 correspond to Gly41 and Met59, respectively, in zebrafish CYB5b.
Our experiments of Cygb reduction show that CYB5 and CYB5R are able to reduce Cygb1 and Cygb2 in the reduced state. This reduction is particularly important for Cygb2, as this protein shows autoxidation rates 5-fold faster than human Cygb.17
A limitation of our previous study20 is the use of the CYB5b isoform, which is mostly present in the mitochondrial outer membrane, thus probably isolated from cytoplasmic cytoglobins. However, the CYB5a isoform is attached to the endoplasmic reticulum, with the heme domain remaining in the cytoplasmic side and thus available for interactions with cytoplasmic molecules.65–67 CYB5R3 is located in the mitochondrial outer membrane and also attached to the endoplasmic reticulum67 and can thus provide electrons to cytosolic cytoglobins through CYB5a. GbX has been found to associate with the plasma membrane when expressed in mammalian cells23. In these conditions a cytosolic, soluble CYB5 could provide electrons to GbX. The presence of soluble CYB5 proteins in zebrafish is not established. However, large scale protein expression studies in human tissues indicate that human CYB5a and CYB5b can be found in the cytosol-dependent or independent of membrane binding-68, 69, Thus, we hypothesize that membrane-bound GbX could be reduced in vivo by the NADH/CYB5/CYB5R system either via CYB5 soluble isoforms present in zebrafish cells – as will be the case in thrombocytes, where GBX has been detected24, or by localizing to the endoplasmic reticulum membrane, where CYB5a and CYB5R are available.
Mammalian Cygb may play an important role in the regulation of NO levels in the vascular wall.19, 21, 70 It is unclear if Cygbs are present in fish blood vessels, but given the important physiological differences between NO metabolism and vascular biology of the fish and mammals, it is conceivable that Cygb in the fish accomplish other unrelated functions. However, we have shown that fish, like mammals, have a very effective Cygb reduction system, suggesting that NO detoxification or other roles requiring the reduced Cygb can be maintained. We observe very low P50 for both Cygbs, indicating that these proteins would be largely saturated with oxygen in vivo and that their biological roles may specifically involve the ferrous-oxy species, at least for Cygb1. Alternatively, Cygb2 can be involved in O2 transport but its P50 may be optimal for oxygen sensing functions. Experiments in different fish species have shown increases of Cygb1 mRNA transcripts under hypoxia – but usually not Cygb2 – which is consistent with this hypothesis71–73. The deoxy Cygb2 species can be formed in hypoxic tissues, where it could fulfill other functions not related to oxygen binding.
ACKNOWLEDGEMENTS
We thank Qin Tong for excellent technical support. S.J.K. was part of the First Experiences in Research program at the University of Pittsburgh, Dietrich School of Arts and Sciences.
Funding Sources
This work was supported by funding from the Institute for Transfusion Medicine and the Hemophilia Center of Western Pennsylvania (to M.T.G.), Ri.MED foundation (to P.C.), National Institutes of Health Grants T32 HL076124 (to M.B.A.), R01 HL098032, R01 HL125886, P01 HL103455, T32 HL110849, and T32 HL007563 (to M.T.G.), R21 ES027390 (to J.T.), K08 HL136857 (to J.J.R.); by a Parker B Francis Foundation Fellowship (to J.J.R.) and by the Independent Research Fund Denmark, Natural Sciences Grant 4181–00094 and the Aarhus University Research Foundation NOVA grant AUFF-E-2016-9-37 (to A.F.).
ABBREVIATIONS
- CYB5
cytochrome b5
- CYB5a
cytochrome b5a
- CYB5b
cytochrome b5b CYB5R, cytochrome b5 reductase 3
- Cygb
cytoglobin
- Cygb1
zebrafish cytoglobin-1
- Cygb2
zebrafish cytoglobin-2
- GbX
Globin X
- NADH
nicotinamide adenine dinucleotide hydrate
Footnotes
Publisher's Disclaimer: This document is confidential and is proprietary to the American Chemical Society and its authors. Do not copy or disclose without written permission. If you have received this item in error, notify the sender and delete all copies.
SEQUENCE INFORMATION
Zebrafish Cytoglobin 1, Uniprot KB Q8UUR3; zebrafish Cytoglobin 2, Uniprot KB Q575S8; zebrafish Cytochrome b5a, Uniprot KB Q7T341; zebrafish Cytochrome b5b, Uniprot KB Q6NY41; zebrafish Cytochrome b5 reductase 3, Uniprot KB Q6NYE6.
CONFLICT OF INTEREST STATEMENT
A.W.D., J.J.R., M.T.G. and J.T. are coinventors of provisional and pending patents for the use of recombinant cytoglobin, neuroglobin and other heme-based molecules as antidotes for carbon monoxide poisoning. Globin Solutions, Inc. has licensed this technology. J.J.R., M.T.G. and J.T. are shareholders and officers in Globin Solutions, Inc. J.J.R. is an officer and director of Globin Solutions, Inc. M.T.G. is a director and advisor of Globin Solutions, Inc. J.T. is an officer of Globin Solutions, Inc. The remaining authors have disclosed that they do not have any potential conflicts of interest.
REFERENCES
- 1.Burmester T, and Hankeln T (2014) Function and evolution of vertebrate globins, Acta Physiol (Oxf) 211, 501–514. [DOI] [PubMed] [Google Scholar]
- 2.Hoogewijs D, Ebner B, Germani F, Hoffmann FG, Fabrizius A, Moens L, Burmester T, Dewilde S, Storz JF, Vinogradov SN, and Hankeln T (2012) Androglobin: a chimeric globin in metazoans that is preferentially expressed in Mammalian testes, Mol Biol Evol 29, 1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardner AM, Cook MR, and Gardner PR (2010) Nitric-oxide dioxygenase function of human cytoglobin with cellular reductants and in rat hepatocytes, J Biol Chem 285, 23850–23857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reeder BJ, Svistunenko DA, and Wilson MT (2011) Lipid binding to cytoglobin leads to a change in haem co-ordination: a role for cytoglobin in lipid signalling of oxidative stress, Biochem J 434, 483–492. [DOI] [PubMed] [Google Scholar]
- 5.Tejero J, Kapralov AA, Baumgartner MP, Sparacino-Watkins CE, Anthonymuthu TS, Vlasova II, Camacho CJ, Gladwin MT, Bayir H, and Kagan VE (2016) Peroxidase Activation of Cytoglobin by Anionic phospholipids: Mechanisms and Consequences, Biochimica et Biophysica Acta-Molecular and Cell Biology of Lipids 1861, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tiso M, Tejero J, Basu S, Azarov I, Wang X, Simplaceanu V, Frizzell S, Jayaraman T, Geary L, Shapiro C, Ho C, Shiva S, Kim-Shapiro DB, and Gladwin MT (2011) Human neuroglobin functions as a redox-regulated nitrite reductase, J Biol Chem 286, 18277–18289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wakasugi K, Nakano T, and Morishima I (2003) Oxidized human neuroglobin acts as a heterotrimeric Galpha protein guanine nucleotide dissociation inhibitor, J Biol Chem 278, 36505–36512. [DOI] [PubMed] [Google Scholar]
- 8.Fago A, Mathews AJ, Moens L, Dewilde S, and Brittain T (2006) The reaction of neuroglobin with potential redox protein partners cytochrome b5 and cytochrome c, FEBS Lett 580, 4884–4888. [DOI] [PubMed] [Google Scholar]
- 9.Kawada N, Kristensen DB, Asahina K, Nakatani K, Minamiyama Y, Seki S, and Yoshizato K (2001) Characterization of a stellate cell activation-associated protein (STAP) with peroxidase activity found in rat hepatic stellate cells, J Biol Chem 276, 25318–25323. [DOI] [PubMed] [Google Scholar]
- 10.Burmester T, Ebner B, Weich B, and Hankeln T (2002) Cytoglobin: a novel globin type ubiquitously expressed in vertebrate tissues, Mol Biol Evol 19, 416–421. [DOI] [PubMed] [Google Scholar]
- 11.Trent JT 3rd, and Hargrove MS (2002) A ubiquitously expressed human hexacoordinate hemoglobin, J Biol Chem 277, 19538–19545. [DOI] [PubMed] [Google Scholar]
- 12.Dewilde S, Kiger L, Burmester T, Hankeln T, Baudin-Creuza V, Aerts T, Marden MC, Caubergs R, and Moens L (2001) Biochemical characterization and ligand binding properties of neuroglobin, a novel member of the globin family, J Biol Chem 276, 38949–38955. [DOI] [PubMed] [Google Scholar]
- 13.Tejero J, Sparacino-Watkins CE, Ragireddy V, Frizzell S, and Gladwin MT (2015) Exploring the mechanisms of the reductase activity of neuroglobin by site-directed mutagenesis of the heme distal pocket, Biochemistry 54, 722–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fago A, Hundahl C, Dewilde S, Gilany K, Moens L, and Weber RE (2004) Allosteric regulation and temperature dependence of oxygen binding in human neuroglobin and cytoglobin. Molecular mechanisms and physiological significance, J Biol Chem 279, 44417–44426. [DOI] [PubMed] [Google Scholar]
- 15.Fuchs C, Luckhardt A, Gerlach F, Burmester T, and Hankeln T (2005) Duplicated cytoglobin genes in teleost fishes, Biochem Biophys Res Commun 337, 216–223. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann FG, Opazo JC, and Storz JF (2011) Differential loss and retention of cytoglobin, myoglobin, and globin-E during the radiation of vertebrates, Genome biology and evolution 3, 588–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corti P, Ieraci M, and Tejero J (2016) Characterization of zebrafish neuroglobin and cytoglobins 1 and 2: Zebrafish cytoglobins provide insights into the transition from six-coordinate to five-coordinate globins, Nitric Oxide 53, 22–34. [DOI] [PubMed] [Google Scholar]
- 18.Fago A (2017) Functional roles of globin proteins in hypoxia-tolerant ectothermic vertebrates, J Appl Physiol (1985) 123, 926–934. [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Follmer D, Zweier JR, Huang X, Hemann C, Liu K, Druhan LJ, and Zweier JL (2012) Characterization of the function of cytoglobin as an oxygen-dependent regulator of nitric oxide concentration, Biochemistry 51, 5072–5082. [DOI] [PubMed] [Google Scholar]
- 20.Amdahl MB, Sparacino-Watkins CE, Corti P, Gladwin MT, and Tejero J (2017) Efficient Reduction of Vertebrate Cytoglobins by the Cytochrome b5/Cytochrome b5 Reductase/NADH System, Biochemistry 56, 3993–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, El-Mahdy MA, Boslett J, Varadharaj S, Hemann C, Abdelghany TM, Ismail RS, Little SC, Zhou D, Thuy LT, Kawada N, and Zweier JL (2017) Cytoglobin regulates blood pressure and vascular tone through nitric oxide metabolism in the vascular wall, Nature communications 8, 14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roesner A, Fuchs C, Hankeln T, and Burmester T (2005) A globin gene of ancient evolutionary origin in lower vertebrates: evidence for two distinct globin families in animals, Mol Biol Evol 22, 12–20. [DOI] [PubMed] [Google Scholar]
- 23.Blank M, Wollberg J, Gerlach F, Reimann K, Roesner A, Hankeln T, Fago A, Weber RE, and Burmester T (2011) A membrane-bound vertebrate globin, PLoS One 6, e25292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corti P, Xue J, Tejero J, Wajih N, Sun M, Stolz DB, Tsang M, Kim-Shapiro DB, and Gladwin MT (2016) Globin X is a six-coordinate globin that reduces nitrite to nitric oxide in fish red blood cells, Proc Natl Acad Sci U S A 113, 8538–8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altuve A, Wang L, Benson DR, and Rivera M (2004) Mammalian mitochondrial and microsomal cytochromes b(5) exhibit divergent structural and biophysical characteristics, Biochem Biophys Res Commun 314, 602–609. [DOI] [PubMed] [Google Scholar]
- 26.Altuve A, Silchenko S, Lee KH, Kuczera K, Terzyan S, Zhang X, Benson DR, and Rivera M (2001) Probing the differences between rat liver outer mitochondrial membrane cytochrome b5 and microsomal cytochromes b5, Biochemistry 40, 9469–9483. [DOI] [PubMed] [Google Scholar]
- 27.McConkey S, Saunders J, and Speare DJ (2013) Comparison of NADH-dependent cytochrome b5 reductase activity and in vitro methemoglobin induction by sodium nitrite in Oncorhynchus mykiss, Salmo salar, and Salvelinus fontinalis, Fish physiology and biochemistry 39, 713–719. [DOI] [PubMed] [Google Scholar]
- 28.Saleh MC, and McConkey S (2012) NADH-dependent cytochrome b5 reductase and NADPH methemoglobin reductase activity in the erythrocytes of Oncorhynchus mykiss, Fish physiology and biochemistry 38, 1807–1813. [DOI] [PubMed] [Google Scholar]
- 29.Scott EM, and Harrington JP (1985) Methemoglobin reductase activity in fish erythrocytes, Comparative biochemistry and physiology. B, Comparative biochemistry 82, 511–513. [DOI] [PubMed] [Google Scholar]
- 30.Freeman L, Beitinger T, and Huey D (1983) Methemoglobin reductase activity in phylogenetically diverse piscine species, Comparative Biochemistry and Physiology Part B: Comparative Biochemistry 75, 27–30. [DOI] [PubMed] [Google Scholar]
- 31.Sparacino-Watkins CE, Tejero J, Sun B, Gauthier MC, Thomas J, Ragireddy V, Merchant BA, Wang J, Azarov I, Basu P, and Gladwin MT (2014) Nitrite reductase and nitric-oxide synthase activity of the mitochondrial molybdopterin enzymes mARC1 and mARC2, J Biol Chem 289, 10345–10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charpilloz C, Veuthey AL, Chopard B, and Falcone JL (2014) Motifs tree: a new method for predicting post-translational modifications, Bioinformatics 30, 1974–1982. [DOI] [PubMed] [Google Scholar]
- 33.Kiemer L, Bendtsen JD, and Blom N (2005) NetAcet: prediction of N-terminal acetylation sites, Bioinformatics 21, 1269–1270. [DOI] [PubMed] [Google Scholar]
- 34.Ren J, Wen L, Gao X, Jin C, Xue Y, and Yao X (2008) CSS-Palm 2.0: an updated software for palmitoylation sites prediction, Protein engineering, design & selection : PEDS 21, 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Efimov I, Parkin G, Millett ES, Glenday J, Chan CK, Weedon H, Randhawa H, Basran J, and Raven EL (2014) A simple method for the determination of reduction potentials in heme proteins, FEBS Lett 588, 701–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santoro MM, and Bolen DW (1988) Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants, Biochemistry 27, 8063–8068. [DOI] [PubMed] [Google Scholar]
- 37.Weber RE (1981) Cationic control of O2 affinity in lugworm erythrocruorin, Nature 292, 386–387. [Google Scholar]
- 38.Janecka JE, Nielsen SS, Andersen SD, Hoffmann FG, Weber RE, Anderson T, Storz JF, and Fago A (2015) Genetically based low oxygen affinities of felid hemoglobins: lack of biochemical adaptation to high-altitude hypoxia in the snow leopard, Journal of experimental biology 218, 2402–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jendroszek A, Malte H, Overgaard CB, Beedholm K, Natarajan C, Weber RE, Storz JF, and Fago A (2018) Allosteric mechanisms underlying the adaptive increase in hemoglobin–oxygen affinity of the bar-headed goose, Journal of Experimental Biology 221, jeb185470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murakami K, Yubisui T, Takeshita M, and Miyata T (1989) The NH2-terminal structures of human and rat liver microsomal NADH-cytochrome b5 reductases, J Biochem 105, 312–317. [DOI] [PubMed] [Google Scholar]
- 41.Rivera M, Barillas-Mury C, Christensen KA, Little JW, Wells MA, and Walker FA (1992) Gene synthesis, bacterial expression, and 1H NMR spectroscopic studies of the rat outer mitochondrial membrane cytochrome b5, Biochemistry 31, 12233–12240. [DOI] [PubMed] [Google Scholar]
- 42.Zhang M, and Scott JG (1996) Purification and characterization of cytochrome b5 reductase from the house fly, Musca domestica, Comp Biochem Physiol B Biochem Mol Biol 113, 175–183. [DOI] [PubMed] [Google Scholar]
- 43.Roma GW, Crowley LJ, Davis CA, and Barber MJ (2005) Mutagenesis of glycine 179 modulates both catalytic efficiency and reduced pyridine nucleotide specificity in cytochrome b 5 reductase, Biochemistry 44, 13467–13476. [DOI] [PubMed] [Google Scholar]
- 44.Kimura S, Emi Y, Ikushiro S, and Iyanagi T (1999) Systematic mutations of highly conserved His49 and carboxyl-terminal of recombinant porcine liver NADH-cytochrome b5 reductase solubilized domain, Biochim Biophys Acta 1430, 290–301. [DOI] [PubMed] [Google Scholar]
- 45.Passon PG, and Hultquist DE (1972) Soluble cytochrome b 5 reductase from human erythrocytes, Biochim Biophys Acta 275, 62–73. [DOI] [PubMed] [Google Scholar]
- 46.Kitajima S, and Minakami S (1983) Human NADH-cytochrome b5 reductases: comparison among those of erythrocyte membrane, erythrocyte cytosol, and liver microsomes, J Biochem 93, 615–620. [DOI] [PubMed] [Google Scholar]
- 47.Roma GW, Crowley LJ, and Barber MJ (2006) Expression and characterization of a functional canine variant of cytochrome b5 reductase, Archives of biochemistry and biophysics 452, 69–82. [DOI] [PubMed] [Google Scholar]
- 48.Iyanagi T (1977) Redox properties of microsomal reduced nicotinamide adenine dinucleotide-cytochrome b5 reductase and cytochrome b5, Biochemistry 16, 2725–2730. [DOI] [PubMed] [Google Scholar]
- 49.Marohnic CC, Bewley MC, and Barber MJ (2003) Engineering and characterization of a NADPH-utilizing cytochrome b 5 reductase, Biochemistry 42, 11170–11182. [DOI] [PubMed] [Google Scholar]
- 50.Reid LS, Taniguchi VT, Gray HB, and Mauk AG (1982) Oxidation-reduction equilibrium of cytochrome b5, Journal of the American Chemical Society 104, 7516–7519. [Google Scholar]
- 51.Walker FA, Emrick D, Rivera JE, Hanquet BJ, and Buttlaire DH (1988) Effect of heme orientation on the reduction potential of cytochrome b5, Journal of the American Chemical Society 110, 6234–6240. [DOI] [PubMed] [Google Scholar]
- 52.Rodgers KK, and Sligar SG (1991) Surface electrostatics, reduction potentials, and the internal dielectric constant of proteins, Journal of the American Chemical Society 113, 9419–9421. [Google Scholar]
- 53.Rivera M, Seetharaman R, Girdhar D, Wirtz M, Zhang X, Wang X, and White S (1998) The reduction potential of cytochrome b 5 is modulated by its exposed heme edge, Biochemistry 37, 1485–1494. [DOI] [PubMed] [Google Scholar]
- 54.Rivera M, Wells MA, and Walker FA (1994) Cation-Promoted Cyclic Voltammetry of Recombinant Rat Outer Mitochondrial Membrane Cytochrome b5 at a Gold Electrode Modified with. beta.-Mercaptopropionic Acid, Biochemistry 33, 2161–2170. [DOI] [PubMed] [Google Scholar]
- 55.Tezcan FA, Winkler JR, and Gray HB (1998) Effects of ligation and folding on reduction potentials of heme proteins, Journal of the American Chemical Society 120, 13383–13388. [Google Scholar]
- 56.Kennedy ML, Silchenko S, Houndonougbo N. v., Gibney BR, Dutton PL, Rodgers KR, and Benson DR (2001) Model hemoprotein reduction potentials: The effects of histidine-to-iron coordination equilibrium, Journal of the American Chemical Society 123, 4635–4636. [DOI] [PubMed] [Google Scholar]
- 57.Hayashi A, Suzuki T, and Shin M (1973) An enzymic reduction system for metmyoglobin and methemoglobin, and its application to functional studies of oxygen carriers, Biochim Biophys Acta 310, 309–316. [DOI] [PubMed] [Google Scholar]
- 58.Hamdane D, Kiger L, Dewilde S, Green BN, Pesce A, Uzan J, Burmester T, Hankeln T, Bolognesi M, Moens L, and Marden MC (2003) The redox state of the cell regulates the ligand binding affinity of human neuroglobin and cytoglobin, J Biol Chem 278, 51713–51721. [DOI] [PubMed] [Google Scholar]
- 59.Lechauve C, Chauvierre C, Dewilde S, Moens L, Green BN, Marden MC, Celier C, and Kiger L (2010) Cytoglobin conformations and disulfide bond formation, FEBS J 277, 2696–2704. [DOI] [PubMed] [Google Scholar]
- 60.Kurian JR, Chin NA, Longlais BJ, Hayes KL, and Trepanier LA (2006) Reductive detoxification of arylhydroxylamine carcinogens by human NADH cytochrome b5 reductase and cytochrome b5, Chem Res Toxicol 19, 1366–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yubisui T, Takeshita M, and Yoneyama Y (1980) Reduction of methemoglobin through flavin at the physiological concentration by NADPH-flavin reductase of human erythrocytes, J Biochem 87, 1715–1720. [DOI] [PubMed] [Google Scholar]
- 62.Westerfield M (1995) The Zebrafish Book, University of Oregon Press, Eugene, OR. [Google Scholar]
- 63.Kroupova H, Machova J, and Svobodova Z (2005) Nitrite influence on fish: a review, Veterinarni Medicina (Praha) 50, 461–471. [Google Scholar]
- 64.Svobodova Z, Machova J, Poleszczuk G, Hůda J, Hamáčková J, and Kroupova H (2005) Nitrite poisoning of fish in aquaculture facilities with water-recirculating systems, Acta Veterinaria Brno 74, 129–137. [Google Scholar]
- 65.Strittmatter P, Rogers MJ, and Spatz L (1972) The binding of cytochrome b 5 to liver microsomes, J Biol Chem 247, 7188–7194. [PubMed] [Google Scholar]
- 66.Mitoma J, and Ito A (1992) The carboxy-terminal 10 amino acid residues of cytochrome b5 are necessary for its targeting to the endoplasmic reticulum, EMBO J 11, 4197–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Borgese N, D’Arrigo A, De Silvestris M, and Pietrini G (1993) NADH-cytochrome b5 reductase and cytochrome b5 isoforms as models for the study of post-translational targeting to the endoplasmic reticulum, FEBS Lett 325, 70–75. [DOI] [PubMed] [Google Scholar]
- 68.Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Bjork L, Breckels LM, Backstrom A, Danielsson F, Fagerberg L, Fall J, Gatto L, Gnann C, Hober S, Hjelmare M, Johansson F, Lee S, Lindskog C, Mulder J, Mulvey CM, Nilsson P, Oksvold P, Rockberg J, Schutten R, Schwenk JM, Sivertsson A, Sjostedt E, Skogs M, Stadler C, Sullivan DP, Tegel H, Winsnes C, Zhang C, Zwahlen M, Mardinoglu A, Ponten F, von Feilitzen K, Lilley KS, Uhlen M, and Lundberg E (2017) A subcellular map of the human proteome, Science 356, eaal3321. [DOI] [PubMed] [Google Scholar]
- 69.The Human Protein Atlas. https://www.proteinatlas.org/
- 70.Liu X, Tong J, Zweier JR, Follmer D, Hemann C, Ismail RS, and Zweier JL (2013) Differences in oxygen-dependent nitric oxide metabolism by cytoglobin and myoglobin account for their differing functional roles, FEBS J 280, 3621–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wawrowski A, Gerlach F, Hankeln T, and Burmester T (2011) Changes of globin expression in the Japanese medaka (Oryzias latipes) in response to acute and chronic hypoxia, J Comp Physiol B 181, 199–208. [DOI] [PubMed] [Google Scholar]
- 72.Roesner A, Hankeln T, and Burmester T (2006) Hypoxia induces a complex response of globin expression in zebrafish (Danio rerio), J Exp Biol 209, 2129–2137. [DOI] [PubMed] [Google Scholar]
- 73.Chao Y, Xia M, Wu R, Chen Q, Zheng Z, and Qi D (2018) Molecular characterization and expression changes of cytoglobin genes in response to hypoxia in a Tibetan schizothoracine fish, Schizopygopsis pylzovi, Fish physiology and biochemistry, 10.1007/s10695-10018-10582-10691. [DOI] [PubMed]